Abstract
Mycobacterium tuberculosis Beijing strains represent targets of special importance for molecular surveillance of tuberculosis (TB), especially because they are associated with spread of multidrug resistance in some world regions. Standard 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing lacks resolution power for accurately discriminating closely related clones that often compose Beijing strain populations. Therefore, we evaluated a set of 7 additional, hypervariable MIRU-VNTR loci for better resolution and tracing of such strains, using a collection of 535 Beijing isolates from six world regions where these strains are known to be prevalent. The typeability and interlaboratory reproducibility of these hypervariable loci were lower than those of the 24 standard loci. Three loci (2163a, 3155, and 3336) were excluded because of their redundant variability and/or more frequent noninterpretable results compared to the 4 other markers. The use of the remaining 4-locus set (1982, 3232, 3820, and 4120) increased the number of types by 52% (from 223 to 340) and reduced the clustering rate from 58.3 to 36.6%, when combined with the use of the standard 24-locus set. Known major clonal complexes/24-locus-based clusters were all subdivided, although the degree of subdivision varied depending on the complex. Only five single-locus variations were detected among the hypervariable loci of an additional panel of 92 isolates, representing 15 years of clonal spread of a single Beijing strain in a geographically restricted setting. On this calibrated basis, we propose this 4-locus set as a consensus for subtyping Beijing clonal complexes and clusters, after standard typing.
INTRODUCTION
Standardized molecular surveillance of infectious diseases is gaining increased importance, especially in the context of globalization and (re-)emergence of some pathogens. This is particularly true for tuberculosis (TB). The complex epidemiology of the disease, linked to the highly variable period of subclinical chronic infection before activation toward contagious states, requires efficient tools to estimate the dynamics of transmission in human populations. The steadily rising threat of multidrug-resistant (MDR) and extremely drug-resistant (XDR) strains of the causative agent, Mycobacterium tuberculosis, in a number of world regions further accentuates the need for more fine-tuned molecular tracing, in order to better detect, prevent, and understand the reasons for the spread of particular clones locally and globally.
Because of its portable data format and relatively high resolution power for many microbial species, multiple-locus variable-number tandem-repeat analysis (MLVA) (1) has become a widely used method for pathogen typing. A format targeting 24 variable-number tandem-repeat (VNTR) markers, including genetic elements called mycobacterial interspersed repetitive units (MIRUs), has been internationally standardized for typing of M. tuberculosis (2). This format, optionally combined with spoligotyping, has been shown to have a predictive value nearly similar to that of standard IS6110 restriction fragment length polymorphism (RFLP)-based typing for tracing TB transmission at population-based and even nationwide levels in several Western European settings (e.g., references 3 to 7). Because of its shorter time-to-result, especially when using marker multiplexing on automated DNA analyzers, standard MIRU-VNTR typing, often complemented by spoligotyping (8), has therefore largely replaced IS6110-RFLP in molecular epidemiological surveillance programs, e.g., at the U.S. CDC (9) and the ECDC (http://www.tuberculosis.rivm.nl/).
However, several studies suggested that 24-locus MIRU-VNTR typing can lack resolution power for discriminating closely related clones of the M. tuberculosis Beijing lineage (5, 10–18). This lineage constitutes the bulk of the East Asian lineage of M. tuberculosis (19). It is of special interest as it is dominant in large regions of the world, including East Asia, Russia and the Commonwealth of Independent States (CIS) countries, and South Africa, and, in the Western world, among TB patients originating from these regions. Beijing strains have been associated with large outbreaks and rapid spread of MDR TB in different geographic regions (reference 20 and references therein). So-called hypervariable MIRU-VNTR loci (2), because of their higher variability than that of the 24 standardized loci, have been tested and used to increase discriminatory power on Beijing strains (5, 10–18, 21). Nonetheless, intra- and interlaboratory analyses suggested lack of reproducibility and robustness of results based on at least some of these hypervariable loci, questioning their informative value and justifying their exclusion from the standard 24-locus set for first-line typing (2). As previous studies evaluated hypervariable loci separately based on isolates from specific areas, and in most cases without parallel estimation of their reproducibility and longitudinal stability to trace transmission chains, no consensus was made on potentially usable loci and on the actual extent and epidemiological relevance of their additional resolution power.
Here, we evaluated the additional discriminatory power provided by a total of 7 candidate hypervariable loci, as compiled from all previous relevant studies, as well as their technical applicability and interlaboratory reproducibility compared to the standard 24-locus set. This evaluation was conducted multicentrically in parallel by Genoscreen and 6 different reference laboratories, on a global sample of more than 500 M. tuberculosis Beijing isolates from six different world regions where this lineage is prevalent. In addition, we measured the clonal stability of the hypervariable and standard markers in a genuine longitudinal outbreak context, by testing 92 isolates from a unique, geographically restricted epidemic, representing 16 years of clonal spread of a single Beijing strain (22). Our results define a consensus subset of hypervariable loci proposed to be used for second-line typing of M. tuberculosis Beijing isolates.
MATERIALS AND METHODS
Isolates and genomic DNA samples.
The origin of the isolates and the corresponding sampling modalities are described in Table 1. Seventy-six to 94 genomic DNA samples, obtained by crude extraction after heat inactivation or by purification (23) from cultured isolates, were initially selected by each of 6 reference laboratories, making a total of 546 samples composing the “reproducibility panel.” The isolates had been identified by the laboratories as Beijing strains, by detection of a typical spoligotype containing at least three of the spacers of the spacers 35 to 43 and none of the spacers 1 to 34 and/or typical IS6110 fingerprints, as defined in references 24 and 25. The corresponding 24-locus MIRU-VNTR genotypes were additionally analyzed on the MIRU-VNTRPlus database as described previously (26, 27), in order to confirm this lineage identification. From this panel, 12 isolates were subsequently excluded after actual identification of another lineage than Beijing, after detection of double alleles in two loci or more of the 24 standard loci (indicative of the presence of two independent strains [28] in the isolate), or after nonamplification of more than two standard loci and/or more than two hypervariable loci suggestive of genomic DNA quality problems. After additional inclusion of one representative of the outbreak panel (see below), a total of 535 M. tuberculosis Beijing isolates were then retained to constitute a second strain panel designated the “global panel,” used to evaluate interstrain marker variability and resolution power. A third panel, designated the “outbreak panel,” included 92 other genomic DNA samples from Beijing isolates all obtained from different patients, covering a 16-year time span (i.e., 1993 to 2008) of a longitudinal outbreak after a single clonal introduction on Gran Canaria Island (22). The clonality of these isolates was previously confirmed by PCR interrogation of a strain-specific IS6110 insertion site, in addition to IS6110 RFLP analysis (29).
TABLE 1.
M. tuberculosis Beijing strain collection
| Geographic origin | No. of isolates | Reproducibility panel (n) | Global panel (n) | Outbreak panel (n) | Sampling |
|---|---|---|---|---|---|
| Northwest Russiaa | 76 | 76 | 76 | Conveniencee | |
| Central Russiab | 94 | 94 | 87 | Convenience | |
| Germany | 94 | 94 | 94 | MDR TB cases | |
| Japan | 94 | 94 | 94 | Randomf | |
| Singapore | 94 | 94 | 93 | Convenience | |
| South Africac | 94 | 94 | 90 | Genotype selectiong | |
| Gran Canaria | 92 | 1d | 92d | Outbreak relatedh | |
| Total | 638 | 546 | 535 | 92 |
Saint Petersburg and Kaliningrad regions.
Samara Oblast.
Cape Town region.
One representative sample of the outbreak panel was included in the global panel.
From Beijing isolates identified in strain collections described in references 39 (n = 41) and 40 (n = 35).
Random selection from 228 Beijing isolates, identified among 325 isolates selected from 3,122 isolates collected for a drug resistance survey from the whole of Japan in 2002.
Selection based on IS6110 RFLP profiles identified in reference 15.
Genotyping.
MIRU-VNTR typing was performed at Genoscreen on a total of 638 isolates and in parallel by each reference center on its own isolate set (except for the outbreak panel, for which typing was performed only at Genoscreen). Typing at Genoscreen was performed using capillary electrophoresis on a 3730-XL DNA analyzer (ABI) and under the conditions of a commercially available typing service and kit, optimized from reference 2. Typing with the standard markers in the reference laboratories was performed using capillary electrophoresis on a 3130-XL DNA analyzer (ABI) and the commercially available kit (n = 2), or in-house protocols adapted from reference 2, combined with capillary electrophoresis on a Beckman Coulter CEQ8000 DNA analyzer (n = 1) on the SV1210 system (Hitachi) (n = 1), or electrophoresis on agarose gels (n = 2). For the analysis of the hypervariable loci, the same electrophoresis platforms were used by the respective laboratories, using either previously published primers and protocols (14, 15, 30) (n = 3) or protocols and primers used or newly designed by Genoscreen (n = 3) (Table 2). To homogenize the counting of repeat units especially for loci containing partial repeats (e.g., locus 0580, alias MIRU 04/ETR-D, or locus 3232, alias QUB-3232), a standard allele calling system (2) was used for the 24 standard markers, while the system used for the hypervariable loci was based on analysis of H37Rv genome sequence and conventions described in Table 2. The corresponding conventional genotype of the reference strain M. tuberculosis H37Rv was sent to each laboratory to verify and ensure consistent allele calling. Genotyping at Genoscreen was initially performed blinded to the results obtained by the respective participating laboratories. Discordant genotyping results between Genoscreen and these laboratories were subsequently reviewed in parallel, and consensus corrections were performed, when necessary.
TABLE 2.
Locus designations and primer sequences used in this study for the hypervariable loci
| Markera | Alias | Consensus panel | Primer pair, 5′–3′ (dye) | Reference | Amplicon size (bp) in H37Rv | Repeat no. × length (bp) in H37Rv |
|---|---|---|---|---|---|---|
| 1982 | QUB-18 | Included | CCGGAATCTGCAATGGCGGCAAATTAAAAG | 2 | 620 | 5 × 78 |
| TGATCTGACTCTGCCGCCGCTGCAAATA (NED) | ||||||
| 3232 | QUB-3232 | Included | CACTAGTTGTTGCGGCGATGG | 41 | 421 | 3 × 56b |
| AAGGGCGGCATTGTGTTC (FAM) | ||||||
| 3820 | Included | ACCTTCATCCTTGGCGAC (NED) | 42 | 444 | 3 × 57 | |
| TGCGCGGTGAATGAGACG | ||||||
| 4120 | Included | GTTCACCGGAGCCAACC (FAM) | 42 | 447 | 2 × 57 | |
| GAGGTGGTTTCGTGGTCG | ||||||
| 2163a | QUB-11a | Excluded | TGTTGAAGAAGCCCGAAGAC (PET) | This study | 451 | 3 × 69 |
| CCGTTTAACGTCGCTGGTAT | ||||||
| 3155 | QUB-15 | Excluded | AGGGGTTCTCGGTCACCC (VIC) | 43 | 252 | 3 × 54 + 1 × 45c |
| TACATTCGCGGCCAAAGG | ||||||
| 3336 | QUB-3636 | Excluded | GATCGGGTGCAGTGGTTTCAGGTG | This study | 571 | 5 × 59 |
| GGGCGGCCAGCGGTGTC (VIC) |
Markers are coded by conventional numbers indicating their position (in kbp) on the M. tuberculosis H37Rv chromosome.
Additional incomplete repeat unit of 48 bp not taken into consideration for allele reporting, by convention.
Repeat units of 54 and 45 bp are jointly considered for allele reporting (i.e., 3 × 54 + 1 × 45 = allele 4), by convention.
Genotyping data analysis.
Unweighted pair group method with arithmetic mean (UPGMA)-based cluster analysis, minimum spanning tree analysis, and determination of the frequencies of single-locus variations (SLVs) were performed using Bionumerics v6.5 (Applied Maths, Belgium) and a categorical coefficient. For samples with missing alleles (at most 2; see above), loci with missing information were not considered for cluster analysis. Assignment to MLVA Mtbc 15-9 genotypes was based only on complete 24-locus genotypes. The MIRU-VNTR allelic diversity (H) at a given locus was calculated as H = 1 − ∑xi2 [(n/n − 1)], where xi is the frequency of the ith allele in the population and n is the number of isolates.
RESULTS
Typeability and reproducibility.
The typeability and reproducibility of standard and hypervariable markers were analyzed on the reproducibility strain panel of 546 isolates, by comparing the results respectively obtained by the reference centers on their own isolate set with those obtained at Genoscreen. The average typeability (as defined by detection of an allele after PCR amplification) per marker ranged from 96.9 (2163a) to 99.7% (3336 and 3155) for the hypervariable markers (average of 98.8%), slightly lower than the 97.6 (3192) to 100% (10 out of 24 markers) range of the 24 standard markers (average, 99.3%) (see Fig. S1a in the supplemental material). Locus 2163a was particularly prone to lack of amplification; amplification of large, nonsizable fragments (i.e., >1,400 bp); or a noninterpretable multibanded pattern with no clear, typical stutter peak pattern (31) (see Table S1). For the 24 standard markers, these limited typeability problems were discernibly skewed by results obtained by one particular laboratory, accounting for 53 of the 75 PCR failures detected among 9,936 individual amplifications (total failure rate of 0.8%). Amplification problems tended to be more equally distributed among laboratories for the hypervariable loci, although the same laboratory encountered more frequent problems (12 failures out of a total of 31, for 2,694 amplifications; total failure rate of 1.2%) (see Fig. S1b).
The average reproducibility per locus reached 98.6% for the standard 24 loci, ranging from 90.9% (locus 0577) to 100% (three loci) (see Fig. S1c in the supplemental material). The average reproducibility of the 7 hypervariable loci was substantially lower (91.9%), ranging from 68.8% (locus 3820) to 98.6% (loci 1982 and 3155). However, for both the standard and hypervariable loci, the discordances were again clearly more frequent for one laboratory, where 86 of the 138 and 106 of the 217 deviating results were obtained for the 24 standard and the 7 hypervariable markers, respectively (see Fig. S1d).
Marker resolution power and variability.
The variability and resolution power of the standard and hypervariable marker sets were evaluated on the global strain panel that included a total of 535 isolates, resulting from exclusion of 12 problematic samples from the reproducibility panel and inclusion of one representative of the outbreak panel (see Materials and Methods).
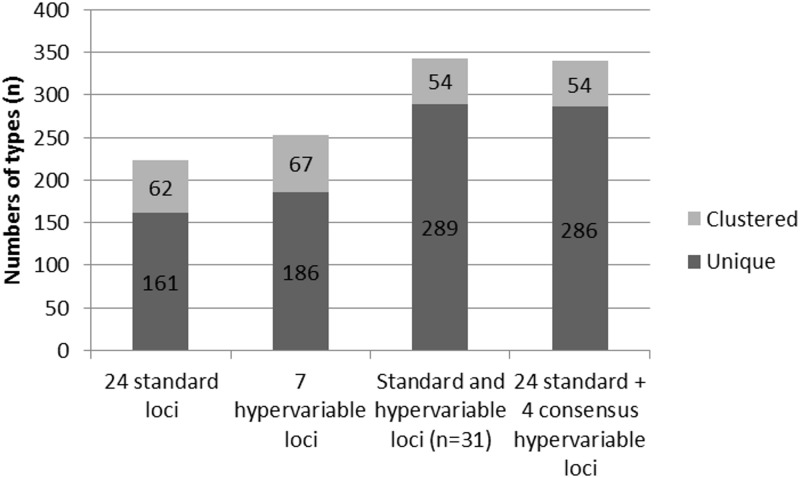
In this panel, the 24 standard markers resolved 223 types; 161 were unique and 62 were in clusters, of which 7 included from 10 to 75 isolates. In comparison, 253 types were detected using the full hypervariable set alone, including 186 unique and 67 clustered types. Similarly to the results obtained for the standard markers, 7 clusters included more than 10 isolates and up to 69 for the largest cluster. Combining the 31 markers from both sets resulted in the maximal apparent resolution power, as reflected both by the highest number of types (n = 343) and unique types (n = 289) and by the lowest number of clustered types (n = 54) (Fig. 1).
FIG 1.
Resolution power of the different marker sets on the global strain panel of 535 Beijing isolates. Histograms indicate numbers of unique (dark gray) and clustered (light gray) genotypes, as well as total numbers of types (unique plus clustered).
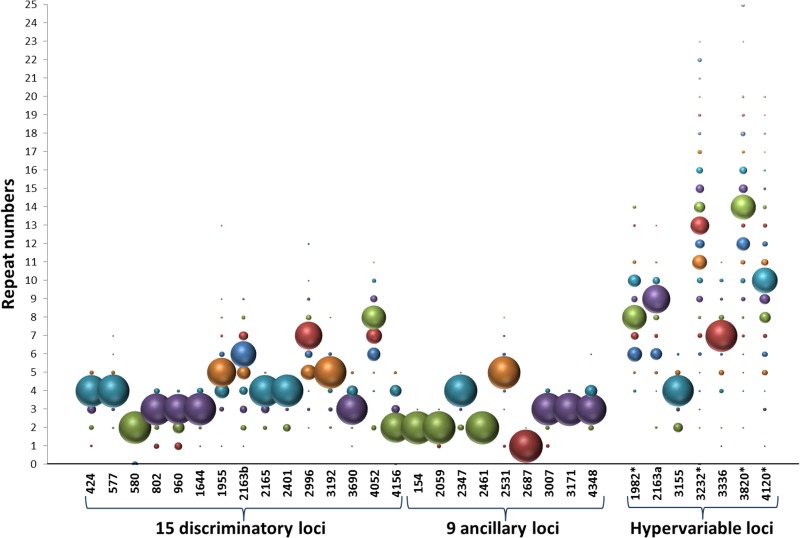
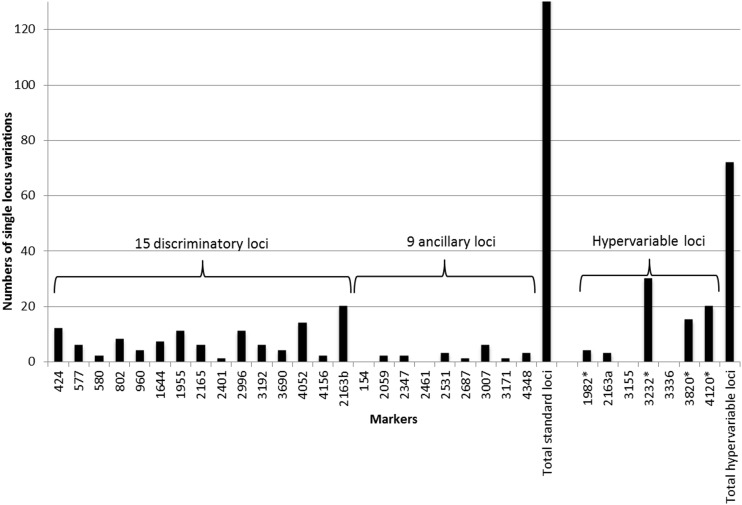
We examined the contribution of individual loci to the total discriminatory power, by looking at their respective allelic diversities (as visualized by the allelic distributions represented on Fig. 2 and as calculated in Table S1 in the supplemental material) and at frequencies of single-locus variations (SLVs) (Fig. 3), indicating when the locus considered was indispensable for distinguishing two types differing by only a single marker. Among the standard 24 loci, as expected from results from previous studies (2–4, 6), the 15 loci of the so-called discriminatory subset globally showed the highest diversities and SLV frequencies, with maximal values classically seen for loci 4052, 2613b, 1955, and 2996 (diversities of up to 0.64 and up to 20 SLVs per locus). The ancillary 9-locus subset consistently showed individual diversities ranging from 0.00 to 0.28 at most, with markers rarely to never involved in SLVs (from 0 to 3 events for 8 markers; 6 for locus 3007).
FIG 2.
Allelic distributions of the 31 markers on the global strain panel of 535 isolates. Bubble sizes are proportional to the frequencies of the different alleles detected for each marker. Bubble colors correspond to different alleles observed for each marker, with corresponding repeat numbers shown on the y axis. The 15 discriminatory loci and 9 ancillary loci compose the standard set of 24 markers. Hypervariable loci retained for the consensus set are marked by an asterisk. Markers are coded by conventional numbers indicating their position (in kbp) on the M. tuberculosis H37Rv chromosome.
FIG 3.
Distribution of single-locus variation events in the global strain panel of 535 isolates. See the legend to Fig. 2 for description and coding of markers.
Among the 7 markers of the hypervariable set, according to the same allelic diversity and SLV frequency parameters (Fig. 2 and 3), three markers (3232, 3820, and 4120) were clearly above any of the 24 standard markers, with allelic diversities of up to 0.83 and 30 SLVs (3232). A fourth one (1982) ranked among the most variable standard loci in terms of allelic diversity (0.65), and 4 SLVs based on this marker were detected. In contrast, the diversity of 2163a (0.42) was not superior to average values seen among the 15 standard discriminatory markers; 3 SLVs based on this marker were detected. Loci 3155 and 3336 displayed low diversities of 0.22 and 0.18, respectively, and were not involved in any single-locus variation over the global strain panel.
We analyzed the frequencies of double alleles detected in a single marker in some isolates, as a reflection of the marker variability by microevolution in a clonal subpopulation (11, 32). Consistently, double alleles were observed substantially more often among the hypervariable loci (33 occurrences among 5 of the 7 loci) than among the standard loci (16 occurrences among 7 of the 24 loci, which were all part of the discriminatory subset of 15 loci) (see Fig. S2 in the supplemental material).
We then examined in more detail the resolution power provided by the combination of the 24 markers with the hypervariable set restricted to 4 loci (1982, 3232, 3820, and 4120), after subtracting loci 3155, 3336, and 2163a, which show very limited to no contribution to the total resolution power (and, for 2163a, also showing more frequent amplification problems; see above). As expected, the total numbers of types (n = 340) and numbers of unique (n = 286) and clustered (n = 54) types were almost identical to the same parameters measured for the 31 loci (Fig. 1).
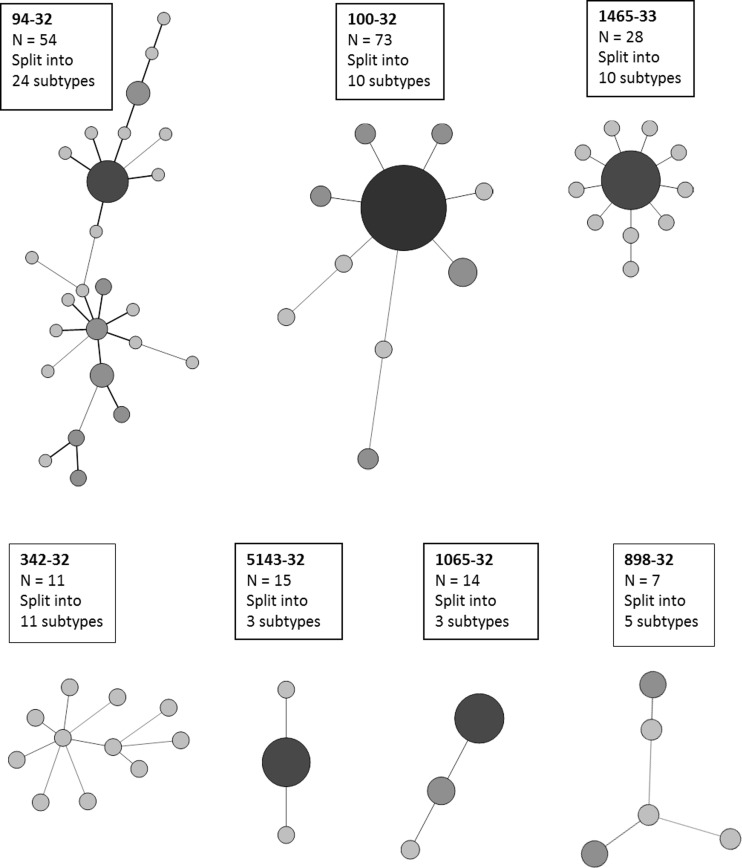
Finally, we evaluated the capacity of this 4-locus hypervariable set to subdivide some of the major Beijing clusters and genotypes, as defined by the standard 24 loci and the MIRU-VNTRPlus MLVA Mtbc 15-9 database nomenclature (26, 27) and containing from 7 to 73 isolates (note that a few samples with one or two missing alleles were not included in this analysis) in this global panel. Among the 7 largest clusters involved, 5 (i.e., 100-32, 94-32, 1065-32, 342-32, and 898-32) were of clear supranational distribution, with at least three collection sites represented in each case. Of these, clusters of 54 “94-32” and 7 “898-32” isolates were largely subdivided by the 4-locus hypervariable set into 24 and 5 subtypes, respectively, and a cluster of 11 “342-32” isolates was fully split (Fig. 4). In contrast, clusters of 73 “100-32” and of 14 “1065-32” isolates were subdivided to a lesser extent into 10 and 3 subtypes, respectively. The two remaining largest clusters, 1465-33 and 5143-32, were mostly of regional prevalence, representing essentially South Africa and including each a single isolate from another site. The cluster of 28 “1465-33” isolates was relatively efficiently subdivided into 10 subtypes, though without separation of the single isolate from Central Russia from those of South Africa. The cluster of 15 “5143-32” isolates was split into only three subtypes, but with separation of the single isolate from Singapore.
FIG 4.
Major 24-locus-based Beijing genotypes subtyped by the 4 consensus hypervariable loci. MLVA Mtbc 15-9 nomenclature codes of the corresponding clonal complexes (e.g., 100-32) are boxed, as well as the numbers of isolates sharing the same 24-locus genotype (e.g., 73 for 100-32) and the numbers of subtypes obtained by using the 4 consensus hypervariable loci.
Longitudinal stability of markers.
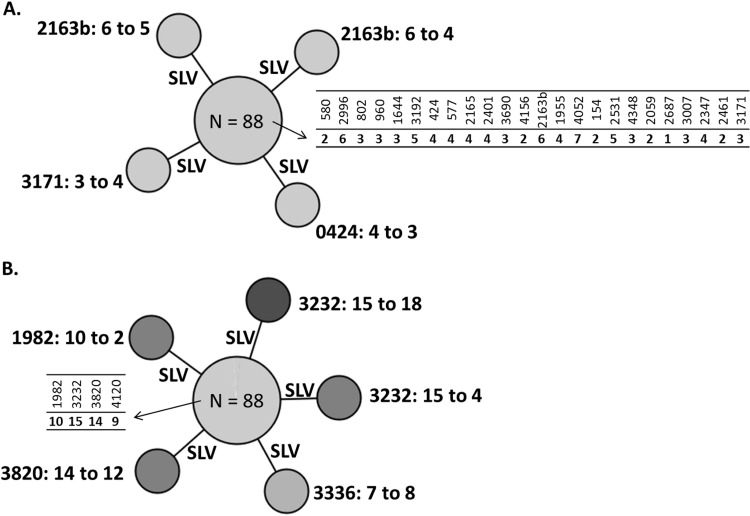
The longitudinal stability of the standard and hypervariable markers was evaluated by typing, at Genoscreen, of the outbreak panel from Gran Canaria. This panel included patient isolates collected at early (1993 to 1996, n = 21), intermediate (1999 to 2005, n = 48), and late (2007 to 2008, n = 23) time points of the outbreak. Among the 92 corresponding isolates, 88 displayed fully identical standard 24-locus genotypes (Fig. 5A). Only four isolates showed a change, restricted to a single-locus variation in each case, involving locus 2163b twice (in one isolate obtained in 2002 and one in 2007–2008, respectively), and 3171 (isolate of 2002) and 0424 (isolate of 2007–2008) once. This corresponded thus to a rate of allele change of 0.2% (4 out of 92 × 24 = 2,208 alleles).
FIG 5.
Longitudinal stability of standard and hypervariable loci evaluated on the outbreak panel, representing 15 years of clonal spread of a single Beijing strain. Minimum spanning trees representing the core genotype and its 4 single-locus variants based on 24 standard loci (A) and the core genotype and its 5 single-locus variants based on 4 consensus hypervariable loci (B), identified on the 92 isolates of the outbreak panel from Gran Canaria.
In comparison, five changes were detected among the hypervariable loci among the same isolates, corresponding to a rate of allele change of 0.7% (5 out of 92 × 7 = 644 alleles) (Fig. 5B). Four changes concerned the consensus set of markers: locus 3232 was involved twice (in one isolate obtained in 1999 and one in 2002, respectively), while 1982 (isolate of 1999) and 3820 (isolate of 2007–2008) were each involved once. The remaining change was detected in locus 3336 (in one isolate of 2007–2008), which is excluded from the consensus set. Of note, the results of locus 2163a could not be interpreted for any of these 92 isolates, as a nonreliably sizable fragment of more than 1,400 bp was amplified in all cases.
Interestingly, the core 24-locus genotype of the Gran Canaria outbreak isolates was shared by two isolates from another and distant site, i.e., Northwest Russia. Both isolates were distinguished from the Gran Canaria group (and from each other) by the 4-locus hypervariable set.
DISCUSSION
Because M. tuberculosis Beijing strains spread in some world regions in association with drug resistance and have been associated with clinical or physiopathological manifestations suggestive of higher virulence in epidemiological or animal model studies (reviewed in reference 24), they represent special targets for epidemiological surveillance and control. However, accurate molecular tracing of Beijing clones and delineation of outbreaks caused by such strains are complicated by the remarkable genetic homogeneity and the widespread geographic distribution of some clonal complexes. Here, based on a multicenter study involving 7 internationally recognized centers, we evaluated and defined a consensus 4-locus set of hypervariable MIRU-VNTR loci (1982, 3232, 3820, and 4120) that, when used in complement to standard 24-locus based MIRU-VNTR typing, (i) substantially improve the resolution power on the Beijing strain population, in particular of some major clonal complexes; (ii) still retain sufficient clonal stability to follow epidemic spread of a particular strain over time; and (iii) have a relatively limited cost in terms of typeability and (inter-)laboratory reproducibility problems, under well-controlled technical conditions.
Crucially, in contrast to previous studies looking at smaller strain populations from single regions, the additional discriminatory power provided by the hypervariable loci was evaluated on a global sample of 535 isolates from different world regions or patient populations with an especially high prevalence of Beijing strains, i.e., mainly from Northwest and Central Russia, Japan, Singapore, South Africa, and MDR-TB patients from Germany. The present panel is known to contain the major genetic branches of the Beijing strain population seen in over more than 50 countries worldwide (T. Wirth, personal communication). On this representative sample, secondary typing using the consensus set of 4 hypervariable loci allowed us to increase the number of types by 52% (from 223 to 340) and accordingly reduced the clustering rate from 58.3 to 36.6%, compared to the standard 24-locus set used alone.
Most of the isolates of this panel were obtained from patients presumed or found to be epidemiologically unlinked, with the exception of 13 isolates in 5 clusters from Singapore. For the latter isolates, genotype identity based on the 24 standard and 4 (or 7) hypervariable loci was confirmed by posterior indication of an epidemiological linkage. A number of other isolates were found to represent the same major Beijing clonal complexes as identified by IS6110 RFLP and 24-locus MIRU-VNTR typing. For instance, the clonal complex defined by 24-locus genotype 100-32 represents the major part of the successful Russian clone B0/W148 (33), while 94-32 (20) is widespread in Central Asia. Clonal complexes represented by genotypes 5143-32 and 1465-33 are prevalent regionally in South Africa (15). The 4-locus hypervariable set specifically subdivided all the corresponding major 24-locus-based clusters in this global panel but to variable extents. Some, including clusters/genotypes 94-32 and 1465-33, were largely or fully subdivided, with, in 3 out of these 4 cases, multiple occurrences of variations in two or more loci in pairwise comparisons between isolates (Fig. 4). In contrast, other clusters, including the most prominent Beijing clonal complex 100-32 and 5143-32, were split into proportionally much fewer subtypes, with mostly single-locus variations between a few individual subtypes and a dominant central subtype.
Not a single instance of double (or more)-locus variations was observed, for both the standard 24-locus and the hypervariable sets, in our outbreak panel of 92 isolates capturing the expansion of a single clone over 16 years. This finding, based on a specific Beijing background, is fully consistent with our analyses of 12- or 24-locus genotypes of >300 isolates from genetically diverse clusters with proven epidemiological links and sets of clonal serial isolates covering time periods of up to 7 years, where neither double-locus nor more-locus variations were observed (2, 34). Thus, although hypervariable loci have a higher evolutionary rate than the standard markers (see below), the same threshold of double-locus (or more) variations can be used for highly reliable exclusion of an epidemiological link between isolates, as for standard 24-locus typing (2). Therefore, patterns of large subdivision with multiple double or more variations among hypervariable 4-locus subtypes, seen, e.g., for 24-locus-based genotype 94-32, represent strong evidence that the detected subtypes reflect in many cases distinct clones with no direct epidemiological links. Such clones are more distant than could be anticipated based on 24-locus data.
Five instances of single-locus variations were nevertheless observed among the 92 isolates of the outbreak panel in the hypervariable 4-locus set, versus 4 among the 24 standard loci (which is also fully in line with the approximately 5% of single-locus variations previously measured among 12- or 24-locus genotypes from epidemiologically proven clusters and serial isolates [2, 34]). As expected for mutations occurring stochastically over such an evolutionary scale, these changes were observed at early, intermediate, or late time points. Although the rate of allele change in the hypervariable set is about three times higher than that of the standard set (taking into account the number of loci in the respective sets), this frequency of single locus variation distributed over a period of 16 years remains relatively low. Hence, even for hypervariable loci, at least in the absence of further specific epidemiological or contact tracing evidence, the definition of molecular clustering should remain restricted to full identity of the markers, as for standard 24-locus typing (2, 35). Indeed, under this definition—calibrated on the actual drift measured during clonal expansion—the cost in terms of false exclusion of a link in the case of a rare single-locus clonal drift will still be far less than the cost of false clustering to expect if single-locus variations were allowed for defining clusters. As additional examples of their probable epidemiological relevance in frequent cases, single-locus variations in the 4-locus set allowed us to distinguish one single isolate from Singapore and another single isolate from Northwest Russia, from the large 24-locus clusters including isolates from South Africa (i.e., 5143-32) and Gran Canaria (i.e., in the outbreak panel), respectively.
Out of the 7 markers tested in total, 3 were excluded from the proposed consensus set, for several reasons. Their variation was fully (for 3155 and 3336) or almost fully (for 2163a) redundant compared to that generated by the consensus set. Their exclusion thus marginally impacted the total number of types obtained among the 535 isolates of the global panel (340, versus 343 before their exclusion). The exclusion of 2163a is further justified by more frequently detected problems, such as lack of amplification; amplification of large, nonsizable fragments (as seen, e.g., for all the 92 isolates from the outbreak panel); or noninterpretable multibanded pattern.
As expected from previous studies (2), the typeability and the interlaboratory reproducibility obtained with the 4 hypervariable loci retained in the consensus were lower than those obtained with the standard 24 loci in this multicentric study. This lower reproducibility reflected the higher frequency of large alleles, which are more difficult to size when using agarose gels or even capillary DNA analyzers without proper calibration. In addition, the frequency of double alleles is higher for these loci, reflecting a higher rate of clonal drift as expected, which might be a source of more frequent mistakes if not properly noticed. For these reasons, we recommend the use of these hypervariable loci as a subtyping method of Beijing clones and clusters, after 24-locus typing. Nevertheless, in order to more reliably exploit the resolution power provided by the consensus set, commercially available kits have been subsequently developed, including specific allelic ladders for proper calibration of the sizing on capillary systems and a robust single 4-plex assay of the 4 target loci. Of note, at least three of the four consensus loci (i.e., 3232, 3820, and 4120) have also been recently recommended for subtyping of Beijing strains in China (36). In principle, these loci could be used as well for subtyping isolates of other M. tuberculosis lineages, although their informative value and the technical ease would remain to be established in these cases.
In conclusion, our study proposes a consensus set of 4 hypervariable MIRU-VNTR loci for epidemiologically relevant subtyping and international tracing of Beijing clonal complexes/clones that are prevalent in and/or across different world regions. This typing tool might further help ensuring the transition until whole-genome sequence analysis might become universally accessible and applicable for molecular biology-guided TB surveillance (e.g., references 37 and 38).
Supplementary Material
ACKNOWLEDGMENTS
I. Millán-Lou is acknowledged for previous analysis of IS6110 insertion sites among isolates from Gran Canaria, and S. X. Wang is thanked for support to the project. Laboratory staff, in particular K. Mak, are thanked for technical assistance.
This work was supported by European Regional Development Fund (ERDF)/Oseo grant A0906016N FS.
C.A.-B. and C.W. are employed by Genoscreen. P.S. is a consultant for the same company. All other authors have declared that no competing interests exist.
Footnotes
Published ahead of print 30 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02519-13.
REFERENCES
- 1.Smith KL, De Vos V, Bryden HB, Hugh-Jones ME, Klevytska A, Price LB, Keim P, Scholl DT. 1999. Meso-scale ecology of anthrax in southern Africa: a pilot study of diversity and clustering. J. Appl. Microbiol. 87:204–207. 10.1046/j.1365-2672.1999.00871.x [DOI] [PubMed] [Google Scholar]
- 2.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510. 10.1128/JCM.01392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allix-Beguec C, Fauville-Dufaux M, Supply P. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398–1406. 10.1128/JCM.02089-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oelemann MC, Diel R, Vatin V, Haas W, Rusch-Gerdes S, Locht C, Niemann S, Supply P. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691–697. 10.1128/JCM.01393-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roetzer A, Schuback S, Diel R, Gasau F, Ubben T, di Nauta A, Richter E, Rusch-Gerdes S, Niemann S. 2011. Evaluation of Mycobacterium tuberculosis typing methods in a 4-year study in Schleswig-Holstein, Northern Germany. J. Clin. Microbiol. 49:4173–4178. 10.1128/JCM.05293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bidovec-Stojkovic U, Zolnir-Dovc M, Supply P. 2011. One year nationwide evaluation of 24-locus MIRU-VNTR genotyping on Slovenian Mycobacterium tuberculosis isolates. Respir. Med. 105(Suppl 1):S67–S73. 10.1016/S0954-6111(11)70014-2 [DOI] [PubMed] [Google Scholar]
- 7.de Beer JL, van Ingen J, de Vries G, Erkens C, Sebek M, Mulder A, Sloot R, van den Brandt AM, Enaimi M, Kremer K, Supply P, van Soolingen D. 2013. Comparative study of IS6110 restriction fragment length polymorphism and variable-number tandem-repeat typing of Mycobacterium tuberculosis isolates in the Netherlands, based on a 5-year nationwide survey. J. Clin. Microbiol. 51:1193–1198. 10.1128/JCM.03061-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moonan PK, Ghosh S, Oeltmann JE, Kammerer JS, Cowan LS, Navin TR. 2012. Using genotyping and geospatial scanning to estimate recent mycobacterium tuberculosis transmission, United States. Emerg. Infect. Dis. 18:458–465. 10.3201/eid1803.111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso M, Alonso Rodriguez N, Garzelli C, Martinez Lirola M, Herranz M, Samper S, Ruiz Serrano MJ, Bouza E, Garcia de Viedma D. 2010. Characterization of Mycobacterium tuberculosis Beijing isolates from the Mediterranean area. BMC Microbiol. 10:151. 10.1186/1471-2180-10-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shamputa IC, Lee J, Allix-Beguec C, Cho EJ, Lee JI, Rajan V, Lee EG, Min JH, Carroll MW, Goldfeder LC, Kim JH, Kang HS, Hwang S, Eum SY, Park SK, Lee H, Supply P, Cho SN, Via LE, Barry CE., III 2010. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J. Clin. Microbiol. 48:387–394. 10.1128/JCM.02167-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwamoto T, Yoshida S, Suzuki K, Tomita M, Fujiyama R, Tanaka N, Kawakami Y, Ito M. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 270:67–74. 10.1111/j.1574-6968.2007.00658.x [DOI] [PubMed] [Google Scholar]
- 13.Mokrousov I, Narvskaya O, Vyazovaya A, Millet J, Otten T, Vishnevsky B, Rastogi N. 2008. Mycobacterium tuberculosis Beijing genotype in Russia: in search of informative variable-number tandem-repeat loci. J. Clin. Microbiol. 46:3576–3584. 10.1128/JCM.00414-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolayevskyy V, Gopaul K, Balabanova Y, Brown T, Fedorin I, Drobniewski F. 2006. Differentiation of tuberculosis strains in a population with mainly Beijing-family strains. Emerg. Infect. Dis. 12:1406–1413. 10.3201/eid1209.041263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, Ndabambi SL, Jordaan AM, Victor TC, van Helden PD, Warren RM. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46:3338–3345. 10.1128/JCM.00770-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wada T, Maeda S, Hase A, Kobayashi K. 2007. Evaluation of variable numbers of tandem repeat as molecular epidemiological markers of Mycobacterium tuberculosis in Japan. J. Med. Microbiol. 56:1052–1057. 10.1099/jmm.0.46990-0 [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama E, Kishida K, Uchimura M, Ichinohe S. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7:499–508. 10.1016/j.meegid.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 18.Kremer K, Au BK, Yip PC, Skuce R, Supply P, Kam KM, van Soolingen D. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43:314–320. 10.1128/JCM.43.1.314-320.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:2869–2873. 10.1073/pnas.0511240103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemann S, Diel R, Khechinashvili G, Gegia M, Mdivani N, Tang YW. 2010. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 48:3544–3550. 10.1128/JCM.00715-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velji P, Nikolayevskyy V, Brown T, Drobniewski F. 2009. Discriminatory ability of hypervariable variable number tandem repeat loci in population-based analysis of Mycobacterium tuberculosis strains, London, UK. Emerg. Infect. Dis. 15:1609–1616. 10.3201/eid1510.090463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Garcia I, Cabrera P, Lafoz C, Samper S, Takiff H, Afonso O, Pavon JM, Torres MJ, van Soolingen D, Enarson DA, Martin C. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164:1165–1170. 10.1164/ajrccm.164.7.2101031 [DOI] [PubMed] [Google Scholar]
- 23.Van Soolingen D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1–26. 10.1046/j.1365-2796.2001.00772.x [DOI] [PubMed] [Google Scholar]
- 24.Parwati I, van Crevel R, van Soolingen D. 2010. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10:103–111. 10.1016/S1473-3099(09)70330-5 [DOI] [PubMed] [Google Scholar]
- 25.Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, Kreiswirth BN, Bifani PJ, van Soolingen D. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040–4049. 10.1128/JCM.42.9.4040-4049.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46:2692–2699. 10.1128/JCM.00540-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. 2010. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38:W326–W331. 10.1093/nar/gkq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, Supply P, Rigouts L. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7:99. 10.1186/1465-9921-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millan-Lou MI, Alonso H, Gavin P, Hernandez-Febles M, Campos-Herrero MI, Copado R, Canas F, Kremer K, Caminero JA, Martin C, Samper S. 2012. Rapid test for identification of a highly transmissible Mycobacterium tuberculosis Beijing strain of sub-Saharan origin. J. Clin. Microbiol. 50:516–518. 10.1128/JCM.06314-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murase Y, Mitarai S, Sugawara I, Kato S, Maeda S. 2008. Promising loci of variable numbers of tandem repeats for typing Beijing family Mycobacterium tuberculosis. J. Med. Microbiol. 57:873–880. 10.1099/jmm.0.47564-0 [DOI] [PubMed] [Google Scholar]
- 31.Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571. 10.1128/JCM.39.10.3563-3571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Hajoj SA, Akkerman O, Parwati I, al-Gamdi S, Rahim Z, van Soolingen D, van Ingen J, Supply P, van der Zanden AG. 2010. Microevolution of Mycobacterium tuberculosis in a tuberculosis patient. J. Clin. Microbiol. 48:3813–3816. 10.1128/JCM.00556-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mokrousov I. 2013. Insights into the origin, emergence, and current spread of a successful Russian clone of Mycobacterium tuberculosis. Clin. Microbiol. Rev. 26:342–360. 10.1128/CMR.00087-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savine E, Warren RM, van der Spuy GD, Beyers N, van Helden PD, Locht C, Supply P. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561–4566. 10.1128/JCM.40.12.4561-4566.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloot R, Borgdorff MW, de Beer JL, van Ingen J, Supply P, van Soolingen D. 2013. Clustering of tuberculosis cases based on variable-number tandem-repeat typing in relation to the population structure of Mycobacterium tuberculosis in the Netherlands. J. Clin. Microbiol. 51:2427–2431. 10.1128/JCM.00489-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo T, Yang C, Gagneux S, Gicquel B, Mei J, Gao Q. 2012. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. J. Clin. Microbiol. 50:633–639. 10.1128/JCM.05539-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roetzer A, Diel R, Kohl TA, Ruckert C, Nubel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rusch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 10:e1001387. 10.1371/journal.pmed.1001387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13:137–146. 10.1016/S1473-3099(12)70277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mokrousov I, Otten T, Zozio T, Turkin E, Nazemtseva V, Sheremet A, Vishnevsky B, Narvskaya O, Rastogi N. 2009. At Baltic crossroads: a molecular snapshot of Mycobacterium tuberculosis population diversity in Kaliningrad, Russia. FEMS Immunol. Med. Microbiol. 55:13–22. 10.1111/j.1574-695X.2008.00470.x [DOI] [PubMed] [Google Scholar]
- 40.Mokrousov I, Otten T, Manicheva O, Potapova Y, Vishnevsky B, Narvskaya O, Rastogi N. 2008. Molecular characterization of ofloxacin-resistant Mycobacterium tuberculosis strains from Russia. Antimicrob. Agents Chemother. 52:2937–2939. 10.1128/AAC.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kam KM, Yip CW, Tse LW, Leung KL, Wong KL, Ko WM, Wong WS. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256:258–265. 10.1111/j.1574-6968.2006.00126.x [DOI] [PubMed] [Google Scholar]
- 42.Smittipat N, Billamas P, Palittapongarnpim M, Thong-On A, Temu MM, Thanakijcharoen P, Karnkawinpong O, Palittapongarnpim P. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 43:5034–5043. 10.1128/JCM.43.10.5034-5043.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D, Hughes SL, Hewinson RG, Neill SD. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519–528 http://mic.sgmjournals.org/content/148/2/519.long [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.