Abstract
Noroviruses are the leading cause of epidemic acute gastroenteritis in the United States. From September 2009 through August 2013, 3,960 norovirus outbreaks were reported to CaliciNet. Of the 2,895 outbreaks with a known transmission route, person-to-person and food-borne transmissions were reported for 2,425 (83.7%) and 465 (16.1%) of the outbreaks, respectively. A total of 2,475 outbreaks (62.5%) occurred in long-term care facilities (LTCF), 389 (9.8%) in restaurants, and 227 (5.7%) in schools. A total of 435 outbreaks (11%) were typed as genogroup I (GI) and 3,525 (89%) as GII noroviruses. GII.4 viruses caused 2,853 (72%) of all outbreaks, of which 94% typed as either GII.4 New Orleans or GII.4 Sydney. In addition, three non-GII.4 viruses, i.e., GII.12, GII.1, and GI.6, caused 528 (13%) of all outbreaks. Several non-GII.4 genotypes (GI.3, GI.6, GI.7, GII.3, GII.6, and GII.12) were significantly more associated with food-borne transmission (odds ratio, 1.9 to 7.1; P < 0.05). Patients in LTCF and people ≥65 years of age were at higher risk for GII.4 infections than those in other settings and with other genotypes (P < 0.05). Phylogeographic analysis identified three major dispersions from two geographic locations that were responsible for the GI.6 outbreaks from 2011 to 2013. In conclusion, our data demonstrate the cyclic emergence of new (non-GII.4) norovirus strains, and several genotypes are more often associated with food-borne outbreaks. These surveillance data can be used to improve viral food-borne surveillance and to help guide studies to develop and evaluate targeted prevention methods such as norovirus vaccines, antivirals, and environmental decontamination methods.
INTRODUCTION
Norovirus is the most common cause of epidemic and endemic gastroenteritis in people of all ages (1, 2). In the elderly, norovirus gastroenteritis is associated with increased hospitalization and mortality rates (3, 4). Recent studies have also shown that, with the increased use of rotavirus vaccine, norovirus has become the primary cause of medically attended acute gastroenteritis in U.S. children (2). Although outbreaks occur throughout the year, 80% occur between November and April (5). Noroviruses are a group of genetically diverse viruses that belong to the family Caliciviridae and that can be classified into 6 different genogroups (6), of which viruses from genogroup I (GI), GII, and GIV are responsible for disease in humans. GI includes nine genotypes based on the complete major capsid protein (VP1) (7), whereas to date, GII contains 22 genotypes, including three genotypes (GII.11, GII.18, and GII.19) that have been uniquely detected in swine (8). Over the past 15 years, GIV viruses have been detected as the cause of only a handful of outbreaks in humans, while other GIV genotypes have been observed in canine species (9). In humans, GII.4 viruses cause the majority of norovirus-related gastroenteritis outbreaks worldwide (10); in the past decade, new GII.4 strains have been emerging every 2 to 3 years (11, 12). The cyclic emergence of new strains and the displacement of previously predominant strains are believed to be driven by genetic drift and population immunity (13). These new strains are often, but not always, associated with increases in the number of outbreaks (14–16).
The norovirus genome consists of a 7.5-kb, single-stranded, positive-sense RNA containing three open reading frames (ORFs) that encode both structural (ORF2, coding for VP1, and ORF3, coding for VP2) and nonstructural (ORF1) proteins. The icosahedral norovirus particles contain 180 copies of VP1, whereas VP2 is incorporated in lower numbers (17). VP1 is divided into two domains, i.e., the shell (S) and protruding (P) domains. The P domain is subdivided into two subdomains, P1 and P2, with P2 being the hypervariable region of the capsid, containing both the antigenic and histo-blood group antigen binding sites. These features make the P2 region one of the best targets to distinguish norovirus strains and to determine if outbreaks are related (11, 18–21). To better understand epidemiologic and genotypic trends of norovirus outbreaks in the United States, we describe and analyze data reported to CaliciNet between September 2009 and August 2013.
MATERIALS AND METHODS
CaliciNet.
Since 2009, laboratory-based outbreak surveillance of norovirus outbreaks in the United States has been conducted using CaliciNet, which is a surveillance network of state and local public health laboratories coordinated by the CDC (11). Nucleotide sequences of small regions of the VP1 gene (regions C and D) (11) and epidemiologic data (outbreak date, city or county, setting, transmission route, number sick, and ages of patients) were downloaded from the national CaliciNet database. All norovirus outbreak data and investigation results were uploaded by certified state or local public health laboratories (Table 1). In 2011, six certified CaliciNet laboratories, including CDC, were established as CaliciNet Outbreak Support Centers (OSCs) to assist in the genotyping of norovirus outbreaks from states not participating in CaliciNet.
TABLE 1.
Norovirus outbreaks reported to CaliciNet in 2009 to 2013
| State, county, or city laboratory | No. of outbreaksf reported from September through August |
|||
|---|---|---|---|---|
| 2009–2010 | 2010–2011 | 2011–2012 | 2012–2013 | |
| Alabamaa | — | 5 | 27 | 15 |
| Alaskab | 2 | 8 | 4 | 4 |
| Arizonab | — | 11 | 28 | 22 |
| Arkansas | — | — | 6 | 7 |
| Californiac | 204 | 82 | 219 | 193 |
| Los Angeles County | — | — | — | 36 |
| Orange County | — | — | — | 12 |
| Colorado | 41 | 33 | 4 | 7 |
| Connecticutb | — | 1 | — | 3 |
| District of Columbia | — | — | 1 | — |
| Delaware | 2 | 16 | 14 | 11 |
| Florida | — | 2 | 25 | 30 |
| Georgiab | 9 | 4 | 19 | 29 |
| Hawaii | 20 | 6 | 10 | 24 |
| Idahoc | 6 | 10 | 11 | 18 |
| Iowab | — | 7 | 13 | 20 |
| Illinoisb | 4 | 9 | 10 | 8 |
| Indiana | — | 3 | 9 | 17 |
| Kansasb | — | 5 | 7 | 3 |
| Kentuckya | — | 2 | 5 | — |
| Louisianab | 9 | 3 | 5 | 5 |
| Mained | 9 | 46 | 5 | 26 |
| Massachusettsa | 1 | 3 | 6 | 6 |
| Maryland | 47 | 53 | 76 | 36 |
| Michigan | 27 | 58 | 52 | 60 |
| Minnesota | 18 | 9 | 17 | 32 |
| Mississippib | — | 1 | 4 | 9 |
| Missourib | 5 | 15 | 9 | 17 |
| Montanab | — | 3 | 4 | 8 |
| North Carolina | 9 | 10 | 28 | 17 |
| New Hampshire | — | 30 | 36 | 3 |
| New Mexico | 5 | 12 | 9 | 13 |
| Nevada | 2 | — | — | 1 |
| New Yorkc | 1 | 15 | 37 | 40 |
| Ohio | 44 | 55 | 80 | 108 |
| Oregon | 70 | 23 | 79 | 102 |
| Rhode Islandb | 14 | 13 | 16 | 15 |
| South Carolina | 1 | 2 | 50 | 35 |
| South Dakotab | — | 4 | — | 2 |
| Tennesseec | 34 | 28 | 23 | 46 |
| Texas | 13 | — | 4 | 8 |
| Utahb | — | 3 | 3 | 6 |
| Virginia | 58 | 63 | 84 | 106 |
| Vermont | 1 | 2 | 13 | 5 |
| West Virginiab | — | 6 | 5 | 2 |
| Wisconsinc | 43 | 61 | 61 | 121 |
| Milwaukee | — | 8 | 11 | 3 |
| Washingtonb | — | 2 | 1 | — |
| Wyominga | — | 5 | 9 | 3 |
| Totale | 699 | 737 | 1,139 | 1,294 |
States that previously submitted data to CaliciNet OSCs that became certified during the study period.
Laboratories that report norovirus outbreaks through a CaliciNet OSC or the National Calicivirus Laboratory at the CDC.
CaliciNet OSCs are regional laboratories that sequence norovirus outbreaks from non-CaliciNet states.
State that is no longer certified but submits outbreak data to a CaliciNet OSC.
Totals do not include 88 cruise ship outbreaks and 3 outbreaks from Puerto Rico. Oklahoma, Nebraska, New Jersey, North Dakota, or Pennsylvania did not submit data to CaliciNet.
“—” indicates that laboratories were not part of CaliciNet during the indicated years.
P2 amplification and sequencing of GI.6 viruses.
Fecal specimens from 43 GI.6 outbreaks were selected based on geographic origin, outbreak date, and availability. The P2 subdomain was amplified using the Qiagen One-Step reverse transcription (RT)-PCR kit (Qiagen, Valencia, CA), in a final reaction volume of 25 μl, according to the manufacturer's recommended reaction conditions. RNase inhibitor (Applied Biosystems, Carlsbad, CA) was added to a final concentration of 15 to 20 U/reaction. The oligonucleotide primers P2I6F (5′-CCC AGA TGT YAA YCA GTC AGT CCA G-3′) and P2I6R (5′-CCC ACA GGC TTR AGT TGA TAR AAT C-3′) were added at a final concentration of 0.6 μM. RT-PCR cycling conditions included reverse transcription at 50°C for 30 min, denaturation at 95°C for 15 min, 40 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 2 min, and a final extension step at 68°C for 10 min. GI.6 P2 amplicons were purified using QIAquick gel extraction or PCR purification kits (Qiagen) and cycle sequenced using a BigDye Terminator kit (version 1.1; Applied Biosystems). Unincorporated fluorescent nucleotides were removed using a BigDye Xterminator kit (Applied Biosystems), and sequences were analyzed with a 3130XL automated sequencer (Applied Biosystems).
Data analysis.
All outbreak and specimen information was downloaded from the CaliciNet database and imported into R (http://www.r-project.org/) for data formatting, statistical analysis, and graphing. State, county, and city data from GI.6-positive outbreaks were matched with U.S. Geological Survey data (http://geonames.usgs.gov/domestic) for latitude and longitude coordinates. Cruise ship outbreaks were excluded from the geographic analysis. For statistical analyses, we used the Kruskal-Wallis test to compare patient ages by outbreak setting, the Mann-Whitney test to determine differences between outbreak sizes for person-to-person versus food-borne transmission outbreaks, and pairwise comparison of patient ages by setting with the Bonferroni correction. Odds ratios (ORs) were calculated to determine the odds of transmission route by genotype, while Pearson's r and linear regression were used to determine the linearity of GI.6 dispersion by distance and time. The χ2 test was used to determine significant differences between GII.4 outbreaks by setting, numbers of outbreaks in elderly patients by genotype, differences in transmission routes by genotype, and seasonal differences between transmission routes. Phylogenetic analysis of the GI.6 P2 region was performed with PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/versions.php) using the approximate likelihood ratio test for branch support with the best model, TIM3ef+I, as determined by jModelTest (http://code.google.com/p/jmodeltest2) using the corrected Akaike information criterion.
RESULTS
Epidemiologic characteristics of norovirus outbreaks in the United States in 2009 to 2013.
The number of states that participated in CaliciNet increased from 10 states in 2009 to 28 states and four local public health laboratories in 2013 (Table 1). In addition, at least 16 states submitted outbreak specimens to one of the six CaliciNet OSCs. Between September 2009 and August 2013, a total of 3,960 norovirus outbreaks were reported, of which 2,254 (57%) occurred in seven states in the Pacific (California and Oregon), Midwest (Wisconsin, Michigan, and Ohio), and South (Tennessee and Virginia) regions. No outbreak information from Oklahoma, Nebraska, New Jersey, North Dakota, or Pennsylvania was available for analysis. A total of 367 outbreaks (9.3%) were reported via the OSCs.
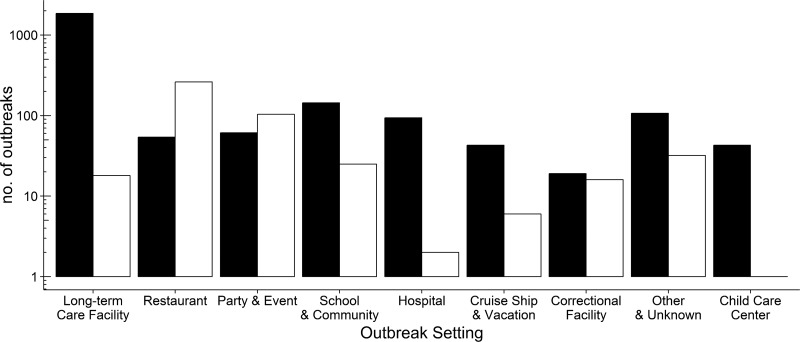
Information on the number of sick people was available for 2,712 outbreaks (68.5%) and ranged from 3 to 1,090 people per outbreak, with a median of 18 people (Table 2). A total of 2,475 norovirus outbreaks (62.5%) occurred in long-term care facilities (LTCF), 389 (9.8%) in restaurants, 227 (5.7%) in schools and communities (schools), 213 (5.4%) at parties or events (parties), and 144 (3.6%) in hospitals (Table 3). The transmission route was reported for 2,895 outbreaks (73%); person-to-person transmission and food-borne transmission were involved in 2,425 (83.7%) and 465 (16.1%) of the outbreaks, respectively. Waterborne transmission was involved in 5 outbreaks (0.1%), and no transmission route was reported for 1,065 (27.0%) outbreaks. A total of 407 food-borne outbreaks (87.5%) were reported for restaurants, parties, schools, and correctional facilities (Fig. 1). The median numbers of people affected were 21 for person-to-person outbreaks and 11 for food-borne outbreaks (P < 0.001).
TABLE 2.
Numbers of people with norovirus gastroenteritis per outbreak according to genotype
| Genotype | No. of ill people (range) | No. of outbreaks | Median no. of ill people per outbreak |
|---|---|---|---|
| GI.1 | 8–19 | 2 | 13.5 |
| GI.2 | 3–18 | 7 | 12 |
| GI.3 | 3–1,090 | 75 | 19 |
| GI.4 | 4–134 | 27 | 18 |
| GI.5 | 3–121 | 21 | 31 |
| GI.6 | 3–1,029 | 161 | 19 |
| GI.7 | 3–74 | 12 | 23.5 |
| GI.9 | 8–21 | 3 | 11 |
| GII.1 | 3–778 | 159 | 19 |
| GII.2 | 3–250 | 49 | 15 |
| GII.3 | 3–60 | 13 | 16 |
| GII.4 Den Haag 2006ba | 3–126 | 81 | 16 |
| GII.4 Osaka 2007a | 8–48 | 5 | 26 |
| GII.4 New Orleans 2009 | 3–698 | 1,025 | 20 |
| GII.4 Sydney 2012 | 3–660 | 802 | 17 |
| GII.4 | 3–698 | 1,913 | 18 |
| GII.5 | 4–307 | 9 | 30 |
| GII.6 | 3–250 | 110 | 19 |
| GII.7 | 3–106 | 39 | 18 |
| GII.12 | 3–235 | 42 | 17.5 |
| GII.13 | 3–89 | 19 | 18 |
| GII.14 | 7–69 | 3 | 15 |
| GII.15 | 109 | 1 | 109 |
| GII.16 | 16–77 | 3 | 26 |
| GII.17 | 5–23 | 4 | 11.5 |
GII.4 Den Haag 2006b and GII.4 Osaka 2007 viruses are referred to as GII.4 Minerva and GII.4 Riviera, respectively, in CaliciNet.
TABLE 3.
Genotype distribution of norovirus outbreaks from 2009 to 2013 according to setting
| Genotypea | Total no. of outbreaks | No. (%) of outbreaks in: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Child care centers | Correctional facilities | Cruise ships and vacation locales | Hospitals | Long-term care facilities | Parties and events | Restaurants | Schools and communities | Unknown settings | ||
| GI.1 | 5 (0.1) | 0 | 0 | 0 | 1 (0.7) | 3 (0.1) | 0 | 0 | 0 | 1 (0.3) |
| GI.2 | 10 (0.3) | 0 | 0 | 0 | 0 | 3 (0.1) | 0 | 4 (1.0) | 2 (0.9) | 1 (0.3) |
| GI.3 | 107 (2.7) | 1 (1.6) | 0 | 2 (2.0) | 2 (1.4) | 28 (1.1) | 7 (3.3) | 26 (6.7) | 21 (9.3) | 20 (6.5) |
| GI.4 | 43 (1.1) | 1 (1.6) | 0 | 1 (1.0) | 0 | 23 (0.9) | 3 (1.4) | 4 (1.0) | 5 (2.2) | 6 (2.0) |
| GI.5 | 24 (0.6) | 0 | 0 | 4 (4.0) | 0 | 11 (0.4) | 2 (0.9) | 4 (1.0) | 2 (0.9) | 1 (0.3) |
| GI.6 | 216 (5.5) | 4 (6.6) | 1 (2.3) | 5 (5.0) | 1 (0.7) | 83 (3.4) | 21 (9.9) | 34 (8.7) | 33 (14.5) | 34 (11.1) |
| GI.7 | 26 (0.7) | 1 (1.6) | 0 | 2 (2.0) | 1 (0.7) | 5 (0.2) | 1 (0.5) | 9 (2.3) | 3 (1.3) | 4 (1.3) |
| GI.8 | 1 (<0.1) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) | 0 | 0 |
| GI.9 | 3 (0.1) | 0 | 0 | 0 | 0 | 3 (0.1) | 0 | 0 | 0 | 0 |
| GII.1 | 214 (5.4) | 5 (8.2) | 1 (2.3) | 1 (1.0) | 1 (0.7) | 90 (3.6) | 26 (12.2) | 35 (9) | 42 (18.5) | 13 (4.2) |
| GII.2 | 69 (1.7) | 6 (9.8) | 0 | 0 | 2 (1.4) | 4 (0.2) | 15 (7.0) | 13 (3.3) | 25 (11.0) | 4 (1.3) |
| GII.3 | 23 (0.6) | 3 (4.9) | 0 | 1 (1.0) | 1 (0.7) | 6 (0.2) | 3 (1.4) | 6 (1.5) | 2 (0.9) | 1 (0.3) |
| GII.4 | 2,853 (72.0) | 21 (34.4) | 40 (93.0) | 76 (75.2) | 125 (86.8) | 2,081 (84.1) | 99 (46.5) | 193 (49.6) | 39 (17.2) | 179 (58.3) |
| GII.4 2006bb | 152 (3.8) | 2 (3.3) | 0 | 1 (1.0) | 6 (4.2) | 109 (4.4) | 2 (0.9) | 18 (4.6) | 1 (0.4) | 13 (4.2) |
| GII.4 2007c | 10 (0.3) | 0 | 0 | 0 | 0 | 6 (0.2) | 1 (0.5) | 1 (0.3) | 1 (0.4) | 1 (0.3) |
| GII.4 2009d | 1,737 (43.9) | 11 (18.0) | 20 (46.5) | 52 (51.5) | 80 (55.6) | 1,273 (51.4) | 61 (28.6) | 88 (22.6) | 17 (7.5) | 135 (44) |
| GII.4 2012e | 954 (24.1) | 8 (13.1) | 20 (46.5) | 23 (22.8) | 39 (27.1) | 693 (28) | 35 (16.4) | 86 (22.1) | 20 (8.8) | 30 (9.8) |
| GII.5 | 11 (0.3) | 0 | 0 | 1 (1.0) | 0 | 3 (0.1) | 0 | 4 (1.0) | 2 (0.9) | 1 (0.3) |
| GII.6 | 151 (3.8) | 10 (16.4) | 0 | 3 (3.0) | 5 (3.5) | 57 (2.3) | 13 (6.1) | 29 (7.5) | 27 (11.9) | 7 (2.3) |
| GII.7 | 64 (1.6) | 5 (8.2) | 0 | 2 (2.0) | 1 (0.7) | 23 (0.9) | 10 (4.7) | 7 (1.8) | 12 (5.3) | 4 (1.3) |
| GII.12 | 98 (2.5) | 3 (4.9) | 0 | 1 (1.0) | 4 (2.8) | 35 (1.4) | 7 (3.3) | 17 (4.4) | 11 (4.8) | 20 (6.5) |
| GII.13 | 30 (0.8) | 1 (1.6) | 1 (2.3) | 2 (2.0) | 0 | 12 (0.5) | 3 (1.4) | 1 (0.3) | 1 (0.4) | 9 (2.9) |
| GII.14 | 3 (0.1) | 0 | 0 | 0 | 0 | 1 (<0.1) | 1 (0.5) | 1 (0.3) | 0 | 0 |
| GII.15 | 1 (<0.1) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.3) | 0 | 0 |
| GII.16 | 3 (0.1) | 0 | 0 | 0 | 0 | 1 (<0.1) | 2 (0.9) | 0 | 0 | 0 |
| GII.17 | 4 (0.1) | 0 | 0 | 0 | 0 | 2 (0.1) | 0 | 0 | 0 | 2 (0.7) |
| GII.21 | 1 (<0.1) | 0 | 0 | 0 | 0 | 1 (<0.1) | 0 | 0 | 0 | 0 |
| Total | 3,960 | 61 (1.5) | 43 (1.0) | 101 (2.6) | 144 (3.6) | 2,475 (62.5) | 213 (5.4) | 389 (9.8) | 227 (5.7) | 307 (7.7) |
Different GII.4 variants.
GII.4 Den Haag 2006b.
GII.4 Osaka 2007.
GII.4 New Orleans 2009.
GII.4 Sydney 2012.
FIG 1.
Norovirus outbreaks by setting and transmission route. The numbers of norovirus outbreaks in different settings are shown for food-borne (□) and person-to-person (■) transmissions.
Person-to-person transmission was reported in 86% of the outbreaks with known transmission routes that occurred during the fall-winter season (fourth and first quarters). During the spring and summer seasons, significantly fewer person-to-person outbreaks occurred than in the expected distribution (P = 0.001), whereas food-borne outbreaks occurred more frequently (P < 0.001) during these months.
Genotypes.
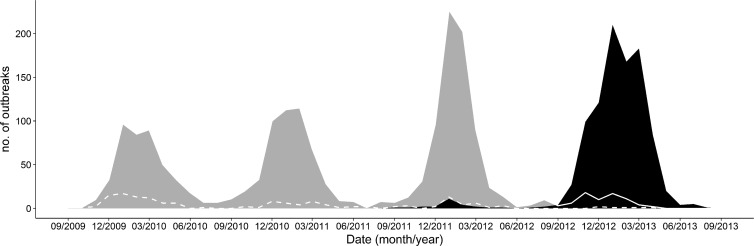
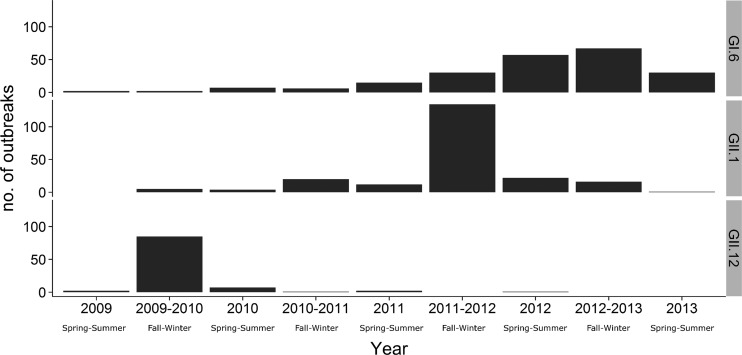
GI and GII norovirus types were detected as the cause of 435 (11%) and 3,525 (89%) of the outbreaks, respectively. Overall, all nine recognized GI genotypes and 14 of the 19 human GII genotypes were detected (Table 3). Most GI outbreaks were caused by three genotypes, i.e., GI.6 (n = 216 [49.7%]), GI.3 (n = 107 [24.6%]), and GI.4 (n = 43 [9.9%]) (Table 3). In all, 2,853 outbreaks (72%) were caused by GII.4 viruses (Den Haag 2006b, Osaka 2007, New Orleans 2009, and Sydney 2012). GII.4 New Orleans 2009 caused 1,737 outbreaks (44%) (Table 3) and was the predominant virus during three consecutive years (2009 to 2012), with the greatest numbers of outbreaks occurring in January 2010 (n = 97), February 2011 (n = 119), and January 2012 (n = 227) (Fig. 2). GII.4 Den Haag 2006b was detected throughout all four winter seasons, albeit at low levels, with a gradually decreasing number of outbreaks, i.e., from 61 outbreaks between October 2009 and March 2010 to six outbreaks between October 2012 and April 2013. GII.4 Sydney 2012 was first detected in September 2011 (Fig. 2) and became the predominant virus in the next winter season (2012 to 2013), causing 896 (72.8%) of the 1,098 outbreaks between September 2012 and April 2013, compared with 74 outbreaks (6.2%) caused by GII.4 New Orleans 2009 (Fig. 2). In addition to the GII.4 viruses, three non-GII.4 genotypes were observed, including a novel GII.12 virus that emerged during 2009-2010 (Fig. 3) (21), a novel GII.1 virus that emerged during 2011-2012, and a new GI.6 virus that gradually increased until the 2012-2013 fall-winter season, causing 67 outbreaks (Fig. 3). Unlike outbreaks caused by GII.12 and GII.1 viruses, GI.6 outbreaks did not exhibit winter seasonality (22). GI.1, GI.2, GI.5, GI.7, GI.8, GII.5, GII.13, GII.14, GII.15, GII.16, GII.17, and GII.21 viruses each caused less than 1% of all outbreaks (Table 3). GII.8, GII.9, GII.10, GII.20, and GII.22 viruses were not detected.
FIG 2.
Distribution of norovirus GII.4 variants from September 2009 through August 2013. Area graphs represent GII.4 Sydney 2012 (black) and GII.4 New Orleans 2009 (gray). The lines track GII.4 Den Haag 2006b (dashed line) from 2009 to 2012 and GII.4 New Orleans 2009 (solid line) from September 2012 through August 2013.
FIG 3.
Norovirus GII.12, GII.1, and GI.6 outbreaks reported to CaliciNet from September 2009 through August 2013. The numbers of outbreaks are shown for spring through summer (April through September) or fall through winter (October through March).
Correlation of genotypes with transmission route.
To determine whether certain genotypes were more often associated with food-borne outbreaks, we analyzed the transmission route of outbreaks caused by 11 different genotypes (GI.3, GI.4, GI.6, GI.7, GII.1, GII.2, GII.3, GII.4, GII.6, GII.7, and GII.12), each having ≥5 outbreaks per transmission route (Table 4). GII.4 outbreaks were more likely to involve person-to-person transmission than food-borne transmission (OR, 3.0; P < 0.05). Of the non-GII.4 outbreaks, GI.4, GII.1, GII.2, and GII.7 outbreaks were not associated with a particular transmission route (P ≥ 0.05), whereas GI.3, GI.6, GI.7, GII.3, GII.6, and GII.12 outbreaks were more likely to be associated with food-borne transmission (OR, 1.93 to 7.1; P < 0.05). Of these, GI.7 and GII.12 were 7 and 4 times more likely, respectively, to be associated with food-borne outbreaks than with person-to-person outbreaks.
TABLE 4.
Genotype frequency distributions of 3,616 norovirus outbreaks reported in 2009 to 2013
| Genotype | No. of outbreaks of the indicated transmission type: |
P (χ2 test) | Odds ratio (95% CI)a | ||
|---|---|---|---|---|---|
| Food-borne | Person-to-person | Unknown | |||
| GI.3 | 20 | 56 | 30 | 0.01 | 1.93 (1.12–3.21) |
| GI.4 | 7 | 18 | 17 | 0.1 | 2.1 (0.8–4.9) |
| GI.6 | 49 | 109 | 55 | <0.01 | 2.5 (1.8–3.6) |
| GI.7 | 8 | 6 | 12 | <0.01 | 7.1 (2.41–22.1) |
| GII.1 | 36 | 141 | 36 | 0.1 | 1.38 (0.93–2.0) |
| GII.2 | 14 | 41 | 14 | 0.05 | 1.84 (0.96–3.3) |
| GII.3 | 8 | 10 | 5 | <0.01 | 4.3 (1.6–11.1) |
| GII.4 | 239 | 1,838 | 764 | <0.01 | 0.33 (0.27–0.40) |
| GII.6 | 34 | 85 | 32 | <0.01 | 2.2 (1.45–3.30) |
| GII.7 | 12 | 36 | 16 | 0.08 | 1.79 (0.88–3.40) |
| GII.12 | 22 | 31 | 45 | <0.01 | 3.89 (2.20–6.98) |
The reference for odds ratios is food-borne transmission. CI, confidence interval.
Correlations of genotypes with outbreak setting, season, and age.
A total of 2,081 (84%) of the 2,475 LTCF outbreaks were caused by one of four GII.4 variants (Den Haag 2006b, Osaka 2007, New Orleans 2009, or Sydney 2012), whereas GI viruses caused 159 (6.4%) of the outbreaks in this setting (Table 3). GII.4 food-borne outbreaks were underrepresented (12.3%; P = 0.006) during the spring-summer seasons, based on the overall number of outbreaks. Non-GII.4 person-to-person outbreaks occurred significantly less in the fall and winter seasons (17%; P < 0.001) and significantly more in the spring-summer seasons (7.3%; P < 0.001).
Because GII.4 was the only genotype significantly associated with person-to-person transmission, we decided to investigate the roles of setting and age in the transmission of viruses of this genotype. All outbreaks with known transmission routes (n = 2,909) were grouped into GII.4 and non-GII.4 outbreaks and stratified by setting. The number of GII.4 outbreaks was dependent on the setting (P < 0.001), and outbreaks occurred more often in LTCF and hospitals (P < 0.001) than in other settings such as schools. Non-GII.4 outbreaks were significantly associated with child care centers, parties, restaurants, and schools (P < 0.001). Patient age information was available for 7,519 patients from 3,593 outbreaks (90.7%). Overall, the patient age range was 0.3 to 111 years, with a median age of 77 years. There was a significant difference in the median ages of the patients from several different outbreak settings (P < 0.001). The median patient ages in LTCF (85 years) and child care centers (4 years) were significantly different from those in all other settings (P < 0.001). For norovirus outbreaks in hospitals and cruise ships, the median ages of patients were 66 years and 61 years, respectively, statistically different (P < 0.001) from the median ages of patients in outbreaks in restaurants (46 years), parties (44 years), and correctional facilities (41 years). To further investigate a possible association between patient age and genotype, genotype-specific outbreak counts for patients ≥65 years and <65 years of age were compared. Overall, there was a direct correlation between certain genotypes and patient ages (P < 0.001). For example, GI.2, GI.3, GI.6, and GI.7 outbreaks affected proportionally fewer patients ≥65 years of age (P, <0.01 to <0.001), whereas GI.4 and GI.5 outbreaks were not significant in the ≥65-year-old age group. Except for GII.4, GII outbreaks had proportionally fewer outbreaks in the ≥65-year-old group (P values of <0.04 to <0.001). GII.4 was the only genotype with proportionally more outbreaks in the ≥65-year-old group. Furthermore, analysis of χ2 results show a linear relationship between the median ages of patients in outbreak settings and deviation from expected GII.4 outbreak counts, indicating that older age is associated with GII.4 outbreaks in schools, parties, restaurants, cruise ships, hospitals, and long-term care facilities.
Phylogeography of GI.6 outbreaks.
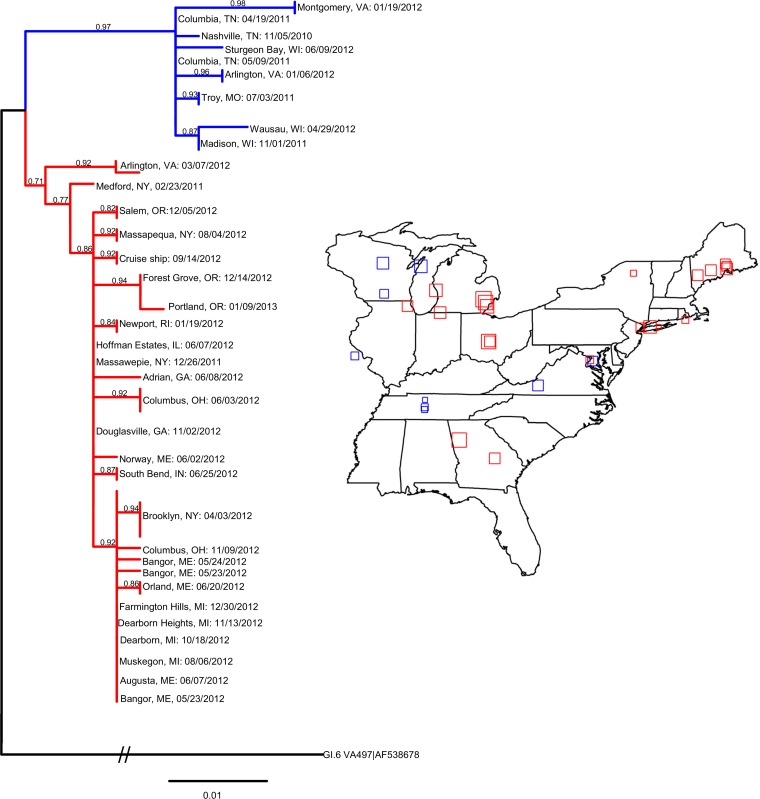
Of the three emerging non-GII.4 strains, GI.6 was further analyzed because it is a rare genotype, had atypical seasonality, and had not been analyzed previously, unlike GII.12 (21). We decided to amplify the P2 region of this virus and to compare the sequences to investigate whether outbreaks could be directly linked or were closely related. P2 sequences from 43 GI.6 outbreaks that spread throughout the United States in 2011 to 2012 could be grouped into southern (Tennessee) and northeastern (New York) subclusters, based on the earliest observed outbreak for each phylogenetic cluster (Fig. 4). The Tennessee cluster had a simple dispersal trend, with a linear relationship between the distance and time from the first outbreak (Pearson's r = 0.92, R2 = 0.85). The New York subcluster was more complex, with at least two dispersions, from Massawepie, NY, to Portland, OR (Pearson's r = 0.85, R2 = 0.72), and from Brooklyn, NY, to Farmington Hills, MI (Pearson's r = 0.86, R2 = 0.73), and two outbreaks (in New York in February 2011 and in Virginia in March 2012) that were not directly related (by date and phylogenetic distance) to the New York subcluster. In summary, phylogeographic analysis of GI.6 outbreaks indicated time and distance dependence of GI.6 outbreaks.
FIG 4.
Time-ordered geographic distribution of GI.6 outbreaks in the eastern half of the United States by P2 subdomain phylogenetic analysis. The phylogenetic nucleotide tree of the GI.6 P2 subdomain was run in PhyML using the custom model TIM3ef+I, as determined by jModelTest using the corrected Akaike information criterion. Branch support was provided by the approximate likelihood ratio test. The distance of the root GI.6 VA497 (GenBank accession no. AF538678) is 1.46 nucleotides between the node of the two GI.6 clusters and reference GI.6. The truncated P2 tree shows the two main clusters of GI.6 circulating primarily in the eastern United States. The complete phylogenetic tree and corresponding outbreaks can be provided upon request. Outbreak information was downloaded from CaliciNet, and outbreaks from the eastern United States were mapped according to the latitude and longitude coordinates of the outbreak (squares). Overlapping outbreaks were jittered for clarity. The outbreak dates were time ordered, and the sizes of the squares indicate the order of the outbreaks within the cluster of the same color as the phylogenetic tree. Smaller squares indicate earlier outbreaks and larger squares indicate later outbreaks. The dates (month/day/year) and cities of the mapped outbreaks are listed in the tree.
DISCUSSION
From September 2009 through August 2013, epidemiologic and virologic data from 3,960 norovirus outbreaks in the United States were reported to CaliciNet. A total of 16.1% of the known outbreaks were reported to have resulted from food-borne transmission. Outbreaks affected a median of 18 people overall, 21 people in person-to-person outbreaks, and 11 people in food-borne outbreaks. A total of 2,475 (63%) of all norovirus outbreaks occurred in LTCF, 389 (9.8%) in restaurants, 227 (5.7%) in schools, 2,131 (5.3%) at parties, and 144 (3.6%) in hospitals.
Two new GII.4 variants, i.e., GII.4 New Orleans 2009 in October 2009 (11) and GII.4 Sydney 2012 in October 2012, replaced previously predominant GII.4 strains. Although certain states reported increased outbreak activity associated with these new variants, such increases could not be confirmed nationally (14, 23–26). In addition, three non-GII.4 strains (GII.12, GII.1, and GI.6) emerged successively and caused between 11 and 15% of all outbreaks for their respective time periods. Using phylogeographic analysis, we identified that the GI.6 strains consisted of two related genetic clusters that spread throughout the United States from two distinct geographic locations. We further observed that outbreaks caused by GI.3, GI.6, GI.7, GII.3, GII.6, and GII.12 were more often associated with food-borne outbreaks, whereas GII.4 was primarily associated with person-to-person transmission.
GII.4 outbreaks occurred disproportionally among older patients in settings such as LTCF, hospitals, and cruise ships, compared with outbreak settings with younger patients (<65 years old) such as schools, which had proportionally fewer GII.4 outbreaks. Moreover, the settings with the fewest GII.4 outbreaks were schools and child care centers, both settings populated nearly exclusively by younger people. Increasing age has been reported as a risk factor for nosocomial gastrointestinal infections (27). Why age is a risk factor is not fully known, but waning immunity or immunosenescence in the elderly population has been suggested to play an important role for viral infections (28, 29). Age by itself may not fully explain why the majority of outbreaks (72%) were caused by GII.4 viruses. Aside from patient age, GII.4 viruses may contain a currently unknown virulence factor that is not present in other norovirus genotypes. Modern GII.4 noroviruses have an amino acid insertion in epitope D, which has been present in GII.4 noroviruses only since the Farmington Hills 2002 variant, and this epitope has been shown to be a blockade epitope for post-2002 GII.4 noroviruses (30). The presence of epitope D in GII.4 noroviruses, along with the inability of aging immune systems to mount robust immune responses (29), may favor GII.4 noroviruses over non-GII.4 noroviruses in people >65 years of age. Antigenic changes over time have been shown to be specifically associated with GII.4 blockade epitopes (30), which supports the hypothesis that the emergence of new GII.4 strains, including GII.4 Sydney 2012, is driven by evolutionary escape from herd immunity (30, 31).
Over the 4 years of data, 16.1% of all outbreaks were epidemiologically linked to the consumption of food. This percentage is in the same range as that reported from a comprehensive study in Europe (32) and slightly lower than that reported by the National Outbreak Reporting System (NORS), which is an Internet-based system for local, state, and territorial U.S. health departments to report all enteric disease outbreaks (33). We identified several genotypes that were more frequently associated with food-borne transmission, including GI.3, GI.6, GI.7, GII.3, GII.6, and GII.12. Our data confirm recent findings that certain non-GII.4 genotypes are more frequently detected in food-borne outbreaks in Europe (34). Interestingly, our data did not show an association between GI.2 or GI.4 viruses and food-borne outbreaks (34). These different findings could reflect temporal variations in the circulation of non-GII.4 genotypes in the population, with specific genotypes emerging only in certain years, as we reported for GII.12 and GI.6 in the United States (21, 22). Interestingly, of the three emergent non-GII.4 genotypes (GII.12, GII.1, and GI.6), GII.1 caused the greatest number of outbreaks, whereas GII.12 and GI.6 outbreaks were 2 to 4 times more likely to be caused by contaminated food than by person-to-person transmission.
Phylogeographic analyses indicated that, in 2011 to 2012, GI.6 viruses spread from the first reported outbreaks in Tennessee and New York. P2 sequences have been used to trace norovirus outbreaks in hospitals and nursing homes (18–20, 35), as well as to establish the number of mutations over the course of a norovirus infection (20). Within an outbreak, sequence diversity between subjects is small (0 to 0.3%); however, viruses collected later in an infection of the same person accumulated several mutations, compared to viruses shed at the onset of the infection (20). P2 sequence data for the GI.6 outbreaks in our study differed 0 to 2%, which could indicate that at least several outbreaks might have had a common source or, alternatively, had minimal circulation. Mutations in the P2 subdomain can affect both antigenic and histo-blood group antigen binding sites and, if favorable for the virus, are likely to be sustained (30, 36, 37). GI.6 outbreaks that do not have 100% identical sequences may still be related, and therefore, the slightly different P2 sequences of the outbreaks that occurred in the southern and northeastern United States might have had a single food-borne source.
From 2009 through 2013, three non-GII.4 viruses (GII.12, GII.1, and GI.6) caused 13% of all outbreaks. These novel strains likely emerged after recombination events (5, 11, 38). Although molecular determinants of virulence are difficult to discern, the increase in the number of outbreaks reported after the emergence of a recombinant norovirus strain (21, 39) suggests that nonstructural proteins may be involved. Recent reports have also indicated that the emergence of the GII.4 pandemic variants are derived from recombination events (40) and the current major variant, GII.4 Sydney 2012, has already started to recombine with other GII.4 variants (24). This would suggest that the major capsid protein is not the sole virulence factor in norovirus disease and that population immunity and genetic drift of the major capsid protein are not the only factors involved in norovirus immune evasion (38, 41). Complete norovirus genome sequence data have shown that changes of several unique amino acid residues within nonstructural protein 4 (NS4) have occurred over time (6, 41), indicating that NS4 may be important in immune evasion. The emergence of GII.12 in 2009 to 2010, GII.1 in 2010 to 2011, and most recently GI.6 in 2011 to 2012 indicates that a combination of capsid and nonstructural proteins may be responsible for a surge of outbreaks from year to year in addition to the major GII.4 variant outbreaks, which occur mainly in the winter. Sequence data clearly show the increase and decrease of these variants in the last 4 years and confirm that the cyclic emergence of new norovirus strains is a global phenomenon (10, 23–27).
Our study has several limitations. First, the epidemiologic information that is entered into CaliciNet for the outbreaks may be preliminary in some cases, as the outbreak investigation might not have been finalized. Final information, including additional epidemiologic, demographic, and clinical details, is captured by the NORS (33), and efforts are under way to integrate NORS and CaliciNet. Second, norovirus genotyping is performed by amplification of a small region of ORF2 (11). However, amplification of the P2 subdomain has been shown to have better resolution to distinguish between norovirus strains (18–21, 24). Unfortunately, because of the high level of genetic variation in ORF2 of noroviruses, P2 assays that detect all norovirus genotypes are difficult to implement in CaliciNet. An additional limitation is the entanglement of age and setting data. Although every effort was made to analyze patient age and outbreak setting data independently, there is a strong correlation between the two, making separate analysis of the parameters difficult. Third, we could not detect differences between genotype profiles of the rarer genotypes. However, rare strains are of interest because they may have the potential to emerge quickly with public health consequences, as was reported for the recombinant ORF1 GII.b viruses, which were reported first after a large waterborne outbreak (42) and then were transmitted through different routes, including food-borne distribution, throughout Europe, causing up to 13% of the outbreaks between 1999 and 2004 (34). Lastly, P2 sequences were obtained from 34% of the reported GI.6 outbreaks, which may not have been representative, and links to a possible seeding event may have been missed.
Since 2009, CaliciNet has captured data from 45 states, allowing for comprehensive national norovirus outbreak surveillance. Information on circulating norovirus genotypes in the United States will provide much-needed data for the development and potential reformulation of a norovirus vaccine (43). Improved understanding of which genotypes are preferentially associated with food-borne outbreaks, as well as the continued tracking of newly emerging strains, will facilitate better understanding of norovirus disease. Because food-borne disease may be associated with certain genotypes, rapid typing of outbreaks will allow for quick implementation of control measures specific for food-borne disease, to limit the spread of norovirus. The data provided in our study will also help to provide enhanced disinfection and prevention strategies, such as focusing on early control and intervention measures in LTCF, the single most important setting for GII.4 norovirus outbreaks. Although it is currently not feasible for routine analysis, future typing methods may include sequencing of the complete genome, which will allow better resolution of outbreaks and detection of emerging variants (44). In addition to tracking of norovirus genotypes based on the viral capsid, the continuous emergence of new recombinant norovirus strains will provide an impetus to identify which nonstructural proteins are important in norovirus virulence and possible immune evasion. Continuous surveillance of changing trends, norovirus strains, and genotype distributions will be helpful in continued efforts to reduce the burden of norovirus gastroenteritis.
ACKNOWLEDGMENTS
This study was supported by the intramural food safety program at the Centers for Disease Control and Prevention and in part by Agriculture and Food Research Initiative Competitive Grant 2011-68003-30395 from the U.S. Department of Agriculture, National Institute of Food and Agriculture.
We thank Brenda Brown and Applied Maths (Austin, TX) for their continued support of CaliciNet and Verónica Costantini, Ben Lopman, Aron Hall, and the anonymous reviewers for their critical review of the manuscript. We gratefully acknowledge the CaliciNet members who contributed to the manuscript, including Courtney Chesnutt and Nicholas Switzer (Alabama Department of Public Health, Bureau of Clinical Laboratories), Cheng Yang (Arkansas Department of Health, Public Health Laboratory), Chao-Yang Pan and Tasha Padilla (California Department of Public Health, Viral and Rickettsial Disease Laboratory), Julia Wolfe (Orange County [California] Public Health Laboratory), Eduardo Ramos (Los Angeles County [California] Public Health Laboratory), Justin Nucci and Mary-Kate Cichon (Colorado Department of Public Health and Environment), Horng-Yuan Kan (Washington, DC, Public Health Laboratory), Gregory Hovan and Jordan Estes Hudson (Delaware Public Health Laboratories), Lea A. Heberlein-Larson and Marshall Cone (Florida Department of Health, Bureau of Public Health Laboratories-Tampa), Precilia Calimlim and Cheryl-Lynn Daquip (Hawaii Department of Health), Amanda Bruesch (Idaho Bureau of Laboratories), Edward Simpson and Nikail Collins (Indiana State Department of Health Laboratories), Erika Buzby (Massachusetts Department of Public Health), Heather Grieser, John Martha, and Brian Bernier (State of Maine, Health and Environmental Testing Laboratory), Jonathan Johnston, Julie Haendiges, and Eric Keller (Maryland Department of Health and Mental Hygiene, Laboratories Administration), Laura Mosher, Victoria Vavricka, and Kevin Rodeman (Michigan Department of Community Health, Bureau of Laboratories), Elizabeth Cebelinski, Ginette Dobbins, and Mary Elizabeth Horn (Minnesota Department of Health, Infectious Disease Laboratory), Shadia Rath (North Carolina State Laboratory of Public Health), Alisha M. Nadeau, Fengxiang Gao, and Jennifer Mahoney (New Hampshire Public Health Laboratories, Department of Health and Human Services), Frederick Gentry (New Mexico Department of Health, Scientific Laboratory Division), Sergey P. Morzunov (Nevada State Public Health Laboratory), Gino Battaglioli and Daryl M. Lamson (New York State Department of Health, Wadsworth Center), Rebekah Carman, Rosemary Hage, Lai Ming Woo, and Eric Brandt (Ohio Department of Health Laboratory), James M. Terry and Laura J. Tsaknaridis (Oregon State Public Health Laboratory), Andrea Maloney (South Carolina Department of Health and Environmental Control), Amy M. Woron and Christina Moore (Tennessee Department of Health, Laboratory Services), Chun Wang and Jenny Zhang (Texas Department of State Health Services), Leigh-Emma Lion (Virginia Division of Consolidated Laboratory Services), Valarie Devlin and Jessica Chenette (Vermont Department of Health Laboratory), Tim Davis, T. J. Whyte, and Tonya Danz (Wisconsin State Laboratory of Hygiene), Jose Navidad and David Bina (City of Milwaukee [Wisconsin] Health Department), and Rob Christensen (Wyoming Public Health Laboratory).
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 30 October 2013
REFERENCES
- 1.Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785. 10.1056/NEJMra0804575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne DC, Vinjé J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. 2013. Norovirus and medically attended gastroenteritis in U.S. children. N. Engl. J. Med. 368:1121–1130. 10.1056/NEJMsa1206589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopman BA, Hall AJ, Curns AT, Parashar UD. 2011. Increasing rates of gastroenteritis hospital discharges in US adults and the contribution of norovirus, 1996–2007. Clin. Infect. Dis. 52:466–474. 10.1093/cid/ciq163 [DOI] [PubMed] [Google Scholar]
- 4.Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. 2012. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin. Infect. Dis. 55:216–223. 10.1093/cid/cis386 [DOI] [PubMed] [Google Scholar]
- 5.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, Vinjé J, Parashar UD. 2013. Norovirus disease in the United States. Emerg. Infect. Dis. 19:1198–1205. 10.3201/eid1908.130465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green K. 2013. Caliciviridae: the noroviruses, p 583–609 In Knipe DM, Howley PM. (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 7.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323. 10.1016/j.virol.2005.11.015 [DOI] [PubMed] [Google Scholar]
- 8.Kroneman A, Vega E, Vennema H, Vinjé J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. 2013. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 158:2059–2068. 10.1007/s00705-013-1708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martella V, Decaro N, Lorusso E, Radogna A, Moschidou P, Amorisco F, Lucente MS, Desario C, Mari V, Elia G, Banyai K, Carmichael LE, Buonavoglia C. 2009. Genetic heterogeneity and recombination in canine noroviruses. J. Virol. 83:11391–11396. 10.1128/JVI.01385-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebenga JJ, Vennema H, Zheng DP, Vinjé J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O'Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 200:802–812. 10.1086/605127 [DOI] [PubMed] [Google Scholar]
- 11.Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinjé J. 2011. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg. Infect. Dis. 17:1389–1395. 10.3201/eid1708.101837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng DP, Widdowson MA, Glass RI, Vinjé J. 2010. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J. Clin. Microbiol. 48:168–177. 10.1128/JCM.01622-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debbink K, Lindesmith LC, Donaldson EF, Baric RS. 2012. Norovirus immunity and the great escape. PLoS Pathog. 8:e1002921. 10.1371/journal.ppat.1002921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leshem E, Wikswo M, Barclay L, Brandt E, Storm W, Salehi E, Desalvo T, Davis T, Saupe A, Dobbins G, Booth HA, Biggs C, Garman K, Woron AM, Parashar UD, Vinjé J, Hall AJ. 2013. Effects and clinical significance of GII.4 Sydney norovirus, United States, 2012–2013. Emerg. Infect. Dis. 19:1231–1238. 10.3201/eid1908.130458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention 2007. Norovirus activity—United States, 2006–2007. MMWR Morb. Mortal. Wkly. Rep. 56:842–846 [PubMed] [Google Scholar]
- 16.Yen C, Wikswo ME, Lopman BA, Vinjé J, Parashar UD, Hall AJ. 2011. Impact of an emergent norovirus variant in 2009 on norovirus outbreak activity in the United States. Clin. Infect. Dis. 53:568–571. 10.1093/cid/cir478 [DOI] [PubMed] [Google Scholar]
- 17.Vongpunsawad S, Prasad BV, Estes MK. 2013. Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J. Virol. 87:4818–4825. 10.1128/JVI.03508-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xerry J, Gallimore CI, Iturriza-Gómara M, Allen DJ, Gray JJ. 2008. Transmission events within outbreaks of gastroenteritis determined through analysis of nucleotide sequences of the P2 domain of genogroup II noroviruses. J. Clin. Microbiol. 46:947–953. 10.1128/JCM.02240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukhrie FH, Beersma MF, Wong A, van der Veer B, Vennema H, Bogerman J, Koopmans M. 2011. Using molecular epidemiology to trace transmission of nosocomial norovirus infection. J. Clin. Microbiol. 49:602–606. 10.1128/JCM.01443-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukhrie FH, Teunis P, Vennema H, Bogerman J, van Marm S, Beersma MF, Koopmans M. 2013. P2 domain profiles and shedding dynamics in prospectively monitored norovirus outbreaks. J. Clin. Virol. 56:286–292. 10.1016/j.jcv.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 21.Vega E, Vinjé J. 2011. Novel GII.12 norovirus strain, United States, 2009–2010. Emerg. Infect. Dis. 17:1516–1518. 10.3201/eid1708.110025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leshem E, Barclay L, Wikswo M, Vega E, Gregoricus N, Parashar UD, Vinjé J, Hall AJ. 2013. Genotype GI.6 norovirus, United States, 2010–2012. Emerg. Infect. Dis. 19:1317–1320. 10.3201/eid1908.130445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett S, MacLean A, Miller RS, Aitken C, Gunson RN. 2013. Increased norovirus activity in Scotland in 2012 is associated with the emergence of a new norovirus GII.4 variant. Euro Surveill. 18:pii=20349 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20349 [PubMed] [Google Scholar]
- 24.Fonager J, Hindbæk LS, Fischer TK. 2013. Rapid emergence and antigenic diversification of the norovirus 2012 Sydney variant in Denmark, October to December, 2012. Euro Surveill. 18:pii=20413 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20413 [PubMed] [Google Scholar]
- 25.van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinjé J, White PA, Koopmans M, NoroNet 2013. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 18:pii=20345 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20345 [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention 2013. Emergence of new norovirus strain GII.4 Sydney—United States, 2012. MMWR Morb. Mortal. Wkly. Rep. 62:55. [PMC free article] [PubMed] [Google Scholar]
- 27.Spackova M, Altmann D, Eckmanns T, Koch J, Krause G. 2010. High level of gastrointestinal nosocomial infections in the German surveillance system, 2002–2008. Infect. Control Hosp. Epidemiol. 31:1273–1278. 10.1086/657133 [DOI] [PubMed] [Google Scholar]
- 28.Reber AJ, Chirkova T, Kim JH, Cao W, Biber R, Shay DK, Sambhara S. 2012. Immunosenescence and challenges of vaccination against influenza in the aging population. Aging Dis. 3:68–90 [PMC free article] [PubMed] [Google Scholar]
- 29.Paulke-Korinek M, Kundi M, Laaber B, Brodtraeger N, Seidl-Friedrich C, Wiedermann U, Kollaritsch H. 2013. Factors associated with seroimmunity against tick borne encephalitis virus 10 years after booster vaccination. Vaccine 31:1293–1297. 10.1016/j.vaccine.2012.12.075 [DOI] [PubMed] [Google Scholar]
- 30.Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. 2012. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 8:e1002705. 10.1371/journal.ppat.1002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinjé J, Baric RS. 1 August 2013. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J. Infect. Dis. [Epub ahead of print.] 10.1093/infdis/jit370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Böttiger B, Falkenhorst G, Johnsen C, von Bonsdorff CH, Maunula L, Kuusi M, Pothier P, Gallay A, Schreier E, Höhne M, Koch J, Szücs G, Reuter G, Krisztalovics K, Lynch M, McKeown P, Foley B, Coughlan S, Ruggeri FM, Di Bartolo I, Vainio K, Isakbaeva E, Poljsak-Prijatelj M, Grom AH, Mijovski JZ, Bosch A, Buesa J, Fauquier AS, Hernandéz-Pezzi G, Hedlund KO, Koopmans M. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959–2965. 10.1128/JCM.00499-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall AJ, Wikswo ME, Manikonda K, Roberts VA, Yoder JS, Gould LH. 2013. Acute gastroenteritis surveillance through the National Outbreak Reporting System, United States. Emerg. Infect. Dis. 19:1305–1309. 10.3201/eid1908.130482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhoef L, Vennema H, van Pelt W, Lees D, Boshuizen H, Henshilwood K, Koopmans M, Food-Borne Viruses in Europe Network 2010. Use of norovirus genotype profiles to differentiate origins of foodborne outbreaks. Emerg. Infect. Dis. 16:617–624. 10.3201/eid1604.090723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xerry J, Gallimore CI, Iturriza-Gómara M, Gray JJ. 2010. Genetic characterization of genogroup I norovirus in outbreaks of gastroenteritis. J. Clin. Microbiol. 48:2560–2562. 10.1128/JCM.00798-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindesmith LC, Costantini V, Swanstrom J, Debbink K, Donaldson EF, Vinjé J, Baric RS. 2013. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J. Virol. 87:2803–2813. 10.1128/JVI.03106-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. 2011. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J. Virol. 85:8635–8645. 10.1128/JVI.00848-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bull RA, Eden JS, Rawlinson WD, White PA. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. 10.1371/journal.ppat.1000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iritani N, Kaida A, Abe N, Sekiguchi J, Kubo H, Takakura K, Goto K, Ogura H, Seto Y. 2012. Increase of GII.2 norovirus infections during the 2009–2010 season in Osaka City, Japan. J. Med. Virol. 84:517–525. 10.1002/jmv.23211 [DOI] [PubMed] [Google Scholar]
- 40.Eden JS, Tanaka MM, Boni MF, Rawlinson WD, White PA. 2013. Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 87:6270–6282. 10.1128/JVI.03464-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motomura K, Yokoyama M, Ode H, Nakamura H, Mori H, Kanda T, Oka T, Katayama K, Noda M, Tanaka T, Takeda N, Sato H, Norovirus Surveillance Group of Japan 2010. Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J. Virol. 84:8085–8097. 10.1128/JVI.02125-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallay A, De Valk H, Cournot M, Ladeuil B, Hemery C, Castor C, Bon F, Mégraud F, Le Cann P, Desenclos JC, Outbreak Investigation Team 2006. A large multi-pathogen waterborne community outbreak linked to faecal contamination of a groundwater system, France, 2000. Clin. Microbiol. Infect. 12:561–570. 10.1111/j.1469-0691.2006.01441.x [DOI] [PubMed] [Google Scholar]
- 43.Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. 2011. Norovirus vaccine against experimental human Norwalk virus illness. N. Engl. J. Med. 365:2178–2187. 10.1056/NEJMoa1101245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundu S, Lockwood J, Depledge DP, Chaudhry Y, Aston A, Rao K, Hartley JC, Goodfellow I, Breuer J. 2013. Next-generation whole genome sequencing identifies the direction of norovirus transmission in linked patients. Clin. Infect. Dis. 57:407–414. 10.1093/cid/cit287 [DOI] [PMC free article] [PubMed] [Google Scholar]