Abstract
A commercial PCR assay of perirectal swab specimens detected 17 (68%) of 25 asymptomatic carriers of toxigenic Clostridium difficile, including 93% with skin and/or environmental contamination. A clinical prediction rule, followed by PCR screening, could be used to identify carriers at high risk of C. difficile shedding.
TEXT
Clostridium difficile is the most important cause of health care-associated infectious diarrhea. In addition, many hospitalized patients and long-term care facility (LTCF) residents are asymptomatic carriers of toxigenic C. difficile (1–9). Although infection control efforts typically focus on patients with C. difficile infection (CDI), there is some evidence that asymptomatic carriers might play an underappreciated role in the transmission of C. difficile. Clabots et al. (7) found that most episodes of nosocomial acquisition of CDI on a hospital ward were epidemiologically linked to asymptomatic new admissions. Riggs et al. (6) demonstrated that more than half of the asymptomatic carriers in an LTCF had skin and environmental spore contamination. Finally, on the basis of molecular typing, Curry et al. (9) demonstrated that incident CDI cases were as frequently linked to asymptomatic carriers as to symptomatic patients. In order to develop control strategies that address asymptomatic carriage, there is a need for simple and effective methods to rapidly identify carriers, particularly those at high risk of spore shedding. Because C. difficile culture is not routinely available and may require several days for processing, we evaluated the utility of a commercial PCR assay of perirectal swab specimens and a previously derived clinical prediction rule for the detection of asymptomatic carriers of toxigenic C. difficile among LTCF residents (6).
This study was conducted at the Cleveland and Hines (Chicago) Department of Veterans Affairs LTCFs in conjunction with the initial phase of a 6-month prospective cohort study evaluating the asymptomatic carriage of C. difficile. Both facilities have ∼150 beds housing a mixture of residential and post-acute-phase residents. Current residents of the facilities and new admissions were eligible for enrollment. Subjects were cultured for C. difficile within 24 h of admission or upon enrollment and then every 2 weeks during their LTCF stay. With BD BBL CultureSwab (Becton Dickinson, Cockeysville, MD), cultures were collected in a standardized manner from the perirectal area and ∼5- by 10-cm areas of the groin, chest and abdomen, and environment (bed rail and bedside table combined). Additional swabs were collected from the perirectal and groin sites of subsets of consecutive subjects at each study site for PCR testing with a commercial assay (Xpert C. difficile, Cepheid, Sunnyvale, CA).
Swabs were directly plated onto selective media for culture of toxigenic C. difficile as previously described (6). The number of C. difficile colonies per swab was determined. The commercial PCR was processed according to the manufacturer's protocol. A medical record review was conducted to obtain demographic information and to apply a previously derived prediction rule (6). The prediction rule included two factors, antibiotic use in the past 3 months and CDI in the past year (6). Molecular typing by restriction endonuclease analysis was performed as previously described (7). Environmental isolates were considered to represent environmental shedding only if the environmental isolate matched concurrent perirectal and/or skin isolates.
We calculated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the prediction rule and of perirectal PCR for the detection of asymptomatic carriers (i.e., positive perirectal culture) and asymptomatic carriers at increased risk of transmission (i.e., positive perirectal culture plus positive skin and/or environmental cultures). We evaluated the potential to use the prediction rule as an initial screen to rule out patients at low risk of asymptomatic carriage, followed by the use of perirectal PCR to evaluate carriage in the subset of patients at increased risk on the basis of the prediction rule. Fisher's exact test was used to compare the percentages of positive skin, environmental, and perirectal PCR results for carriers with 1 to 25 versus ≥26 colonies of C. difficile per perirectal swab. Data were analyzed with SPSS statistical software version 10.0 (SPSS Inc., Chicago, IL). The institutional review boards of both hospitals approved the study protocol.
From 141 subjects (88 from Cleveland and 53 from Chicago), 204 sets of matched PCR and culture specimens were collected (mean, 1.4 sets per patient; range, 1 to 4). The mean age of the participants was 70 (range, 41 to 98) years. One-hundred thirty-six (97%) of the subjects were men, and 81 (57%) were clinical prediction rule positive on one or more occasions.
Of the 141 subjects, 21 (15%) were asymptomatic carriers of toxigenic C. difficile on the basis of positive perirectal cultures; 19 (22%) of 88 in Cleveland and 2 of 53 (4%) at Hines. Of the 21 asymptomatic carriers, 17 (81%) had received antibiotics within the past 3 months and 6 (29%) had CDI within the past year; only 2 (10%) carriers had neither previous antibiotic exposure nor a previous CDI. Fifteen (71%) of the 21 carriers had 2 or more sets of cultures collected, and 12 (80%) of the 15 remained positive for all subsequent cultures. Only one asymptomatic carrier subsequently developed a CDI.
Of the total of 204 sets of specimens, 25 (12%) were positive for toxigenic C. difficile by perirectal culture. None of the patients had positive groin swab PCR results. Tables 1 and 2 show the sensitivity, specificity, PPV, and NPV of the perirectal swab PCR results and of the clinical prediction rule for the detection of asymptomatic carriers (Table 1) and for the detection of carriers with positive skin and/or environmental cultures (Table 2). Perirectal PCR was positive for 17 (68%) of 25 positive perirectal culture specimens and 13 (93%) of 14 carrier specimens with positive skin and/or environmental cultures.
TABLE 1.
Performance of perirectal swab testing by PCR and a clinical prediction rulea in comparison with that of perirectal swab toxigenic culture for detection of asymptomatic carriers of C. difficile
| Testing method | No. positive/no. toxigenic culture positive | No. negative/no. toxigenic culture negative | % Sensitivity (no. positive/total; 95% CI)b | % Specificity (no. negative/total; 95% CI) | % PPV (no. positive/total; 95% CI) | % NPV (no. negative/total; 95% CI) |
|---|---|---|---|---|---|---|
| Perirectal swab PCR | 17/25 | 179/179 | 68 (17/25; 46–84) | 100 (179/179; 97–100) | 100 (17/17; 77–100) | 96 (179/187; 91–98) |
| Prediction rule | 23/25 | 89/179 | 92 (23/25; 77–99) | 50 (89/179; 42–57) | 20 (23/113; 14–29) | 98 (89/91; 92–100) |
| Prediction rule screen and then perirectal PCR | 16/25 | 179/179 | 64 (16/25; 43–81) | 100 (179/179; 97–100) | 100 (16/16; 76–100) | 95 (179/188; 91–98) |
Prediction rule, previous CDI in thepast year and/or antibiotic use in the previous 3 months.
CI, confidence interval.
TABLE 2.
Performance of perirectal swab testing by toxigenic culture or PCR and a clinical prediction rule for detection of asymptomatic carriers of C. difficile with skin and/or environmental contamination
| Testing method | No. positive/no. skin or environment positive | No. negative/no. skin or environment negative | % Sensitivity (no. positive/total; 95% CI)b | % Specificity (no. negative/total; 95% CI) | % PPV (no. positive/total; 95% CI) | % NPV (no. negative/total; 95% CI) |
|---|---|---|---|---|---|---|
| Perirectal toxigenic culture | 14/14 | 179/190 | 100 (14/14; 73–100) | 94 (179/190; 90–97) | 56 (14/25; 35–75) | 100 (179/179; 97–100) |
| Perirectal swab PCR | 13/14a | 186/190 | 93 (13/14; 64–100) | 98 (186/190; 94–99) | 76 (13/17; 50–92) | 99 (186/187; 97–100) |
| Prediction rule | 14/14 | 91/190 | 100 (14/14; 73–100) | 48 (91/190; 41–55) | 12 (14/113; 7–20) | 100 (91/91; 95–100) |
| Prediction rule screen and then perirectal PCR | 13/14 | 187/190 | 93 (13/14; 64–100) | 98 (187/190; 95–100) | 81 (13/16; 54–95) | 99 (187/188; 97–100) |
The one patient with positive skin and/or environmental cultures but negative perirectal PCR had positive perirectal culture, positive groin culture, and positive groin PCR.
CI, confidence interval.
The clinical prediction rule was positive at the time of collection for 112 (55%) of 204 sets of specimens. The prediction rule detected 23 (92%) of 25 positive perirectal culture specimens and 14 (100%) of 14 asymptomatic carrier specimen sets with positive skin and/or environmental cultures (Tables 1 and 2). However, the prediction rule had suboptimal specificity. Combined use of the prediction rule as an initial sensitive screen, followed by the use of perirectal PCR only for prediction rule-positive subjects, would have decreased the number of PCR tests required from 204 to 112 but reduced the sensitivity of the screening procedure from 92% on the basis of the prediction rule alone to 64% for the detection of asymptomatic carriers (Table 1). However, the combined use of the prediction rule and then perirectal PCR provided excellent sensitivity and specificity for the detection of the subset of asymptomatic carriers with skin and/or environmental contamination (Table 2). Analysis of the data including only the initial set of culture and PCR results for each patient yielded similar results (data not shown).
As shown in Fig. 1, the likelihood of carriers having positive skin or environmental cultures or positive perirectal PCR results increased as the number of colonies per swab increased. In comparison to carriers with 1 to 25 colonies per swab, those with ≥26 colonies per swab had significantly higher frequencies of skin shedding (1 [10%] of 10 versus 10 [67%] of 15; P = 0.012) and environmental shedding (0 of 10 versus 9 [60%] of 15; P = 0.003) and were more likely to have positive perirectal PCR results (3 [30%] of 10 versus 14 [93%] of 15; P = 0.002).
FIG 1.
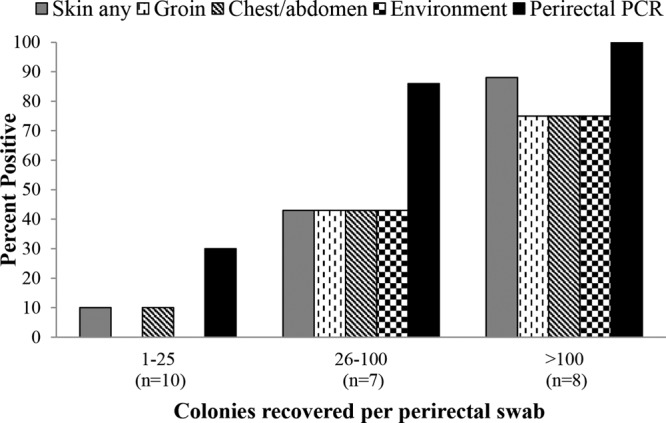
Percentages of positive perirectal PCR results and skin and environment cultures from LTCF residents with asymptomatic carriage of C. difficile on the basis of positive perirectal cultures, stratified by the number of C. difficile colonies recovered per swab. The skin sites cultured included the groin and the combined chest and abdomen. The environmental sites cultured included the bed rail and bedside table. Twenty-five sets of specimens were collected from 21 asymptomatic carriers.
Our findings suggest that a commercial PCR assay of perirectal swab specimens could provide a sensitive and rapid means to identify asymptomatic carriers of C. difficile in LTCFs at high risk of spore shedding. The number of colonies recovered from perirectal swabs varied widely, and a low burden of colonization was associated with a low risk of skin or environmental shedding or a positive perirectal PCR result. Previous studies have demonstrated that asymptomatic carriers typically have significantly lower concentrations of C. difficile in their stool than CDI patients do (6, 8, 9). Our results are consistent with a recent report that a commercial PCR assay of perirectal swab specimens was not sufficiently sensitive to detect carriers with a low burden of C. difficile (limit of detection, ∼4 log10 CFU) (10).
Our study has some limitations. The study population included mostly men, and the number of asymptomatic carriers was small. Additional studies of other health care populations are needed. We tested only one commercial PCR assay, and there may be variability in the sensitivity of nucleic acid amplification tests (11). We tested perirectal and groin CultureSwabs (Becton Dickinson) with the Cepheid Xpert C. difficile kit, which has been validated only with other types of swabs and for stool specimens. However, we have previously demonstrated similar performance of the Xpert C. difficile assay for the testing of CDI patients with perirectal samples collected with CultureSwabs and concurrent stool samples processed with the Cepheid sample collection device (12). Although the difference in the frequency of asymptomatic carriage between the study sites may reflect different carriage patterns in the facilities, it is possible that there were differences in swab collection or culturing methods between the sites. Routine screening for asymptomatic carriage may be useful as a control strategy only in facilities with a relatively high prevalence of carriage. Although it is plausible that carriers with positive perirectal PCR results are at increased risk of transmission because of increased skin and environmental shedding, further studies are needed to evaluate the transmission risk of PCR-negative versus PCR-positive carriers. The swab culture method that was used is less sensitive than broth enrichment cultures collected with moistened gauze (6), and therefore, our findings may underestimate the frequency of skin and environmental contamination. Although the clinical prediction rule is simple to apply, many LTCFs may not use PCR for CDI testing or the results of testing may not be available in a timely manner. Finally, although there is increasing evidence that asymptomatic carriers may contribute to the transmission of C. difficile (7, 9), current guidelines do not recommend screening for carriage (13).
Our results validate the previously derived clinical prediction rule as a sensitive method to detect asymptomatic carriers of C. difficile in LTCF populations but confirm that it does not have sufficient specificity to be useful alone as a screening test. However, our findings suggest that the prediction rule might be very useful as an initial simple and sensitive screen to identify a subset of high-risk patients who could then have perirectal swabs collected for PCR or culture. The use of the prediction rule as an initial screen would significantly reduce the cost of screening, and the combined strategy could identify the carriers that are most likely to serve as a reservoir for transmission. Because contact precautions may have adverse effects, there is a need for the development of strategies to reduce the risk of transmission from asymptomatic carriers that will not require isolation (e.g., enhanced environmental disinfection, bathing to reduce the burden of spores on the skin [14]).
ACKNOWLEDGMENTS
This study was supported by the Centers for Disease Control and Prevention and by the Department of Veterans Affairs.
C. J. Donskey is a consultant for GOJO and 3M and has received research grants from ViroPharma, Pfizer, and Cubist. D.N.G. holds patents for the treatment and prevention of CDI licensed to ViroPharma; is a consultant for Sanofi Pasteur, Merck, ViroPharma, GSK, Roche, Novartis, Cubist, Cangene, and Actelion; and holds a research grant from GOJO.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, Brassard P, Dendukuri N, Béliveau C, Oughton M, Brukner I, Dascal A. 2011. Host and pathogen factors for Clostridium difficile infection and colonization. N. Engl. J. Med. 365:1693–1703. 10.1056/NEJMoa1012413 [DOI] [PubMed] [Google Scholar]
- 2.Bender BS, Bennett R, Laughon BE, Greenough WB, III, Gaydos C, Sears SD, Forman MS, Bartlett JG. 1986. Is Clostridium difficile endemic in chronic-care facilities? Lancet ii:11–13 [DOI] [PubMed] [Google Scholar]
- 3.Thomas DR, Bennett RG, Laughon BE, Greenough WB, III, Bartlett JG. 1990. Postantibiotic colonization with Clostridium difficile in nursing home patients. J. Am. Geriatr. Soc. 38:415–420 [DOI] [PubMed] [Google Scholar]
- 4.Walker KJ, Gilliland SS, Vance-Bryan K, Moody JA, Larsson AJ, Rotschafer JC, Guay DR. 1993. Clostridium difficile colonization in residents of long-term care facilities: prevalence and risk factors. J. Am. Geriatr. Soc. 41:940–946 [DOI] [PubMed] [Google Scholar]
- 5.Simor AE, Yake SL, Tsimidis K. 1993. Infection due to Clostridium difficile among elderly residents of a long-term-care facility. Clin. Infect. Dis. 17:672–678. 10.1093/clinids/17.4.672 [DOI] [PubMed] [Google Scholar]
- 6.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. 2007. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin. Infect. Dis. 45:992–998. 10.1086/521854 [DOI] [PubMed] [Google Scholar]
- 7.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. 1992. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J. Infect. Dis. 166:561–567. 10.1093/infdis/166.3.561 [DOI] [PubMed] [Google Scholar]
- 8.Guerrero DM, Becker JC, Eckstein EC, Kundrapu S, Deshpande A, Sethi AK, Donskey CJ. 2013. Asymptomatic carriage of toxigenic Clostridium difficile by hospitalized patients. J. Hosp. infect. 85:155–158. 10.1016/j.jhin.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 9.Curry SR, Muto CA, Schlackman JL, Pasculle AW, Shutt KA, Marsh JW, Harrison LH. 2013. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin. Infect. Dis. 57:1094–1102. 10.1093/cid/cit475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curry SR, Schlackman JL, Hamilton TM, Henderson TK, Brown NT, Marsh JW, Shutt KA, Brooks MM, Pasculle AW, Muto CA, Harrison LH. 2012. Perirectal swab surveillance for Clostridium difficile by use of selective broth preamplification and real-time PCR detection of tcdB. J. Clin. Microbiol. 50:4078–4082. 10.1128/JCM.00679-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gyorke CE, Wang S, Leslie JL, Cohen SH, Solnick JV, Polage CR. 2013. Evaluation of Clostridium difficile fecal load and limit of detection during a prospective comparison of two molecular tests, the illumigene C. difficile and Xpert C. difficile/Epi tests. J. Clin. Microbiol. 51:278–280. 10.1128/JCM.02120-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kundrapu S, Sunkesula VC, Jury LA, Sethi AK, Donskey CJ. 2012. Utility of perirectal swab specimens for diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 55:1527–1530. 10.1093/cid/cis707 [DOI] [PubMed] [Google Scholar]
- 13.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455. 10.1086/651706 [DOI] [PubMed] [Google Scholar]
- 14.Nerandzic MM, Rackaityte E, Jury LA, Eckart K, Donskey CJ. 2013. Novel strategies for enhanced removal of persistent Bacillus anthracis surrogates and Clostridium difficile spores from skin. PLoS One 8:e68706. 10.1371/journal.pone.0068706 [DOI] [PMC free article] [PubMed] [Google Scholar]


