Abstract
The complement system is an important first line of defense against the human pathogen Haemophilus influenzae. To survive and propagate in vivo, H. influenzae has evolved mechanisms for subverting this host defense, most of which have been shown to involve outer surface structures, including lipooligosaccharide glycans and outer surface proteins. Bacterial defense against complement acts at multiple steps in the pathway by mechanisms that are not fully understood. Here we identify outer membrane protein P5 as an essential factor in serum resistance of both H. influenzae strain Rd and nontypeable H. influenzae (NTHi) clinical isolate NT127. P5 was essential for resistance of Rd and NT127 to complement in pooled human serum. Further investigation determined that P5 expression decreased cell surface binding of IgM, a potent activator of the classical pathway of complement, to both Rd and NT127. Additionally, P5 expression was required for NT127 to bind factor H (fH), an important inhibitor of alternative pathway (AP) activation. Collectively, the results obtained in this work highlight the ability of H. influenzae to utilize a single protein to perform multiple protective functions for evading host immunity.
INTRODUCTION
Haemophilus influenzae is a pathogenic Gram-negative bacterium that colonizes the human nasopharynx and can invade the mucosal epithelium or disseminate to other sites, causing otitis media, upper and lower respiratory tract infections, and meningitis. A vaccine targeting the polyribosylribitol phosphate capsule of the most invasive serotype, H. influenzae type b (Hib), was introduced in the early 1990s, effectively reducing the incidence of Hib disease (1), although it remains significant in countries lacking vaccine coverage. Nontypeable Haemophilus influenzae (NTHi) strains lack an outer surface capsule and are therefore unaffected by the Hib vaccine (2). NTHi strains are important causes of sinusitis, conjunctivitis, and pneumonia (3, 4) and are the second most common cause of bacterial otitis media behind Streptococcus pneumoniae (3). NTHi strains are also among the most prevalent organisms found in the lungs of patients with exacerbations of chronic obstructive pulmonary disease (COPD) (5–8) and cystic fibrosis (CF) (9–11). Although NTHi strains are infrequently associated with invasive disease, and most instances of bacteremia occur in children with underlying medical issues (3, 12), emerging evidence suggests that healthy individuals are also at risk of invasive NTHi infection (13–17).
To survive in the host and cause disease, NTHi must defend itself against immune mechanisms. The complement system is an important first line of defense against invading pathogens that mediates lysis of Gram-negative bacteria through terminal complement, targets microbes for phagocytosis by opsonization, and stimulates the inflammatory response (18). Invasive NTHi strains are likely to encounter complement in blood, whereas in noninvasive infections, they are likely to be exposed to complement in the middle ear exudates during otitis media (19, 20), the nasopharyngeal mucosa during inflammation (21, 22), and the lungs during exacerbation of COPD and asthma (23). Moreover, recent evidence indicates that the ability of NTHi strains to resist killing by complement correlates with the severity of pulmonary and invasive disease (24). Thus, bacterial defense against complement appears to be an important feature of both invasive and noninvasive NTHi infections.
Complement activation on a pathogen may proceed through one or more of three pathways: the classical pathway, the mannose-binding lectin (MBL) pathway, or the alternative pathway (AP). All three pathways lead to the deposition of complement protein C3 on the microbial surface and subsequent clearance through phagocytosis of pathogens opsonized with C3 or lytic pathway activation (18). Classical pathway activation is initiated by immunoglobulin (select IgG subclasses or IgM) or C-reactive protein (CRP), bound to the surface of a pathogen (18, 25), whereas the lectin pathway is activated through binding of MBL or ficolins to select surface carbohydrates on microbes. Both pathways lead to the assembly of the classical C3 convertase C4bC2a, which cleaves C3 and promotes downstream activation of the lytic pathway. The AP is activated by the cleavage of C3, which can be initiated through the action of the classical and lectin C3 convertases or by spontaneous hydrolysis of C3 (26). The C3b fragment released from the cleavage of C3 associates with a cleavage product of factor B, Bb, generating the AP C3 convertase. C3b generated by the C3 convertases can stimulate the production of more C3 convertases, effectively amplifying the pathway. Much like the classical and MBL pathways, AP activation results in downstream lytic pathway effects and clearance of pathogens (18).
The mechanisms by which NTHi defends itself against host complement are not fully understood; however, current evidence implicates multiple cell surface structures. Lipooligosaccharide (LOS) glycans are essential for mediating this function, as mutations that truncate the LOS lead to severe defects in complement resistance and virulence in animal models (27–30). Surface proteins have also been shown to be involved, including P6, via an unknown mechanism, and proteins E and F, which were shown to bind the host complement regulator vitronectin (31, 32). Importantly, other complement regulators, such as factor H (fH), factor HL-1, and C4-binding protein (C4BP), are also bound by NTHi as mechanisms of complement defense (31); however, the specific surface structures that mediate these interactions have not been elucidated (31).
We recently identified a role for the periplasmic disulfide oxidoreductase DsbA, an enzyme critical for maturation and stability of proteins exported to the cell surface containing disulfide bonds (33), in the resistance of H. influenzae to complement in human serum (34). Bioinformatic identification of putative DsbA substrates revealed a subset with potential roles in complement resistance (35); the outer membrane protein P5 was selected from this list as a candidate because it is ∼50% identical to Escherichia coli outer membrane protein A (OmpA) (36), a factor previously shown to be important for complement resistance in E. coli (37–39). NTHi P5, a β-barrel protein with eight predicted transmembrane spans, four outer surface loops (40), and a predicted disulfide between C323 and C335, was shown to be required for virulence in a chinchilla ear infection model (41) and has also been implicated in adhesion of H. influenzae to various mucosal surface structures (42–46). However, a role for P5 in complement resistance has not been previously reported. In this study, we elucidate the mechanism of NTHi complement resistance mediated by P5.
MATERIALS AND METHODS
Strains and culture conditions.
H. influenzae RdAW (referred to here as Rd) (GenBank accession no. NZ_ACSM00000000), a capsule-deficient serotype d derivative (47), and nontypeable H. influenzae strain NT127 (GenBank accession no. NZ_ACSL01000014.1), originally isolated from the blood of a child with meningitis (27, 48, 49), were grown in brain heart infusion (BHI) broth supplemented with 10 μg/ml hemin and 10 μg/ml NAD (sBHI) or on sBHI agar plates at 35°C. Development of competence for transformation of H. influenzae was accomplished as previously described (50). For selection of Rd- and NTHi-derived strains, the following antibiotics were used: 8 μg/ml tetracycline (Tc), 20 μg/ml kanamycin (Km), and 10 μg/ml gentamicin (Gm). For strain generation, plasmids and PCR products were constructed by using standard molecular biology techniques (51). For complementation of mutants, DNA fragments were amplified by PCR, cloned between adjacent SapI restriction sites of the chromosomal delivery vector pXT10, linearized, and used to transform H. influenzae strains, as previously described (47).
P5 mutant strain construction.
P5 mutant strains RP5G and NTP5V were constructed by replacement of the coding sequences of P5 (HI1164 in Rd and HIAG_00526 in NT127, respectively) with the gentamicin resistance gene from the aacC1 Gm resistance cassette via PCR “stitching.” First, three overlapping fragments were generated, representing the 1,008-bp region immediately 5′ of the P5 translational start codon (primers 5omp1 [5′-TGCTACTCTCACTTAATTCAAGCGCAT-3′] and 3omp1 [5′-TGCTGCTGCGTAACATTTTGATGTCCTCTATTTAGTGATCGAATAGT-3′]), the 537-bp coding region of the gentamicin resistance gene (primers 5gent2 [5′-ATGTTACGCAGCAGCAACGATGTT-3′] and 3gent2 [5′-TTAGGTGGCGGTACTTGGGTCGAT-3′]), and a 1,477-bp region immediately 3′ of the P5 translational termination codon (primers 5omp2 [5′-AAGTACCGCCACCTAATTTTAGTATTTGTTTAACGAAAGATTAAATACAGCA-3′] and 3omp2 [5′-TTAGATAAACTAACTCGTTATCCAGATGCGA-3′]). Subsequently, these fragments were assembled by using overlap extension PCR with primers 5omp1 and 3omp2. The resulting 2,990-bp exchange fragment was transformed into competent cells of strain RdAW or strain NTV (a version of NT127 carrying a modified xylose locus for efficient recombination with plasmid pXT10 and its derivatives [48]) and selected on medium containing Gm to create strains RP5G and NTP5V, respectively.
Complementation of the mutations in RP5G and NTP5V was performed by generating a 1,503-bp fragment containing the Rd P5 coding region and 441 bp of sequence immediately upstream of the P5 translational start site using primers 5pOmpAHA (5′-AAAGCTCTTCAATGAAAAAAACTGCAATCGCATTAGTAGT-3′) and 3OmpAS (5′-TTTGCTCTTCTTTATTTAGTACCGTTTACCGCGATTTCTACA-3′), which introduce SapI sites in the termini of the fragments. The resulting 1,458-bp fragment was digested with SapI and ligated between SapI restriction sites of the chromosomal delivery vector pXT10, which does not replicate in H. influenzae (47). Ligated products were used to amplify 1,373-bp fragments including 932 bp of the 5′ xylA-flanking sequence fused to the 441-bp putative promoter region of P5 (primers PXT10thyAF [5′-AGGGCTTGAATCGCACCTCCA-3′] and 3P51 [5′-TTTGATGTCCTCTATTTAGTGATCGAATAGT-3′]). Next, PCR stitching (primers pXT10thyAF and 3revRfaD1 [5′-AACAGGCTACGATAAACCATTCAAAACAGT-3′]) was used to join the 1,373-bp fragments with 1,063-bp fragments containing the P5 coding sequence amplified from either Rd or NT127 (primers p5switch [5′-ACTATTCGATCACTAAATAGAGGACATCAAAATGAAAAAAACTGCAATCGCATTAGTAGT-3′] and 3ompkan1 [5′-CATCAGAGATTTTGAGACACGGGCCTCTTATTTAGTACCGTTTACCGCGATTTCTACA-3′]) and a 2,716-bp PCR product containing the Km resistance (Kmr) gene and homology to xylB amplified from a kanamycin-marked derivative of pXT10 (primers 5pkan1 [5′-GAGGCCCGTGTCTCAAAATCTCTGATG-3′] and 3revRfaD1). The resulting 5,072-bp fragments were introduced into strain RP5G (Rd P5-containing fragment) or NTP5V (NT127 P5-containing fragment), and transformants were selected on Km, generating strains RP5X and NTP5X, respectively.
Growth analysis.
Strains were cultured in triplicate in sBHI broth at 35°C for 16 h (starting inoculum of an optical density at 600 nm [OD600] of 0.01) in a Versamax microplate reader (Molecular Devices, Sunnyvale, CA) set to read the absorbance at 600 nm every 10 min. Growth yields were obtained by calculating the averages and standard deviations of the final readings of each triplicate set of wells. Doubling times were determined by using nonlinear regression analysis with an R2 value of >0.995 (Prism 5.03; GraphPad Software, La Jolla, CA) and are reported as the averages and standard deviations of each triplicate set of wells.
Serum bactericidal assay.
The sensitivity of P5 mutants to serum was determined as previously described (52). Briefly, strains from log-phase cultures were diluted in Hanks' balanced salt solution (HBSS) with 0.15 mM calcium and 1 mM magnesium (HBSS++) to 1.3 × 104 CFU/ml and incubated at 37°C for 30 min with or without pooled normal human serum (NHS) from healthy anonymous donors aged 18 to 65 years (final concentrations are specified in the figures) (Innovative Research, Novi, MI) and plated onto sBHI agar for CFU enumeration. The reaction was also performed in the presence or absence of 10 mM Mg2+ EGTA to block the classical and lectin pathways and selectively activate the alternative pathway. Heat-inactivated serum used in this assay was generated by incubation of NHS at 56°C for 30 min. C1q-depleted sera and purified C1q were obtained from Complement Technologies, Inc. (Tyler, TX). Results of the serum bactericidal assay are reported as percent survival, which was calculated by dividing the CFU recovered from serum-treated samples by the CFU recovered from the sample that lacked serum. Statistical analyses were performed by using one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test (Prism 5.03; GraphPad Software, La Jolla, CA).
SDS and polymyxin B assays.
Sensitivity to SDS was determined by plating 1:10 serial dilutions (five total) of triplicate log-phase cultures of each strain onto sBHI agar containing 20, 50, 75, or 100 μg/ml of SDS. After a 24-h incubation at 35°C, growth was evaluated by comparing colony formation of strains grown on sBHI agar with that of strains grown on sBHI agar supplemented with SDS. Equivalent CFU numbers between normal sBHI agar and SDS-containing agar were scored as “normal growth,” whereas reduced CFU numbers (sensitivity of detection was 1 colony) on SDS-containing agar compared with those on sBHI agar were scored as “minimal growth.” An absence of CFU on SDS plates was scored as “growth inhibited.” For evaluating sensitivity to polymyxin B, strains were inoculated into sBHI broth containing polymyxin B at final concentrations of 100, 200, 300, 400, 500, and 600 nM or with sBHI alone. Cultures were incubated at 35°C for 16 h in a Versamax microplate reader (Molecular Devices, Sunnyvale, CA) set to read the absorbance at 600 nm at the end of the incubation period. Sensitivity was scored as a relative growth yield, as assessed by OD600 values.
Complement binding.
Western blotting for assessment of binding of C4 and iC3b was performed as previously described (30, 53). Purified iC3b was purchased from Complement Technologies (Tyler, TX). Briefly, log-phase cultures of each strain were washed and suspended in HBSS++ (final reaction mixture volume, 0.5 ml). NHS was added to a final concentration of 10% and incubated for 30 min at 37°C. Bacteria were lysed in 1× SDS-PAGE sample buffer, and lysates were separated on 4 to 12% SDS-PAGE gels for immunoblotting with primary antibodies (Abs) to human iC3b (monoclonal Ab [MAb] G-3E, a kind gift of Kyoko Iida [54]) and C4 (sheep polyclonal anti-human C4; Biodesign/Meridian Life Science, Inc., Memphis, TN) and alkaline phosphatase-conjugated secondary antibodies, as described previously (53, 55). Band densities were calculated by densitometry using ImageJ (National Institutes of Health, Bethesda, MD).
Flow cytometry.
Measurement of complement component C4, fH, IgG, or IgM binding was performed as described previously (56). Briefly, log-phase bacteria were washed and suspended in HBSS++ to a density of 108 CFU/ml. The bacteria were then incubated with 5% NHS for 30 min at 37°C. Bacterium-bound C4 fragments and human Abs were detected with anti-human C4 (Biodesign/Meridian), IgM (Sigma), and IgG (Sigma) fluorescein isothiocyanate (FITC)-conjugated polyclonal antibodies (Sigma). fH was detected by using an anti-fH MAb (MAb 90X, catalog no. A254; Quidel Corporation) followed by anti-mouse IgG FITC (Sigma).
RESULTS
Construction and characterization of P5 mutants.
P5 mutants were constructed by replacing their entire protein-coding regions with the protein-coding region of aacC1, encoding gentamicin resistance, to generate nonpolar deletions in Rd and in NT127, a clinical NTHi strain isolated from the cerebrospinal fluid of a patient with meningitis (48). The amino acid sequence of P5 varies between strains (see Fig. S1 in the supplemental material). Therefore, complementation was achieved by expressing each strain's respective P5 allele at the xylose locus, as previously described (47). The set of isogenic strains comprised the parent strain (Rd), an Rd P5 mutant (RP5G), a complemented Rd P5 mutant (RP5X), the NT127 parent strain carrying the “empty vector” (NTV), an NT127 P5 mutant carrying the empty vector (NTP5V), and a complemented NT127 P5 mutant (NTP5X) (Table 1).
TABLE 1.
Strains used in this study
| Strain | Genotype, description, and/or relevant feature(s) | Reference |
|---|---|---|
| NT127 | Nontypeable H. influenzae clinical isolate | 48 |
| NTV | NT127 xylAΔ4–804::tetAR; tetAR sequence from pXT10 replaces xylA | 48 |
| NTP5V | NT127 ΔP5::aacC1 xylAΔ4–804::tetAR; P5 deletion mutant with tetAR Tetr cassette replacing xylA | This study |
| NTP5X | NT127 ΔP5::aacC1 xylAΔ4–804::P5; P5 deletion mutant complemented with P5 expressed via the P5 promoter in place of xylA | This study |
| Rd | RdAW; wild type; H. influenzae capsule-deficient type d | 77 |
| RP5G | Rd ΔP5::aacC1; P5 deletion mutant | This study |
| RP5X | Rd ΔP5::aacC1 xylAΔ4–804::P5; P5 deletion mutant complemented with Rd P5 expressed via the Rd P5 promoter in place of xylA | This study |
The strains were evaluated for in vitro growth in rich media. Rd P5 mutant strain RP5G exhibited generation times and growth yields similar to those of parent strain Rd or complemented strain RP5X (Table 2). However, NT127 P5 mutant strain NTP5V exhibited 57% and 50% increases in generation time and 21% and 26% decreases in growth yield compared with parent strain NTV and complemented strain NTP5X, respectively (Table 2). These data suggest that P5 is important for optimal growth of NTHi strains but not for growth of Rd.
TABLE 2.
Growth phenotypes of P5 mutants
| Strain | Mean growth rate (min) ± SD | Mean growth yield (OD600) ± SD |
|---|---|---|
| Rd | 43.3 ± 2.0 | 0.63 ± 0.05 |
| RP5G | 47.4 ± 1.0 | 0.60 ± 0.02 |
| RP5X | 46.4 ± 0.3 | 0.69 ± 0.03 |
| NTV | 39.4 ± 1.3 | 0.78 ± 0.03 |
| NTP5V | 62.1 ± 3.5a | 0.61 ± 0.03a |
| NTP5X | 41.2 ± 0.4 | 0.83 ± 0.01 |
Statistical comparison determined by using the Kruskal-Wallis test (P = 0.0273) with Dunn's posttest (P < 0.05) (significant between NTV and NTP5V for growth rate and between NTP5V and NTP5X for yield).
In considering P5 as a candidate mediator of complement resistance, it was important to evaluate potential indirect effects on cell surface composition and stability. The outer surface LOS structures are critical mediators of serum resistance of H. influenzae that could potentially be altered by P5 (27, 28, 57, 58). By silver staining of SDS-PAGE gels, the LOS bands were found to have similar mobility between Rd, RP5G, and RP5X or between NTV and NTP5V (see Fig. S2 in the supplemental material), suggesting that P5 does not mediate structural changes in the LOS. To evaluate potential effects of P5 on membrane stability, we examined whether the loss of P5 resulted in enhanced sensitivity to detergents. H. influenzae strains exposed to a range of SDS concentrations exhibited similar sensitivities at all doses (see Table S1 in the supplemental material). Similarly, no differences were detected in sensitivity to polymyxin B between strains Rd and RP5G (data not shown). Thus, P5 mutants resist membrane disruption by both a negatively and a positively charged detergent to the same extent as their wild-type counterparts, suggesting that P5 mutants are not defective for outer membrane stability.
P5 mutants exhibit increased sensitivity to killing by human serum.
To investigate a potential role of P5 in complement resistance of H. influenzae, strains were assayed for survival in the presence of normal human serum (NHS). To exclude potential effects of variable growth rates between strains, serum bactericidal assays were performed with bacteria resuspended in HBSS++, which prevents replication, and viability of all strains was unaffected by incubation in HBSS++ alone (data not shown). A range of serum concentrations was established for wild-type strains Rd and NTV. The average percent survival of Rd was 92.8% ± 21.0% in 1% NHS across 3 independent experiments, including the biological replicates shown in Fig. 1A as well as data from two other experiments (not shown) (n = 9), with an interquartile range of 72.6% to 114.3%. Rd yielded no colonies after incubation in serum at concentrations of 2% or higher. The average percent survival of parent strain NTV was 18.3% ± 8.1% in 3% NHS across 3 independent experiments, including the biological replicates shown in Fig. 1B as well as data from two other experiments (not shown) (n = 9), with an interquartile range of 12.0% to 26.3%.
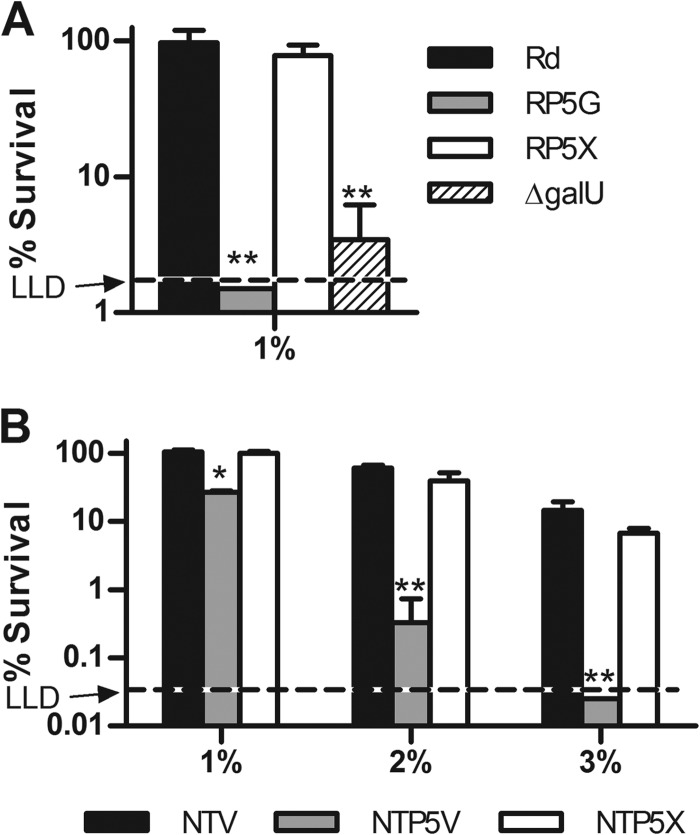
FIG 1.
Effect of the P5 mutation on resistance of H. influenzae to human serum. Strains were treated with NHS for 30 min at 37°C and plated for survivors. (A) Parent strain Rd, Rd P5 mutant strain RP5G, complemented Rd P5 mutant strain RP5X, and the Rd ΔgalU mutant were treated with 1% NHS. The lower limit of detection (LLD) was 1.5%. (B) NTHi parent strain NTV, P5 mutant strain NTP5V, and complemented P5 mutant strain NTP5X were treated with 1%, 2%, or 3% NHS. The LLD was 0.05%. Columns represent the mean percent survival (CFU of treated samples/CFU of untreated samples) of 3 replicates, and error bars indicate standard deviations. Statistical comparisons between parent strains and mutants were done by ANOVA (P = 0.0002 for panel A and P < 0.0001 for panel B) with Tukey's multiple-comparison test (*, P < 0.01; **, P < 0.001). (Survival of Rd after exposure to 2% or 3% NHS was below the LLD [not shown].)
P5 mutant strains were then evaluated for survival in the serum concentrations described above. When incubated in 1% NHS, survival of Rd P5 mutant strain RP5G was reduced to levels below the lower limit of detection (LLD) of the assay, whereas survival of parent strain RDV and complemented strain RP5X was unaffected (Fig. 1A). For comparison, the viability of an H. influenzae Rd mutant carrying a disruption mutation in the coding region of galU (59), which encodes UDP-glucose pyrophosphorylase, an enzyme essential for bacteria to synthesize the LOS outer core (60), was significantly reduced in this assay (survival, 3.5% ± 1.9%). Heat inactivation abrogated the bactericidal effect of serum on the P5 and galU mutants, consistent with an essential role for complement in killing of bacteria in this assay (data not shown).
NTHi parent strain NTV was more serum resistant than Rd, as NTV could partially resist 2% and 3% serum (with survival rates of 61.0% ± 6.0% and 14.6% ± 4.7%, respectively) (Fig. 1B), whereas no colonies were recovered for Rd at the latter two serum concentrations (data not shown). In the presence of 1% serum, survival of NT127 P5 mutant strain NTP5V was reduced by ∼70% compared with parent strain NTV or complemented P5 mutant strain NTP5X, which were unaffected (Fig. 1B). Treatment with 2% NHS reduced survival of NTP5V by ∼99% compared with that of the parent and complemented strains, which were both reduced by ∼40%. Incubation with 3% NHS reduced the survival of the P5 mutant to less than the LLD of 0.25%, which was an average of 584-fold lower than that of the parent strain and 272-fold lower than that of the complemented strain at this concentration. Again, heat inactivation eliminated the bactericidal activity of NHS on all NTHi strains (data not shown). Together, these results indicate that P5 is required for complement resistance of both Rd and an NTHi clinical isolate.
C3 and C4 deposition on P5 mutants.
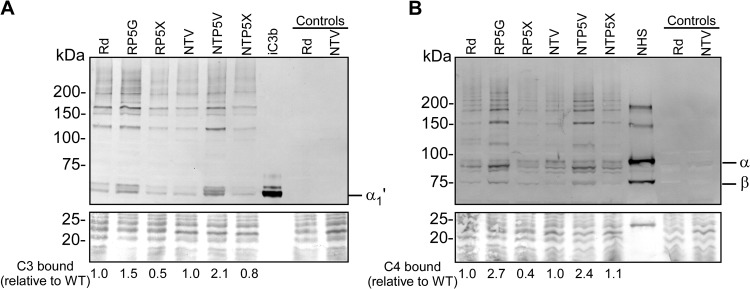
The strains were next evaluated for complement C3 fragment deposition. Activation of C3 results in covalent binding of C3b to bacterial targets; C3b is then converted to iC3b by the actions of factor H and factor I. The amount of iC3b covalently bound to bacteria was measured by incubating strains in 5% NHS and performing Western blotting with a monoclonal antibody directed against a neoepitope on the α1′ chain of iC3b, a cleavage product of C3b (54, 55). Detection of the α1′ chain of iC3b covalently bound to bacterial targets (the 68-kDa α1′ chain of iC3b migrates as a covalently linked complex with its target) by Western blotting permitted us to determine whether targets for C3b/iC3b deposition were altered by the loss of P5. The amount of C3 deposition on each mutant relative to that on the wild-type strain was evaluated by determining the total amount of iC3b bound by using densitometric analysis of visible bands in each lane. As expected, nonopsonized strain Rd or strain NTV controls did not produce detectable iC3b signals on Western blots (Fig. 2A). Deposition of iC3b on RP5G was increased by 1.5-fold compared to that on parent strain Rd and by 3-fold relative to that on complemented P5 mutant strain RP5X (Fig. 2A). It is not clear why complementation reduced iC3b binding to a level somewhat below that of the wild type; however, insertion of the gene encoding P5 (HI1164) at the xyl locus may lead to increased expression as a result of the change in genomic location. Similar to data obtained for Rd strains, iC3b deposition on NTP5V was increased 2.1-fold compared to that on its parent strain, NTV, and was increased 6.8-fold compared to that on complemented strain NTP5X (Fig. 2A). The targets for C3 fragments on the P5 knockout mutants were similar to those on the wild-type and complemented strains.
FIG 2.
Effect of P5 mutation on binding of complement components C3 and C4. The strains listed were incubated with 5% NHS for 30 min at 37°C. Controls are nonopsonized Rd and NTV for specificity and purified iC3b or NHS to visualize complement fragments. (A) Western blot with primary anti-human anti-iC3b antibody and secondary anti-human alkaline phosphatase-conjugated antibody. The position of the 68-kDa α1′ iC3b fragment is denoted α1′. (B) Western blot with anti-human C4 polyclonal antibody and secondary anti-human alkaline phosphatase-conjugated antibody. NHS alone shows the 95-kDa α chain (α), the 75-kDa β chain (β), and the 33-kDa γ chain (γ is not present, as the lower section of the blot was used for protein staining); higher-molecular-mass bands in this lane represent unreduced or partially reduced precursors. The bottom portion of each gel was stained with Coomassie blue to serve as a loading control. C3 or C4 binding relative to the wild type was determined by using densitometry of total visible bands in each lane.
The classical pathway is important to initiate killing of NTHi (61). C4b is an essential component of the classical pathway C3 convertase, and increased C4b deposition on the surface of H. influenzae results in greater bactericidal activity (28). To determine if mutation of P5 affects C4b deposition onto H. influenzae, strains were incubated in 5% NHS and evaluated for total C4b deposition by Western blotting with an anti-C4 polyclonal antibody. NHS alone served as a positive control for the ∼95-kDa and ∼75-kDa α and β chains, respectively (Fig. 2B). Activation of C4 results in cleavage of its α chain to the ∼87-kDa α′ chain, which binds covalently (through either ester or amide linkages) and migrates as a complex with its bacterial targets. C4b binding relative to the wild type was determined by using densitometry of visible bands in each lane. RP5G was found to bind 2.7-fold more C4b than Rd and 6.75-fold more C4 than complemented strain RP5C (Fig. 2B). Strain NTP5V bound 2.4-fold more C4b than NTV and 2.2-fold more C4b than NTP5X (Fig. 2B). As seen with iC3b, targets for C4b on the wild type and the P5 deletion mutants were similar. Taken together with results shown in Fig. 2A, the data indicate that P5 plays a role in the inhibition of classical pathway activation on the surface of H. influenzae strains. The increased C3 fragment deposition that accompanies the loss of P5 may result from increased classical pathway activation and/or independently from increased alternative pathway activation and is examined below.
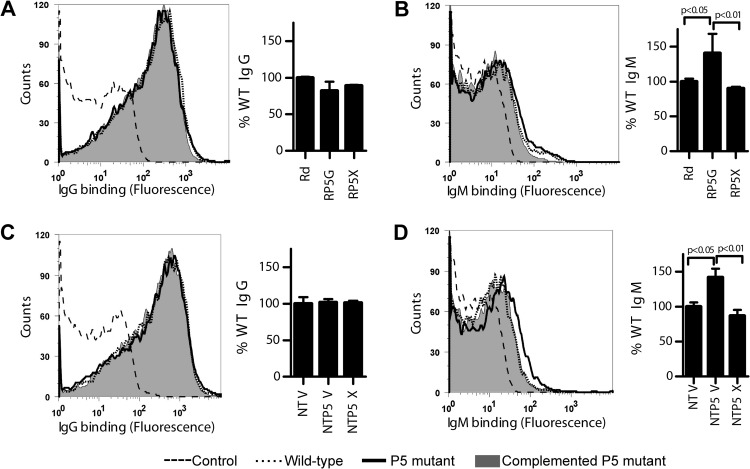
Immunoglobulin binding to P5 mutants.
Antibody binding to the surface of a pathogen initiates complement activation via the classical pathway. We evaluated the effects of P5 on antibody binding to the surface of H. influenzae. Strains were incubated with NHS, and binding of IgG and IgM was measured by flow cytometry. Data are represented as the percentage of wild-type levels (strain Rd or NTV). Levels of IgG binding were similar between the parent strains, P5 mutants, and complemented strains for both Rd and NT127 (Fig. 3A and C). In contrast, IgM binding to the P5 mutant was increased by ∼50% over that of the Rd and NT127 parent and complemented strains (Fig. 3B and D), and these differences were statistically significant. The observed increase in binding of IgM to the surface of P5 mutants could contribute to their enhanced killing by normal human serum.
FIG 3.
Effect of P5 mutation on binding of serum antibodies. Parent strain Rd, P5 mutant strain RP5G, and complemented P5 mutant strain RP5X (A and B) or NTHi parent strain NTV, NTHi P5 mutant strain NTP5V, and complemented NTHi P5 mutant strain NTP5X (C and D) were incubated with 5% NHS for 30 min at 37°C and assayed for binding of IgG or IgM by flow cytometry. Histograms are representative of flow cytometry data from one of the replicates for each strain. On bar graphs, % WT indicates the median fluorescence of each strain relative to that of the wild type (Rd or NTV). Bars represent the means of three independent replicates, and error bars represent standard deviations. Statistical comparisons were done by ANOVA (P = 0.0005 for panel B and P = 0.0008 for panel D) with Tukey's multiple-comparison test (posttest results are shown on graphs).
Resistance to alternative pathway activation.
The P5 mutants were next examined for their sensitivity to the AP alone. Strains were treated with NHS in buffer in the presence or absence of 10 mM Mg2+ EGTA, which inhibits classical/MBL pathway activation, and assayed for survival. Incubation with Mg2+ EGTA-containing buffer alone did not decrease the viability of any of the strains (data not shown). When strains were incubated in 20% NHS in the presence of Mg2+ EGTA, survival of the Rd P5 mutant was similar to that of the parent strain (Fig. 4A). In contrast, the NT127 P5 mutant exhibited a statistically significant 2.2-fold decrease in survival compared to that of the parent strain (Fig. 4A). Strains treated with 20% Mg2+ EGTA serum were next analyzed for C3 binding by Western blotting. Consistent with the survival data, C3 binding to the Rd P5 mutant and wild-type strain Rd were similar, whereas the NT127 P5 mutant exhibited a 1.6-fold increase in C3 binding compared to parent strain NTV (Fig. 4B). The bactericidal results obtained with Mg2+ EGTA serum were confirmed by using 20% C1q-depleted serum; C1q is required for classical pathway activation but does not participate in the lectin pathway. C1q-depleted serum (final concentration of 4%) supplemented with C1q at a physiological concentration (70 μg/ml) was used as a control in which all three pathways were intact. Depletion of C1q did not restore survival of the NT127 P5 deletion mutant to wild-type levels (Fig. 4C). Similar survival rates of the NT127 P5 mutant in Mg2+ EGTA serum (Fig. 4A, white bar) (only the AP is functional) and in C1q-depleted serum (Fig. 4C, white bar) (AP and lectin pathways are functional) suggested that the lectin pathway did not contribute to increased killing of the P5 deletion mutant. Supplementation of C1q-depleted serum with purified C1q restored >99% killing of both the wild type and the P5 deletion mutant even in 4% serum, confirming that an intact classical pathway was required for killing at low serum concentrations (Fig. 4C). At higher (20%) serum concentrations, the AP alone could compromise the survival of the P5 deletion mutant for only NT127 and not Rd. Taken together, these data strongly suggest that P5 is important for interfering with AP activity on select strains of H. influenzae.
FIG 4.
Effect of P5 mutation on resistance of H. influenzae to alternative pathway-mediated killing. (A) Parent strain Rd, Rd P5 mutant strain RP5G, NTHi parent strain NTV, and NTHi P5 mutant strain NTP5V were treated with NHS in the presence of 20 mM Mg2+ EGTA for 30 min at 37°C and plated for survivors. The LLD was 0.15%. (B) Western blot with primary anti-human iC3b antibody and secondary anti-human alkaline phosphatase-conjugated antibody. Controls are nonopsonized wild-type strains (Rd and NTV) and purified iC3b for visualization of the 68-kDa α1′ iC3b fragment. The position of the 68-kDa α1′ iC3b fragment is denoted α1′. The bottom portion of the gel was stained with Coomassie blue as a loading control. C3 binding relative to the wild type was determined by using densitometry of total visible bands in each lane. (C) The strains indicated were assayed for survival in either 20% NHS depleted of C1q (−C1q) or 4% C1q-depleted serum supplemented with C1q (+C1q). The LLD was 0.15%. Bars represent the mean percent survival (CFU of treated samples/CFU of untreated samples) of 3 replicates. (D and E) The strains listed were incubated with 10 μg/ml purified fH and assayed for fH binding by flow cytometry using anti-fH monoclonal antibody 90X. Histograms are representative of flow cytometry data from one of the replicates of each strain. On bar graphs, % WT fH indicates averages of the median fluorescence values (background subtracted) of each strain relative to that of the wild type. Error bars indicate the standard deviations, and statistical comparisons were done by a t test (A and D) or ANOVA (P = 0.0005 for panel C) with Tukey's multiple-comparison test (posttest results are shown on graphs).
Inhibition of the AP mediated by NT127 P5 could be the result of binding of AP inhibitor factor H (fH). The NT127 P5 mutant (NTP5V) bound barely detectable amounts of purified fH in a flow cytometry assay compared with the parent strain (NTV) (Fig. 4D). In contrast, binding of fH to the Rd P5 mutant was equivalent to that of the parental strain (Fig. 4E), consistent with the negligible effect of Rd P5 on survival and iC3b binding in serum possessing only the AP (Fig. 4A and B). These results indicate that P5 is required for fH binding to NT127, which constitutes a probable mechanism by which P5 variants can contribute to NTHi AP evasion.
DISCUSSION
Complement is a major effector of the innate immune response and is present in an increased abundance at mucosal surfaces in the context of infection and other inflammatory conditions (31). The association between complement deficiencies and increased susceptibility to infection has long been recognized for invasive Haemophilus infections (62, 63). Evidence that colonization by NTHi at the mucosal surface requires evasion of complement was obtained in a chinchilla model of middle ear infection. Specifically, depletion of complement using cobra venom factor restored virulence to a serum-sensitive NTHi mutant (which could not sialylate its LOS as a result of a deletion of the siaB gene, encoding cytidinemonophospho-N-acetylneuraminic acid synthetase) that was otherwise avirulent in complement-sufficient animals (30). Recently, strains isolated from pulmonary infections were shown to exhibit higher levels of serum resistance than nasopharyngeal isolates (64, 65). While the mechanism of complement-mediated defense against NTHi in the lung is not fully understood, defects in complement-mediated phagocytosis of NTHi have been identified with macrophages isolated from patients with COPD in comparison to those of healthy nonsmokers (66), suggesting the importance of opsonophagocytosis in controlling NTHi in pulmonary infections. For these reasons, in this work, we sought to elucidate new factors involved in complement evasion by NTHi.
We previously found that H. influenzae mutants deficient in periplasmic disulfide bond formation as a result of a mutation in the dsbA-encoded disulfide oxidoreductase were killed more readily by serum complement (34). By informatics-based approaches, outer membrane protein P5 was identified as a candidate DsbA-dependent protein with a potential role in this phenotype (34, 35). In this report, we show that P5 in both Rd and an NTHi strain are required for full serum resistance. P5 regulated the classical pathway, and loss of P5 was associated with increased IgM binding and C4 deposition, with no apparent change in levels of IgG binding (Fig. 2 and 3). Although the increase in IgM binding to P5 mutants was only 50% over the level of binding to wild-type strains, IgM is very efficient at activating complement compared with IgG, as a single IgM molecule is sufficient to engage the C1 complex and initiate the classical pathway (67). Moreover, surveys of clinical NTHi isolates have revealed a correlation between higher levels of IgM binding and decreased serum resistance (64, 65). The epitope targeted by serum IgM on P5 mutants is currently not known; however, IgM that is bactericidal for NTHi in normal human serum is directed primarily against the LOS (68). Thus, the loss of P5 may increase the exposure of IgM-binding epitopes on the LOS, leading to decreased serum resistance of the strain.
It is unclear why P5 mutants bind increased amounts of IgM while IgG levels remain equal between the mutant and wild-type strains. However, a similar observation has been made with Haemophilus ducreyi mutants deficient in an outer surface protein, DsrA (69). dsrA mutants exhibit increased binding of IgM, which was found to be responsible for the increased activation of classical pathway components on the surface of this bacterium, but levels of bound IgG were equivalent between the mutant and wild-type strains (69). It was suggested that DsrA may physically exclude IgM from the surface of this bacterium or that the loss of dsrA results in the upregulation of a novel IgM epitope-containing factor on the surface of the bacterium (69). These scenarios are also plausible in the case of P5 mutants; however, further investigation will be necessary to determine exactly how P5 participates in limiting surface IgM deposition on H. influenzae strains. Potentially, these examples are indicative of a general strategy by which Gram-negative pathogens utilize outer surface proteins to exclude IgM from their surfaces and avoid complement activation.
A protein similar to P5 in E. coli, OmpA, was implicated previously in serum resistance. OmpA has been suggested to bind complement regulatory factor C4BP (38, 39), a host protein that normally functions to limit inappropriate classical pathway activation. Preliminarily studies showed that P5 mutants and wild-type strains bound similar levels of C4BP (S. Ram and C. V. Rosadini, unpublished data). Thus, P5 is unlikely to play a role in the binding of C4BP to our strains. However, we did find that P5 of NT127 is required for defense against alternative pathway activation via its ability to promote the binding of another complement-inhibitory molecule, fH (Fig. 4D). H. influenzae strains that bind fH were shown previously to be more sensitive to NHS when fH was depleted (70). Thus, the significant decrease in fH binding to NTHi P5 mutants strongly suggests a mechanism for their loss of resistance to the AP. Interestingly, P5 was not required for Rd strains to bind fH (Fig. 4E), suggesting that this strain binds fH via an alternative mechanism. Furthermore, outer surface loops of P5, which are likely to be involved in fH binding, are different between Rd and NT127 (see Fig. S1 in the supplemental material), which likely accounts for the difference in function. Of note, in a survey of the serum resistance of 18 clinical NTHi isolates, Marti-Lliteras et al. identified a strain with a predicted truncation of P5 that exhibited a moderate level of serum resistance albeit a level lower than that of 16 of the 17 other strains tested (71). Therefore, it is possible that some clinical isolates may possess an alternative fH-binding mechanism contributing resistance analogous to that of Rd, and it will be of interest to evaluate this possibility with isogenic mutants.
Evidence for alternative fH-binding mechanisms within the species raises the possibility that H. influenzae strains capable of high levels of fH binding may possess more than one binding mechanism, a strategy common to many other organisms, including Neisseria meningitidis (72, 73), Streptococcus pneumoniae (74), Borrelia burgdorferi (75), and Candida albicans (76). Importantly, Hallstrom et al. found that among clinical isolates of NTHi from cases of sepsis, disease severity was correlated with increased serum resistance and binding of complement-inhibitory proteins, including fH (24). An understanding of how pathogenic NTHi strains bind fH and whether differences in this ability involve the acquisition of multiple binding mechanisms is therefore of potential clinical relevance to the severity of invasive infection.
Previously, P5 was implicated in the pathogenesis of H. influenzae as an adherence factor for attachment of H. influenzae to host mucosal structures (42–46). The work presented here describes new functional roles for this abundant outer membrane protein, including limiting the binding of IgM to the bacterial surface and participating in the binding of fH. Overall, P5's role in resistance to both the classical and AP complement pathways highlights the ability of H. influenzae to utilize a single protein to perform diverse virulence-associated functions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health grants AI-095740 and AI-049437 to B.J.A.
Footnotes
Published ahead of print 25 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01224-13.
REFERENCES
- 1.CDC 2002. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children—United States, 1998-2000. JAMA 287:2206–2207. 10.1001/jama.1994.03510400017006 [DOI] [PubMed] [Google Scholar]
- 2.Poolman J, Bakaletz L, Cripps A, Denoel P, Forsgren A, Kyd J. 2000. Developing a nontypeable Haemophilus influenzae (NTHi) vaccine. Vaccine 19:S108–S115. 10.1016/S0264-410X(00)00288-7 [DOI] [PubMed] [Google Scholar]
- 3.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr. Infect. Dis. J. 28:43–48. 10.1097/INF.0b013e318184dba2 [DOI] [PubMed] [Google Scholar]
- 4.Murphy TF. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16:129–134. 10.1097/00001432-200304000-00009 [DOI] [PubMed] [Google Scholar]
- 5.Mannino DM, Kiriz VA. 2006. Changing the burden of COPD mortality. Int. J. Chron. Obstruct. Pulmon. Dis. 1:219–233 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2707151/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. 2004. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 170:266–272. 10.1164/rccm.200403-354OC [DOI] [PubMed] [Google Scholar]
- 7.Murphy TF, Sethi S. 2002. Chronic obstructive pulmonary disease: role of bacteria and guide to antibacterial selection in the older patient. Drugs Aging 19:761–775. 10.2165/00002512-200219100-00005 [DOI] [PubMed] [Google Scholar]
- 8.Sethi S, Murphy TF. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336–363. 10.1128/CMR.14.2.336-363.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saiman L. 2004. Microbiology of early CF lung disease. Paediatr. Respir. Rev. 5(Suppl A):S367–S369. 10.1016/S1526-0542(04)90065-6 [DOI] [PubMed] [Google Scholar]
- 10.Moller LV, Regelink AG, Grasselier H, Dankert-Roelse JE, Dankert J, van Alphen L. 1995. Multiple Haemophilus influenzae strains and strain variants coexist in the respiratory tract of patients with cystic fibrosis. J. Infect. Dis. 172:1388–1392. 10.1093/infdis/172.5.1388 [DOI] [PubMed] [Google Scholar]
- 11.Gilligan PH. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 4:35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Neill JM, St Geme JW, III, Cutter D, Adderson EE, Anyanwu J, Jacobs RF, Schutze GE. 2003. Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas. J. Clin. Microbiol. 41:3064–3069. 10.1128/JCM.41.7.3064-3069.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Shibata H, Nakazawa M, Myokai M, Ikegaya K, Tsuchiya K, Kamimaki T. 2011. Meningitis and septicemia caused by nontypeable Haemophilus influenzae in a previously healthy 2-year-old girl. J. Infect. Chemother. 17:559–562. 10.1007/s10156-011-0213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuthill SL, Farley MM, Donowitz LG. 1999. Nontypable Haemophilus influenzae meningitis. Pediatr. Infect. Dis. J. 18:660–662. 10.1097/00006454-199907000-00024 [DOI] [PubMed] [Google Scholar]
- 15.Nizet V, Colina KF, Almquist JR, Rubens CE, Smith AL. 1996. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173:180–186. 10.1093/infdis/173.1.180 [DOI] [PubMed] [Google Scholar]
- 16.Campos J, Hernando M, Roman F, Perez-Vazquez M, Aracil B, Oteo J, Lazaro E, de Abajo F. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42:524–529. 10.1128/JCM.42.2.524-529.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerquetti M, Ciofi degli Atti ML, Renna G, Tozzi AE, Garlaschi ML, Mastrantonio P. 2000. Characterization of non-type B Haemophilus influenzae strains isolated from patients with invasive disease. The HI Study Group. J. Clin. Microbiol. 38:4649–4652 http://jcm.asm.org/content/38/12/4649.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walport MJ. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058–1066. 10.1056/NEJM200104053441406 [DOI] [PubMed] [Google Scholar]
- 19.Narkio-Makela M, Hellwage J, Tahkokallio O, Meri S. 2001. Complement-regulator factor H and related proteins in otitis media with effusion. Clin. Immunol. 100:118–126. 10.1006/clim.2001.5043 [DOI] [PubMed] [Google Scholar]
- 20.Narkio-Makela M, Teppo AM, Meri S. 2000. Complement C3 cleavage and cytokines interleukin-1beta and tumor necrosis factor-alpha in otitis media with effusion. Laryngoscope 110:1745–1749. 10.1097/00005537-200010000-00035 [DOI] [PubMed] [Google Scholar]
- 21.Van Zele T, Coppieters F, Gevaert P, Holtappels G, Van Cauwenberge P, Bachert C. 2009. Local complement activation in nasal polyposis. Laryngoscope 119:1753–1758. 10.1002/lary.20484 [DOI] [PubMed] [Google Scholar]
- 22.Andersson M, Michel L, Llull JB, Pipkorn U. 1994. Complement activation on the nasal mucosal surface—a feature of the immediate allergic reaction in the nose. Allergy 49:242–245. 10.1111/j.1398-9995.1994.tb02656.x [DOI] [PubMed] [Google Scholar]
- 23.Marc MM, Korosec P, Kosnik M, Kern I, Flezar M, Suskovic S, Sorli J. 2004. Complement factors c3a, c4a, and c5a in chronic obstructive pulmonary disease and asthma. Am. J. Respir. Cell Mol. Biol. 31:216–219. 10.1165/rcmb.2003-0394OC [DOI] [PubMed] [Google Scholar]
- 24.Hallstrom T, Resman F, Ristovski M, Riesbeck K. 2010. Binding of complement regulators to invasive nontypeable Haemophilus influenzae isolates is not increased compared to nasopharyngeal isolates, but serum resistance is linked to disease severity. J. Clin. Microbiol. 48:921–927. 10.1128/JCM.01654-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Clos TW. 2000. Function of C-reactive protein. Ann. Med. 32:274–278. 10.3109/07853890009011772 [DOI] [PubMed] [Google Scholar]
- 26.Pangburn MK, Muller-Eberhard HJ. 1983. Initiation of the alternative complement pathway due to spontaneous hydrolysis of the thioester of C3. Ann. N. Y. Acad. Sci. 421:291–298. 10.1111/j.1749-6632.1983.tb18116.x [DOI] [PubMed] [Google Scholar]
- 27.Wong SM, St Michael F, Cox A, Ram S, Akerley BJ. 2011. ArcA-regulated glycosyltransferase Lic2B promotes complement evasion and pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 79:1971–1983. 10.1128/IAI.01269-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho DK, Ram S, Nelson KL, Bonthuis PJ, Smith AL. 2007. lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J. Immunol. 178:1002–1012 http://www.jimmunol.org/content/178/2/1002.long [DOI] [PubMed] [Google Scholar]
- 29.Hood DW, Makepeace K, Deadman ME, Rest RF, Thibault P, Martin A, Richards JC, Moxon ER. 1999. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33:679–692. 10.1046/j.1365-2958.1999.01509.x [DOI] [PubMed] [Google Scholar]
- 30.Figueira MA, Ram S, Goldstein R, Hood DW, Moxon ER, Pelton SI. 2007. Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect. Immun. 75:325–333. 10.1128/IAI.01054-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallstrom T, Riesbeck K. 2010. Haemophilus influenzae and the complement system. Trends Microbiol. 18:258–265. 10.1016/j.tim.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 32.Su YC, Jalalvand F, Morgelin M, Blom AM, Singh B, Riesbeck K. 2013. Haemophilus influenzae acquires vitronectin via the ubiquitous protein F to subvert host innate immunity. Mol. Microbiol. 87:1245–1266. 10.1111/mmi.12164 [DOI] [PubMed] [Google Scholar]
- 33.Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589. 10.1016/0092-8674(91)90532-4 [DOI] [PubMed] [Google Scholar]
- 34.Rosadini CV, Wong SM, Akerley BJ. 2008. The periplasmic disulfide oxidoreductase DsbA contributes to Haemophilus influenzae pathogenesis. Infect. Immun. 76:1498–1508. 10.1128/IAI.01378-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosadini CV, Gawronski JD, Raimunda D, Arguello JM, Akerley BJ. 2011. A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infect. Immun. 79:3366–3376. 10.1128/IAI.05135-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munson RS, Jr, Grass S, West R. 1993. Molecular cloning and sequence of the gene for outer membrane protein P5 of Haemophilus influenzae. Infect. Immun. 61:4017–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiser JN, Gotschlich EC. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasadarao NV, Blom AM, Villoutreix BO, Linsangan LC. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352–6360 http://www.jimmunol.org/content/169/11/6352.long [DOI] [PubMed] [Google Scholar]
- 39.Wooster DG, Maruvada R, Blom AM, Prasadarao NV. 2006. Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunology 117:482–493. 10.1111/j.1365-2567.2006.02323.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb DC, Cripps A. 1998. Secondary structure and molecular analysis of interstrain variability in the P5 outer-membrane protein of non-typable Haemophilus influenzae isolated from diverse anatomical sites. J. Med. Microbiol. 47:1059–1067. 10.1099/00222615-47-12-1059 [DOI] [PubMed] [Google Scholar]
- 41.Sirakova T, Kolattukudy PE, Murwin D, Billy J, Leake E, Lim D, DeMaria T, Bakaletz L. 1994. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect. Immun. 62:2002–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy MS, Bernstein JM, Murphy TF, Faden HS. 1996. Binding between outer membrane proteins of nontypeable Haemophilus influenzae and human nasopharyngeal mucin. Infect. Immun. 64:1477–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Z, Nagata N, Molina E, Bakaletz LO, Hawkins H, Patel JA. 1999. Fimbria-mediated enhanced attachment of nontypeable Haemophilus influenzae to respiratory syncytial virus-infected respiratory epithelial cells. Infect. Immun. 67:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto N, Bakaletz LO. 1996. Selective adherence of non-typeable Haemophilus influenzae (NTHi) to mucus or epithelial cells in the chinchilla eustachian tube and middle ear. Microb. Pathog. 21:343–356. 10.1006/mpat.1996.0067 [DOI] [PubMed] [Google Scholar]
- 45.Hill DJ, Toleman MA, Evans DJ, Villullas S, Van Alphen L, Virji M. 2001. The variable P5 proteins of typeable and non-typeable Haemophilus influenzae target human CEACAM1. Mol. Microbiol. 39:850–862. 10.1046/j.1365-2958.2001.02233.x [DOI] [PubMed] [Google Scholar]
- 46.Bookwalter JE, Jurcisek JA, Gray-Owen SD, Fernandez S, McGillivary G, Bakaletz LO. 2008. A carcinoembryonic antigen-related cell adhesion molecule 1 homologue plays a pivotal role in nontypeable Haemophilus influenzae colonization of the chinchilla nasopharynx via the outer membrane protein P5-homologous adhesin. Infect. Immun. 76:48–55. 10.1128/IAI.00980-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong SM, Akerley BJ. 2003. Inducible expression system and marker-linked mutagenesis approach for functional genomics of Haemophilus influenzae. Gene 316:177–186. 10.1016/S0378-1119(03)00762-5 [DOI] [PubMed] [Google Scholar]
- 48.Harrington JC, Wong SM, Rosadini CV, Garifulin O, Boyartchuk V, Akerley BJ. 2009. Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated ytfE gene. Infect. Immun. 77:1945–1958. 10.1128/IAI.01365-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U. S. A. 106:16422–16427. 10.1073/pnas.0906627106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321–342 [DOI] [PubMed] [Google Scholar]
- 51.Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K. (ed). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc. New York, NY [Google Scholar]
- 52.Ram S, Ngampasutadol J, Cox AD, Blom AM, Lewis LA, St Michael F, Stupak J, Gulati S, Rice PA. 2007. Heptose I glycan substitutions on Neisseria gonorrhoeae lipooligosaccharide influence C4b-binding protein binding and serum resistance. Infect. Immun. 75:4071–4081. 10.1128/IAI.01109-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ram S, Cox AD, Wright JC, Vogel U, Getzlaff S, Boden R, Li J, Plested JS, Meri S, Gulati S, Stein DC, Richards JC, Moxon ER, Rice PA. 2003. Neisserial lipooligosaccharide is a target for complement component C4b. Inner core phosphoethanolamine residues define C4b linkage specificity. J. Biol. Chem. 278:50853–50862. 10.1074/jbc.M308364200 [DOI] [PubMed] [Google Scholar]
- 54.Iida K, Mitomo K, Fujita T, Tamura N. 1987. Characterization of three monoclonal antibodies against C3 with selective specificities. Immunology 62:413–417 [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis LA, Ram S, Prasad A, Gulati S, Getzlaff S, Blom AM, Vogel U, Rice PA. 2008. Defining targets for complement components C4b and C3b on the pathogenic neisseriae. Infect. Immun. 76:339–350. 10.1128/IAI.00613-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ram S, Cullinane M, Blom AM, Gulati S, McQuillen DP, Monks BG, O'Connell C, Boden R, Elkins C, Pangburn MK. 2001. Binding of C4b-binding protein to porin. J. Exp. Med. 193:281–295. 10.1084/jem.193.3.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erwin AL, Bonthuis PJ, Geelhood JL, Nelson KL, McCrea KW, Gilsdorf JR, Smith AL. 2006. Heterogeneity in tandem octanucleotides within Haemophilus influenzae lipopolysaccharide biosynthetic gene losA affects serum resistance. Infect. Immun. 74:3408–3414. 10.1128/IAI.01540-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffin R, Bayliss CD, Herbert MA, Cox AD, Makepeace K, Richards JC, Hood DW, Moxon ER. 2005. Digalactoside expression in the lipopolysaccharide of Haemophilus influenzae and its role in intravascular survival. Infect. Immun. 73:7022–7026. 10.1128/IAI.73.10.7022-7026.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong SM, Alugupalli KR, Ram S, Akerley BJ. 2007. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol. Microbiol. 64:1375–1390. 10.1111/j.1365-2958.2007.05747.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Priebe GP, Dean CR, Zaidi T, Meluleni GJ, Coleman FT, Coutinho YS, Noto MJ, Urban TA, Pier GB, Goldberg JB. 2004. The galU gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 72:4224–4232. 10.1128/IAI.72.7.4224-4232.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams BJ, Morlin G, Valentine N, Smith AL. 2001. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69:695–705. 10.1128/IAI.69.2.695-705.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ram S, Lewis LA, Rice PA. 2010. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin. Microbiol. Rev. 23:740–780. 10.1128/CMR.00048-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkelstein JA, Moxon ER. 1992. The role of complement in the host's defense against Haemophilus influenzae. J. Infect. Dis. 165(Suppl 1):S62–S65 [DOI] [PubMed] [Google Scholar]
- 64.Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. 2011. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 7:e1001247. 10.1371/journal.ppat.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langereis JD, Stol K, Schweda EK, Twelkmeyer B, Bootsma HJ, de Vries SP, Burghout P, Diavatopoulos DA, Hermans PW. 2012. Modified lipooligosaccharide structure protects nontypeable Haemophilus influenzae from IgM-mediated complement killing in experimental otitis media. mBio 3(4):e00079-12. 10.1128/mBio.00079-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berenson CS, Kruzel RL, Eberhardt E, Sethi S. 2013. Phagocytic dysfunction of human alveolar macrophages and severity of chronic obstructive pulmonary disease. J. Infect. Dis. 208:2036–2045. 10.1093/infdis/jit400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Janeway CA, Travers P, Walport M, Shlomchik MJ. 2001. Immunobiology: the immune system in health and disease, 5th ed. Garland Science, New York, NY [Google Scholar]
- 68.Gnehm HE, Pelton SI, Gulati S, Rice PA. 1985. Characterization of antigens from nontypable Haemophilus influenzae recognized by human bactericidal antibodies. Role of Haemophilus outer membrane proteins. J. Clin. Invest. 75:1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdullah M, Nepluev I, Afonina G, Ram S, Rice P, Cade W, Elkins C. 2005. Killing of dsrA mutants of Haemophilus ducreyi by normal human serum occurs via the classical complement pathway and is initiated by immunoglobulin M binding. Infect. Immun. 73:3431–3439. 10.1128/IAI.73.6.3431-3439.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hallstrom T, Zipfel PF, Blom AM, Lauer N, Forsgren A, Riesbeck K. 2008. Haemophilus influenzae interacts with the human complement inhibitor factor H. J. Immunol. 181:537–545 http://www.jimmunol.org/content/181/1/537.long [DOI] [PubMed] [Google Scholar]
- 71.Marti-Lliteras P, Lopez-Gomez A, Mauro S, Hood DW, Viadas C, Calatayud L, Morey P, Servin A, Linares J, Oliver A, Bengoechea JA, Garmendia J. 2011. Nontypable Haemophilus influenzae displays a prevalent surface structure molecular pattern in clinical isolates. PLoS One 6:e21133. 10.1371/journal.pone.0021133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177:501–510 http://www.jimmunol.org/content/177/1/501.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lewis LA, Ngampasutadol J, Wallace R, Reid JE, Vogel U, Ram S. 2010. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 6:e1001027. 10.1371/journal.ppat.1001027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zipfel PF, Hallstrom T, Hammerschmidt S, Skerka C. 2008. The complement fitness factor H: role in human diseases and for immune escape of pathogens, like pneumococci. Vaccine 26(Suppl 8):I67–I74. 10.1016/j.vaccine.2008.11.015 [DOI] [PubMed] [Google Scholar]
- 75.Kraiczy P, Stevenson B. 2013. Complement regulator-acquiring surface proteins of Borrelia burgdorferi: structure, function and regulation of gene expression. Ticks Tick Borne Dis. 4:26–34. 10.1016/j.ttbdis.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo S, Hoffmann R, Skerka C, Zipfel PF. 2013. Glycerol-3-phosphate dehydrogenase 2 is a novel factor H-, factor H-like protein 1-, and plasminogen-binding surface protein of Candida albicans. J. Infect. Dis. 207:594–603. 10.1093/infdis/jis718 [DOI] [PubMed] [Google Scholar]
- 77.Wong SM, Akerley BJ. 2005. Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol. Microbiol. 55:724–738. 10.1111/j.1365-2958.2004.04439.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.