Abstract
New variants of the influenza A(H1N1)pdm09 and A(H3N2) viruses were detected in Taiwan between 2012 and 2013. Some of these variants were not detected in clinical specimens using a common real-time reverse transcription-PCR (RT-PCR) assay that targeted the conserved regions of the viral matrix (M) genes. An analysis of the M gene sequences of the new variants revealed that several newly emerging mutations were located in the regions where the primers or probes of the real-time RT-PCR assay bind; these included three mutations (G225A, T228C, and G238A) in the A(H1N1)pdm09 virus, as well as one mutation (C163T) in the A(H3N2) virus. These accumulated mismatch mutations, together with the previously identified C154T mutation of the A(H1N1)pdm09 virus and the C153T and G189T mutations of the A(H3N2) virus, result in a reduced detection sensitivity for the real-time RT-PCR assay. To overcome the loss of assay sensitivity due to mismatch mutations, we established a real-time RT-PCR assay using degenerate nucleotide bases in both the primers and probe and successfully increased the sensitivity of the assay to detect circulating variants of the human influenza A viruses. Our observations highlight the importance of the simultaneous use of different gene-targeting real-time RT-PCR assays for the clinical diagnosis of influenza.
INTRODUCTION
Influenza A viruses belonging to family Orthomyxoviridae are classified into various subtypes based on their two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). So far, 17 HA (H1 to H17) and 10 NA (N1 to N10) subtypes have been identified, and all except H17 and N10 have been found to circulate in waterfowl (1, 2). Since 1918, three subtypes (H1N1, H2N2, and H3N2) have established stable lineages and have caused sustained epidemics in the human population (3). In the 21st century, the influenza A(H1N1) virus [now termed A(H1N1)pdm09], originally found in swine, caused human infections in Mexico and the United States during March 2009 (4) and resulted in a global pandemic. To date, the virus circulated across four influenza seasons (2009 to 2013), and most countries have experienced multiple waves of infection (5–8). In the early stage of the 2009 H1N1 pandemic, between April and July 2009, the virus had begun to evolve and diversified into at least 7 clades (clades 1 to 7) with particular spatial and geographic patterns (9). In Taiwan, the imported clade 7 viruses first became predominant in August 2009. The new variants continued to emerge and replaced the old ones (5), indicating that the A(H1N1)pdm09 viruses had evolved in a manner similar to those of the human seasonal H1N1 viruses. The currently circulating human influenza A(H3N2) viruses descended from the 1968 pandemic virus. The continued mutation of the virus persistently results in the generation of novel antigenic strains that will replace the current ones and can cause annual epidemics. The substitution rate of the influenza A(H3N2) virus was estimated to be 5.7 nucleotides per year for the HA1 genes (10), with an average of 3.6 amino acid changes per year (11). The constant and rapid evolution of this virus is also reflected in the frequency of updates made by the WHO as to the recommended vaccine strains (12).
Molecular techniques for detecting influenza viruses exhibit high sensitivity and specificity. The real-time reverse transcription-PCR (RT-PCR) method, which meets the requirements for rapid and accurate influenza virus identification and subtyping, has been recommended for use in detecting the human influenza A(H1N1)pdm09 virus and the highly pathogenic avian influenza A(H5N1) virus for influenza surveillance, outbreak management, diagnosis, and treatment (13, 14). To develop an influenza A genus-specific real-time RT-PCR assay that can detect all subtypes, primers and probe are usually designed to detect the highly conserved regions of the matrix (M) genes. Two commonly used real-time RT-PCR assays recommended by the WHO (13–15) were designed to target the M gene region 144 to 251 (the start codon of the M1 gene was designated 1), which contained contiguous residues with low entropy among all influenza A subtypes. However, given the constant and rapid evolution of influenza A viruses, it is worth determining whether gene mutations located in these primer- and probe-binding sites can affect the abilities of these assays to detect influenza viruses. Such an effect would necessitate a continuous updating of the sequences of the primers and probes used in the real-time RT-PCR assays according to the gene sequences of the circulating viruses in order to maintain an optimal level of detection.
New variants of the influenza A(H1N1)pdm09 and A(H3N2) viruses were detected in Taiwan from October 2012 to August 2013. These new variants carried mutations in the M genes that were located within the highly conserved regions targeted by the commonly used real-time RT-PCR assays. These newly emerging mismatch mutations, together with the previously identified ones, comprise four nucleotide mutations (C154T, G225A, T228C, and G238A) in the A(H1N1)pdm09 viruses that were located within the forward and reverse primer-binding regions and three nucleotide mutations (C153T, C163T, and G189T) in the A(H3N2) viruses that were located within the forward primer-binding region and the probe-binding region; these mutations have been found to decrease the detection sensitivity of the PCR due to mismatches in primer or probe binding. We also established a modified real-time RT-PCR assay to improve the detection sensitivity and overcome the hurdle of detecting these mutant viruses.
MATERIALS AND METHODS
Collection of clinical specimens and virus isolates.
Laboratory influenza surveillance in Taiwan was conducted through a national influenza surveillance network coordinated by the Taiwan Centers for Disease Control (CDC). Throat swabs were collected from hospitalized patients, outpatients in the community, and cluster patients who exhibited influenza-like illnesses, and the samples were transported to the laboratories of the influenza surveillance network in Taiwan for diagnosis. The specimens from the hospitalized and cluster patients were tested using real-time RT-PCR and virus culture, and those from outpatients in the community were tested using virus culture. The real-time RT-PCR assays and virus culture using Madin-Darby canine kidney (MDCK) cells were described previously (16, 17). All of the isolated influenza viruses were sent to the Taiwan CDC, and 30 to ∼50% of the isolates received from each type or subtype were randomly selected and analyzed for their partial HA genes using conventional RT-PCR followed by Sanger sequencing for viral genetic characterization (5, 18).
Genetic analyses of the influenza A viruses.
Genetic analyses of the influenza A viruses were performed using freshly cultured isolates from the clinical specimens rather than serially passaged isolates. To analyze the phylogenetic relationships between the HA and M genes of the A(H1N1)pdm09 and A(H3N2) viruses isolated in Taiwan from 2012 to 2013, the HA and M genes were first examined using conventional RT-PCR using the primers and protocol published by the WHO (19). Viral RNA was extracted from the clinical specimens using the automated TANBead (Taiwan Advanced Nanotech, Inc., Taiwan) or the MagNA Pure LC (Roche, Penzberg City, Germany) extraction systems, according to the manufacturer's instructions. The nucleotide sequences of the amplified PCR products were then determined and translated into amino acid residues. The sequencing reactions were performed bidirectionally on the 3730 DNA analyzer (Applied Biosystems, Life Technologies, USA). The resultant sequence contigs were assembled using the Sequencher version 5.0 sequence analysis software (Gene Codes Corporation, Ann Arbor, MI, USA), and the assembled complete viral sequences were used in the phylogenetic analyses. Multiple sequence alignments, protein translation, and phylogenetic analyses were performed on the basis of the nucleotide sequences using the MEGA4 (20) and BioEdit software programs (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A phylogenetic tree was constructed using the neighbor-joining method, and 1,000 bootstrap replications were performed to evaluate its reliability.
Modified real-time RT-PCR for the broad detection of new influenza A variants.
In this study, the real-time RT-PCR assay designed for the detection of the new A(H1N1)pdm09 and A(H3N2) variants circulating in 2012 to 2013 was established by the Taiwan CDC. The sequences of the primers and probe were modified from those described by Ward et al. (15) but targeted the same regions of the genome. For the broad detection of each variant using a single assay, several degenerate nucleotide bases were included in the modified primers and probe; these degenerate sequences were based on the analyzed M gene sequences of both the A(H1N1)pdm09 and A(H3N2) viruses. The modified primer sequences of the new assay (designated the At assay) were FluA-144Ft (5′-AAGACCAATYYTGTCACCTYTGA-3′) and FLUA-214Rt (5′-GGACTGCARCGYAGACGCTTTA-3′), and the modified probe sequence was FluA-184Pt (5′-FAM-TTTGTNTTCACGCTCACCGT-BBQ-3′) (FAM, 6-carboxyfluorescein; BBQ, BlackBerry quencher). The Roche LightCycler 480 RNA master hydrolysis probe system (Roche, Penzberg City, Germany) was used. Real-time RT-PCR was performed in a 25-μl reaction mixture containing 5 μl of RNA template, 3.8 μl water, 7.4 μl enzyme mix, a final concentration of 1 μM each primer, 250 nM probe, 1.3 μl of the activator [in the final concentration of 3.25 mM Mn(OAC)2], and 1 μl of the enhancer. The reaction mixtures were incubated at 63°C for 3 min, followed by 95°C for 30 s. The reaction mixtures were then subjected to 45 cycles of 95°C for 10 s, 53°C for 30 s, and 72°C for 3 s.
Nucleotide sequence accession numbers.
The nucleotide sequences of the influenza viruses included in this study have been submitted to GenBank under the accession no. KF495607 to KF495656.
RESULTS
Characteristics of the influenza A viruses circulating in Taiwan in 2012 to 2013.
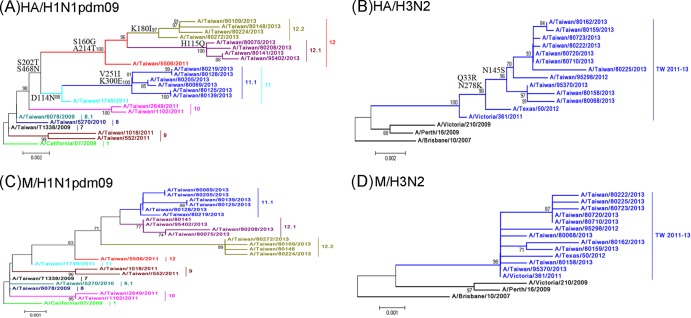
From October 2012 to August 2013 in the influenza laboratory surveillance network of Taiwan, a total of 783, 953, and 33 cases tested positive for the influenza A (H1N1)pdm09, A(H3N2), or influenza B viruses, respectively, using real-time RT-PCR and/or virus isolation. The influenza epidemic of 2012 to 2013 in Taiwan was caused by the cocirculation of influenza A(H1N1)pdm09 and A(H3N2) viruses. Among the isolated viruses, 378 A(H1N1)pdm09, 537 A(H3N2), and 14 influenza B viruses were randomly selected to determine their partial HA sequences. To further genetically characterize these influenza viruses, 14 A(H1N1)pdm09 and 11 A(H3N2) viruses were selected as representative isolates, and their full-length HA and M genes were analyzed by DNA sequencing and compared with those of previous circulating viruses. Based on the results of the HA phylogeny (Fig. 1A), the A(H1N1)pdm09 viruses that reemerged in 2012 to 2013 were classified as three novel variants, clades 11.1, 12.1, and 12.2, which branched from the previously predominant clade 11 and 12 viruses that circulated in 2010 to 2011, which had the amino acid signatures D114N-S202T-S468N and S160G-S202T-A214T-S468N, respectively. Viruses of the clade 11.1 harbored the additional V251I-K300E substitutions, and viruses of the clades 12.1 and 12.2 harbored the additional substitutions H155Q and K180I, respectively (Fig. 1A). Regarding the A(H3N2) viruses found in 2012 to 2013, most of these viruses clustered into one large TW2011-13 clade, which first appeared in Taiwan in 2011, and carried the amino acid signature Q33R-N145S-N278K, along with several additional mutations (Fig. 1B). The various clade variants of the A(H1N1)pdm09 viruses were antigenically similar to the vaccine strains of A/California/07/2009, and the A(H3N2) viruses of the TW2011-13 clade were antigenically similar to the A/Victoria/361/2011 (H3N2) viruses (data not shown).
FIG 1.
Phylogenetic relationships of the full-length HA gene (A and B) and the M gene (C and D) sequences of the influenza A(H1N1)pdm09 and A(H3N2) viruses in Taiwan. The phylogenetic trees were constructed using the neighbor-joining method, with 1,000 bootstrap replications. Branch values of >70 are indicated. The sequences of the vaccine strains, including that of A/California/7/2009 of the A(H1N1)pdm09 subtype, as well as A/Texas/50/2012, A/Victoria/361/2011, A/Victoria/210/2009, A/Perth/16/2009, and A/Brisbane/10/2007 of the A(H3N2) subtype, were downloaded from the NCBI database and are included as references. The classification of the specific evolutionary clades is defined. The major amino acid signatures of the virus clades are indicated, including D114N-S202T-S468N for clade 11, V251I-K300E for clade 11.1, S160G-S202T-A214T-S468N for clade 12, H155Q for clade 12.1, and K180I for clade 12.2 of the A(H1N1)pdm09 viruses and Q33R-N145S-N278K for clade TW2010-13 of the A(H3N2) viruses.
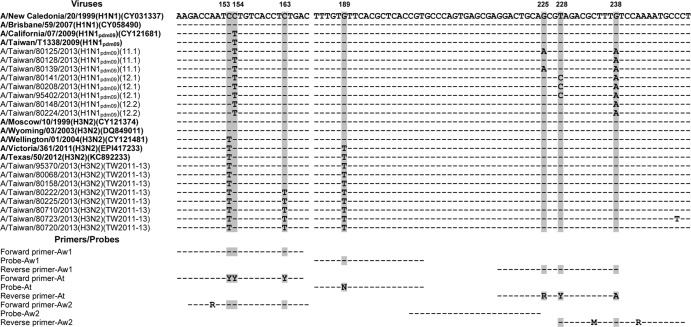
Phylogenetic analysis of the viral M genes showed that the M genes of clades 11.1, 12.1, and 12.2 of the A(H1N1)pdm09 viruses (classified by the HA genes) were located in clades that were distinct from those of the previous clade 1 to 10 viruses in Taiwan (Fig. 1C). The M gene phylogeny of the A(H3N2) viruses also showed that the M genes of the circulating A(H3N2) viruses fell into the same clades as the vaccine strain A/Victoria/361/2011 and diverged from the vaccine strains isolated prior to 2009 (Fig. 1D). These results indicate that the new variants with the mutated M genes emerged after 2011. Among the changed nucleotides, some of the mutations were located between positions 144 and 251, which were designed for the primer- and probe-binding sites of commonly used real-time RT-PCR assays. Of the 14 tested A(H1N1)pdm09 viruses, some carried the G225A (n = 2), T228C (n = 4), and G238A (n = 14) mutations, and of 11 tested A(H3N2) viruses, some carried the C153T (n = 11), C163T (n = 5), and G189T (n = 11) mutations in the M genes (Fig. 2). These mutations were generated sequentially. The M genes of the clade 11 and 12 A(H1N1)pdm09 viruses in 2010 to 2011 had the G238A mutation (Fig. 2). The G225A and T228C mutations emerged in 2012 to 2013 in Taiwan and appeared in some of the clade 11.1 viruses and all of the clade 12.1 viruses, respectively. Regarding the A(H3N2) viruses, the C163T mutations in the M genes emerged in the TW2011-13 clade strains that circulated beginning in 2011 in Taiwan. Because positions 144 to 251 were usually selected as the targets of molecular detection assays for the broad detection of the influenza A viruses, we investigated the influence of these M gene mutations on the widely used M gene real-time RT-PCRs recommended by the WHO (designated Aw1 and Aw2 in this study). The alignment between the nucleotide sequences of the viral M genes and the primers and probes used in the Aw1 and Aw2 assays are shown in Fig. 2. The C154T, G225A, T228C, and G238A mutations of the A(H1N1)pdm09 viruses can cause mismatches with the forward and reverse primers of both the Aw1 and Aw2 assays (Fig. 2); the C153T, C163T, and G189T mutations of the A(H3N2) viruses can result in mismatches with the forward primers of the Aw1 and Aw2 assays, as well as the probe of the Aw1 assay (Fig. 2).
FIG 2.
Nucleotide sequence alignments between the M genes of the A(H1N1)pdm09 and A(H3N2) viruses and the primer and probe sequences of the three real-time RT-PCR assays. The M gene sequence of the A/New Caledonia/20/1999 strain was used as a reference, and the identical nucleotide bases of the other viruses and the primer/probe sequences are indicated by dashed lines. Different nucleotide bases, compared to the reference, are directly indicated. Positions that contain modified nucleotide bases in the primer-probe sequences of the At assay established in this study, including positions 153, 154, 163, 189, 225, 228, and 238, are indicated by gray shading. Reference viruses of either A(H1N1)pdm09 or A(H3N2) subtypes included in the alignments are shown in bold along with their sequence accession numbers. The evolutionary clades (based on the HA sequences) of the viruses are also shown in parentheses in the nomenclature.
Modification of the real-time RT-PCR assay for the broad detection of newly evolved influenza A viruses.
The mismatches found in the M genes of the new A(H1N1)pdm09 and A(H3N2) variants potentially affect the detection ability of the Aw1 and Aw2 real-time RT-PCR assays. To test this hypothesis and to overcome the impact of these mismatch mutations, we established a real-time RT-PCR assay (the At assay) using degenerate nucleotide bases; the At assay described here was modified from the Aw1 assay. We compared the performances of the three Aw1, Aw2, and At real-time RT-PCR assays. The modified primer and probe sequences were designed to completely match the currently circulating A(H1N1)pdm09 viruses of clades 11, 11.1, 12, 12.1, and 12.2, as well as the A(H3N2) viruses of the TW2011-13 clade. To examine whether the modified At assay could improve the detection sensitivity, a subset of clinical specimens that were positive for influenza viruses from each evolutionary clade were randomly selected and simultaneously analyzed using the Aw1 and At assays. Some problematic specimens that had positive results in the real-time RT-PCR subtyping assays (targeting the HA gene) but were undetectable by the Aw1 assay were also selected and reanalyzed using the At assay. In the detection of A(H1N1)pdm09 specimens, 6 of 40 samples tested negative in the Aw1 assay and positive in the At assay, and overall, the threshold cycle (CT) values of all tested samples shortened 3.1 to 14.25 cycles (Table 1). For the detection of A(H3N2) specimens, 10 of 40 samples were missed using the Aw1 assay, and overall, the CT values of all tested samples shortened 2.24 to 12.74 cycles (Table 2). The results indicate that the At assay is more sensitive for detecting the newly emerging A(H1N1)pdm09 and A(H3N2) viruses than the Aw1 assay. To further determine the detection sensitivity of the assay, 10-fold serial dilutions were made of DNA templates from the A(H1N1)pdm09 viruses belonging to various HA clades, as well as A(H3N2) viruses of the TW2011-13 clade, and these dilutions were analyzed using the At assay. The results were then compared with those obtained from the Aw1 and Aw2 assays. The calculated detection limits of the At, Aw1, and Aw2 assays are shown in Table 3. Based on these results, the detection sensitivity of the At assay was improved 10- to 20- and 200-fold compared to that of Aw1 assay for detecting the new variants of the A(H1N1)pdm09 and A(H3N2) viruses, respectively (Table 3); this indicates that the At real-time RT-PCR assay, which uses degenerate nucleotide bases in its primers and probe, can overcome the impact of mismatch mutations and can be used for the broad detection of human influenza A viruses that emerged in 2012 to 2013.
TABLE 1.
CT values of the A(H1N1)pdm09-positive clinical specimens given by two real-time RT-PCR assays
| Specimen no. |
CT values from the indicated assay |
ΔCTc | HA clade A(H1N1)pdm09 | Collection date (in 2013) | |
|---|---|---|---|---|---|
| Aw1a | Atb | ||||
| 95,316 | 34.92 | 31.00 | 3.92 | 12.2 | 28 Decemberd |
| 95,380 | UDe | 32.62 | NDf | 7 March | |
| 95,402 | 32.85 | 25.74 | 7.11 | 12.1 | 14 March |
| 95,409 | 34.53 | 30.12 | 4.41 | ND | 15 March |
| 95,411 | UD | 34.95 | ND | 15 March | |
| 95,460 | 34.55 | 27.17 | 7.38 | 12.1 | 3 April |
| 95,507 | 34.53 | 25.36 | 9.17 | 12.1 | 25 April |
| 80,070 | 33.98 | 30.08 | 3.9 | 12.2 | 24 January |
| 80,075 | 28.56 | 21.29 | 7.27 | 12.1 | 25 January |
| 80,079 | 33.46 | 27.27 | 6.19 | 12.2 | 26 January |
| 80,109 | 32.20 | 28.98 | 3.22 | 12.2 | 4 February |
| 80,125 | 35.73 | 29.64 | 6.09 | 11.1 | 8 February |
| 80,128 | 32.70 | 28.21 | 4.49 | 11.1 | 8 February |
| 80,148 | 35.00 | 29.47 | 5.53 | 12.2 | 15 February |
| 80,205 | 32.30 | 26.18 | 6.12 | 11.1 | 27 February |
| 80,208 | 29.04 | 22.48 | 6.56 | 12.1 | 28 February |
| 80,210 | 32.64 | 23.8 | 8.84 | 12 | 28 February |
| 80,219 | 27.14 | 21.35 | 5.79 | 11.1 | 1 March |
| 80,224 | 32.82 | 26.56 | 6.26 | 12.2 | 4 March |
| 80,243 | 33.45 | 30.34 | 3.11 | 12.2 | 7 March |
| 80,261 | 30.77 | 23.46 | 7.31 | 11.1 | 11 March |
| 80,272 | 22.94 | 17.11 | 5.83 | 12.2 | 13 March |
| 80,310 | 37.52 | 30.63 | 6.89 | 12.1 | 20 March |
| 80,426 | 31.08 | 23.88 | 7.20 | 12.1 | 6 April |
| 80,462 | 34.57 | 27.03 | 7.54 | 12.1 | 10 April |
| 80,470 | 32.41 | 25.95 | 6.46 | 12.2 | 11 April |
| 80,515 | 34.98 | 26.97 | 8.01 | 12.1 | 18 April |
| 80,981 | 38.24 | 29.97 | 8.27 | 11.1 | 21 June |
| 80,993 | 33.82 | 28.23 | 5.59 | 11.1 | 26 June |
| 81,041 | 31.46 | 23.39 | 8.07 | 11.1 | 8 July |
| 81,049 | 34.80 | 29.52 | 5.28 | 11.1 | 10 July |
| 81,085 | 37.92 | 26.26 | 11.66 | 11.1 | 23 July |
| 81,087 | 33.85 | 29.17 | 4.68 | 11.1 | 24 July |
| 81,109 | 35.73 | 27.80 | 7.93 | 11.1 | 2 August |
| 81,110 | UD | 32.94 | ND | 3 August | |
| 81,135 | UD | 27.76 | 11.1 | 17 August | |
| 81,157 | UD | 33.54 | ND | 23 August | |
| 77,757 | 34.58 | 27.68 | 6.90 | 12.1 | 17 June |
| 78,049 | UD | 34.16 | ND | 1 August | |
| 78,123 | 37.81 | 23.56 | 14.25 | 11 | 18 August |
Assay recommended by the WHO for the detection of all influenza A viruses.
Modified assay established in this study.
Values = (CT of Aw1) − (CT of At).
In 2012.
UD, M gene of the influenza A virus was undetectable.
ND, unavailable.
TABLE 2.
CT values of the A(H3N2)-positive clinical specimens given by two real-time RT-PCR assays
| Specimen no. |
CT values from the indicated assay |
ΔCTc | HA clade [A(H3N2)] | Collection date (in 2013) | |
|---|---|---|---|---|---|
| Aw1a | Atb | ||||
| 95,298 | 29.34 | 23.74 | 5.60 | TW2011-13 | 9 Octoberd |
| 95,370 | 29.19 | 23.92 | 5.27 | TW2011-13 | 2 March |
| 95,537 | 35.68 | 25.08 | 10.60 | TW2011-13 | 8 May |
| 95,638 | 38.98 | 26.24 | 12.74 | TW2011-13 | 6 July |
| 95,666 | 35.33 | 24.33 | 11.00 | TW2011-13 | 16 August |
| 95,679 | UDe | 30.89 | TW2011-13 | 23 August | |
| 80,068 | 30.72 | 25.81 | 4.91 | TW2011-13 | 24 January |
| 80,158 | 28.29 | 23.05 | 5.24 | TW2011-13 | 18 February |
| 80,159 | 29.09 | 23.72 | 5.37 | TW2011-13 | 18 February |
| 80,162 | 25.01 | 20.22 | 4.79 | TW2011-13 | 18 February |
| 80,222 | 33.45 | 22.64 | 10.81 | TW2011-13 | 2 March |
| 80,225 | 32.09 | 24.05 | 8.04 | TW2011-13 | 4 March |
| 80,710 | 31.35 | 21.16 | 10.19 | TW2011-13 | 10 May |
| 80,720 | 32.17 | 20.79 | 11.38 | TW2011-13 | 13 May |
| 80,723 | 35.80 | 26.79 | 9.01 | TW2011-13 | 14 May |
| 80,231 | UD | 33.21 | NDf | 5 March | |
| 80,270 | UD | 34.54 | ND | 13 March | |
| 80,636 | 35.35 | 33.11 | 2.24 | ND | 1 May |
| 80,665 | UD | 36.15 | ND | 5 May | |
| 80,683 | 36.54 | 29.56 | 6.98 | TW2011-13 | 7 May |
| 80,864 | 31.77 | 23.71 | 8.06 | TW2011-13 | 3 June |
| 80,889 | 37.31 | 28.09 | 9.22 | TW2011-13 | July 6 |
| 80,911 | 37.90 | 29.41 | 8.49 | TW2011-13 | 10 June |
| 80,950 | 35.04 | 27.02 | 8.02 | TW2011-13 | 15 June |
| 80,978 | 24.52 | 20.57 | 3.95 | TW2011-13 | 22 June |
| 81,007 | 36.95 | 29.09 | 7.86 | TW2011-13 | 29 June |
| 81,013 | UD | 27.52 | TW2011-13 | 28 June | |
| 81,018 | 21.88 | 19.12 | 2.76 | TW2011-13 | 1 July |
| 81,024 | UD | 32.54 | ND | 3 July | |
| 81,083 | 37.23 | 27.69 | 9.54 | TW2011-13 | 22 July |
| 81,111 | UD | 31.32 | ND | 4 August | |
| 81,130 | 34.31 | 22.58 | 11.73 | TW2011-13 | 13 August |
| 81,139 | 32.68 | 21.86 | 10.82 | TW2011-13 | 16 August |
| 81,156 | 39.78 | 29.33 | 10.45 | TW2011-13 | 23 August |
| 77,163 | 38.67 | 27.00 | 11.67 | TW2011-13 | 1 May |
| 77,917 | 35.08 | 24.27 | 10.81 | TW2011-13 | 16 July |
| 78,001 | 37.90 | 29.29 | 8.61 | TW2011-13 | 29 July |
| 78,056 | UD | 36.00 | ND | 3 August | |
| 78,093 | UD | 34.61 | ND | 13 August | |
| 78,128 | UD | 28.93 | TW2011-13 | 19 August | |
Assay recommended by the WHO for the detection of all influenza A viruses.
Modified assay established in this study.
Values = (CT of Aw1) − (CT of At).
In 2012.
UD, M gene of the influenza A virus was undetectable.
ND, unavailable.
TABLE 3.
Detection limits of the three real-time RT-PCR assays on the different clades of the influenza A(H1N1)pdm09 and A(H3N2) viruses
| Virus strain | Detection limit (M gene copies per reaction) of the indicated assay |
M gene variations at nucleotide positiona: |
HA clade | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aw1 | Aw2b | At | 153 | 154 | 163 | 189 | 225 | 228 | 238 | ||
| A/TW/80125/2013 | 300 | 300 | 30 | —c | T | — | — | A | — | A | 11.1 |
| A/TW/80128/2013 | 750 | 75 | 37.5 | — | T | — | — | — | — | A | 11.1 |
| A/TW/80208/2013 | 315 | 315 | 15.8 | — | T | — | — | — | C | A | 12.1 |
| A/TW/80720/2013 | 3,500 | 350 | 17.5 | T | — | T | T | — | — | — | TW2011-13 |
The variations at the indicated positions were compared to the sequences of the primers and the probe used in the Aw1 assay.
Assay recommended by the WHO for the detection of A(H1N1)pdm09 viruses.
—, nucleotides that were identical to the sequences of the primers and the probe used in the Aw1 assay.
DISCUSSION
Influenza A viruses exhibit rapid evolution that may be attributed to the lack of proofreading abilities of the viral RNA polymerases, strong selection pressure to evade the immune system, and frequent genome segment rearrangement. In the experimental virus-infected MDCK cells that did not have immune pressure, the mutation rates of the nonstructural (NS) genes in influenza A(H1N1) viruses were determined to be 2.0 × 10−6 mutations per site per infectious cycle and 2.6 × 10−3 mutations per site per year (21). For circulating influenza viruses in humans, which were under selection pressure, the fixation rates were estimated to be 3.7 × 10−3 and 5.72 × 10−3 nucleotide substitutions per site per year for the HA genes of influenza A(H3N2) viruses; these rates were based on the estimated accumulation of adaptive substitutions in human influenza A(H3N2) viruses during the periods 1983 to 1997 and 1992 to 2005, respectively (22, 23). For the influenza A(H1N1)pdm09 virus, the overall substitution rate was estimated to be 3.2 × 10−3 mutations per site per year during 2009 to 2010. Each of the influenza virus genome segments may evolve differently because of different selection pressures. Comparing the rate differences of the various segments, the substitution rates of the surface proteins, HA and NA, were thought to be higher than those of the internal proteins (24). However, the M and NS segments of the influenza A(H3N2) viruses during 1992 to 2005 evolved at a rate of 5.2 × 10−3 nucleotide substitutions per site per year, which was comparable to the rate of 5.72 × 10−3 nucleotide substitutions per site per year for the HA gene (23). Because of the high mutation rates of the HA, NA, and M segments, which are used for the identification and subtyping of influenza A viruses, constant real-time gene sequencing is required to maintain a high level of sensitivity in the detection assays.
A quantitative real-time RT-PCR assay developed by Ward et al. (15) (Aw1 in this study) targeted the M gene of influenza A virus and has been used in >100 studies; additionally, this assay has been recommended by the WHO to be used for the detection of all influenza A viruses (13). This assay is highly sensitive, rapid, and accurate. At the time of that study's publication, the detection limit was estimated to be 2,138 viral cDNA copies/ml of virus transport medium and 10 viral RNA copies/PCR (15). Due to these advantages, we selected the Aw1 assay for routine influenza A surveillance and integrated it with several HA gene-targeting real-time RT-PCR assays to detect and determine the subtypes of the A(H1N1)pdm09 and A(H3N2) viruses in the same run (17). In general, the CT value given by the Aw1 assay was comparable to that of the HA assays for the same sample. Notably, during the 2012-2013 influenza season, we found that the CT values of most clinical specimens determined by the Aw1 assay were higher than those of the HA assays for both the A(H1N1)pdm09 and A(H3N2) viruses. These delayed M gene CT values sometimes caused problems, as clinical specimens may have undetectable CT values of influenza A virus while having a positive CT value for either the A(H1N1)pdm09 or A(H3N2) subtype. To investigate these irrational data, we performed a sequence analysis of the viral M genes and found that some nucleotide mutations of the circulating A(H1N1)pdm09 and A(H3N2) viruses occurred in the regions targeted by the Aw1 assay. These observed sequence mismatches between the viral templates and the primers and probe may affect the detection ability of the Aw1 assay and may explain the delay in the CT values of the Aw1 assay. The M gene mutations of A(H1N1)pdm09 viruses were also reported to be problematic with some commercial assays, causing a diagnostic conundrum for clinical microbiology labs (25–27). This experience shows that the simultaneous use of different gene-targeting assays is appropriate for correctly identifying influenza viruses, and it also provides some evidence for the existence of newly evolved variants.
When tracing the M gene sequences of previous isolates, it was shown that the primer and probe sequences of the Aw1 assay matched well with those of the old human seasonal influenza A(H1N1) and A(H3N2) viruses circulating before 2003 (Fig. 2). However, variations at positions 153, 154, 163, 189, 225, 228, and 238 gradually accumulated. Three nucleotide positions (153, 163, and 189) were altered in the A(H3N2) variants, whereas four positions (154, 225, 228, and 238) were mutated in the A(H1N1)pdm09 viruses (Fig. 2). Based on a total of 4,543 M gene sequences (available from the National Center for Biotechnology Information [NCBI]) from influenza A(H3N2) viruses that globally circulated between 1968 and 2012, the C153T mutation first appeared and began to spread in 2001 and reached 100% penetrance in 2005 (28). This was also the first fixed mutation and resulted in a mismatch between the A(H3N2) virus and the forward primer of the Aw1 assay. Next, the G189T mutation appeared in 2010. This mutation also became fixed and caused a mismatch with the Aw1 probe sequence. The C163T mutation emerged in 2012 to 2013 and resulted in another mismatch with the Aw1 forward primer (Fig. 2). For the influenza A(H1N1)pdm09 viruses, the C154T mismatch appeared in 2009 when the virus was first found to infect humans. Based on a total of 5,353 M gene sequences collected globally during 2009 to 2012 (available at NCBI), the G238A mutation, which results in a mismatch with the Aw1 reverse primer sequence, first appeared in 2010 and reached 100% prevalence by 2012. In the 2012-2013 influenza season, we observed the newly emerged G225A and T228C mutations in different clade viruses; the former was observed in some of the HA clade 11.1 A(H1N1)pdm09 viruses, and the latter was found in all of the HA clade 12.1 viruses. Both mutations caused mismatches to the reverse primer sequences of the Aw1 assay. In general, the M genes of influenza viruses are more conserved than the HA and NA genes. The regions designed for the primer or probe binding of real-time RT-PCR assays were rarely found to be mutated. For example, for human seasonal H1N1 viruses, the M gene sequences of the viruses isolated from 1999 to 2009, as well as those of the human A(H3N2) viruses circulating before 2003, were well matched to the binding regions of the primers and probe in the Aw1 real-time RT-PCR assay. It is unexpected that the mutations in the M gene sequences of the A(H1N1)pdm09 and A(H3N2) viruses accumulated rapidly from 2009 to 2013. Taken together, four mismatch mutations in the A(H1N1)pdm09 virus and three mutations in A(H3N2) viruses have occurred and may hamper the detection abilities of the Aw1 assays.
In this study, we modified the primer and probe sequences of the Aw1 real-time RT-PCR assay in response to the newly emerging human influenza A viruses detected in 2012 to 2013. Using degenerate sequences at the positions of the mutations, including 153Y (C/T), 154Y, and 163Y in the forward primer, 189N (A/T/C/G) in the probe, and 225 R (A/G) and 228Y in the reverse primer (Fig. 2), all of the modified primers and probe can recognize the circulating A(H1N1)pdm09 and A(H3N2) viruses. The modified At assay was able to overcome the impact of the mismatch mutations and improved the detection sensitivity 10- to 20- and 200-fold in the detection of new A(H1N1)pdm09 and A(H3N2) variants, respectively, in 2012 to 2013 (Table 3). Although we did not test the effects of these mutations on the other Aw2 real-time RT-PCR assay, the C153T, C154T, C163T, T228C, and G238A mutations located at the forward and reverse primer sites may also influence the performance of the Aw2 real-time RT-PCR assay. In conclusion, given the increasing genetic diversity of influenza viruses, a highly sensitive degenerate real-time RT-PCR system can be used for the fast and accurate identification of any novel influenza A variants and can also minimize the need to frequently update the assay parameters. The simultaneous use of real-time RT-PCR assays targeting different genes is also appropriate to ensure accurate identification of the influenza virus.
ACKNOWLEDGMENTS
We thank the members of Taiwan CDC contracted virology laboratories in the Influenza Surveillance Network of Taiwan.
This study was supported by grants from Centers for Disease Control, Ministry of Health and Welfare, Taiwan (grant no. DOH102-DC-2601 and DOH102-DC-2213).
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822. 10.1128/JVI.79.5.2814-2822.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox NJ, Subbarao K. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51:407–421. 10.1146/annurev.med.51.1.407 [DOI] [PubMed] [Google Scholar]
- 4.CDC 2009. Outbreak of swine-origin influenza A (H1N1) virus infection–Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467–470 [PubMed] [Google Scholar]
- 5.Yang JR, Huang YP, Chang FY, Hsu LC, Lin YC, Su CH, Chen PJ, Wu HS, Liu MT. 2011. New variants and age shift to high fatality groups contribute to severe successive waves in the 2009 influenza pandemic in Taiwan. PLoS One 6:e28288. 10.1371/journal.pone.0028288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandaranayake D, Jacobs M, Baker M, Hunt D, Wood T, Bissielo A, Macfarlane M, Lopez L, Mackereth G, Huang Q. 2011. The second wave of 2009 pandemic influenza A(H1N1) in New Zealand, January-October 2010. Euro Surveill. 16:pii: 19788 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19788 [PubMed] [Google Scholar]
- 7.CDC 2010. Update: influenza activity—United States, August 30, 2009-March 27, 2010, and composition of the 2010-11 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 59:423–430 [PubMed] [Google Scholar]
- 8.Mytton OT, Rutter PD, Mak M, Stanton EA, Sachedina N, Donaldson LJ. 2012. Mortality due to pandemic (H1N1) 2009 influenza in England: a comparison of the first and second waves. Epidemiol. Infect. 140:1533–1541. 10.1017/S0950268811001968 [DOI] [PubMed] [Google Scholar]
- 9.Nelson M, Spiro D, Wentworth D, Beck E, Fan J, Ghedin E, Halpin R, Bera J, Hine E, Proudfoot K, Stockwell T, Lin X, Griesemer S, Kumar S, Bose M, Viboud C, Holmes E, Henrickson K. 2009. The early diversification of influenza A/H1N1pdm. PLoS Curr. 1:RRN1126. 10.1371/currents.RRN1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bush RM, Fitch WM, Bender CA, Cox NJ. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457–1465. 10.1093/oxfordjournals.molbev.a026057 [DOI] [PubMed] [Google Scholar]
- 11.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. 10.1126/science.1097211 [DOI] [PubMed] [Google Scholar]
- 12.Hay AJ, Gregory V, Douglas AR, Lin YP. 2001. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1861–1870. 10.1098/rstb.2001.0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO 2007. Recommendations and laboratory procedures for detection of avian influenza A(H5N1) virus in specimens from suspected human cases. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/resources/documents/RecAIlabtestsAug07.pdf [Google Scholar]
- 14.WHO 2009. WHO information for laboratory diagnosis of pandemic (H1N1) 2009 virus in humans–revised. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/WHO_Diagnostic_RecommendationsH1N1_20090521.pdf [Google Scholar]
- 15.Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, Snowden BW, Tisdale M. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29:179–188. 10.1016/S1386-6532(03)00122-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih SR, Chen GW, Yang CC, Yang WZ, Liu DP, Lin JH, Chiu SC, Chen HY, Tsao KC, Huang CG, Huang YL, Mok CK, Chen CJ, Lin TY, Wang JR, Kao CL, Lin KH, Chen LK, Eng HL, Liu YC, Chen PY, Lin JS, Wang JH, Lin CW, Chan YJ, Lu JJ, Hsiung CA, Chen PJ, Su IJ. 2005. Laboratory-based surveillance and molecular epidemiology of influenza virus in Taiwan. J. Clin. Microbiol. 43:1651–1661. 10.1128/JCM.43.4.1651-1661.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang JR, Lo J, Liu JL, Lin CH, Ho YL, Chen CJ, Wu HS, Liu MT. 2009. Rapid SYBR green I and modified probe real-time reverse transcription-PCR assays identify influenza H1N1 viruses and distinguish between pandemic and seasonal strains. J. Clin. Microbiol. 47:3714–3716. 10.1128/JCM.01646-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jian JW, Chen GW, Lai CT, Hsu LC, Chen PJ, Kuo SH, Wu HS, Shih SR. 2008. Genetic and epidemiological analysis of influenza virus epidemics in Taiwan during 2003 to 2006. J. Clin. Microbiol. 46:1426–1434. 10.1128/JCM.01560-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO 2009. Sequencing primers and protocol. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 21.Nobusawa E, Sato K. 2006. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 80:3675–3678. 10.1128/JVI.80.7.3675-3678.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson NM, Galvani AP, Bush RM. 2003. Ecological and immunological determinants of influenza evolution. Nature 422:428–433. 10.1038/nature01509 [DOI] [PubMed] [Google Scholar]
- 23.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. 10.1038/nature06945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Todd KM, Yen-Lieberman B, Kaul K, Mangold K, Shulman ST. 2010. Unique finding of a 2009 H1N1 influenza virus-positive clinical sample suggests matrix gene sequence variation. J. Clin. Microbiol. 48:665–666. 10.1128/JCM.02318-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhiman N, Espy MJ, Irish CL, Wright PA, Smith TF, Pritt BS. 2010. Evidence for amino acid changes in a 57-base-pair region of the highly conserved matrix gene of pandemic (H1N1) 2009 influenza A virus. J. Clin. Microbiol. 48:3817–3819. 10.1128/JCM.01237-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binnicker MJ, Baddour LM, Grys TE, Espy MJ, Hata DJ, Wotton JT, Patel R. 2013. Identification of an influenza A H1N1/2009 virus with mutations in the matrix gene causing a negative result by a commercial molecular assay. J. Clin. Microbiol. 51:2006–2007. 10.1128/JCM.00446-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601. 10.1128/JVI.02005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]