Abstract
Enterotoxigenic Escherichia coli (ETEC) is a leading cause of death due to diarrheal illness among young children in developing countries, and there is currently no effective vaccine. Many elements of ETEC pathogenesis are still poorly defined. Here we demonstrate that YghJ, a secreted ETEC antigen identified in immunoproteomic studies using convalescent patient sera, is required for efficient access to small intestinal enterocytes and for the optimal delivery of heat-labile toxin (LT). Furthermore, YghJ is a highly conserved metalloprotease that influences intestinal colonization of ETEC by degrading the major mucins in the small intestine, MUC2 and MUC3. Genes encoding YghJ and its cognate type II secretion system (T2SS), which also secretes LT, are highly conserved in ETEC and exist in other enteric pathogens, including other diarrheagenic E. coli and Vibrio cholerae bacteria, suggesting that this mucin-degrading enzyme may represent a shared virulence feature of these important pathogens.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) bacteria are a diverse group of organisms that share the ability to secrete and effectively deliver heat-stable toxin (ST) and/or heat-labile toxin (LT) enterotoxins (1). Collectively, these organisms cause millions of infections and are one of the leading pathogens associated with death following moderate to severe diarrhea in young children (2–4). In the classical paradigm for ETEC pathogenesis, these organisms adhere to the small intestinal mucosa via plasmid-encoded antigens known as colonization factors (CFs). At this site, ETEC bacteria deliver toxins to receptors on epithelial cells. Bound toxin increases host cell cyclic nucleotide concentrations, activating the cystic fibrosis transmembrane regulatory channel (CFTR) and ultimately culminating in fluid efflux into the intestinal lumen (1).
Most ETEC vaccine development has focused on a subset of antigens, including CFs and LT (5). Unfortunately, lack of ST immunogenicity, incomplete protection afforded by LT immunization, and substantial antigenic heterogeneity of the CFs (6) have hindered the development of a broadly protective ETEC vaccine. However, the recent discovery of novel ETEC antigens (7, 8), the complex nature of immune responses to infection (9), and the modulation of many bacterial genes during infection (10) suggest that neither molecular pathogenesis nor the nature of the protective immune response following infection with these pathogens is completely understood.
For instance, while direct engagement of intestinal epithelial cells is essential for efficient toxin delivery by ETEC (11), the virulence features contributing to this process are still poorly defined. Interestingly, in the intestine, access of pathogens to enterocytes is largely limited by intestinal mucins, including MUC2 (12), the major mucin secreted into the lumen of the intestine, as well as cell-surface-bound mucins, including MUC3 (13). While many enteric pathogens, including Vibrio cholerae and other diarrheagenic E. coli, secrete enzymes that degrade mucin, these enzymes had not previously been identified in ETEC.
The present studies suggest that YghJ, a conserved (14, 15) secreted molecule highlighted in recent immunoproteome (9) and transcriptome (10) analysis of ETEC, is a mucin-binding metalloprotease that degrades intestinal mucins, including MUC2 and MUC3 to promote access of ETEC to enterocytes, thereby accelerating efficient delivery of the heat-labile toxin to cognate receptors on the epithelial surface.
MATERIALS AND METHODS
Maintenance and propagation of human cell lines.
LS174T cells (American Type Culture Collection [ATCC] CL-188) were propagated in Eagle's minimum essential medium supplemented with 10% (final concentration) of fetal bovine serum (FBS). Caco-2 intestinal epithelial cells were maintained in Eagle's minimum essential medium containing FBS (20% [vol/vol]). Jurkat cells were propagated in RPMI 1640 medium supplemented with FBS (10%). All cells were maintained at 37°C in 5% CO2.
Bacterial strains and growth conditions.
Bacterial strains used are listed in Table 1. In most experiments, ETEC bacteria were grown in Luria broth at 37°C overnight from frozen glycerol stocks. To optimize secretion of YghJ via the type II secretion system (T2SS), bacteria were grown in Casamino Acids-yeast extract medium (CAYE) (11).
TABLE 1.
Bacterial strains and plasmids used in these studies
| Strain or plasmida | Descriptionb | Reference(s) or source |
|---|---|---|
| Strains | ||
| H10407 | Wild-type ETEC strain O78:H11; CFA/1; LT+/ST+; EtpA+ | 56, 57 |
| E24377A | WT ETEC strain O139:H28 | 58 |
| B7A | WT ETEC strain O148:H28 | 59 |
| Nissle 1917 | O6:H1 probiotic strain originally isolated from an asymptomatic German soldier | 32 |
| HS | Isolated from a healthy adult scientist at WRAIR; O9:H4; motile | 31 |
| jf1123 | H10407 derivative with isogenic deletion of gspE | 11 |
| jf1124 | H10407 derivative with isogenic deletion of gspM | 11 |
| jf1121 | H10407 derivative with isogenic deletion of gspG | 11 |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) | Invitrogen |
| yghJ mutant | Isogenic insertion in yghJ; Kmr Cmr | 14 |
| Plasmids | ||
| pBAD/Myc-HisA | Arabinose-inducible expression plasmid | Invitrogen |
| pQL001 | yghJ cloned into XhoI and HindIII sites of pBAD/myc-HisA in frame with myc-His tags | This study |
| pQL117* | Site-directed mutation of yghJ in codon corresponding to E1309(D) | This study |
| pQL114* | Site-directed mutation of yghJ in codon corresponding to E1309(A) | This study |
| pQL150* | Site-directed mutation of yghJ in codon corresponding to H1308(A) | This study |
| pQL152* | Site-directed mutation of yghJ in codon corresponding to H1312(A) | This study |
| pQL124* | 66-nucleotide in-frame deletion in yghJ | This study |
Mutagenized yghJ plasmids (indicated by asterisks) used plasmid pQL001 as a template for mutagenesis.
WRAIR, Walter Reed Army Institute of Research; Strr, streptomycin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant.
Cloning, expression, and purification of recombinant YghJ.
yghJ was amplified from E. coli H10407 genomic DNA using primers jf031912.1 (5′-tccgagctcgagatccTTGTCACTTGCGTTATTAATGAATAAG-3′ [native ETEC sequence is capitalized; XhoI site underlined]) and jf031912.2 (5′-cccaagcttcgaattCTCGGCAGACATCTTATGCTC-3′ [HindIII site underlined]) for directional cloning into XhoI and HindIII sites in pBAD/myc-HisA to create pQL001. E. coli TOP10(pQL001) was then induced with arabinose to express recombinant YghJ tagged with myc and polyhistidine (rYghJ-myc-His6), and the fusion protein was purified from clarified lysates to homogeneity by nickel affinity chromatography, followed by gel filtration chromatography (HiLoad 16/60 Superdex 200 prep grade; GE Life Sciences) (10). Recombinant proteins with mutations in the putative metalloprotease domain were purified similarly. Chelation of metal ions from the recombinant protein to produce the apoenzyme was performed by dialyzing 500 μg of rYghJ overnight at 4°C against 1 liter of phosphate-buffered saline (PBS) containing 10 mM EDTA, 1 mM 1,10-phenanthroline, and 1 g Chelex 100 resin (Na+ form). Phenanthroline was then removed by dialysis for 3 h against 1 liter of PBS containing 1 mM EDTA and 1 g Chelex resin. YghJ holoenzyme was then reconstituted by adding 2 mM ZnSO4, NiSO4, or MgSO4 and incubating (5 min) at room temperature.
Bioinformatic analysis of YghJ and T2SS sequences.
Domain enhanced lookup time accelerated BLAST (Delta-BLAST) searches were used to identify potential functional domains within YghJ (16). Protein homology/analogy recognition engine (Phyre2) was then used to compare regions of YghJ with libraries of known protein structures to further elucidate potential functional residues (17).
BLASTN was used to query multiple draft ETEC genomes emerging from an ongoing project to sequence multiple clinical isolates (http://gscid.igs.umaryland.edu/wp.php?wp=comparative_genome_analysis_of_enterotoxigenic_e._coli_isolates_from_infections_of_different_clinical_severity).
Mutagenesis of the putative M60/pf13402 metallopeptidase domain.
Site-directed mutagenesis of the native yghJ gene fragment carried on pQL001 was performed by high-fidelity PCR using mutagenic primer sets as indicated in Table 2. Mutagenized plasmids were then used to transform E. coli TOP10 to ampicillin resistance. Mutations were confirmed by sequencing using primers jf101812.3 (5′-CCGAAGAAGAACCTGAATGC-3′) and jf101812.4 (5′-TTACCGTTGGATTCAGCACA-3′).
TABLE 2.
Oligonucleotides used in these studies
| Primer | Sequence (5′→3′)a | Description |
|---|---|---|
| jf031912.1 | TCCGAGCTCGAGATCCTTGTCACTTGCGTTATTAATGAATAAG (XhoI site underlined) | yghJ forward cloning primer |
| jf031912.2 | CCCAAGCTTCGAATTCTCGGCAGACATCTTATGCTC (HindIII site underlined) | yghJ reverse cloning primer |
| jf082312.1 | GCTGATCTGGCATGATGTCGGTCATAACGCC (GAA→GAT mutation underlined) | Site-directed mutagenesis (E1309D) forward |
| jf082312.2 | GGCGTTATGACCGACATCATGCCAGATCAGC | Site-directed mutagenesis (E1309D) reverse |
| jf082312.3 | GGCTGATCTGGCATGCAGTCGGTCATAACGC (GAA→GCA mutation underlined) | Site-directed mutagenesis (E1309A) forward |
| jf082312.4 | GCGTTATGACCGACTGCATGCCAGATCAGCC | Site-directed mutagenesis (E1309A) reverse |
| jf112612.1 | ACGACTGGCTGATCTGGGCTGAAGTCGGTCATAACG (CAT→GCT mutation underlined) | Site-directed mutagenesis (H1308A) forward |
| jf112612.2 | CGTTATGACCGACTTCAGCCCAGATCAGCCAGTCGT | Site-directed mutagenesis (H1308A) reverse |
| jf112612.3 | CTGGCATGAAGTCGGTGCTAACGCCGCAGAAACG (CAT→GCT mutation underlined) | Site-directed mutagenesis (H1312A) forward |
| jf112612.4 | CGTTTCTGCGGCGTTAGCACCGACTTCATGCCAG | Site-directed mutagenesis (H1312A) reverse |
| jf101712.1 | GACTGGCTGATCTGG(−66)AACGTGCTGGCGCTG | Mutagenesis primer to construct 22-amino-acid internal deletion of YghJ residues H1308 to N1329 |
| jf101712.2 | CAGCGCCAGCACGTT(−66)CCAGATCAGCCAGTC | Reverse complement of jf101712.1 |
| jf101812.3 | CCGAAGAAGAACCTGAATGC | Forward sequencing primer yghJ from nt 3616 to 3635 |
| jf101812.4 | TTACCGTTGGATTCAGCACA | Reverse sequencing primer yghJ from nt 4319 to 4300 |
(−66) indicates the position of the 66-nucleotide in-frame deletion in yghJ extending from nucleotide (nt) 3904 to 3969.
Production and purification of polyclonal antibody.
Polyclonal antisera against rYghJ-myc-His6 were produced in rabbits as previously described (8). Antibodies to YghJ were prepared by cross absorption of resulting antisera against an immobilized E. coli lysate column (Thermo Scientific) followed by affinity purification using the recombinant protein immobilized on nitrocellulose membranes (18, 19).
Purification of MUC2.
MUC2 was purified from supernatants of tissue culture medium from LS174T cells (ATCC CL-188), a goblet cell-like adenocarcinoma line that makes abundant MUC2 (20, 21). Briefly, LS174T cells were grown as described above; conditioned medium was recovered, concentrated by ultrafiltration using a 100-kDa-molecular-weight-cutoff filter (MWCO), and then buffer exchanged with 10 mM Tris-HCl and 250 mM NaCl (pH 7.4) prior to size exclusion chromatography using Sepharose CL-2B resin (22). Fractions were checked for MUC2 by anti-MUC2 dot immunoblotting. MUC2-positive fractions, corresponding to a protein peak in the column void volume, were separated on 3 to 8% Tris-acetate gradient gels, stained with Sypro Ruby to check purity, and immunoblotted using anti-MUC2 to verify the identity of the protein. Fractions containing intact, full-length MUC2 were then pooled and saved at −80°C for subsequent assays.
Protein interaction studies.
To examine interaction of YghJ with the human intestinal mucin MUC3, lysate from Caco-2 cells containing MUC3 was separated by SDS-PAGE as described above and transferred to nitrocellulose membranes. To examine interaction with MUC2, purified protein was spotted on nitrocellulose membranes. Far Western analysis was then performed with purified rYghJ-myc-His6. Briefly, nitrocellulose membranes with immobilized mucins were blocked for 1 h with 1% bovine serum albumin (BSA) in PBS before incubating with 50 μg/ml of purified rYghJ-myc-His6 overnight at 4°C. Proteins were detected by immunoblotting using antimucin antibodies as described below or anti-YghJ antibody affinity purified from rabbit serum.
Mucin degradation studies.
To examine the activity of YghJ against the cell-associated mucin MUC3, we followed a protocol similar to that described by Szabady et al. (23). Briefly, Caco-2 epithelial cell monolayers were grown in 96-well tissue culture plates for 48 to 72 h postconfluence to optimize MUC3 expression (20) on the epithelial surface. Supernatant was removed and replaced with 100 μl of minimum essential medium (MEM) containing rYghJ-myc-His6 at a final concentration of 50 μg/ml. Following overnight treatment of the cell monolayers at 37°C and 5% CO2, the medium was removed, and the monolayers were lysed in 20 μl of lysis buffer (50 mM sodium phosphate, 250 mM NaCl, 0.1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and complete EDTA-free protease inhibitor cocktail [Roche]). Following incubation on ice for 30 min and repeated freeze (dry ice)-thaw (37°C) cycles, the lysates were centrifuged at 10,000 × g (4°C) to pellet debris. Clarified lysates were then separated on gradient (3 to 8% Tris-acetate; Invitrogen) PAGE. Following transfer to nitrocellulose membranes, Caco-2 lysates were immunoblotted with anti-MUC3A/B goat polyclonal IgG antibodies (F-19 [catalog no. sc-13314; Santa Cruz]) that recognize an internal region of mucin 3A of human origin (gene identification [ID] 4584).
To examine degradation of purified MUC2, 0.1 μg of protein was treated for at least 30 min with 5 μg of rYghJ-myc-His6 at 37°C. Reaction products were resolved by SDS-PAGE, and MUC2 digests were examined with anti-MUC2 rabbit polyclonal (IgG) (H-300 [catalog no. sc-15334; Santa Cruz]) that recognizes an epitope corresponding to amino acids 4880 to 5179 at the C terminus of human mucin 2 (gene ID 4583).
Degradation of MUC2 by LS174T cells was examined by incubating cells grown on coverslips with purified recombinant YghJ for 3 h. MUC2 remaining after treatment was detected by immunofluorescence confocal microscopy as described below, and images were saved as z-stacks. Image data were then analyzed using Volocity three-dimensional (3D) image analysis software (version 6.2; PerkinElmer, Inc.).
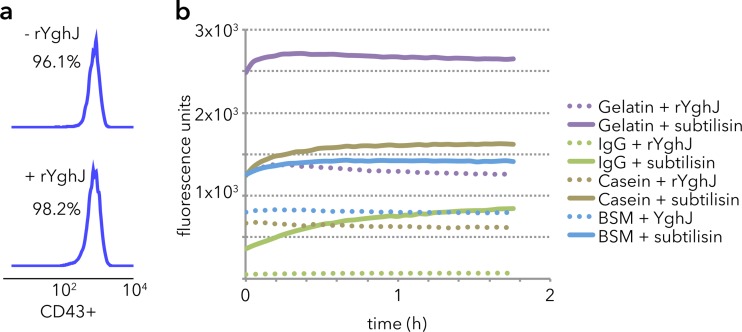
To determine whether YghJ degrades the mucin-like CD43 glycoprotein on the surfaces of human T lymphocytes, we followed a protocol similar to that outlined by Szabady et al. (23). Briefly, 1 × 106 Jurkat cells were incubated for 3 h at 37°C and 5% CO2 with 5 μg of YghJ suspended in a total volume of 100 μl of RPMI 1640 medium. The cells were pelleted at 400 × g for 5 min, washed with PBS, and blocked with 1% BSA in PBS, and then cell surface CD43 was labeled with phycoerythrin-conjugated anti-CD43 (mouse anti-human CD43 monoclonal clone L10; Invitrogen) and analyzed by flow cytometry.
To test the activity of YghJ in degrading other substrates, we examined the hydrolysis of fluorescein-labeled proteins (24). Quenched substrates, including IgG, gelatin, and bovine submaxillary mucin (BSM) were prepared by labeling with fluorescein isothiocyanate (FITC) (Sigma) as previously described (24). Casein-FITC was purchased from Sigma. Briefly, 10 μl of YghJ (50 μg) was added to 90 μl of 0.1 mg/ml substrate in 100 mM Tris (pH 7.5) in black 96-well U-bottom polypropylene plates. The plates were sealed with optically clear film (TempPlate; USA Scientific) and incubated at 37°C, and fluorescence was measured every 5 min for several hours on a Tecan F200 plate reader using an excitation filter at 485 nm and an emission filter at 535 nm. Increase in fluorescence due to release of quenched FITC upon proteolysis was tested using the protease subtilisin as a positive control (0.1 to 1 mg/ml stock).
In vitro assessment of toxin delivery and binding.
Caco-2 epithelial cell monolayers were infected with ETEC H10407, yghJ mutants, or complemented mutants at multiplicities of infection of approximately 100 (bacteria/cell). Cultures of bacteria were grown overnight in Luria broth from frozen glycerol stocks, diluted 1:100, and grown for 1 h. One microliter of each culture with or without antibodies as indicated in the figures was then added to confluent Caco-2 monolayers seeded into 96-well plates. Cultures of mutants complemented with pBAD-based expression plasmids were supplemented with 0.0002% arabinose. Two hours after inoculation, the monolayers were washed 3 times with tissue culture medium, and the medium was replaced with 100 μl of fresh medium/well and returned to the incubator (37°C, 5% CO2) for 2.5 h. As previously described, we used a cyclic AMP (cAMP) enzyme immunoassay (EIA) (Arbor Assays, Ann Arbor, MI) (11) to examine the efficiency of toxin delivery.
To examine the effect of YghJ on toxin access to cell surface receptors on enterocytes, cells were treated with fluorophore-conjugated cholera toxin B subunit. Briefly, Caco-2 or LS174T cells grown on coverslips were treated with YghJ at a final concentration of 25 μg/ml for 3 h at 37°C and 5% CO2. Following treatment, cell membranes were stained with CellMask (catalog no. C10046; Life Technologies) and then fixed with 2% paraformaldehyde for 10 min. After the cells were blocked with 1% BSA, they were labeled with cholera toxin subunit B conjugate (catalog no. C34775; Life Technologies). Images were acquired by fluorescence confocal microscopy with z-stacks, and signal data were then analyzed using Volocity 3D image analysis software (version 6.2; PerkinElmer, Inc.).
Mouse intestinal colonization and competition assays.
Colonization of the ileum of infected mice was assessed using the CD-1 mouse intestinal colonization model (25). Briefly, mice were pretreated with streptomycin; the mice were given streptomycin (5 g/liter) in drinking water for 24 h, followed by drinking water alone for 12 h. After the administration of famotidine (50 mg/kg of body weight) to reduce gastric acidity, mice were challenged with approximately 105 CFU of either the lacZYA::Kmr strain or the yghJ::Kmr mutant by oral gavage. Twenty-four hours later, the mice were sacrificed, and dilutions of saponin intestinal lysates were plated onto Luria agar plates containing kanamycin (25 μg/ml). Competition assays were performed in a similar manner by simultaneous challenge with approximately 104 CFU of each strain. After 24 h, lysates were plated onto agar containing kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to identify wild-type (WT) (lacZYA::Kmr) (white) or yghJ mutant (blue) colonies. In mouse intestinal colonization studies to examine penetration of the mucous layer, mice were inoculated with 108 CFU of either WT H10407 or the yghJ mutant, and after 24 h, mice were sacrificed, and samples were fixed in formalin or Carnoy's solution to preserve the mucin layer.
Immunohistochemistry.
Sections of small intestine were preserved in formalin, embedded in paraffin, and cut into 4-μm sections for further processing. Following removal of paraffin, sections were immunostained overnight at 4°C with anti-MUC2 rabbit polyclonal antibody (ab76774; Abcam) (1:100), followed by goat anti-rabbit biotinylated secondary antibody (Jackson ImmunoResearch Laboratories; 1:800) and streptavidin-horseradish peroxidase (HRP) conjugate (catalog no. 016-030-084; Jackson ImmunoResearch Laboratories) (1:1,600). Samples were then developed with 3,3′-diaminobenzidine (Betazoid DAB; Biocare) and counterstained with hematoxylin. Light microscopic images were acquired using a Zeiss Axioscope microscope fitted with an Axiocam HRm camera or a Zeiss Axioskop 2 MOT microscope fitted with a CRI Nuance FX multispectral imaging system (Cambridge Research and Instrumentation).
Immunofluorescence microscopy.
To identify ETEC H10407 (serotype O78:H11) in the intestine, sections were incubated with anti-O78 antisera obtained from the E. coli Reference Center at Pennsylvania State University, followed by fluorophore Alexa Fluor goat anti-rabbit conjugates. MUC2 in mouse intestinal sections was identified using anti-MUC2 mouse monoclonal antibody (clone 966/1) (ab11197; Abcam). Confocal microscopy images were obtained in the Molecular Microbiology Imaging Facility using a Zeiss LSM 510 Meta confocal laser scanning microscope.
To identify cAMP in sections of mouse small intestine, sections were first stained with CellMask orange (catalog no. C10045; Life Technologies) diluted 1:2,000 for 5 min at 37°C, blocked in 1% BSA for 30 min at 37°C, then labeled with anti-cAMP rabbit polyclonal antibody (catalog no. 07-1497; Millipore) diluted 1:100 for 1 h. After the sections were washed, goat anti-rabbit–Alexa Fluor 488 conjugate was added at 1:200 for 30 min. After the sections were washed and incubated with TO-PRO-3 nucleus counterstain (1:250) for 15 min at room temperature, coverslips were mounted with Prolong gold. To quantify fluorescence, intensity ImageJ (v1.47) was used to measure the mean fluorescence per pixel (http://rsbweb.nih.gov/ij/docs/menus/analyze.html#measure). These values were then normalized for the number of epithelial cells present in the image.
FACS analysis and mucin accumulation studies.
Fluorescence-activated cell sorting (FACS) analysis was used to determine the presence of YghJ on the surfaces of ETEC (26). Briefly, bacteria suspended in phosphate-buffered saline (pH 7.2) were first fixed with 2% paraformaldehyde for 15 min, washed twice in PBS, and blocked with 1% BSA in PBS for 30 min. The resulting cell suspensions were incubated with affinity-purified anti-YghJ antibody in blocking solution for 1 h at room temperature, washed three times, and then incubated with anti-rabbit antibody–Alexa Fluor 594 conjugate.
To study binding of MUC2 to the surfaces of ETEC, supernatants from overnight cultures of LS174T cells grown in confluent monolayers were sterilized by filtration through a 0.22-μm filter and then concentrated through a 100-kDa-molecular-weight-cutoff centrifugal filter to concentrate MUC2 secreted into culture supernatants. Following inoculation with bacteria, the infected MUC2-positive (MUC2+) retentate was incubated at 37°C and 5% CO2, and aliquots were retrieved at various time points. Following centrifugation at 10,000 × g and 4°C, the bacterial pellets were washed with PBS, fixed with 2% paraformaldehyde for 15 min, washed again in PBS to remove the fixative, blocked with 1% BSA in PBS, incubated with rabbit polyclonal anti-MUC2 (1:50), and detected with goat anti-rabbit fluorescent (Alexa Fluor 488; Invitrogen) antibody conjugate. MUC2 binding to bacteria was then examined by flow cytometry (26). Flow cytometry data were processed in FlowJo v10.0.5 (Tree Star, Inc.).
RESULTS
YghJ is required for efficient delivery of heat-labile toxin.
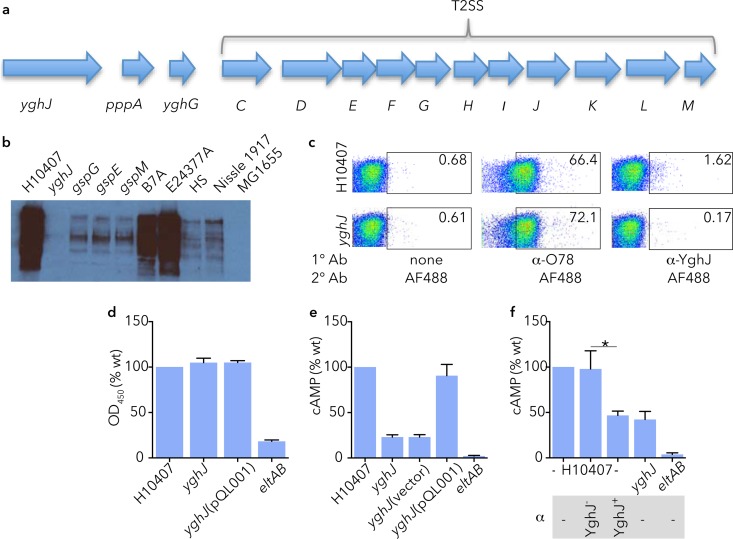
YghJ, encoded immediately upstream of the genes encoding the type II secretion system (T2SS) that is responsible for export of the heat-labile toxin (Fig. 1a) (27) was found in abundance in culture supernatants of several ETEC strains, including H10407, B7A, and E24377A (Fig. 1b). However, in keeping with earlier studies suggesting that YghJ is secreted by the T2SS in other E. coli pathotypes (28), we did not find YghJ in culture supernatants of ETEC strains with mutations in T2SS genes gspE, gspG, and gspM or in cultures of E. coli MG1655 that lacks an intact T2SS (27). YghJ and the T2SS proteins were also found on the chromosomes of nonpathogenic E. coli strains (29, 30) (see supplemental material) including HS, an intestinal commensal strain previously isolated from a healthy adult (31), and Nissle 1917, originally obtained from an asymptomatic German soldier (32). However, relative to ETEC strains, the amount of YghJ secreted by these strains was quite small. Because we had previously detected YghJ in association with outer membrane vesicles (9), we also examined the surface of ETEC for YghJ by flow cytometry (Fig. 1c), revealing that some YghJ also remains associated with the surface of wild-type strain H10407. While YghJ export is dependent on T2SS, conversely, mutation of yghJ had no impact on secretion of LT (14), and likewise, trans-complementation of the mutant with a recombinant YghJ expression plasmid (pQL001) did not affect toxin secretion (Fig. 1d), suggesting that this molecule does not compete with LT or facilitate its secretion. However, we noted that the yghJ mutant was appreciably deficient in the ability to deliver LT as determined by the diminished activation of cAMP in target epithelial cells (Fig. 1e). Likewise, affinity-purified antibodies against YghJ significantly impaired delivery of LT by ETEC (Fig. 1f). Together, these studies suggest that these type II secretion effectors, YghJ and LT, act in concert to intoxicate enterocytes, leading to activation of cyclic nucleotides that drive chloride secretion that results in diarrhea.
FIG 1.
YghJ is a highly conserved type II secretion system (T2SS) effector molecule that facilitates delivery of heat-labile toxin (LT). (a) The product of the yghJ gene, located upstream of the T2SS genes responsible for secretion of the heat-labile toxin as shown in panel a is secreted by ETEC strains with an intact T2SS. (b) Immunoblot demonstrating YghJ in trichloroacetic acid (TCA) precipitates of overnight cultures of ETEC strains H10407, B7A, and E24377A, but not the yghJ mutant, T2SS mutants (gspG, gspE, and gspM), or strain MG1655 (E. coli K-12), which has a truncated version of yghJ and lacks most T2SS genes. Commensal strains HS and Nissle 1917 secrete comparatively little YghJ. (c) Flow cytometry data demonstrating that some YghJ remains associated with the surface of strain H10407 (serotype O78:H11; top right panel) but not with the yghJ mutant (bottom right panel). The regions enclosed in rectangles show the areas gated for surface expression, and the number in the top right-hand corner of each panel represents the percentage of positive cells for the respective antigen under consideration. Anti-O78 (α-O78) data are included as positive controls for surface expression. 1° Ab, primary antibody; 2° Ab, secondary antibody; α-YghJ, anti-YghJ antibody; AF488, Alexa Fluor 488. (d) Secretion of heat-labile toxin by ETEC is not affected by yghJ. GM1 ganglioside enzyme-linked immunosorbent assay (ELISA) data for wild-type (wt) H10407 ETEC, yghJ mutant, and complemented yghJ mutant (pQL001) relative to the LT mutant (eltAB) are shown. OD450, optical density at 450 nm. (e) cAMP production by target Caco-2 cells following infection with wild-type ETEC H10407, yghJ mutant, or complemented mutant. The LT deletion mutant (eltAB) mutant is included here as a negative control. (f) Affinity-purified polyclonal antibodies (α) against YghJ (YghJ+), but not antibody preabsorbed with recombinant YghJ (YghJ−) impair delivery of LT to target epithelial cells. Values that are significantly different (P < 0.05) from each other are indicated by a bar and asterisk (n = 4; two-tailed Mann-Whitney analysis).
Intimate enterocyte engagement requires YghJ.
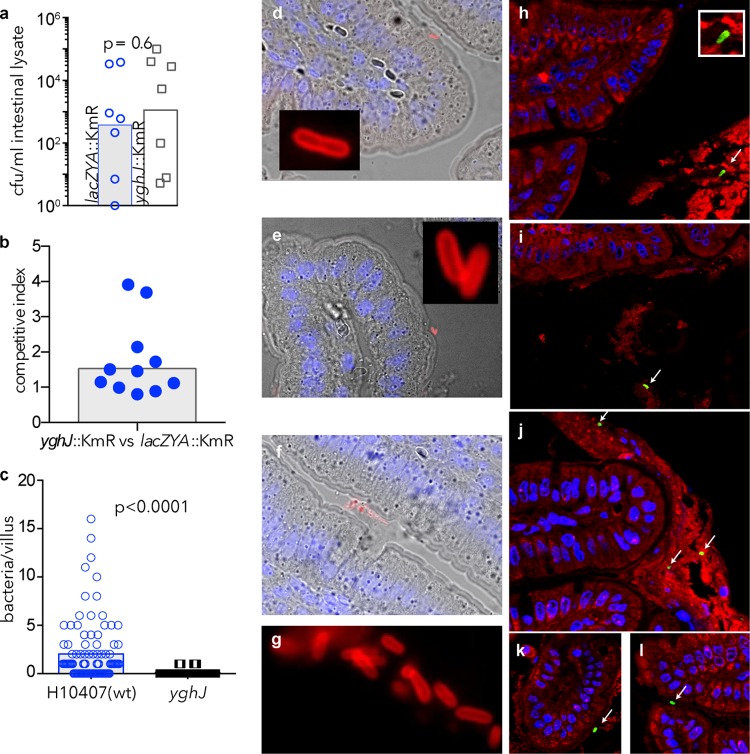
YghJ shares homology with accessory colonization factor D (AcfD), a lipoprotein (33) important for intestinal colonization of Vibrio cholerae (34). Curiously, relative to ETEC (H10407) carrying a mutation in the lacZYA locus, which does not affect intestinal colonization (25), we actually recovered slightly more yghJ mutant organisms from small intestinal lysates regardless of whether mice were challenged with the individual strains (Fig. 2a) (P = 0.6) or together in competition assays (Fig. 2b). Interestingly, however, microscopic examination of ileal sections from mice challenged with either strain H10407 or the yghJ mutant demonstrated many WT organisms intimately associated with the surfaces of enterocytes (Fig. 2c and Fig. 2d to g), while we could identify few if any yghJ mutant organisms, suggesting that YghJ is required for bacteria to gain efficient access to the intestinal epithelial cells (P < 0.0001).
FIG 2.
YghJ is required for efficient access to the surfaces of enterocytes. (a) Total ETEC recovered from intestinal lysates following infection with either the wild-type (wt) strain bearing a Kmr cassette insertion in the lacZYA locus (lacZYA::Kmr) or the yghJ::Kmr mutant. The bars show the geometric mean number of CFU/ml. Each symbol shows the value for an individual mouse. (b) Competition assay between the lacZYA mutant and the yghJ mutant showing total bacteria recovered from intestinal lysates (the bar represents the average competitive index). (c) Bacteria intimately associated with the surfaces of intestinal villi (n = 100) from intestinal segments of mice (n = 2) following challenge with either H10407 (wt) (blue open circles), or the yghJ mutant (open squares) (P values reflect two-tailed Mann-Whitney nonparametric analysis). (d to f) ETEC H10407 (serotype O78:H11) intimately associated with enterocyte surfaces of intestinal villi. Bacteria identified by anti-O78 (red) immunofluorescence imaging are shown in the insets and in panel g. (h and i) Trapping of the yghJ mutant in luminal mucus (MUC2 is shown in red; bacteria identified by anti-O78 antibodies appear green). H10407 also appears in the lumen (j), but it is also present in close association with the enterocyte surface (k and l).
To gain access to the enterocyte surface, enteric pathogens must transit through complex glycoproteins known as mucins. Because there were few yghJ mutant organisms on the enterocyte surface, we questioned whether these organisms have lost the ability to traverse the mucous layer in the small intestine. In the small intestine, the major luminal mucin is MUC2 (13). In sections fixed to retain luminal mucus (Fig. 2h to l), we identified many yghJ mutant bacteria within the MUC2+ luminal mucous layer (Fig. 2h and i), while WT organisms appeared both in the lumen (Fig. 2j) and at the enterocyte surface (Fig. 2k and l).
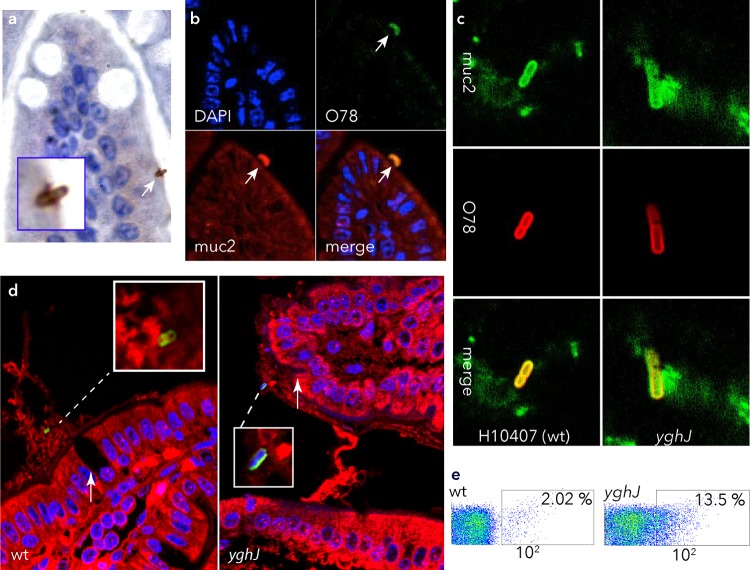
Deletion of yghJ accelerates accumulation of mucin on the surface of ETEC.
In sections of intestine from mice challenged with WT ETEC, we identified MUC2-coated bacteria intimately associated with the mucosal surface by immunohistochemistry (Fig. 3a) or by confocal microscopy where organisms appeared labeled with both MUC2 and anti-O78 (Fig. 3b). Similarly, for both the H10407 strain and the yghJ mutant, luminal bacteria could be identified where MUC2 appeared in close association with the surfaces of organisms (Fig. 3c). In intestinal sections where efforts were made to preserve luminal mucin, we observed both wild-type and yghJ mutant bacteria (Fig. 3d) in plumes of MUC2 deposited over goblet cells. Because the yghJ mutant appeared to have limited access to the mucosal surface, we questioned whether the kinetics of ETEC association with MUC2 might differ. Following short incubation (1 h) with MUC2-containing conditioned medium, the yghJ mutant was more frequently coated with MUC2 relative to the wild-type ETEC strain (Fig. 3e). Collectively, these data suggest that ETEC colonization of the small intestine relates in part to its interactions with intestinal mucins and that YghJ modulates these interactions.
FIG 3.
Mucin accumulation on the surface of ETEC is accelerated in yghJ mutant bacteria. (a) Light microscopy image of MUC2 immunostained ETEC intimately attached to the enterocyte surface following wild-type H10407 oral challenge. The image was acquired following immunostaining with MUC2 polyclonal antibody, biotinylated secondary antibody, streptavidin-HRP conjugate, and 3,3′-diaminobenzidine (Betazoid DAB; Biocare), counterstained with hematoxylin. (b) Confocal microscopic images of ETEC H10407-infected mouse intestine demonstrating colocalization of MUC2 (red) with O78 antigen (green) on the surface of ETEC. (c) MUC2 identified on the surfaces of luminal bacteria, in frozen sections of small intestine (green, MUC2; red, anti-O78; yellow, merged images) following intestinal infection. (d) Wild-type (wt) ETEC H10407 (anti-O78, green) and yghJ mutant (anti-O78, green; nucleic acid, blue) in plumes of MUC2+ mucin (red) deposited over the surfaces of goblet cells (white arrows). (e) yghJ mutants exhibit excess accumulation of MUC2 relative to wild-type H10407. Data represent MUC2+ bacteria determined by flow cytometry following incubation in MUC2+ supernatants from LS174T cells. The regions enclosed in rectangles show the areas gated for surface expression, and the number in the top right-hand corner shows the percentage of MUC2+ bacteria.
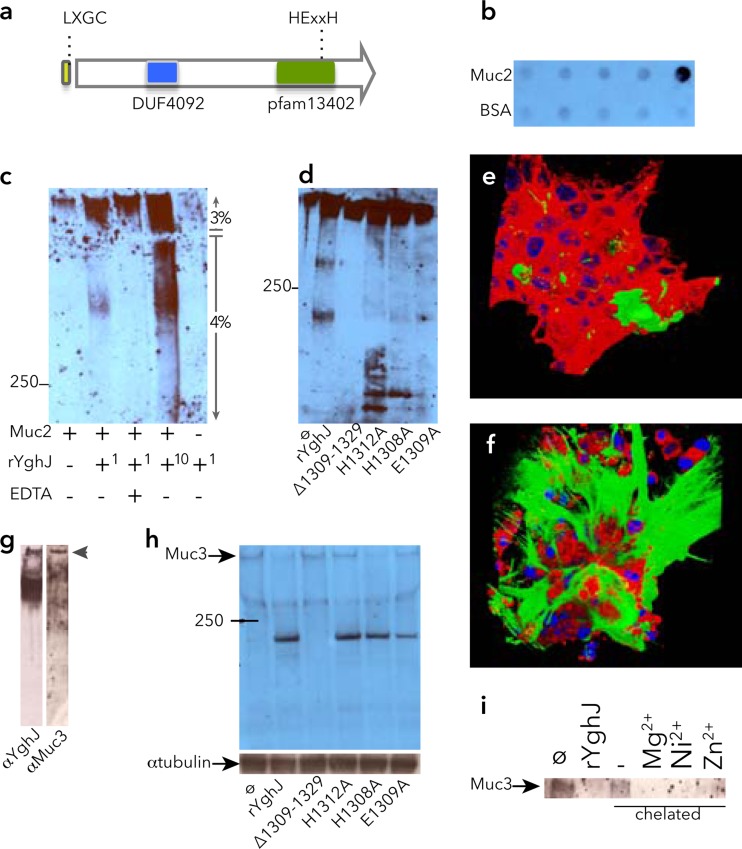
YghJ specifically binds and degrades intestinal mucins.
Comparison of the predicted YghJ protein sequence to known proteins demonstrated several potential functional domains or motifs (Fig. 4a) including a N-terminal signal peptide, followed by a putative lipoprotein (L-X-G-C) motif, a domain of unknown function found in Proteobacteria (DUF4092) located at amino acid residues 422 to 591 of YghJ, and finally an enhancin/M60-like peptidase domain (pfam13402) at residues 1090 to 1386. Included within the predicted peptidase domain is a putative HEXXH metalloprotease motif at positions 1308 to 1312. Enhancin (35) and other members of this family bind to and degrade mucins or mucin-like glycoproteins; therefore, we examined the ability of YghJ to degrade mucins. YghJ bound to purified MUC2 (Fig. 4b), and YghJ degraded this intestinal mucin in a dose-dependent fashion (Fig. 4c). As predicted by the putative metalloprotease motif within the peptidase domain, this activity was at least partially inhibited by EDTA. Similarly, we found that YghJ degradation of MUC2 was affected by mutagenizing the canonical metalloprotease motif. Further comparisons (17) suggested that a 22-amino-acid region extending from H1308 to N1329 contained a potential active site within YghJ, and mutant recombinant protein YghJ with the region from H1308 to N1329 deleted [YghJ(ΔH1309-N1329)] demonstrated greatly reduced MUC2 mucinase activity in vitro (Fig. 4d), while mutagenesis of individual residues within the HEXXH motif partially prevented mucin degradation. To further examine activity of YghJ, we studied LS174T cells, which make abundant MUC2 (20). Treatment of cells with rYghJ significantly reduced the amount of extracellular MUC2 (Fig. 4e) relative to mock-treated controls (Fig. 4f). YghJ also appeared to interact with the predominant cell-bound mucin in the small intestine, MUC3 (Fig. 4g). In addition, YghJ degraded MUC3 on the surfaces of intestinal epithelial cells in vitro (Fig. 4h), and degradation at least partly depended on an intact HEXXH motif. Similarly, chelation of metal from the recombinant protein by dialysis against phenanthroline abolished mucinase activity, while supplementation of divalent cations restored activity to the chelated protein (Fig. 4i). Together, these data suggested that YghJ is a member of a family of metalloproteases that degrade mucins.
FIG 4.
YghJ is a metalloprotease that binds and degrades secreted and cell-bound intestinal mucins. (a) Potential motifs and domains within YghJ include a potential LXGC lipoprotein motif at the amino-terminal end following the signal peptide, a domain of unknown function (DUF4092), and a M60-like peptidase domain (pfam13402) at the carboxy-terminal end of the protein. Within this domain lies a putative HEXXH metalloprotease motif. (b) YghJ interacts with purified MUC2. Shown in the anti-YghJ immunoblot are 10-fold dilutions of the bait proteins (MUC2 in the top row and the BSA control in the bottom row) immobilized on nitrocellulose membranes prior to incubation with rYghJ-myc-His6. (c) YghJ degrades purified MUC2 as shown in the anti-MUC2 immunoblot. In the rYghJ row below the blot, (+1 indicates ∼5 μg of rYghJ-myc-His6, while +10 indicates ∼50 μg rYghJ-myc-His6). The numbers to the right of the blot indicate 3% stacking gel and 4% resolving gel. The position of the molecular mass marker (in kilodaltons) is indicated to the left of the blot. (d) Mutations in the canonical metalloprotease motif located between H1308 and H1312 affect degradation of MUC2 as shown in the anti-MUC2 immunoblot. From left to right, the lanes contain untreated MUC2 (ø) and MUC2 treated with either rYghJ or mutant proteins. (e and f) Composite images of MUC2-expressing LS174T cells following treatment with rYghJ (e) or mock treatment control (f). MUC2 is shown in green, the membrane is shown in red, and the nuclei are shown in blue. (g) Far Western immunoblots demonstrating interaction of rYghJ with MUC3 in Caco-2 cell intestinal lysates. Both lanes contain Caco-2 cell lysates. The left strip was probed with rYghJ and developed with anti-YghJ antibody, while the right strip was developed with anti-MUC3 antibody. (h) Degradation of MUC3 on the surfaces of Caco-2 cells by rYghJ or rYghJ with mutations in the putative metalloprotease motif. (i) YghJ exhibits features of metalloproteases. MUC3 signal from untreated Caco-2 cells is shown in the leftmost lane of the immunoblot (ø lane). YghJ degradation of MUC3 (rYghJ lane) was prevented when equal amounts of recombinant YghJ chelated with phenanthroline to remove metals was used to treat cells (− lane), while supplementation of chelated YghJ with divalent cations (Zn2+, Ni2+, and Mg2+) restored protease activity.
In contrast, YghJ was inactive against the mucin-like CD43 molecule expressed by Jurkat cells (Fig. 5a), bovine submaxillary mucin, gelatin, or IgG (Fig. 5b). Collectively, these data suggest that YghJ has specific activity for the major mucins in the vertebrate lumen and on the surfaces of cells in the small intestine.
FIG 5.
rYghJ does not degrade other substrates. (a) YghJ does not degrade the mucin-like CD43 glycoprotein on the surfaces of Jurkat cells as shown in CD43 flow cytometry. The two numbers are the percentages of CD43+ cells in the two populations. (b) Fluorophore release assays using fluorescently labeled quenched protein substrates demonstrate an increase in fluorescence following treatment with the general protease subtilisin but not with YghJ.
Conservation of YghJ features in peptides from E. coli and V. cholerae.
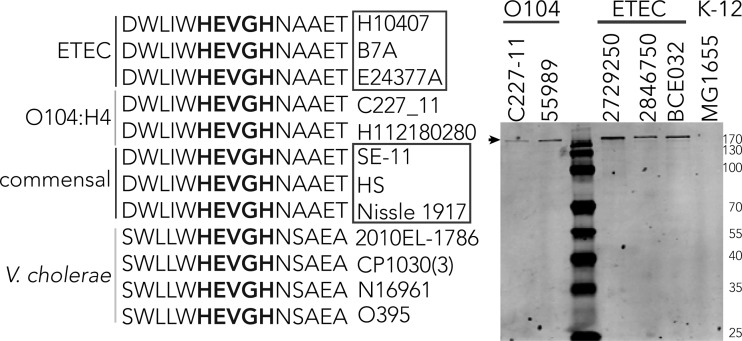
Interestingly, we found that YghJ and genes encoding the downstream T2SS were highly conserved in ETEC and commensal strains that have been sequenced (see Table S1 in the supplemental material), as well as more recently sequenced O104:H4 Shiga toxin-producing enteroaggregative E. coli (36) (see Fig. S1a and S1b in the supplemental material). In contrast to commensal strains however, both the ETEC strains and the O104 strains secreted significant amounts of YghJ (Fig. S1c). The YghJ peptide is currently annotated as a homologue of the Vibrio cholerae protein AcfD. Alignments of YghJ and AcfD from Vibrio cholerae classical 6th pandemic and El Tor 7th pandemic strains demonstrated extensive homology (Fig. S1a and S1b); multiple common features include the putative lipoprotein (LXGC) motif at their amino termini, the domain of unknown function, and the M60-like peptidase domain where the metalloprotease motif sequence HEVGH was completely conserved (Fig. 6).
FIG 6.
Alignment of sequences surrounding the putative metalloprotease motifs (shown in bold type) of YghJ from ETEC and similar proteins from other enteric organisms. Individual strain designations are shown to the right of the sequences. These strains include Shiga toxin-producing E. coli serotype O104:H4 and commensal E. coli strains HS, SE-11, and Nissle 1917. AcfD proteins from V. cholerae O1 strains include strain 2010 EL-1786 (El Tor biotype, 2010 Haiti isolate), CP1030 (3) (Mexico), N16961 (prototypical 7th pandemic El Tor biotype), O395 classical O1 serotype (prototypical 6th pandemic, Ogawa biotype). (b) YghJ is secreted by ETEC and E. coli O104 isolates, as shown by the immunoblot of TCA precipitates. The small black arrow at the top indicates the migration of YghJ. The positions of molecular mass markers (in kilodaltons) are shown to the right of the gel.
YghJ is required for efficient toxin delivery to intestinal epithelia.
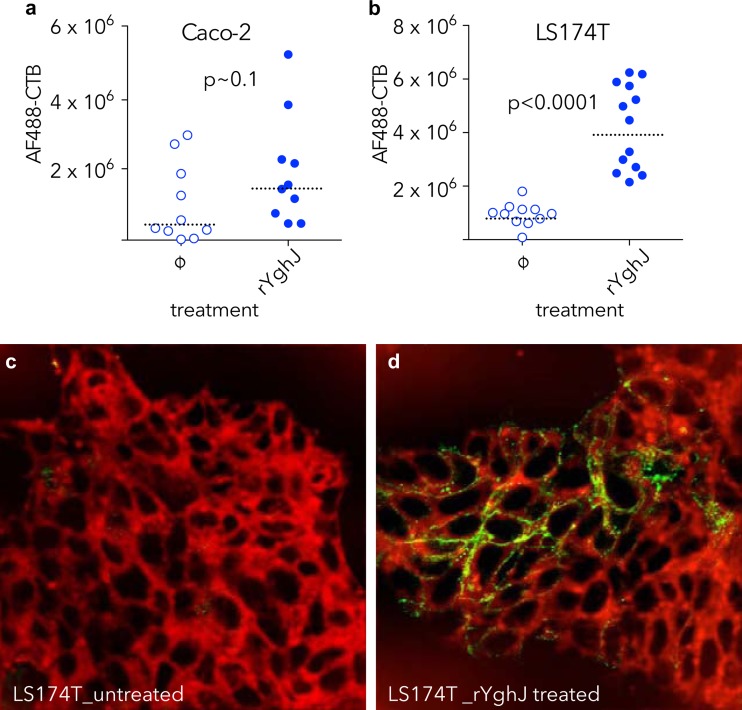
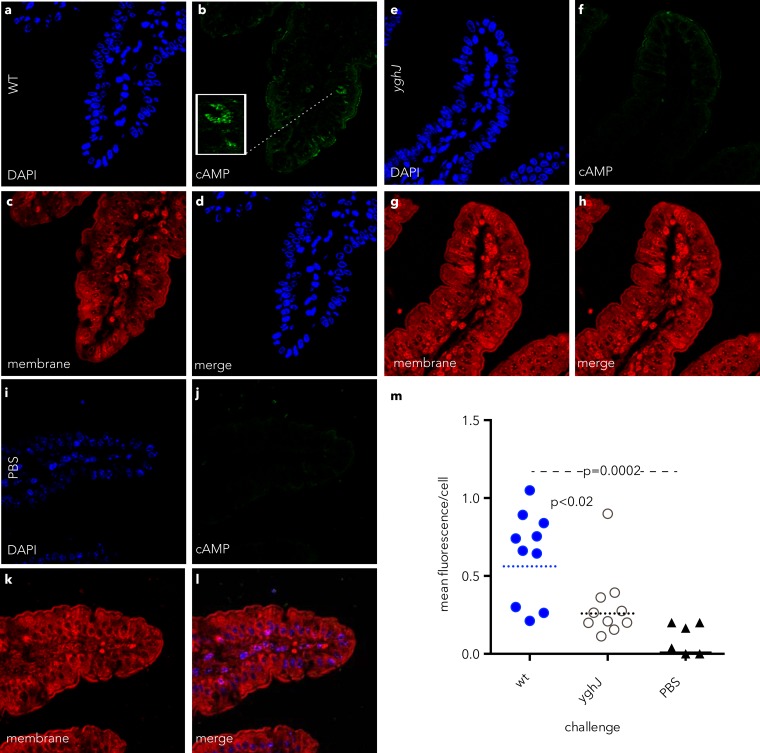
Because earlier studies showed that direct interaction with epithelial cells is required for efficient delivery of heat-labile toxin by ETEC (11), we postulated that degradation of mucins would promote access of toxin to surface receptors. Accordingly, we found that treatment of epithelial cells with YghJ significantly enhanced binding of labeled cholera toxin to the epithelial surface (Fig. 7). Likewise, we reasoned that degradation of mucin in the intestinal lumen would permit direct access of the bacteria to enterocytes and enhance LT-stimulated cAMP activation in target epithelial cells. Indeed, we found substantial increases in cAMP content of small intestinal epithelial cells following infection with wild-type ETEC relative to those infected with the YghJ mutant or unchallenged (PBS-challenged) control mice (Fig. 8).
FIG 7.
YghJ enhances toxin access to enterocytes. (a) Quantitative immunofluorescence data demonstrating binding of fluorescently labeled cholera toxin B-subunit (AF488-CTB) obtained following treatment of Caco-2 cells with rYghJ compared to mock-treated (ø) cells. (b) AF488-CTB binding to LS174T cells is enhanced following rYghJ treatment. P < 0.0001 by Mann-Whitney nonparametric comparison of treated and untreated cell populations. The fluorescence data are normalized to the number of cell nuclei present in each field. (c and d) Images of CTB binding to LS174T cells are shown in panels c (untreated) and d (YghJ treated). Figures represent merged images of membrane (CellMask [red]), and Alexa Fluor 488-conjugated CTB-green. Membrane images were deconvolved in ImageJ, and brightness and contrast were adjusted for clarity, while AF-CTB images were not enhanced following acquisition.
FIG 8.
YghJ leads to enhanced cAMP activation in target intestinal epithelial cells. (a to l) Confocal immunofluorescence images of cAMP produced in small intestinal epithelial cells of mice challenged with wild-type (wt) ETEC H10407 (a to d), the yghJ mutant (e to h), or mock-infected, PBS-challenged control mice (i to l). (m) Quantification of cAMP fluorescence per epithelial cell. The dotted horizontal lines and the solid horizontal line represent geometric mean levels. The values for the wt and yghJ groups were statistically significant by Mann-Whitney (two-tailed) nonparametric comparison (P < 0.02).
As MUC2 forms a critical barrier that limits access of both pathogens and commensal organisms to the epithelial surface of the intestine (13), it has been suggested that MUC2 contributes to elimination of pathogens loosely associated with tissue, as they become trapped in luminal mucin that is removed by peristaltic flow (12). Therefore, we reasoned that in the absence of MUC2 mucinase activity provided by YghJ, mutant bacteria would be more rapidly shed in stools. However, we did not observe consistent differences in fecal shedding following oral challenge with wild-type or yghJ mutant bacteria (data not shown), perhaps suggesting that YghJ plays a more complex role than simply degrading luminal gel-forming mucins or that it shares this ability with other enzymes.
Collectively, however, these studies support the concept that YghJ contributes significantly to the pathogenesis of ETEC, in part by degrading intestinal mucin, thereby permitting these organisms to overcome this important host defense mechanism. By degrading mucin, these organisms gain direct access to epithelial cells required for efficient delivery of their toxin payload.
DISCUSSION
Fundamentally, the ETEC pathovar is defined by the ability to produce and effectively deliver heat-labile toxin (LT) and/or heat-stable toxin (ST) enterotoxins to epithelial receptors where they then cause net fluid secretion and diarrhea (1). Most pathogenic Escherichia coli, including ETEC, have relatively few pathovar-specific genes (30). Consequently, it has been suggested that little more than acquisition of genes encoding the known toxins LT and ST, which are invariably carried on plasmids, may be sufficient for E. coli to effectively deliver these toxins to enterocytes (37). Similarly, it has been suggested that commensal E. coli may act as “genetic sinks” for pathogen evolution (30). The results of the present studies support these hypotheses.
By assimilating the yghJ gene and downstream elements encoding the type II secretion system (T2SS) from commensal strains, ETEC strains have ensured both the secretion of LT via the T2SS and its ultimate delivery to epithelial receptors through the mucinase activity of a second T2SS effector, YghJ. Interestingly, while commensal strains produce and secrete small amounts of YghJ, only ETEC exported this protein in abundance. Therefore, ETEC might gain competitive access to enterocytes by optimizing export of this mucin-degrading protein.
YghJ belongs to a large and diverse family of eukaryotic and prokaryotic proteins containing putative metalloprotease domains (38). Interestingly, enhancin, the prototype molecule of the family of proteases most closely related to YghJ, was isolated from an insect virus and targets insect intestinal mucins (35, 39). It bears a canonical HEXXH metalloprotease motif within a domain (M60-like pfam13402) that is strongly associated with pathogens and commensal organisms that colonize mucosal surfaces (40).
The significant homology between YghJ and V. cholerae AcfD (34) is worthy of comment. AcfD has antivibriocidal activity (33) and is required for efficient intestinal colonization of Vibrio cholerae (34). Given the presence of an enhancin-like protease domain, it has been suggested that AcfD is a mucinase (40). To our knowledge, this has not been tested experimentally. AcfD along with two other lipoproteins encoded by V. cholerae, TcpC, and ToxR-activated gene A (TagA), are regulated by the ToxR virulence regulon (33). Interestingly, TagA (23), encoded on the same V. cholerae pathogenicity island (VPI) that encodes AcfD (41), also is a secreted mucinase. Notably, the hemagglutinin protease of V. cholerae is also a mucinase that is secreted via the T2SS (42, 43), responsible for secretion of cholera toxin (CT). Therefore, V. cholerae appears to be equipped with multiple enzymes with the ability to degrade intestinal mucins, and similar to ETEC, it uses the T2SS to export both an enterotoxin and one or more mucinases that facilitate the delivery of toxin.
The identification of YghJ homologues in a variety of enteric pathogens with known mucinases might suggest that multiple mucin-degrading enzymes could contribute cooperatively to virulence. A variety of different diarrheagenic E. coli produce mucinases (44–46), and the recent O104:H4 outbreak strain, also carries the gene that encodes Pic, an established mucinase shared by enteroaggregative E. coli and Shigella flexneri (44, 47, 48). Interestingly, mucinase activity of Pic and other molecules was established initially with readily available bovine submaxillary mucin (44). However, YghJ had no demonstrable activity against this material, suggesting structural and functional differences in mucinases. Finding other mucinases with different substrate specificities in these pathotypes raises the possibility that other molecules in addition to YghJ that degrade mucin exist in ETEC.
This concept is supported by our accompanying paper (60), which demonstrates that many ETEC strains which secrete the YghJ metalloprotease also secrete EatA, a plasmid-encoded serine protease (7) with MUC2-degrading enzymatic activity.
These enzymes are part of a dynamic complex between ETEC and the mucosa. Interestingly, elevations in intracellular cAMP have been associated with commensurate increases in mucin secretion by enterocytes (49), suggesting that LT, which exerts its toxic effect by stimulating production of this cyclic nucleotide, may itself be a mucin secretagogue. In addition, a wide variety of microbial products or pathogen-associated molecular patterns stimulate production, release, or altered glycosylation of mucins (50). Therefore, it is not surprising that some enteric pathogens make multiple enzymes to subvert this dynamic barrier.
Both intestinal commensal organisms and enteric pathogens must interact with mucins to thrive in their intestinal niche. E. coli (51) and other enteric pathogens such as Yersinia enterocolitica (52) can bind to intestinal mucus and isolated mucus-derived proteins (53), and this may be mediated in part by lipopolysaccharide (LPS) on the surfaces of the organisms. The present studies suggest that ETEC bacteria interact specifically with MUC2, the major intestinal mucin, and that MUC2 appears to colocalize with oligosaccharide of LPS on ETEC. The apparent coating of organisms with MUC2 raises questions regarding the role it could play in modulating both innate and adaptive immune responses to these organisms. YghJ and other mucin-degrading enzymes may allow ETEC and other pathogens to exploit these interactions.
As our understanding of ETEC pathogenesis evolves, new approaches to vaccine development emerge (5, 54, 55). Likewise, current information regarding the nature of E. coli commensalism in the intestine is inadequate, illustrated by the very limited number of “commensal” genomes presently available. Given the apparent paucity of pathotype-specific vaccine targets, it will be important to determine whether highly conserved antigens such as YghJ could be effectively targeted as vaccine candidates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the Department of Veterans Affairs and grant R01AI89894 from the National Institutes of Health. The Washington University School of Medicine Digestive Disease Research Core Center was supported by grant P30DK052574 from the National Institute of Diabetes and Digestive and Kidney Disease, and the Genome Sequencing Center for Infectious Diseases is supported by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272200900009C.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID, NIH, or the VA.
We thank Peter Howard for supplying the yghJ mutant of strain H10407 and the laboratories of Phillip Tarr, Lothar Wieler, and Robyn Klein for supplying cells used in these studies. Finally, we thank Phillip Tarr for reviewing the manuscript and Wandy Beatty for assistance with image processing.
Footnotes
Published ahead of print 18 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01106-13.
REFERENCES
- 1.Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12:89–98. 10.1016/j.micinf.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm AM. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect. Immun. 75:3961–3968. 10.1128/IAI.00459-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qadri F, Svennerholm AM, Faruque AS, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465–483. 10.1128/CMR.18.3.465-483.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Sack DA. 2012. Progress and hurdles in the development of vaccines against enterotoxigenic Escherichia coli in humans. Expert Rev. Vaccines 11:677–694. 10.1586/erv.12.37 [DOI] [PubMed] [Google Scholar]
- 6.Isidean SD, Riddle MS, Savarino SJ, Porter CK. 2011. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29:6167–6178. 10.1016/j.vaccine.2011.06.084 [DOI] [PubMed] [Google Scholar]
- 7.Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786–1794. 10.1128/IAI.72.3.1786-1794.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245–2258. 10.1128/IAI.74.4.2245-2258.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy K, Bartels S, Qadri F, Fleckenstein JM. 2010. Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect. Immun. 78:3027–3035. 10.1128/IAI.00264-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kansal R, Rasko DA, Sahl JW, Munson GP, Roy K, Luo Q, Sheikh A, Kuhne KJ, Fleckenstein JM. 2013. Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect. Immun. 81:259–270. 10.1128/IAI.00919-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsey FC, Fischer JF, Fleckenstein JM. 2006. Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell. Microbiol. 8:1516–1527. 10.1111/j.1462-5822.2006.00736.x [DOI] [PubMed] [Google Scholar]
- 12.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 6:e1000902. 10.1371/journal.ppat.1000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9:265–278. 10.1038/nrmicro2538 [DOI] [PubMed] [Google Scholar]
- 14.Strozen TG, Li G, Howard SP. 2012. YghG (GspSbeta) is a novel pilot protein required for localization of the GspSbeta type II secretion system secretin of enterotoxigenic Escherichia coli. Infect. Immun. 80:2608–2622. 10.1128/IAI.06394-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahl JW, Steinsland H, Redman JC, Angiuoli SV, Nataro JP, Sommerfelt H, Rasko DA. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79:950–960. 10.1128/IAI.00932-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol. Direct 7:12. 10.1186/1745-6150-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D, Harlow E. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J. Biol. Chem. 286:29771–29779. 10.1074/jbc.M111.251546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Klinken BJ, Oussoren E, Weenink JJ, Strous GJ, Buller HA, Dekker J, Einerhand AW. 1996. The human intestinal cell lines Caco-2 and LS174T as models to study cell-type specific mucin expression. Glycoconj. J. 13:757–768. 10.1007/BF00702340 [DOI] [PubMed] [Google Scholar]
- 21.Bu XD, Li N, Tian XQ, Huang PL. 2011. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell 43:201–206. 10.1016/j.tice.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 22.Davies JR, Carlstedt I. 2000. Isolation of large gel-forming mucins. Methods Mol. Biol. 125:3–13 [DOI] [PubMed] [Google Scholar]
- 23.Szabady RL, Yanta JH, Halladin DK, Schofield MJ, Welch RA. 2011. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology 157:516–525. 10.1099/mic.0.044529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis WG, Robinson LS, Perry J, Bick JL, Peipert JF, Allsworth JE, Lewis AL. 2012. Hydrolysis of secreted sialoglycoprotein immunoglobulin A (IgA) in ex vivo and biochemical models of bacterial vaginosis. J. Biol. Chem. 287:2079–2089. 10.1074/jbc.M111.278135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen KP, Randolph MM, Fleckenstein JM. 2006. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74:869–875. 10.1128/IAI.74.2.869-875.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris JA, Roy K, Woo-Rasberry V, Hamilton DJ, Kansal R, Qadri F, Fleckenstein JM. 2011. Directed evaluation of enterotoxigenic Escherichia coli autotransporter proteins as putative vaccine candidates. PLoS Negl. Trop. Dis. 5:e1428. 10.1371/journal.pntd.0001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7066–7071. 10.1073/pnas.092152899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077. 10.1073/pnas.0915077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshima K, Toh H, Ogura Y, Sasamoto H, Morita H, Park SH, Ooka T, Iyoda S, Taylor TD, Hayashi T, Itoh K, Hattori M. 2008. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 15:375–386. 10.1093/dnares/dsn026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 32.Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. 2010. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol. Genet. Genomics 283:469–484. 10.1007/s00438-010-0532-9 [DOI] [PubMed] [Google Scholar]
- 33.Parsot C, Taxman E, Mekalanos JJ. 1991. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 88:1641–1645. 10.1073/pnas.88.5.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Granados RR. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. U. S. A. 94:6977–6982. 10.1073/pnas.94.13.6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717. 10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822–5831. 10.1128/JB.00710-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlings ND, Barrett AJ. 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248:183–228 [DOI] [PubMed] [Google Scholar]
- 39.Lepore LS, Roelvink PR, Granados RR. 1996. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebr. Pathol. 68:131–140. 10.1006/jipa.1996.0070 [DOI] [PubMed] [Google Scholar]
- 40.Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP. 2012. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS One 7:e30287. 10.1371/journal.pone.0030287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karaolis DK, Lan R, Kaper JB, Reeves PR. 2001. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect. Immun. 69:1947–1952. 10.1128/IAI.69.3.1947-1952.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbye LJ, Sandkvist M, Bagdasarian M. 1993. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 132:101–106. 10.1016/0378-1119(93)90520-D [DOI] [PubMed] [Google Scholar]
- 43.Sikora AE, Zielke RA, Lawrence DA, Andrews PC, Sandkvist M. 2011. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J. Biol. Chem. 286:16555–16566. 10.1074/jbc.M110.211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, Murphy E, Cross A, Sztein MB, Nataro JP. 2011. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 108:12881–12886. 10.1073/pnas.1101006108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45:277–288. 10.1046/j.1365-2958.2002.02997.x [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez-Jimenez J, Arciniega I, Navarro-Garcia F. 2008. The serine protease motif of Pic mediates a dose-dependent mucolytic activity after binding to sugar constituents of the mucin substrate. Microb. Pathog. 45:115–123. 10.1016/j.micpath.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 48.Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. 2009. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect. Immun. 77:2465–2473. 10.1128/IAI.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradbury NA. 2000. Protein kinase-A-mediated secretion of mucin from human colonic epithelial cells. J. Cell. Physiol. 185:408–415. [DOI] [PubMed] [Google Scholar]
- 50.Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G. 2013. Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 10:211–220. 10.1038/nmeth.2371 [DOI] [PubMed] [Google Scholar]
- 51.Cohen PS, Arruda JC, Williams TJ, Laux DC. 1985. Adhesion of a human fecal Escherichia coli strain to mouse colonic mucus. Infect. Immun. 48:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mantle M, Husar SD. 1994. Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety. Infect. Immun. 62:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen PS, Rossoll R, Cabelli VJ, Yang SL, Laux DC. 1983. Relationship between the mouse colonizing ability of a human fecal Escherichia coli strain and its ability to bind a specific mouse colonic mucous gel protein. Infect. Immun. 40:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darsley MJ, Chakraborty S, DeNearing B, Sack DA, Feller A, Buchwaldt C, Bourgeois AL, Walker R, Harro CD. 3 October 2012. The oral, live attenuated enterotoxigenic Escherichia coli vaccine ACE527 reduces the incidence and severity of diarrhea in a human challenge model of diarrheal disease. Clin. Vaccine Immunol. 10.1128/CVI.00364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svennerholm AM, Lundgren A. 2012. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert Rev. Vaccines 11:495–507. 10.1586/erv.12.12 [DOI] [PubMed] [Google Scholar]
- 56.Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans DJ, Jr, Evans DG. 1973. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect. Immun. 8:322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements ML, Cheney C, Patnaik R. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DuPont HL, Formal SB, Hornick RB, Snyder MJ, Libonati JP, Sheahan DG, LaBrec EH, Kalas JP. 1971. Pathogenesis of Escherichia coli diarrhea. N. Engl. J. Med. 285:1–9. 10.1056/NEJM197107012850101 [DOI] [PubMed] [Google Scholar]
- 60.Kumar P, Luo Q, Vickers TJ, Sheikh A, Lewis WG, Fleckenstein JM. 2014. EatA, an immununogenic protective antigen of enterotoxigenic Escherichia coli, degrades intestinal mucin. Infect. Immun. 82:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.