Abstract
Vibrio parahaemolyticus is the leading cause of bacterial seafood-borne gastroenteritis worldwide, yet little is known about how this pathogen colonizes the human intestine. The alternative sigma factor RpoN/sigma-54 is a global regulator that controls flagellar synthesis, as well as a wide range of nonflagellar genes. We constructed an in-frame deletion mutation in rpoN (VP2670) in V. parahaemolyticus RIMD2210633, a clinical serogroup O3:K6 isolate, and examined the effects in vivo using a streptomycin-treated mouse model of colonization. We confirmed that deletion of rpoN rendered V. parahaemolyticus nonmotile, and it caused reduced biofilm formation and an apparent defect in glutamine synthetase production. In in vivo competition assays between the rpoN mutant and a wild-type RIMD2210633 strain marked with the β-galactosidase gene lacZ (WBWlacZ), the mutant colonized significantly more proficiently. Intestinal persistence competition assays also demonstrated that the rpoN mutant had enhanced fitness and outcompeted WBWlacZ. Mutants defective in the polar flagellum biosynthesis FliAP sigma factor also outcompeted WBWlacZ but not to the same level as the rpoN mutant, which suggested that lack of motility is not the sole cause of the fitness effect. In an in vitro growth competition assay in mouse intestinal mucus, the rpoN mutant also outcompeted the wild type and exhibited faster doubling times when grown in mucus and on individual components of mucus. Genes in the pathways for the catabolism of mucus sugars also had significantly higher expression levels in a ΔrpoN mutant than in the wild type. These data suggest that in V. parahaemolyticus, RpoN plays an important role in carbon utilization regulation, which may significantly affect host colonization.
INTRODUCTION
Vibrio parahaemolyticus is a Gram-negative bacterium ubiquitous in the marine and estuarine environments worldwide (1–4). Vibrio parahaemolyticus is also the leading cause of seafood-associated bacterial gastroenteritis in the United States and Asia (5, 6), which usually stems from the consumption of raw or undercooked shellfish (7, 8). Typically, infection by this organism leads to nausea, vomiting, fever, and a diarrhea distinct from that of the related Vibrio cholerae. Less commonly, infection by V. parahaemolyticus can cause wound infection and septicemia, leading to mortality in immunocompromised individuals (9–12).
Much effort has gone into understanding the mechanisms that contribute to V. parahaemolyticus pathogenesis, with a particular focus on virulence factors produced by this bacterium. Strains that caused disease often possessed either the thermostable direct hemolysin (TDH) or the TDH-related hemolysin (TRH), while nonpathogenic strains typically lacked these two markers (13–15). Additionally, sequence analysis of RIMD2210633, an O3:K6 isolate (TDH+ TRH−), revealed the presence of two type 3 secretion systems (T3SS), one on each chromosome (T3SS-1 and T3SS-2) (16, 17). T3SS-1 is common to both clinical and nonclinical strains of V. parahaemolyticus and is a major contributor to cytotoxicity in in vitro models (18–22). T3SS-1 is the major contributor to lethality in the intraperitoneal mouse model, and it also plays a minor role in virulence in the infant rabbit model (23). The T3SS-2 gene cluster is flanked by a copy of the tdh gene (tdhA and tdhS) within an 80-kb pathogenicity island named VPaI-7, a region found only in clinical isolates (16, 24–26). T3SS-2 plays only a minor role in cytotoxicity but is the major contributing factor toward enterotoxicity (17, 22, 23, 27). Studies have demonstrated that fluid accumulation in the rabbit ileal loop model as well as virulence in the infant rabbit and infant pig models are dependent upon a functional T3SS-2 (17, 22, 23, 27).
Little is known about how V. parahaemolyticus survives within the host gastrointestinal (GI) tract. We recently developed an orogastric adult mouse model that allows for the oral infection and persistent colonization of mice with V. parahaemolyticus (28). In conjunction with this model, we also demonstrated that the Vibrio-specific two-component regulator ToxRS is an important colonization factor and is required for resistance to acid and bile salt stresses (28). We determined that ToxRS contributes to in vivo survival via its regulation of the outer membrane protein OmpU, which is essential for acid and bile salt tolerance (28). It stands to reason that there are a great many other factors that contribute to V. parahaemolyticus colonization and survival within the host that have not yet been identified.
Bacteria employ a housekeeping sigma factor, σ70 (RpoD), that controls most of the cell's gene expression and a number of alternative sigma factors that typically control a smaller subset of genes with different promoter specificities (29). RpoN (σ54) is one such alternative sigma factor, which in Escherichia coli was shown to regulate approximately 30 operons, half of which were involved in nitrogen metabolism (30). The function of RpoN has also been studied in V. cholerae, where it was shown to play a role in the regulation of flagellum synthesis, ammonium assimilation, virulence, dicarboxylic acid transport, type VI secretion systems, and quorum sensing (26, 31–36). Studies demonstrated that deletion of rpoN in V. cholerae leads to aflagellate cells that are nonmotile and are defective for growth on minimal medium where ammonium is the only nitrogen source because of the mutant's inability to express glutamine synthetase (32, 37). Importantly, the rpoN mutant exhibited a defect in colonization in the infant mouse model of cholera, and this defect was glutamine synthetase independent and not entirely due to loss of motility (32). Loss of RpoN in other Vibrio spp. has also been attributed to loss of motility, reduced biofilm production, and, in the case of V. fischeri and V. anguillarum, decreased host colonization (38–40).
To the best of our knowledge, this is the first paper to evaluate the role of RpoN in V. parahaemolyticus host colonization and survival. Here, we demonstrate that an rpoN deletion mutant in V. parahaemolyticus colonizes the streptomycin-treated adult mouse model significantly better than the wild-type strain. To determine whether this enhanced fitness effect is due to a lack of motility alone, we constructed in-frame deletions in the sigma factor-encoding genes fliAP and fliAL and a double deletion in these genes, which produced strains that lacked polar, lateral, or both flagellar systems, respectively. In in vivo long-term persistence competition assays, we found that although the fliAP mutants but not the fliAL mutant outcompeted the wild type, they did not do so to the same level as the rpoN mutant. These data indicate that the rpoN mutant's enhanced fitness was not due to its role in motility alone. In vitro competition assays were performed between wild-type and mutant strains grown on mouse intestinal mucus. In these assays, ΔrpoN and ΔfliAP outcompeted the wild type and these isolates had a faster doubling time on mucus and its components, suggesting that carbon utilization plays a key role in enhanced fitness in vivo.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All strains and plasmids used in this study are listed in Table 1. A previously described streptomycin-resistant V. parahaemolyticus RIMD2210633 O3:K6 clinical isolate and a streptomycin-resistant β-galactosidase-positive RIMD2210633 isolate named WBWlacZ were used in this study (28, 41). Unless stated otherwise, all V. parahaemolyticus strains were grown aerobically in LB broth (Fisher Scientific, Pittsburgh, PA) containing 3% NaCl at 37°C. For growth studies of all mutants, LB was used as a nutritionally complex medium, while M9 minimal medium (Sigma-Aldrich, St. Louis, MO) was used as a simple medium consisting of salts to which amino acids and carbon sources can be added. M9 medium contains no carbon source but does contain ammonium as the sole nitrogen source that can be converted to glutamine by glutamine synthetase. For experiments analyzing growth characteristics of the ΔrpoN strain in M9 medium, the medium was supplemented with 2 mM glutamine unless otherwise stated. Genetic manipulations utilized the E. coli diaminopimelic acid (DAP) auxotroph β2155λ pir. The E. coli β2155 λpir strain was cultured on medium supplemented with 0.3 mM DAP (Sigma). When required, antibiotics were added to LB broth at the following concentrations: streptomycin, 200 μg/ml; chloramphenicol, 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| Vibrio parahaemolyticus | ||
| RIMD2210633 | O3:K6 clinical isolate, Strr | 16, 41 |
| WBWlacZ | RIMD2210633, Strr, lacZ | 28 |
| ΔrpoN | RIMD2210633 ΔrpoN (VP2670), Strr | This study |
| ΔfliAP | RIMD2210633 ΔfliAP (VP2232), Strr | This study |
| ΔfliAL | RIMD2210633 ΔfliAL (VPA1555), Strr | This study |
| ΔfliAP ΔfliAL | RIMD2210633 ΔfliAP ΔfliAL (VP2232 and VPA1555), Strr | This study |
| ΔrpoNprpoN | ΔrpoN harboring pBBRrpoN | This study |
| ΔfliAPpfliAP | ΔfliAP harboring pBBRfliAP | This study |
| ΔfliAP ΔfliALpfliAP | ΔfliAP ΔfliAL harboring pBBRfliAP | This study |
| Escherichia coli | ||
| β2155 λpir | ΔdapA::erm pir, for bacterial conjugation | |
| Plasmids | ||
| pDS132 | Suicide plasmid, Cmr, SacB | 69 |
| pDSΔrpoN | pDS132 harboring truncated rpoN (VP2670) gene | This study |
| pDSΔfliAP | pDS132 harboring truncated fliAP (VP2232) gene | This study |
| pDSΔfliAL | pDS132 harboring truncated fliAL (VPA1555) gene | This study |
| pBBR1MCS | Expression vector, lacZ promoter, Cmr | |
| pBBRrpoN | pBBR1MCS harboring full-length rpoN | This study |
| pBBRfliAP | pBBR1MCS harboring full-length fliAP | This study |
Construction of V. parahaemolyticus RIMD2210633 rpoN, fliAP, fliAL, and fliAP fliAL deletion mutants.
In-frame nonpolar deletions were constructed in the sigma factors encoded by VP2670 (rpoN), VP2232 (fliAP), and VPA1555 (fliAL), and double deletion mutations in fliAP and fliAL were constructed using splice overlapping extension (SOE) PCR and an allelic exchange method as previously described (28, 41). Briefly, primers were designed to open reading frame (ORF) VP2670, which encodes the alternate sigma factor RpoN, using the V. parahaemolyticus RIMD2210633 genome sequence as the template, and primers were purchased from Integrated DNA Technologies (Coralville, IA) (Table 2). These primers were used to perform SOE PCR and to obtain a 465-bp truncated version of the 1,467-bp rpoN gene. The ΔrpoN mutant PCR fragment was cloned into the suicide vector pDS132, which was designated pDSΔrpoN. pDSΔrpoN was subsequently transformed into the E. coli strain β2155 λpir. pDSΔrpoN was conjugated into V. parahaemolyticus RIMD2210633 via cross-streaking on LB plates containing 0.3 mM DAP. Growth from these plates was transferred to LB plates containing 3% NaCl, streptomycin (200 μg/ml), and chloramphenicol (25 μg/ml). The 3% NaCl allowed for optimal V. parahaemolyticus growth. The absence of DAP from these plates, plus the addition of chloramphenicol and streptomycin, only selected for V. parahaemolyticus cells that harbored pDSΔrpoN. Exconjugate colonies were cultured overnight in LB-3% NaCl without chloramphenicol and were subsequently serially diluted and plated on LB-3% NaCl with 10% sucrose to select for cells which had lost pDSΔrpoN. Double-crossover deletion mutants were screened by PCR using SOEFLrpoNF and SOEFLrpoNR primers and sequenced for verification. The same protocol was used to create in-frame deletion mutations in the alternative sigma factor genes fliAP and fliAL, as well as fliAP fliAL, a double mutation of these genes. We constructed 84-bp and 294-bp truncated versions of the fliAP and fliAL genes, respectively. All deletions were confirmed to be in-frame via gene sequencing. To ensure the phenotypes observed in this study were not due to disruption of downstream genes, the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains were complemented as described previously (28). Briefly, a full-length copy of the rpoN gene was PCR amplified and cloned into the expression vector pBBR1MCS and transferred into the ΔrpoN strain, designated ΔrpoNprpoN. Assays utilizing the ΔrpoNprpoN strain were routinely supplemented with 100 mM isopropyl-β-d-thiogalactopyranoside (IPTG). This process was repeated for the ΔfliAP and ΔfliAP ΔfliAL strains. Complementation primers can be found in Table 2.
TABLE 2.
Primers used in this study
| Primer name and use | Sequence (5′–3′)a | Melting temp (°C) | Product (bp) |
|---|---|---|---|
| Splice overlap extension PCR | |||
| SOEvp2670A | TCT AGA TTA TCT CGG CGA ACA ATT CC | 56 | 315 |
| SOEvp2670B | GAT TTC TGT TGG CTC GTT GG | 43 | |
| SOEvp2670C | CCA ACG AGC CAA CAG AAA TCT TCT CCA GCC ACG TCA GTA C | 67 | 302 |
| SOEvp2670D | GAT ATC TGC ATT GAA TCG GTA AGA TCA | 55 | |
| SOEvp2670FlFor | GAC TCA AAC CCG CTA TTG GA | 55 | 1,325 |
| SOEvp2670FlRev | TGA CTT GAA GGG GCA ATA CC | 55 | |
| SOEvp2232A | TCT AGA GCG TTG ATC GAG GTT GAA CT | 59 | 756 |
| SOEvp2232B | AAC GTT TAC GCA CCA AGC TC | 56 | |
| SOEvp2232C | GAG CTT GGT GCG TAA ACG TTT AAA CTT GCC ATG CTG GTC | 66 | 609 |
| SOEvp2232D | GAG CTC AAC TTC TGG CAC ACA AAG CA | 61 | |
| SOEvp2232FlFor | ACG CGA CGA ATA ATT TGA CC | 53 | 2,831 |
| SOEvp2232FlRev | TGC CAG TGG CTT ACA TAG CA | 57 | |
| SOEvpa1555A | AAT CTA GAC AGA CGC ACT TTG TCA TGC T | 60 | 411 |
| SOEvpa1555B | ATG AAC GCG AAG TTG GTT AA | 53 | |
| SOEvpa1555C | TTA ACC AAC TTC GCG TTC ATC GAC TGT CAA AGC GAG ATC A | 66 | 453 |
| SOEvpa1555D | AAG AGC TCA ACT TGA AAC CAC GGC TAC G | 62 | |
| SOEvpa1555FlFor | GGT GAA ACC AGC TCA CTG CT | 58 | 1,450 |
| SOEvpa1555FlRev | TTC TGG TGC TTC GAT GTG AG | 55 | |
| Complement primers | |||
| rpoNforward | GGT ACC TGA GCA TTA CAA GGT AAG TAA CAC TG | 60 | 1,500 |
| rpoNreverse | TCT AGA AGT GCC TTA AAG TAG GCG TTT | 58 | |
| fliAPforward | CCC GGG TCG CAG AGG ACC CTT TTG | 66 | 753 |
| fliAPreverse | TCT AGA TTA GTC ATT TTG TGT CCA CGC AC | 59 | |
| RT-PCR primers | |||
| VP0063For | TCA ACT CCA TAT CGC CAT CA | 54 | 219 |
| VP0063Rev | CAC GAG CGA ACA TTC AGA AA | 53 | |
| VPA1674For | ATG CCT GGT TCA AAA ACG T | 53 | 211 |
| VPA1674Rev | CCT TTA AGA CGC TGG TTT GC | 55 | |
| VPA1083For | CCT TTT GCA CCG AGA GTG AT | 55 | 222 |
| VPA1083Rev | TGT GGC AAA AGA TGC AAA AA | 52 | |
| VPA1425For | TGA GAT CAT GGC GAA CTC AG | 55 | 210 |
| VPA1425Rev | GTT CGT TGG TTG GCT TGT TT | 54 |
Underlined base pairs indicate restriction sites.
Motility assay.
Motility assays were conducted on either LB containing 2% NaCl and 0.3% agar for swimming motility assays or heart infusion (HI) (Remel, Lenexa, KS) plates containing 2% NaCl and 1.5% agar for swarming motility assays. For swim assays, a single colony was picked from a fresh plate using a sterile pipette tip and stabbed into LB containing 0.3% agar. Plates were incubated overnight at 37°C, and images of each plate were taken the following day. Swarming assays were performed similarly, with the exception that the strains were inoculated only onto the surface of the HI-1.5% agar plates using sterile pipette tips. These plates were incubated at 30°C for 60 h before images were taken.
Electron microscopy.
The wild-type and ΔrpoN strains were analyzed for their ability to produce flagella using transmission electron microscopy. Strains were grown either in LB-3% NaCl broth or on an HI plate with 2% NaCl. Cells were adhered to a Formvar/carbon grid (Electron Microscopy Sciences, Hatfield, PA) and negatively stained with 1% phosphotungstate. Images were taken using a Zeiss LIBRA transmission electron microscope.
Biofilm assay.
Vibrio parahaemolyticus wild-type and mutant strains were grown overnight at 37°C aerobically in LB broth with 3% NaCl. Overnight cultures were diluted 1:40 in fresh LB-3% NaCl and allowed to grow statically in a 96-well plate for 24 h. After static incubation, the culture was decanted and each well was washed once with sterile phosphate-buffered saline (PBS) to remove any loosely adherent cells. Crystal violet (Electron Microscopy Sciences) was added to each well at a concentration of 0.1% (wt/vol) and incubated at room temperature for 30 min. The crystal violet was decanted, and each well was washed once with sterile PBS. Crystal violet that stained adhering cells (biofilm) was solubilized with dimethylsulfoxide (DMSO), and the optical density at 595 nm (OD595) of each well was measured using a Tecan Sunrise microplate reader and Magellan plate reader software (Tecan Systems Inc., San Jose, CA).
Mouse streptomycin pretreatment and inoculation preparation.
All experiments involving mice were approved by the University of Delaware Institutional Animal Care and Use Committee. Male C57BL/6 mice, aged 6 to 10 weeks, were housed under specific-pathogen-free conditions in standard cages in groups (4 per group) and provided standard mouse feed and water ad libitum. Treatment with streptomycin and inoculations were performed as previously described (28). In essence, mice were fasted for 4 h and then orogastrically administered 20 mg streptomycin per mouse (100 μl of streptomycin at 200 mg/ml). Food and water were returned upon antibiotic treatment. Twenty hours after antibiotic treatment, the mice were fasted for 4 h and then inoculated with a 100-μl bacterial suspension in PBS by gavage. Water was returned immediately upon infection, and food was returned 2 h postinfection. Vibrio parahaemolyticus inocula were prepared from wild-type RIMD2210633 and WBWlacZ, ΔrpoN, ΔfliAP, ΔfliAL, and ΔfliAPΔfliAL mutant cultures grown aerobically overnight in LB supplemented with 3% NaCl and streptomycin (200 μg/ml) at 37°C. The WBWlacZ strain used in competition experiments, as described below, is RIMD2210633 with a β-galactosidase gene knock-in, as previously described (28). This strain is designated WBWlacZ and substitutes for the wild-type strain in coinfections, which allows for color selection of the WBWlacZ strain (blue colonies) against a background of mutants (white colonies). These overnight cultures were diluted 1:50 with LB streptomycin and grown aerobically at 37°C for 4 h. An aliquot of the 4-h bacterial culture was pelleted and washed once with sterile PBS before being resuspended in fresh PBS to a final concentration of ∼1 × 1010 CFU/ml. Each inoculum was serially diluted and the titers determined on LB streptomycin plates to determine the dose administered.
In vivo competition assays.
Inocula for competition assays were prepared as described above, with the following modifications. Four-hour cultures of wild-type, WBWlacZ, ΔrpoN, ΔfliAP, ΔfliAL, and ΔfliAP ΔfliAL strains were pelleted by centrifugation at 4,000 × g, washed with PBS, and centrifuged again. The resulting bacterial pellet was resuspended in PBS to a concentration of approximately 1 × 1010 CFU/ml based on the culture OD600. A 1-ml aliquot of each deletion mutant strain was combined with 1 ml of the WBWlacZ strain, yielding a bacterial suspension of 1 × 1010 CFU/ml with a ratio of 1:1 CFU of mutant to WBWlacZ strain. A bacterial suspension of 1 × 1010 CFU/ml with a ratio of 1:1 CFU of wild-type to WBWlacZ strain was also examined in all experiments. The inoculum was serially diluted and plated on plates of LB agar containing 3% NaCl plus streptomycin and 120 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to determine the exact ratio in the inocula. Mice were prepared by following the streptomycin treatment protocol outlined above. Mice were inoculated with 100 μl of the appropriate bacterial suspension as previously described. In order to evaluate strain persistence within the mouse intestine, fresh fecal pellets (2 to 4 per animal) were collected at daily intervals. The pellets were weighed and then placed in 2 ml of sterile PBS for mechanical homogenization. Diluted samples were plated for CFU on LB agar containing 3% NaCl plus streptomycin and X-Gal for a blue (WBWlacZ) versus white (wild-type or deletion mutant strain) screen of colonies after incubation at 37°C overnight. In a separate experiment, mice were treated and infected as described above and then sacrificed 24 h postinfection, and the entire gastrointestinal tract was harvested from each mouse. Samples were placed in 8 ml of sterile PBS, mechanically homogenized, serially diluted in PBS, and plated on LB-3% NaCl plus streptomycin and X-Gal for CFU determination. In vitro competition assays were performed in tandem with the in vivo assays by diluting the inoculum 1:50 into fresh LB-3% NaCl broth. This culture was incubated at 37°C for 24 h and then serially diluted and plated on LB agar with 3% NaCl and X-Gal. The competitive index (CI) was determined with the following equation: CI = ratio out(mutant/wild-type)/ratio in(mutant/wild-type). A CI of >1 indicates that the test strain has the ability to outcompete the wild-type strain, while a CI of <1 indicates that the test strain is less fit than the wild-type strain. “Ratio out” refers to the ratio of deletion mutant to WBWlacZ colonies recovered from the mouse intestine, while “ratio in” refers to the ratio of the deletion mutant to WBWlacZ colonies enumerated from the inoculum.
Bacterial localization.
To elucidate where in the gastrointestinal tract V. parahaemolyticus was colonizing, the small intestine, cecum, and large intestine were analyzed separately for colonizing bacteria. Mice and inocula were prepared as outlined above. Streptomycin-treated mice were infected with either the wild-type strain or the ΔrpoN strain. Twenty-four hours postinfection, mice were sacrificed by CO2 asphyxiation, and the entire gastrointestinal tract was removed and separated into the small intestine, cecum, and large intestine. Individual organs were placed into 4 ml of sterile PBS, mechanically homogenized, and serially diluted in PBS. Serially diluted samples were plated for CFU on LB-streptomycin selection plates and incubated at 37°C overnight.
Mucus extraction.
Intestinal mucus was extracted from the gastrointestinal tract of mice as follows. Mice were orally treated with streptomycin as outlined above to mimic the in vivo mouse experiments. Twenty-four hours after antibiotic treatment, mice were sacrificed by CO2 asphyxiation and their GI tracts were dissected. Mucus was collected by flushing the cecum with PBS to remove its contents and then by gently scraping the walls of the intestine to remove the mucus. Extracts from small intestine, cecum, or large intestine were also collected. Extracted mucus was stored at −80°C until further use. For growth assays, 200 mg of mucus was diluted in 5 ml of PBS, homogenized, and centrifuged for 10 min at 500 × g to pellet out tissue and fecal materials. To determine the ability of strains to utilize mucus as a sole carbon source, the small intestine, cecum, or large intestine mucus was added to M9 medium at a final concentration of 30 μg of protein/ml (42, 43).
Mucus and carbon utilization.
Vibrio parahaemolyticus strains (wild type and WBWlacZ, ΔfliAP, ΔfliAL, and ΔfliAPΔfliAL mutants) were grown overnight in M9 medium supplemented with 3% NaCl and 0.4% glucose (M9G medium), and the ΔrpoN strain was grown in M9G supplemented with 2 mM glutamine to compensate for the defect in the mutant's ability to utilize ammonium present in M9 as a sole nitrogen source (32). Overnight cultures were pelleted by centrifugation for 10 min at 4,000 × g. Bacterial pellets were washed once with M9 and then resuspended in M9 with no carbon source. Cultures were diluted 1:40 into fresh M9 supplemented with 3% NaCl and either gluconate (0.4%), ribose (0.4%), arabinose (0.4%), mannose (0.4%), glucose (0.4%), or mouse intestinal mucus (30 μg/ml). The rpoN mutant in these experiments was also supplemented with 2 mM glutamine. Diluted cultures were transferred to a 96-well microtiter plate and incubated at 37°C with shaking. Optical densities at 595 nm were taken hourly for a total of 24 h using a Tecan Sunrise microplate reader and Magellan plate reader software. Doubling times for each strain and carbon source were calculated by dividing the time interval used (min) by the number of generations (n): n = ln(OD2/OD1)/ln2, where OD1 is the optical density of the culture at the start of the log phase and OD2 is the optical density of the culture at the end of the log phase. Each experiment was performed in triplicate with at least two biological replicates.
RNA extraction and qPCR.
Vibrio parahaemolyticus strains were grown for 4 h in LB-3% NaCl and then diluted 1:50 into M9 medium containing no ammonium and supplemented with cecal mucus as a sole carbon and nitrogen source. For these experiments, in order to best mimic in vivo conditions, M9 was made without the addition of ammonium. Therefore, in these experiments all bacterial strains tested were forced to utilize the supplemented mucus as the sole carbon and nitrogen source. To determine expression patterns of carbon metabolism genes in mucus, we examined early-exponential-phase cultures; therefore, all cultures were grown to an OD595 of 0.15. Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. The RNA samples were quantified using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA) and subsequently treated with Turbo DNase (Invitrogen) per the manufacturer's instructions. cDNA was synthesized using Superscript II reverse transcriptase (RT) (Invitrogen) according to the manufacturer's protocol, using 500 ng of RNA as a template and priming with 200 ng of random hexamers. cDNA samples were then diluted 1:25 and used for quantitative real-time PCR (qPCR). Real-time PCRs used the HotStart-IT SYBR green qPCR master mix (USB, Santa Clara, CA) and were run on an Applied Biosystems 7500 fast real-time PCR system (Foster City, CA). Gene-specific primers were designed using Primer 3 software according to the real-time PCR guidelines and are listed in Table 2. Data were analyzed using Applied Biosystems 7500 software. Expression levels of each gene, as determined by their cycle threshold (CT) values, were normalized using the 16S rRNA gene to correct for sampling errors. Differences in the ratios of gene expression were determined using the ΔΔCT method (44).
Mucus competition assays.
In vitro competition assays were performed in mouse intestinal cecal mucus. Mixed cultures containing the WBWlacZ strain and test strains were prepared by following the mouse inoculum protocol outlined above. Intestinal mucus was diluted to a final concentration of 30 μg/ml of protein in M9 medium containing no ammonium. Again, for these experiments, the ammonium was removed from the M9 medium so that neither the wild type nor the deletion mutants had an advantage over the rpoN mutant, which is defective in utilizing ammonium as a nitrogen source. Forcing the bacteria to rely solely on the supplied mucus for carbon and nitrogen also closely mimics the conditions experienced by the strain in the mouse intestine. Mixed cultures of wild-type and deletion mutant strains were then diluted 1:50 into the M9 (no ammonium) medium supplemented with mucus and incubated aerobically overnight at 37°C. After 24 h, cultures were serially diluted and plated on LB-3% NaCl plus X-Gal, and the CI values were calculated as outlined above. For these experiments the rpoN mutant cultures were not supplemented with glutamine; thus, they were grown similarly to the other deletion mutant strains where the supplied mucus served as the sole carbon and nitrogen sources.
RESULTS
Growth analysis of ΔrpoN strain.
RpoN has been shown to be a global regulator in a number of Gram-negative bacteria, where it is known to regulate motility, nitrogen assimilation, quorum sensing, and virulence (30–33, 37–40, 45, 46). VP2670 encodes the RpoN homologue in V. parahaemolyticus RIMD2210633 and shared 81% identity with RpoN (VC2529) from V. cholerae N16961. To determine the role of this regulator in V. parahaemolyticus pathogenesis, we constructed an in-frame deletion in VP2670. We created an unmarked 465-bp truncated version of the rpoN gene in V. parahaemolyticus RIMD2210633 using SOE PCR and homologous recombination.
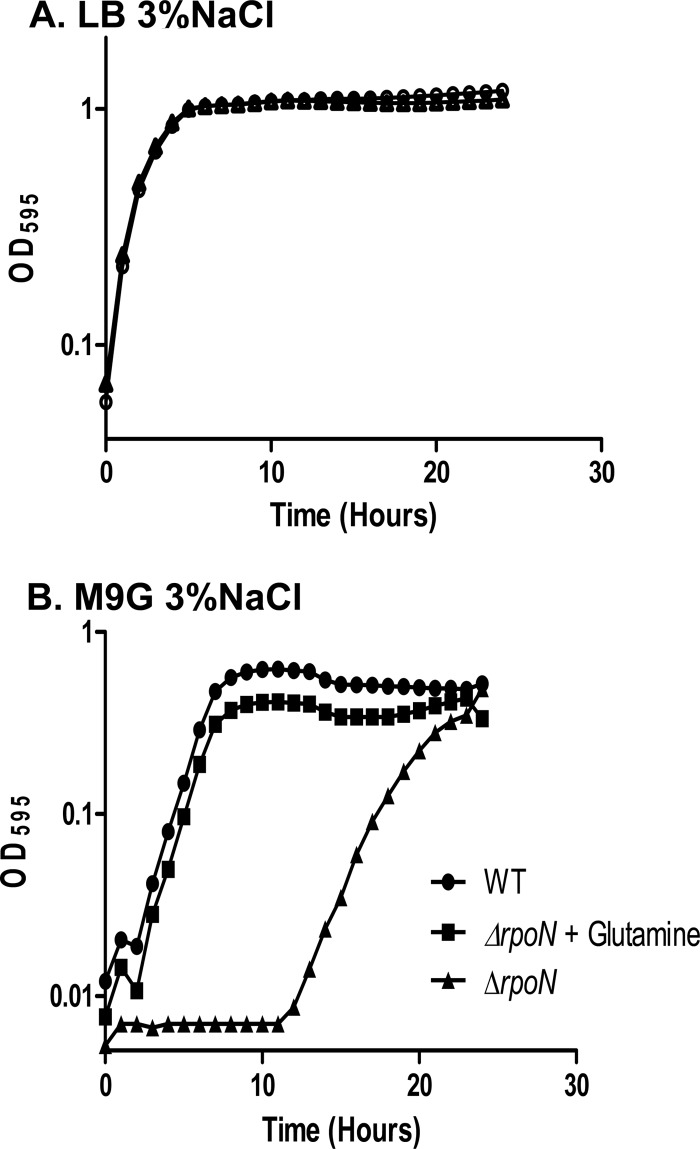
To ensure that the rpoN mutant had no general growth defects, we compared growth of the mutant to that of the wild-type strain grown under standard laboratory conditions (LB-3% NaCl) and found that the ΔrpoN strain exhibited no defects in growth relative to the wild-type parent strain (Fig. 1A). We also compared the growth of the ΔrpoN strain to that of the wild-type strain in an M9G medium and found that the ΔrpoN strain exhibited a substantial lag phase of approximately 11 h compared to the wild-type strain before reaching the same biomass as the parent strain (Fig. 1B). To demonstrate that the growth of the mutant in M9G was not the result of a suppressor mutation, we took cells of the rpoN mutant grown overnight in M9G and inoculated them into new media and found a similar lag time with these cultures. The 11-h lag phase could be reduced to the wild-type level by the addition of 2 mM glutamine to the medium (Fig. 1B). This suggests that the V. parahaemolyticus rpoN mutant has a defect in the synthesis of glutamine synthetase (encoded by VP0121), an enzyme that synthesizes glutamine from the ligation of glutamate and ammonia. While RpoN deletions in some Gram-negative bacteria result in glutamine auxotrophy (46–48), our data suggest that RpoN is important but not absolutely necessary, since delayed growth of the rpoN mutant was observed in minimal medium with ammonium as the sole nitrogen source. This phenotype is also observed in other Vibrio species, such as V. cholerae, V. fischeri, and V. alginolyticus (32, 38, 40), which implies that in Vibrio spp., glutamine synthetase is basally transcribed independent of RpoN. However, under certain conditions, such as growth in M9G medium containing ammonium as a sole nitrogen source, RpoN is required for maximal expression of glutamine synthetase.
FIG 1.
Growth characteristics of wild-type and ΔrpoN strains. (A) Both the wild-type (circle) and ΔrpoN (triangle) strains were grown aerobically at 37°C in LB medium with 3% NaCl. (B) Both the wild-type (circle) and ΔrpoN (triangle) strains were grown aerobically at 37°C in M9 minimal medium with 3% NaCl and 0.4% glucose (M9G). Additionally, the ΔrpoN strain was also grown in M9G plus 2 mM glutamine (square). All cultures were grown in triplicate, and each experiment was performed at least twice. Error bars, indicating standard deviations, are too small to be observed.
RpoN controls swimming and swarming motility in V. parahaemolyticus RIMD2210633.
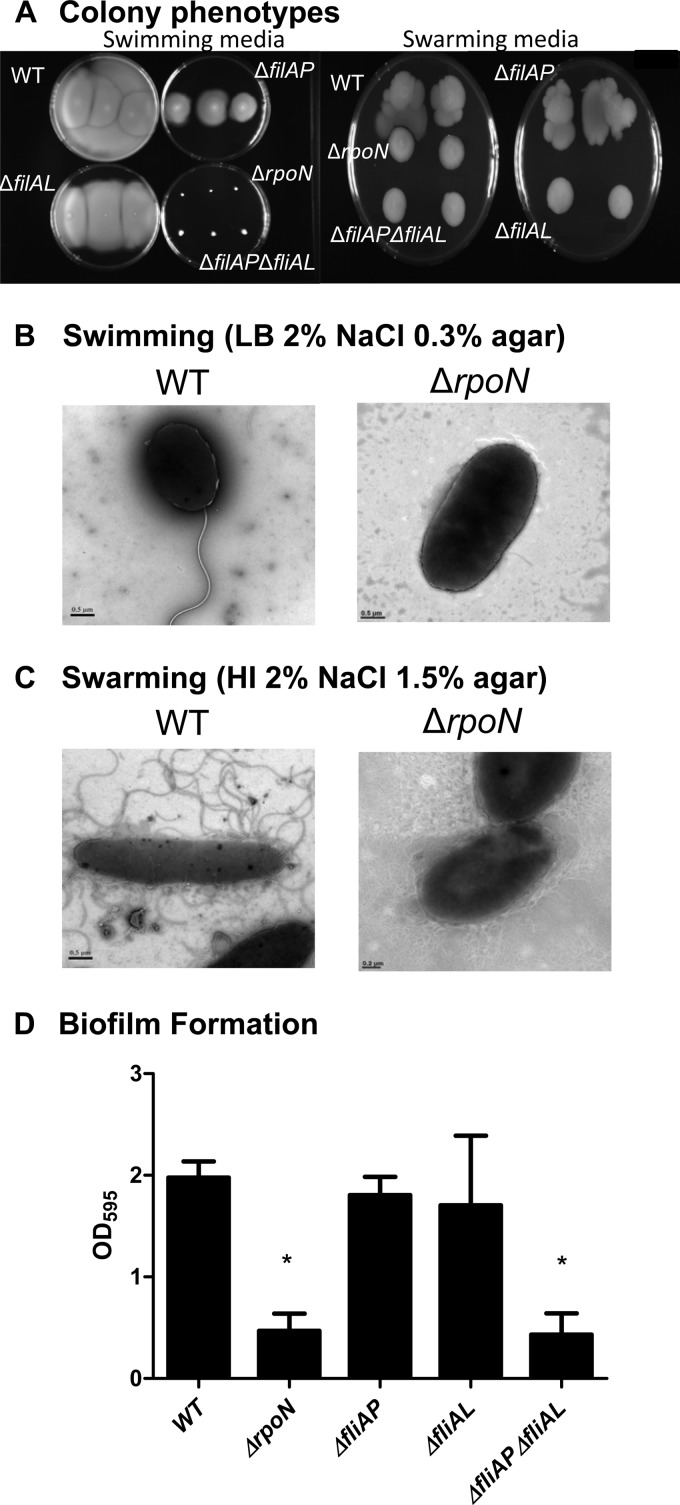
The role of RpoN in the regulation of flagellar synthesis has been studied in a number of Vibrio species (32, 38–40, 49, 50). Vibrio parahaemolyticus produces both a polar flagellum for movement through liquid medium (swimming) and a lateral flagellar system for movement over solid surfaces (swarming) (49, 50). We compared the ability of the wild-type and the rpoN mutant strains to swim and swarm. A colony of the wild-type strain stabbed onto LB plates containing 0.3% agar and incubated overnight showed the initial inoculum spreading throughout the plate, which is indicative of swimming (Fig. 2A). However, when a colony of the ΔrpoN strain was examined under the same conditions, there was no movement beyond the initial inoculum site (Fig. 2A). These data confirm that the rpoN mutant does not have the ability to swim in this medium. Similarly, when a colony of the wild-type strain was inoculated onto HI-1.5% agar plates and incubated, the colony growth expanded in an asymmetrical pattern, indicative of swarming behavior. In contrast, when the ΔrpoN strain was examined under the same conditions, the ΔrpoN colonies that formed were round and symmetrical, which indicates that this strain does not have the ability to swarm (Fig. 2A). Additionally, we compared the ability of the wild-type and ΔrpoN strains to produce flagella under both swimming (Fig. 2B) and swarming (Fig. 2C) conditions and confirmed that no flagella were produced in the rpoN mutant. The defects in swimming and swarming in the ΔrpoN mutant are due to the loss of polar and lateral flagella, respectively, as indicated by transmission electron microscopy (Fig. 2B and C).
FIG 2.
Examination of motility, flagellar synthesis, and biofilm production of V. parahaemolyticus RIMD2210633 and an rpoN deletion mutant. (A) To examine the motility phenotypes of the ΔrpoN, ΔfliAP, ΔfliAL, and ΔfliAP ΔfliAL strains, bacterial colonies were stabbed onto an LB plate containing 2% NaCl and 0.3% agar to assess swimming. Plates were incubated at 37°C, and photographs were taken after 24 h. To examine swarming, strains were grown on the surface of a heart infusion plate containing 2% NaCl and 1.5% agar. The plates were incubated at 30°C, and photographs were taken after 60 h. Electron microscopy was used to examine the production of polar and lateral flagella in wild-type and ΔrpoN mutant strains grown in swimming (B) and swarming (C) media. (D) To examine biofilm production, the wild-type strain and sigma factor deletion mutants were grown overnight aerobically in LB supplemented with 3% NaCl at 37°C. Overnight cultures were diluted into fresh LB-3% NaCl and allowed to grow statically for 24 h at 37°C in a 96-well plate. After such time, the culture was removed and adherent cells were stained with crystal violet. The crystal violet was solubilized in DMSO, and the OD595 was determined. Bars represent the OD595 of each strain. All cultures were grown in triplicate, and each experiment was performed at least twice. Error bars indicate standard errors. P values were calculated using an unpaired Student t test with a 95% confidence interval. Asterisks denote significant differences between biofilm production of the mutant strains and that of the wild-type strain. *, P < 0.01.
We also examined the effects of deletion of the rpoN gene on the bacterium's ability to form biofilms. We found that the ΔrpoN and ΔfliAP ΔfliAL strains produced significantly less (P < 0.01) biofilm than the wild-type strain after static growth for 24 h in LB-3% NaCl (Fig. 2D).
Loss of RpoN enhances V. parahaemolyticus RIMD2210633 colonization.
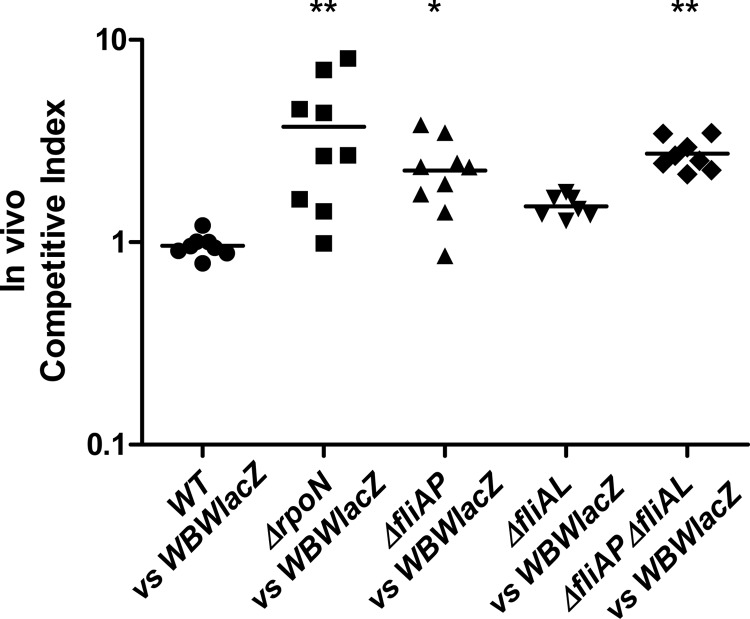
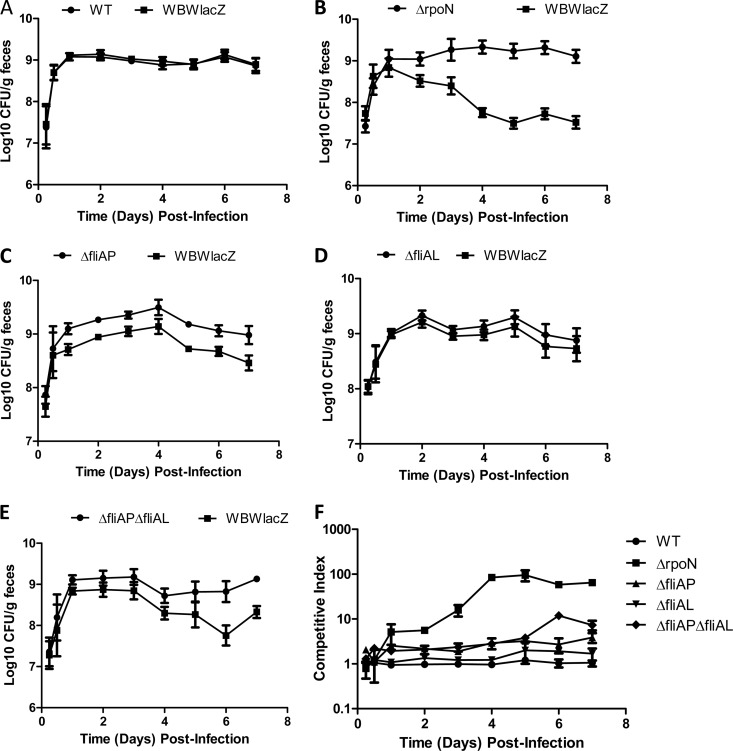
Studies on a number of enteric pathogens have shown that motility is an important phenotype for colonization and infection (32, 33, 39, 40, 51–54). To determine whether RpoN was essential for colonization in V. parahaemolyticus pathogenesis, we used the streptomycin-treated mouse model of colonization (28). For this analysis, a β-galactosidase-positive RIMD2210633 strain named WBWlacZ was used, which was previously demonstrated to behave similarly to the wild type in vitro and in vivo (28). We examined the ability of WBWlacZ and the rpoN mutant to cocolonize the intestinal tract of streptomycin-pretreated adult mice. An in vivo competition assay in adult C57BL/6 mice was performed by pretreating mice with an orogastric dose of streptomycin (20 mg/mouse) 24 h prior to orogastric coinoculation with a mixture of 5 × 108 CFU V. parahaemolyticus WBWlacZ and 5 × 108 CFU of either the wild-type strain (n = 8) or the ΔrpoN mutant (n = 9). The wild-type and WBWlacZ strains did not outcompete each other in vivo (CI = 0.96) (Fig. 3), indicating that insertion of the lacZ gene is neither deleterious nor beneficial, confirming previously reported values (28). In in vitro assays, the wild-type versus WBWlacZ strain comparison had a CI of 1, as previously shown (28). Conversely, the rpoN mutant significantly (P < 0.01) outcompeted the WBWlacZ strain by 3.8-fold in vivo (Fig. 3). This indicates that the ΔrpoN strain has increased fitness in vivo compared to the wild-type strain.
FIG 3.
In vivo competition assays. A 1:1 mixed culture of WBWlacZ and test strain (wild type or deletion mutants) was used to orogastrically infect streptomycin-treated adult mice. CFU were calculated 24 h postinfection from the entire gastrointestinal tracts using blue/white colony selection. Data are pooled from two separate experiments and reported as competitive index (CI), which is calculated as CI = ratio out(test strain/WBWlacZ)/ratio in(test strain/WBWlacZ). The solid lines indicate the means. P values were calculated using a Kruskal-Wallis one-way ANOVA followed by a Dunn multiple-comparison posttest. Asterisks denote significant differences between the CI of the mutant strains and that of the wild-type strain. *, P < 0.05; **, P < 0.01.
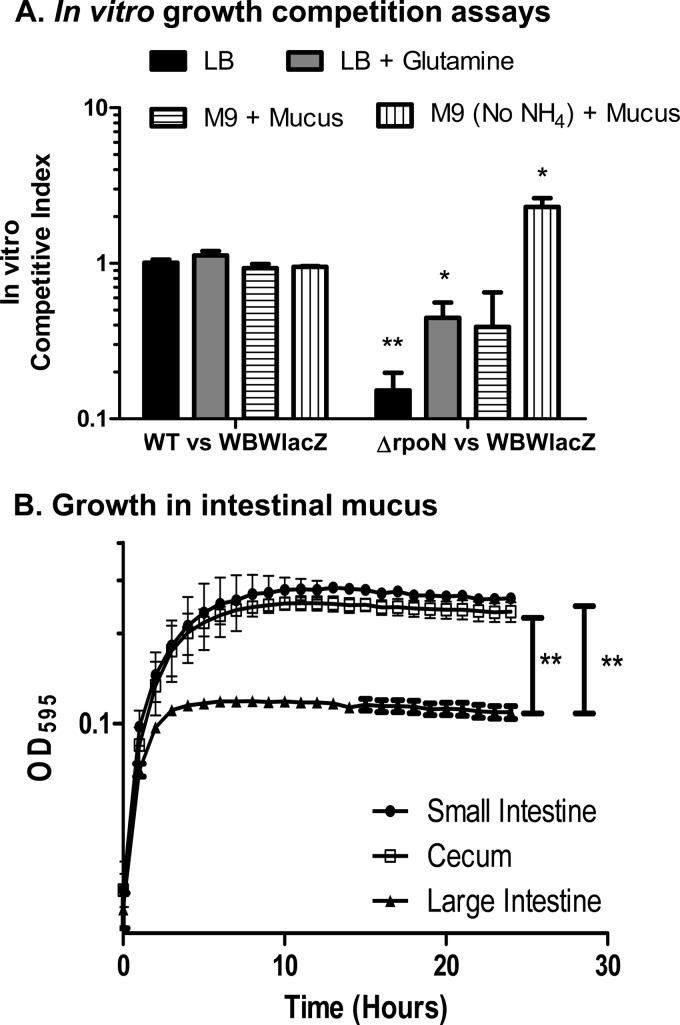
In vitro competition assays were performed between these strains in LB broth, and in these assays WBWlacZ outcompeted the rpoN mutant with a CI of 0.25 (Fig. 4A). We believe that this defect in vitro most likely is a result of the ΔrpoN strain's defect in glutamine synthetase and that this defect is exacerbated when it must compete with WBWlacZ under in vitro conditions. To examine this further, in vitro competition assays were performed in LB supplemented with glutamine. In this assay, the mutant made a recovery but was still outcompeted by the WBWlacZ strain (Fig. 4A). To mimic in vivo conditions more closely, in vitro competition assays were performed in M9 (with and without ammonium) supplemented with intestinal mucus. For these assays, the ability of V. parahaemolyticus to grow on mouse intestinal mucus as a sole nutrient source was first assessed. Mucus is the main component of mucous membranes and is the primary source of nutrients for most intestinal microbiota (55, 56). We collected mucus from streptomycin-treated mice from the small intestine, cecum, and large intestine to mimic in vivo conditions used in this study. In in vitro competition assays, V. parahaemolyticus was grown overnight in M9 supplemented with small intestine, cecum, or large intestine mucus (30 μg/ml of mucus protein) as a sole carbon source at 37°C. Growth occurred on all three mucus extracts, but the bacterium grew to significantly higher optical densities on M9 supplemented with either small intestine mucus or cecal mucus compared to its growth on large intestine mucus (P < 0.001) (Fig. 4B). In M9 supplemented with mucus, the rpoN mutant was outcompeted but not to the same extent as in LB (Fig. 4A). In M9 medium (no ammonium) supplemented with mucus, the rpoN mutant was able to outcompete the WBWlacZ strain with a CI of 2.3 (Fig. 4A). These results suggest that both strains obtain nitrogen from mucus and that the glutamine synthetase defect in the ΔrpoN strain does not play a significant role in vivo.
FIG 4.
V. parahaemolyticus in vitro competition assays and growth in mouse intestinal mucus. (A) In vitro competition assays showing competitive indices in LB-3% NaCl broth with and without 2 mM added glutamine or in M9 3% NaCl medium supplemented with mucus either with or without ammonium as the nitrogen source. (B) Wild-type V. parahaemolyticus was grown overnight at 37°C aerobically in M9 3% NaCl supplemented with either small intestine mucus (circles), cecal mucus (squares), or large intestine mucus (triangle), and growth was determined for 24 h at OD595. All mucus samples were adjusted to contain 30 μg/ml of protein. The final OD595 was used to compare the growth potential on each substrate. Error bars indicate standard errors. P values were calculated using an unpaired Student t test with a 95% confidence interval. Asterisks denote significant differences between the competitive index of the ΔrpoN strain and the competitive index of the wild-type strain (A) or the final OD595 of the wild-type strain grown utilizing mucus isolated from the small intestine, cecum, or large intestine (B). *, P < 0.05; **, P < 0.001.
To begin to determine how the rpoN mutant outcompeted the wild-type strain in vivo, we performed single-infection in vivo mouse colonization experiments to compare the localization of the ΔrpoN mutant to that of the wild-type strain. Streptomycin-treated mice were given an oral dose of 1 × 109 CFU of either the wild-type or ΔrpoN strain. Twenty-four hours postinfection, the mice were sacrificed and their small intestine, cecum, and large intestine were harvested separately and plated for CFU. We found that the ΔrpoN strain colonized the mouse small intestine at an approximately 2.5-fold higher level than the wild type (P < 0.05). However, there was no significant difference (P > 0.05) between the amount of ΔrpoN and WBWlacZ strains recovered from the cecum and large intestine (1.2- and 1.6-fold, respectively) (data not shown).
We next examined whether the superior ability of the ΔrpoN strain to colonize the mouse intestine was dependent upon the inability of the ΔrpoN strain to synthesize polar, lateral, or both flagellar systems. In V. parahaemolyticus, the sigma factor FliAP is required for the synthesis of the single-sheath flagellum and requires sodium motive force to swim in liquid media (49, 57). FliAL, encoded by lafS, regulates the synthesis of the lateral flagella, which requires energy from the proton motive force to drive swarming motility on solid media (50, 58, 59). We examined the expression level of fliAP in the rpoN mutant background and, as expected, we found highly reduced expression (data not shown). In order to test whether a lack of polar or lateral flagella alone played a role in the superior colonization of the rpoN mutant, we constructed single deletion mutations in fliAP (VP2232) and fliAL (VPA1555) sigma factor genes and in a double mutant lacking fliAP and fliAL genes. As expected, we confirmed that the ΔfliAP strain did not produce a polar flagellum and was defective in swimming motility but exhibited the same phenotype as the wild-type strain on swarming plates (Fig. 2A). The ΔfliAL strain exhibited wild-type levels of swimming motility but was unable to swarm, whereas the double deletion mutant showed defects in both swim and swarm motility (Fig. 2A). Neither the ΔfliAP nor ΔfliAL strain exhibited defects in the ability to produce biofilm. However, as previously mentioned, the ΔrpoN and ΔfliAP ΔfliAL strains were defective in biofilm production compared with the wild-type strain (P < 0.01) (Fig. 2D), indicating that motility, either by polar or lateral flagella, is important for biofilm production and that a defect exhibited by the ΔrpoN mutant in biofilm production may be motility related.
The colonization ability of the ΔfliAP, ΔfliAL, and ΔfliAP ΔfliAL strains was determined in vivo via competition assay using our streptomycin-treated adult mouse model. Like the ΔrpoN strain, the ΔfliAP and ΔfliAP ΔfliAL strains were able to significantly outcompete the WBWlacZ strain, with ΔfliAP and ΔfliAP ΔfliAL strains having competitive indices of 2.3 (P < 0.05) and 2.7 (P < 0.001), respectively (Fig. 3). The fliAL mutant was not significantly different from the WBWlacZ strain. Under in vitro conditions, all 3 motility mutant strains were found to have a CI close to 1. Even though the polar mutants outcompeted WBWlacZ in vivo, they did not do so to the same level as the rpoN mutant, suggesting that other factors are involved in its superior colonization.
The RpoN mutant shows superior colonization in long-term persistence competition assays compared to WBWlacZ.
To determine whether the deletion mutants exhibit different phenotypes during the later stages of colonization, in vivo long-term persistence assays in the mouse intestine were performed. To accomplish this, streptomycin-treated mice were coinfected with WBWlacZ and either the wild-type, ΔrpoN, ΔfliAP, ΔfliAL, or ΔfliAP ΔfliAL strain. The Vibrio levels within the intestine were monitored by collecting and subsequently plating feces from infected mice daily for 7 days. This experiment confirmed that the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL mutants (Fig. 5B, C, and E) were able to outcompete the WBWlacZ strain starting at 24 h postinfection and continuing through day 7. There was no significant difference between the wild-type and WBWlacZ strain at any point during the 7 days of fecal monitoring (Fig. 5A and F). Similarly, a comparison of WBWlacZ to ΔfliAL strains exhibited a CI of approximately 1 at 24 h and throughout the 7 days, indicating that in this model, lateral flagella are not required for colonization (Fig. 5D and F). In the 7-day persistence assays, the ΔrpoN strain was significantly more fit in the mouse intestine than any of the other strains tested, exhibiting a CI shift from approximately 2 at 24 h postinfection to approximately 70 at 7 days postinfection (Fig. 5B and F). The ΔfliAP strain outcompeted the WBWlacZ strain with a CI of approximately 3 through the 7 days in the mouse intestine (Fig. 5C and F). The ΔfliAP ΔfliAL strain outcompeted the WBWlacZ strain with a competitive index of approximately 10 by 7 days postinfection (Fig. 5E and F). These results suggest that the lack of FliAP and a polar flagellum gives a competitive advantage in vivo. However, in all in vivo experiments, the ΔrpoN strain outcompeted the WBWlacZ strain at much higher levels than either the ΔfliAP or ΔfliAP ΔfliAL strains, suggesting that while loss of motility likely contributes to the colonization phenotype of all three strains, it is clear that RpoN controls additional factors that are important for host colonization.
FIG 5.
In vivo persistence competition assays. Groups of streptomycin-treated mice were orally dosed with a mixed inoculum consisting of the WBWlacZ strain and either the wild-type (A), ΔrpoN (B), ΔfliAP (C), ΔfliAL (D), or ΔfliAP ΔfliAL (E) strain. At the indicated intervals postinfection, fecal pellets were collected from mice, weighed, and homogenized in PBS. Samples were serially diluted and plated on LB agar containing 3% NaCl and X-Gal to determine the number of V. parahaemolyticus organisms per gram of fecal material. Data are pooled from two separate experiments and reported as log-transformed CFU/gram of feces (A to E) or as the competitive index (F), which is calculated as CI = ratio out(test strain/WBWlacZ)/ratio in(test strain/WBWlacZ). Error bars indicate standard errors.
Growth in mouse intestinal mucus and mucus components.
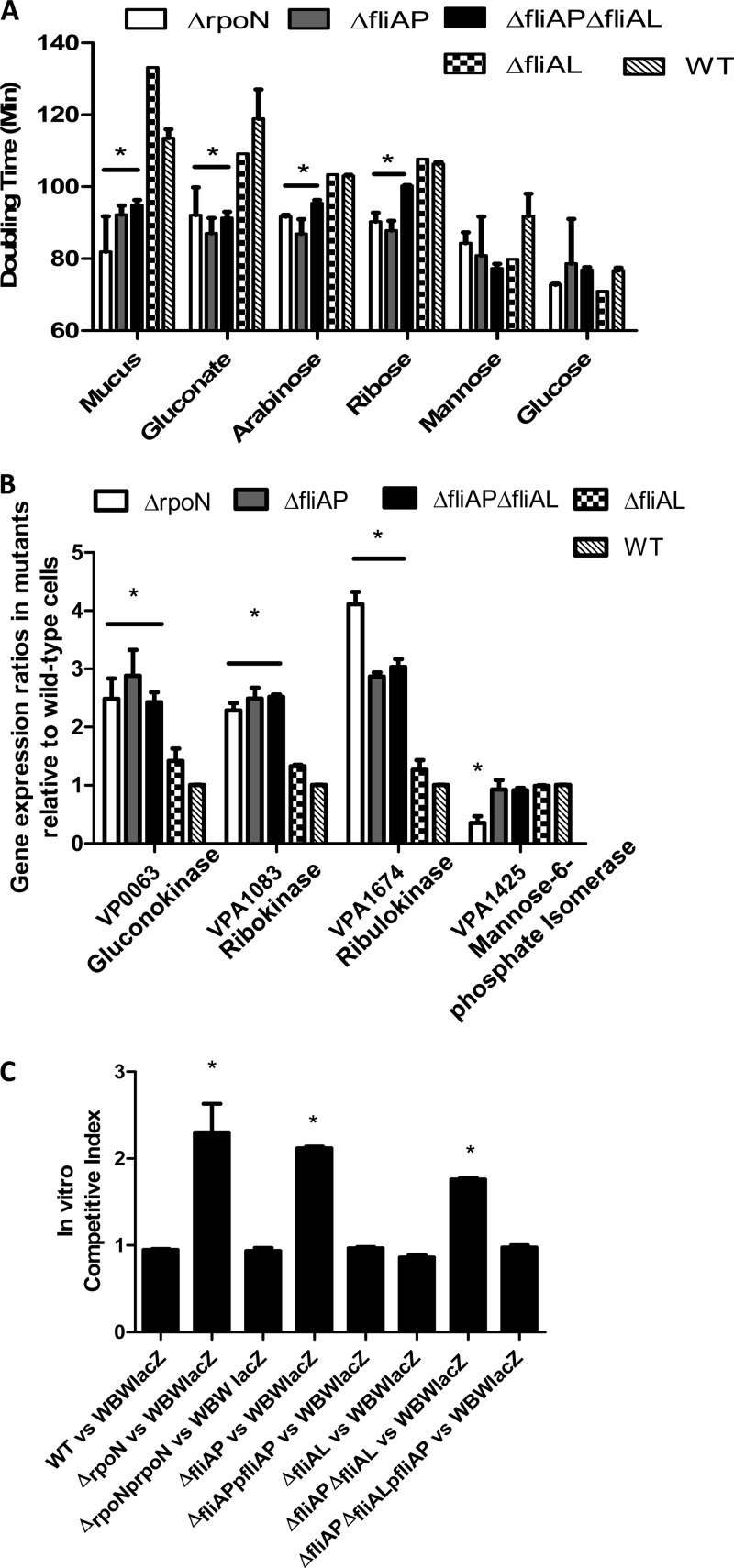
One of the barriers to pathogen colonization of the gut for many species is competition from the resident microbial populations for nutrients (60, 61). We hypothesized that one of the ways our rpoN mutant is outcompeting WBWlacZ is by the more efficient in vivo utilization of carbon sources. To explore this, the doubling time of each of the mutants was examined in M9 supplemented with mucus where, for the rpoN mutant assays, glutamine was also added to M9. The ΔrpoN, ΔfliAP, ΔfliAPΔfliAL, ΔfliAL, and wild-type strains exhibited average doubling times of 81.9 min, 92.2 min, 94.3 min, 130.0 min, and 113.5 min, respectively (Fig. 6A). The doubling times for the ΔrpoN, ΔfliAP, and ΔfliAPΔfliAL strains were significantly faster (P < 0.05) than those for the wild-type strain (Fig. 6A).
FIG 6.
Growth characteristics of V. parahaemolyticus on cecal mucus. (A) Doubling times of wild-type and mutant strains. Overnight cultures of the ΔrpoN, ΔfliAP, ΔfliAPΔfliAL, ΔfliAL, and wild-type strains were grown aerobically in M9 medium supplemented with either cecal mucus (30 μg/ml of protein), gluconate (0.4%), mannose (0.4%), arabinose (0.4%), ribose (0.4%), or glucose (0.4%) for 24 h, and growth was determined at OD595. Doubling times were calculated as mentioned in Materials and Methods. (B) Expression analysis of wild-type and mutant strains. RNA was extracted on two separate occasions from wild-type and mutant ΔrpoN, ΔfliAP, ΔfliAL, and ΔfliAP ΔfliAL cells grown in cecal mucus and analyzed by qPCR in duplicate for each biological replicate. Bars represent the expression of the gluconokinase (VP0063), ribokinase (VPA1083), ribulokinase (VP1674), and mannose-6-phosphate isomerase (VPA1425) normalized to 16S rRNA and are relative to levels in the wild-type cells. (C) In vitro competitive indices of strains grown on mucus. Mixed cultures of WBWlacZ and ΔrpoN, ΔfliAP, ΔfliAL, ΔfliAP ΔfliAL, or complemented strains were grown for 24 h in M9 medium supplemented with cecal mucus. The competitive index for each strain relative to the WBWlacZ strain was calculated. Each experiment was performed in triplicate with at least two replicates. Errors bars indicate standard errors. P values were calculated using an unpaired Student t test with a 95% confidence interval. Asterisks denote significant differences between the doubling times of the mutant strains and the doubling times of the wild-type strain (A), gene expression in the mutant strains relative to the wild-type strain (B), or competitive index of mutant strains compared to the competitive index of the wild-type strain (C). *, P < 0.05.
To ascertain if any of the individual major carbon components in mucus were contributing more than others to the mutants' superior growth, we compared the growth of all strains in M9 medium supplemented with gluconate, arabinose, ribose, mannose, or glucose as the sole carbon source, and for the rpoN mutant assays, glutamine was also added to M9 (Fig. 6A). When cells were grown in M9 medium with gluconate as the sole carbon source, the ΔrpoN, ΔfliAP, and ΔfliAPΔfliAL strains had average doubling times of 92.1 min, 87.0 min, and 91.3 min, respectively, which were significantly faster (P < 0.05) than the 118.9-min doubling time of the wild-type strain (Fig. 6A). More similar to the wild-type strain, the ΔfliAL strain had a doubling time of 107.9 min (P > 0.05). When cells were grown using arabinose as the sole carbon source, the ΔrpoN, ΔfliAP, and ΔfliAPΔfliAL strains had average doubling times of 91.2 min, 86.8 min, and 95.4 min, respectively, which were significantly faster (P < 0.05) than the 104.8-min and 103.2-min doubling times of the ΔfliAL and wild-type strains, respectively (Fig. 6A). With ribose as the sole carbon source, the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains had average doubling times of 90.5, 87.8, and 100.0 min, respectively, which were again significantly shorter (P < 0.05) than those of the ΔfliAL and wild-type strains, with average doubling times of 110.9 and 106.3 min, respectively (Fig. 6A). We found no significant difference between the generation times of the wild-type strain and the mutant strains when grown in minimal medium containing glucose or mannose as sole carbon sources (Fig. 6A). Taken together, these data demonstrate that the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains have higher growth rates than the wild-type strain when grown on mucus, and these strains also exhibit higher growth rates on gluconate, arabinose, and ribose as sole carbon sources, which are the major carbon sources present in mucus.
Loss of RpoN and FliAP derepresses key metabolic genes.
Given that the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains had higher growth rates in mouse intestinal mucus as well as in a number of carbon sources commonly present in mucus, we hypothesized that these sigma factors act as repressors of certain metabolic pathways. Therefore, the expression patterns of genes encoding gluconokinase (VP0063), ribokinase (VPA1083), ribulokinase (VPA1674), and mannose-6-phosphate isomerase (VPA1475), required for gluconate, ribose, arabinose, and mannose catabolism, respectively, were examined during growth in mucus. For these expression experiments, M9 lacking both ammonium and glutamine was used for all strains examined, ensuring the only source of nitrogen for all strains was mucus. Higher expression was found for the genes encoding the metabolic enzymes gluconokinase, ribokinase, and ribulokinase for the hypercolonizing mutants than for the wild type (Fig. 6B). Gluconokinase (encoded by VP0063) catalyzes the conversion of d-gluconate to 6-phospho-d-gluconate, which can then be shuttled to the pentose phosphate pathway or the Entner-Doudoroff pathway. We analyzed expression levels of this gene in both the wild-type strain and the mutant strains grown to an OD of 0.15 in M9 supplemented with mucus. The expression levels were approximately 2.5-, 2.9-, and 2.8-fold higher in the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains, respectively, than in the wild-type strain (P < 0.05) (Fig. 6B). Ribokinase (encoded by VPA1083) is the enzyme responsible for converting d-ribose to d-ribose-5-phosphate, which can be utilized by the cell via the pentose phosphate pathway. The expression levels of this gene were approximately 2.3-, 2.5-, and 2.5-fold higher in the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains, respectively (P < 0.05), compared to the wild-type strain (Fig. 6B). Ribulokinase (encoded by VPA1674) catalyzes the conversion of l-ribulose (which is generated via the isomerization of l-arabinose) into l-ribulose-5-phosphate, which is then converted into an intermediate of the pentose phosphate pathway. Expression of this gene was approximately 4-, 2.9-, and 3-fold higher in the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains, respectively (P < 0.05), than in the wild-type strain (Fig. 6B). These expression data suggest that when grown in intestinal mucus, the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains are exhibiting higher expression of genes in the pathways for the catabolism of gluconate, arabinose, and ribose than the wild-type strain. This could mean that in wild-type cells, either RpoN or FliAP acts, either directly or indirectly, as a repressor for these metabolic pathways. Furthermore, the higher expression patterns seen in these pathways in the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains could explain the higher growth rates (lower doubling times) observed when these strains were grown using mucus, gluconate, arabinose, and ribose as sole carbon sources and why the mutant strains can outcompete the WBWlacZ strain both in vitro and in vivo. Mannose-6-phosphate isomerase (encoded by VPA1425) catalyzes the isomerization of mannose-6-phosphate to fructose-6-phosphate. This allows cells to utilize mannose as a carbon source via glycolysis. We found no significant alteration in expression of this gene in the ΔfliAP and ΔfliAP ΔfliAL strains compared to wild-type expression levels (Fig. 6B). However, expression of VPA1425 was significantly decreased (P < 0.05) in the ΔrpoN strain, indicating that RpoN acts as a positive regulator of mannose catabolism. The ΔfliAL strain, which exhibited no difference in growth rates in mucus, did not exhibit significant differences in metabolic gene expression relative to the wild-type strain (Fig. 6B).
To assess whether the faster doubling time and increased expression of catabolic genes in mucus of the hypercolonizers leads to a competitive advantage, we performed in vitro competition assays in M9 medium supplemented with intestinal mucus. For these experiments, M9 medium lacked ammonium and glutamine for all assays, so that the bacteria were forced to derive nitrogen solely from the mucus. In these experiments, the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains were able to outcompete (P < 0.05) the WBWlacZ strain approximately 2.3-, 2.1-, and 1.8-fold, respectively (Fig. 6C). Furthermore, when the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains were complemented with functional genes, they lost the ability to outcompete the WBWlacZ strain for growth utilizing mucus as the sole nutritional source (Fig. 6C).
DISCUSSION
In an effort to better understand how V. parahaemolyticus colonizes and survives within the host, we examined a strain where the alternative sigma factor RpoN was deleted. We confirmed that loss of RpoN renders V. parahaemolyticus aflagellate and nonmotile, causing a defect in biofilm formation and likely a defect in the synthesis of glutamine from glutamate and ammonium. We also demonstrated that the ΔrpoN strain has the ability to significantly outcompete WBWlacZ in vivo in a streptomycin-treated adult mouse model of colonization. Additionally, other mutant strains defective for polar flagellar synthesis, ΔfliAP and ΔfliAP ΔfliAL strains, also demonstrated enhanced fitness in vivo but not to the same extent as the rpoN mutant. This was also true in persistence colonization assays performed over 7 days, which again showed that the ΔrpoN strain significantly outcompeted WBWlacZ. These results suggested that the ΔrpoN in vivo phenotype is not due in any large part to a lack of motility. In in vitro competition assays in minimal medium using intestinal mucus as the sole nutritional source, the rpoN, fliAP, and fliAP fliAL mutants outcompeted WBWlacZ but not to the same extent as in the in vivo competition assays. These strains also exhibited faster doubling times than the wild-type strain when grown in intestinal mucus, as well as in minimal medium supplemented with the sole carbon sources gluconate, arabinose, and ribose, which are all constituents of mucus. Overall, these results suggest that the ΔrpoN mutant in vivo phenotype is due in part to carbon utilization along with unidentified factors under the control of the RpoN regulon.
The role RpoN plays in gene regulation and host colonization has been studied in V. cholerae as well as in a number of other Vibrio species. In V. cholerae, the RpoN regulon encompasses nearly 500 genes, most of which are independent of flagellar synthesis (33). In addition to its role in the regulation of polar flagellum synthesis in V. cholerae, RpoN has been shown to play a role in growth on ammonium as the sole nitrogen source, the regulation of the type VI secretion system, and the regulation of quorum sensing. In addition, it was found that rpoN deletion mutants exhibited different growth patterns than the wild-type strain on a number of carbon sources, suggesting a role in carbon metabolism (26, 31–33, 35, 36, 62). Deletion of rpoN in V. cholerae also resulted in a strain that exhibited a nearly 30-fold decrease in competitive fitness compared to the wild-type strain using the cholera infant mouse model of colonization (32). FliAP has also been studied in V. cholerae and has been shown to regulate approximately 330 genes (33). The fliAP mutants in V. cholerae were nonmotile, demonstrated an increase in virulence gene expression and decreased biofilm production, and were able to differentially utilize various carbon sources compared to the wild-type strain (33). As with the rpoN deletion strains, V. cholerae that lacks a functional FliAP displays a reduced competitive index in the infant mouse model (54).
We recently introduced a new animal model for the study of V. parahaemolyticus colonization and survival within the host (28). This model relies on the streptomycin pretreatment of adult mice to facilitate the colonization of the mice by V. parahaemolyticus. Through this model, we demonstrated that the two-component system ToxRS and the outer membrane protein OmpU were important survival factors for this organism in vivo, whereas T3SS-1 and T3SS-2 were not required for in vivo colonization (28). Here, we report that deletion of rpoN, fliAP, or a double deletion of fliAP fliAL results in strains that have increased fitness and a significant competitive advantage in vivo. This is in contrast to deletions of rpoN in V. cholerae, V. anguillarum, and V. fischeri, which all have deleterious effects on the ability of these strains to colonize their particular model host organism (32, 39, 40). However, it has been shown that loss of motility in biotype El Tor V. cholerae strains results in decreased CIs in vivo, whereas nonmotile classical biotype strains colonize as well as the wild-type strains (54, 63, 64).
There is evidence that a defect in motility can contribute to in vivo survival. Studies by Leatham et al. and Gauger et al. showed that a streptomycin-treated mouse model for E. coli invasiveness positively selected for motility-defective mutants in E. coli MG1665 (42, 65). Gauger et al. found that 30 to 40% of E. coli cells were nonmotile 3 days postinfection, and that number increased to 80 to 90% nonmotile cells 15 days postinfection (65). When reinfected into streptomycin-treated adult mice, these nonmotile strains were demonstrated to be better colonizers than motile E. coli MG1665 strains. Furthermore, nonmotile strains were able to grow 10 to 20% faster on mouse cecal mucus than their isogenic, motile, parent strains and 15 to 30% faster on sugars that are known to be present in the intestine (42, 65). This is similar to the data we present in this study on V. parahaemolyticus. We have shown that deletion of rpoN only causes a growth defect on minimal medium which lacks a glutamine source. We also demonstrated that the ΔrpoN strain has the ability to outcompete the wild-type strain during coinfections of streptomycin-treated adult mice. With this in vivo phenotype, the ΔrpoN strain exhibited higher growth rates in mouse intestinal mucus as well as in a number of sugars that are known to be present in the mouse intestine. In order to further our understanding of this phenotype, we constructed deletion mutants in the polar and lateral flagellar sigma factors, as well as a double deletion mutation in both sigma factors. These strains would retain the motility-defective phenotype but have a fully functioning RpoN. We found that strains carrying a deletion in the polar flagellum sigma factor, FliAP, or a defect in both flagellar systems were slightly better colonizers than the wild-type parent strains but not to the same extent as the rpoN mutant. These data suggest that loss of motility is not the cause of the increased fitness in vivo. As with the ΔrpoN strain, these strains also had higher growth rates on intestinal mucus and intestinal sugars than the wild-type strain. The ΔfliAL strain defective in lateral flagellum synthesis was the only mutant that could not significantly outcompete the wild type and had a doubling time similar to that of the wild type, suggesting that the fliAL gene plays no role in colonization or growth rates. The fact that ΔfliAP and ΔfliAPΔfliAL strains colonized and doubled at similar rates further supports this hypothesis. It would appear that strains that are defective in the synthesis of the polar flagellum are better suited for an in vivo lifestyle. In E. coli, flagellar synthesis accounts for approximately 2% of the total energy cost of the cell (65, 66). Therefore, nonmotile strains, which no longer have to synthesize or expend energy to rotate the flagella, may use that excess energy for other cellular functions, causing them to grow at higher rates than the motile parent strain. This may be the case for the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains, as these are all defective for motility. In addition, when we analyzed the expression patterns of genes in the pathways for gluconate, ribose, and arabinose metabolism, we found that the three aforementioned mutants exhibited significantly higher expression ratios than the wild-type strain. This indicates that these pathways are repressed by either RpoN or FliAP and in the absence of these sigma factors, the pathways are derepressed, which leads to higher growth rates when these carbon sources are present in the growth medium. Recently, it was shown that lack of a functional polar flagellum does indeed cause global changes in V. parahaemolyticus (58), as cells grown on a surface (conditions where the polar flagellum is inhibited) or in mutant strains with defects in the polar flagellum exhibited alterations in nonmotility genes. However, genes relating to carbon utilization pathways were not found to be altered. It should be noted, however, that they examined expression in cells grown on heart infusion plates. Media such as LB and HI broth/plates typically do not contain fermentable carbon sources, which may have prevented the expression of carbon catabolism pathways (67, 68).
The derepression of the gluconate, ribose, and arabinose pathways exhibited in the ΔrpoN, ΔfliAP, and ΔfliAP ΔfliAL strains likely contributes to the higher growth rates in the aforementioned carbon sources as well as these strains' ability to outcompete the WBWlacZ strain in media containing mucus as the sole nutrition source. However, if this were solely the case, it would be expected that all three strains would outcompete the WBWlacZ strain equally in vivo as well. While the ΔfliAP and ΔfliAP ΔfliAL strains are similar in their ability to outcompete the WBWlacZ strain, the ΔrpoN strain outcompetes the WBWlacZ strain at a much greater magnitude, which indicates that there is more to this phenotype than just a loss of motility and ability to utilize carbon sources more efficiently. As mentioned previously, RpoN is a global regulator that controls the expression of a large number of nonmotility-associated genes (31–33). The ΔrpoN strain most likely is benefiting from some altered expression of these nonmotility genes in addition to the benefit seen in nonmotile but RpoN+ strains (ΔfliAP and ΔfliAP ΔfliAL mutants). It would be of merit to examine the RpoN regulon further in V. parahaemolyticus to examine which other genes that are contributing to host colonization are differentially regulated by RpoN.
This is the first report on the ability of V. parahaemolyticus to utilize intestinal mucus as a sole carbon source. Mucus is a glycoprotein that contains fucose, galactose, gluconate, glucuronate, mannose, N-acetylglucosamine, N-acetylneuraminic acid, N-acetylgalactosamine, ribose, and arabinose as its major components (55, 56). Many of the pathways required to catabolize these compounds are present in V. parahaemolyticus. We have shown that this organism has the ability to grow in intestinal mucus and utilize a variety of sugar sources commonly found in intestinal mucus, specifically gluconate, mannose, ribose, and arabinose. Furthermore, strains that catabolize these nutritional sources at higher rates demonstrated higher fitness within the host. It will be interesting to examine further which catabolic pathways are more important during host survival and to see if this organism has preferred nutritional sources within the host.
ACKNOWLEDGMENTS
We thank Brandy Haines Menges, Sai Siddarth Kalburge, and Nityananda Chowdhury for technical assistance and helpful discussion.
This work was supported in part by National Science Foundation grant IOS-0918429 to E.F.B. Bioimaging work was performed at the Delaware Biotechnology Institute BioImaging Center and was supported by grants from the National Center for Research Resources (5P30RR031160-03) and the National Institute of General Medical Sciences (8 P30 GM103519-03) from the National Institutes of Health.
Footnotes
Published ahead of print 18 November 2013
REFERENCES
- 1.Joseph SW, Colwell RR, Kaper JB. 1982. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10:77–124. 10.3109/10408418209113506 [DOI] [PubMed] [Google Scholar]
- 2.Kaneko T, Colwell RR. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordstrom JL, DePaola A. 2003. Improved recovery of pathogenic Vibrio parahaemolyticus from oysters using colony hybridization following enrichment. J. Microbiol. Methods 52:273–277. 10.1016/S0167-7012(02)00188-4 [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman AM, DePaola A, Bowers JC, Krantz JA, Nordstrom JL, Johnson CN, Grimes DJ. 2007. Variability of total and pathogenic Vibrio parahaemolyticus densities in northern Gulf of Mexico water and oysters. Appl. Environ. Microbiol. 73:7589–7596. 10.1128/AEM.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J. Infect. Dis. 181:1661–1666. 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- 6.Su YC, Liu C. 2007. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24:549–558. 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 7.Blackstone GM, Nordstrom JL, Vickery MC, Bowen MD, Meyer RF, DePaola A. 2003. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 53:149–155. 10.1016/S0167-7012(03)00020-4 [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin JB, DePaola A, Bopp CA, Martinek KA, Napolilli NP, Allison CG, Murray SL, Thompson EC, Bird MM, Middaugh JP. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353:1463–1470. 10.1056/NEJMoa051594 [DOI] [PubMed] [Google Scholar]
- 9.Dabanch PJ, Herrero CD, Pavez AC, Veas PN, Braun JS, Porte TL. 2009. Vibrio parahaemolyticus bacteremia: case report and literature review. Rev. Chilena Infectol. 26:360–362 [PubMed] [Google Scholar]
- 10.Lim TK, Stebbings AE. 1999. Fulminant necrotising fasciitis caused by Vibrio parahaemolyticus. Singapore Med. J. 40:596–597 [PubMed] [Google Scholar]
- 11.Payinda G. 2008. Necrotizing fasciitis due to Vibrio parahaemolyticus. N. Z. Med. J. 121:99–101 [PubMed] [Google Scholar]
- 12.Ralph A, Currie BJ. 2007. Vibrio vulnificus and V. parahaemolyticus necrotising fasciitis in fishermen visiting an estuarine tropical northern Australian location. J. Infect. 54:e111–e114. 10.1016/j.jinf.2006.06.015 [DOI] [PubMed] [Google Scholar]
- 13.DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nordstrom JL, Wells J, Puhr N, Gendel SM. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999–4005. 10.1128/AEM.69.7.3999-4005.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CN, Flowers AR, Young VC, Gonzalez-Escalona N, DePaola A, Noriea NF, III, Grimes DJ. 2009. Genetic relatedness among tdh+ and trh+ Vibrio parahaemolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb. Ecol. 57:437–443. 10.1007/s00248-008-9418-3 [DOI] [PubMed] [Google Scholar]
- 15.Osawa R, Okitsu T, Morozumi H, Yamai S. 1996. Occurrence of urease-positive Vibrio parahaemolyticus in Kanagawa, Japan, with specific reference to presence of thermostable direct hemolysin (TDH) and the TDH-related-hemolysin genes. Appl. Environ. Microbiol. 62:725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- 17.Park KS, Ono T, Rokuda M, Jang MH, Okada K, Iida T, Honda T. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659–6665. 10.1128/IAI.72.11.6659-6665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broberg CA, Zhang L, Gonzalez H, Laskowski-Arce MA, Orth K. 2010. A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 329:1660–1662. 10.1126/science.1192850 [DOI] [PubMed] [Google Scholar]
- 19.Burdette DL, Seemann J, Orth K. 2009. Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol. Microbiol. 73:639–649. 10.1111/j.1365-2958.2009.06798.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdette DL, Yarbrough ML, Orth K. 2009. Not without cause: Vibrio parahaemolyticus induces acute autophagy and cell death. Autophagy 5:100–102. 10.4161/auto.5.1.7264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gode-Potratz CJ, Chodur DM, McCarter LL. 2010. Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J. Bacteriol. 192:6025–6038. 10.1128/JB.00654-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiyoshi H, Kodama T, Iida T, Honda T. 2010. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78:1772–1780. 10.1128/IAI.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, Davis BM, Bronson RT, Waldor MK. 2012. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. 10.1371/journal.ppat.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd EF, Cohen AL, Naughton LM, Ussery DW, Binnewies TT, Stine OC, Parent MA. 2008. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110. 10.1186/1471-2180-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley CC, Quirke A, Reen FJ, Boyd EF. 2006. Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. 10.1186/1471-2164-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izutsu K, Kurokawa K, Tashiro K, Kuhara S, Hayashi T, Honda T, Iida T. 2008. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76:1016–1023. 10.1128/IAI.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineyro P, Zhou X, Orfe LH, Friel PJ, Lahmers K, Call DR. 2010. Development of two animal models to study the function of Vibrio parahaemolyticus type III secretion systems. Infect. Immun. 78:4551–4559. 10.1128/IAI.00461-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitaker WB, Parent MA, Boyd A, Richards GP, Boyd EF. 2012. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80:1834–1845. 10.1128/IAI.06284-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. 1998. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp. Quant. Biol. 63:141–155. 10.1101/sqb.1998.63.141 [DOI] [PubMed] [Google Scholar]
- 30.Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422–444. 10.1128/MMBR.65.3.422-444.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong TG, Mekalanos JJ. 2012. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40:7766–7775. 10.1093/nar/gks567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501–520. 10.1046/j.1365-2958.1998.00809.x [DOI] [PubMed] [Google Scholar]
- 33.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F, Klose KE. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191:6555–6570. 10.1128/JB.00949-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734. 10.1371/journal.pone.0006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams SG, Varcoe LT, Attridge SR, Manning PA. 1996. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515. 10.1111/j.1365-2958.2004.04154.x [DOI] [PubMed] [Google Scholar]
- 37.Klose KE, Novik V, Mekalanos JJ. 1998. Identification of multiple sigma54-dependent transcriptional activators in Vibrio cholerae. J. Bacteriol. 180:5256–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawagishi I, Nakada M, Nishioka N, Homma M. 1997. Cloning of a Vibrio alginolyticus rpoN gene that is required for polar flagellar formation. J. Bacteriol. 179:6851–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole R, Milton DL, Horstedt P, Wolf-Watz H. 1997. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology 143(Part 12):3849–3859. 10.1099/00221287-143-12-3849 [DOI] [PubMed] [Google Scholar]
- 40.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. 2004. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70:2520–2524. 10.1128/AEM.70.4.2520-2524.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. 2010. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl. Environ. Microbiol. 76:4720–4729. 10.1128/AEM.00474-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leatham MP, Stevenson SJ, Gauger EJ, Krogfelt KA, Lins JJ, Haddock TL, Autieri SM, Conway T, Cohen PS. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039–8049. 10.1128/IAI.73.12.8039-8049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen PS, Laux DC. 1995. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 253:309–314 [DOI] [PubMed] [Google Scholar]
- 44.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, Hebraud M, Hechard Y. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581–1590. 10.1099/mic.0.26860-0 [DOI] [PubMed] [Google Scholar]
- 46.Totten PA, Lara JC, Lory S. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klose KE, Mekalanos JJ. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reitzer LJ, Bueno R, Cheng WD, Abrams SA, Rothstein DM, Hunt TP, Tyler B, Magasanik B. 1987. Mutations that create new promoters suppress the sigma 54 dependence of glnA transcription in Escherichia coli. J. Bacteriol. 169:4279–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YK, McCarter LL. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693–3704. 10.1128/JB.182.13.3693-3704.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart BJ, McCarter LL. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508–4518. 10.1128/JB.185.15.4508-4518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eaton KA, Morgan DR, Krakowka S. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123–127. 10.1099/00222615-37-2-123 [DOI] [PubMed] [Google Scholar]
- 52.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614. 10.1078/1438-4221-00173 [DOI] [PubMed] [Google Scholar]
- 53.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644–7656. 10.1128/IAI.73.11.7644-7656.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 105:9769–9774. 10.1073/pnas.0802241105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427–7432. 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peekhaus N, Conway T. 1998. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 180:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarter LL. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445–462. 10.1128/MMBR.65.3.445-462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. 2011. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79:240–263. 10.1111/j.1365-2958.2010.07445.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarter LL, Wright ME. 1993. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J. Bacteriol. 175:3361–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. 10.1371/journal.pone.0053957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stecher B, Hardt WD. 2011. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 14:82–91. 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 62.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186–1202. 10.1111/j.1365-2958.2005.04902.x [DOI] [PubMed] [Google Scholar]
- 63.Gardel CL, Mekalanos JJ. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. U. S. A. 98:6889–6894. 10.1073/pnas.111581598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gauger EJ, Leatham MP, Mercado-Lubo R, Laux DC, Conway T, Cohen PS. 2007. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect. Immun. 75:3315–3324. 10.1128/IAI.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macnab R. 1996. Flagella and motility, p 123–145 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC [Google Scholar]
- 67.Haque RU, Baldwin JN. 1964. Purification and properties of Staphylococcal beta-hemolysin. I. Production of beta-hemolysin. J. Bacteriol. 88:1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749. 10.1128/JB.01368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. 10.1016/j.plasmid.2004.02.003 [DOI] [PubMed] [Google Scholar]