Abstract
Since the establishment of sequence-based typing as the gold standard for DNA-based typing of Legionella pneumophila, the Legionella laboratory at the Centers for Disease Control and Prevention (CDC) has conducted routine sequence-based typing (SBT) analysis of all incoming L. pneumophila serogroup 1 (Lp1) isolates to identify potential links between cases and to better understand genetic diversity and clonal expansion among L. pneumophila bacteria. Retrospective genotyping of Lp1 isolates from sporadic cases and Legionnaires' disease (LD) outbreaks deposited into the CDC reference collection since 1982 has been completed. For this study, we compared the distribution of sequence types (STs) among Lp1 isolates implicated in 26 outbreaks in the United States, 571 clinical isolates from sporadic cases of LD in the United States, and 149 environmental isolates with no known association with LD. The Lp1 isolates under study had been deposited into our collection between 1982 and 2012. We identified 17 outbreak-associated STs, 153 sporadic STs, and 49 environmental STs. We observed that Lp1 STs from outbreaks and sporadic cases are more similar to each other than either group is to environmental STs. The most frequent ST for both sporadic and environmental isolates was ST1, accounting for 25% and 49% of the total number of isolates, respectively. The STs shared by both outbreak-associated and sporadic Lp1 included ST1, ST35, ST36, ST37, and ST222. The STs most commonly found in sporadic and outbreak-associated Lp1 populations may have an increased ability to cause disease and thus may require special attention when detected.
INTRODUCTION
Legionnaires' disease (LD) is a major cause of respiratory disease in the United States, with the reported legionellosis incidence rates having increased almost 3-fold during 2000 to 2009 (1). In addition, LD is the most common waterborne disease in the United States (2). Although 59 species of legionellae have been identified so far (http://old.dsmz.de/microorganisms/bacterial_nomenclature_info.php?genus=Legionella), they possess different capacities to cause legionellosis. More than 90% of the isolates associated with LD are Legionella pneumophila, and up to 84% of these are L. pneumophila serogroup 1 (Lp1) (3, 4). Moreover, the majority of Legionella isolates associated with disease are Lp1 that react positively with monoclonal antibody 2 (MAb2) from the “Joly” MAb panel (5) or MAb3/1 from the “Dresden” MAb panel (6–11).
Sequence-based typing (SBT) is a rapid, highly discriminatory, and reproducible seven-gene molecular typing method that has become an internationally recognized procedure for genotyping L. pneumophila isolates (12–15). Results of recent studies on the distribution of sequence types (STs) of L. pneumophila isolates from culture collections in Europe, Asia, and North America suggested that a select group of STs appeared to be predominant among L. pneumophila isolates causing clinical sporadic cases and outbreaks. Therefore, similar to select Legionella species and serogroups, some L. pneumophila STs appear to have an enhanced ability to cause disease in humans. In 2009, our group reported a comparison of STs among 100 clinical Lp1 isolates and 50 environmental Lp1 isolates that were deposited into the Centers for Disease Control and Prevention (CDC) culture collection between 2001 and 2006 (7). The clinical and environmental isolates analyzed had different sets of STs and had only three STs in common, ST1, ST36, and ST154. The identification of STs specific to clinical cases indicated that, similar to serology-based hierarchy, virulence ranking for legionellae also exists at the genetic level, and there are certain sequence types that appear to have a higher capacity to cause sporadic cases of legionellosis or outbreaks.
To investigate this phenomenon further, we conducted SBT analysis on a larger set of Lp1 isolates from the United States deposited into the CDC Legionella reference collection during the last 30 years. This set was comprised of three distinct source types: (i) isolates thought to have caused outbreaks, based on CDC investigations, in which matching isolates from patients and epidemiologically implicated environmental reservoirs were identified; (ii) clinical isolates from sporadic cases sent to CDC for reference testing from state and local health departments, and (iii) environmental isolates with no known association with cases of legionellosis. Isolates of the last source type were obtained from Legionella eradication and surveillance studies conducted by CDC or sent to us by two commercial companies conducting routine sampling for their clients. We compared STs from these different groups to investigate geographical and temporal distribution of STs in the United States and compare our findings to those from other countries.
(Some of this work was presented previously as posters at the 110th General Meeting of the American Society for Microbiology, San Diego, CA, and the 111th General Meeting of the American Society for Microbiology, New Orleans, LA.)
MATERIALS AND METHODS
Outbreak-associated isolates.
Community and travel-associated LD outbreaks are defined as two or more cases of legionellosis that occurred within a year of each other and associated with the same location. A health care-associated outbreak is defined as either a single case of laboratory-confirmed, definite-case-care-associated LD or two or more cases of laboratory-confirmed, possible-health care-associated LD occurring within 6 months of each other (16). Both clinical and environmental isolates were selected from 26 outbreaks that occurred in North America and the Caribbean and were investigated by the CDC between 1982 and 2012 (Table 1). This represents 21% (26/125) of the total number of outbreaks for which the CDC laboratory was assisting during this time period. These 26 outbreaks were selected based on the availability of matching clinical isolates from patients and environmental isolates from epidemiologically implicated reservoirs. Matches for isolates collected from outbreaks of 2006 to 2012 were made based on SBT data, whereas matches for 1982-2005 outbreaks were initially established using a panel of seven monoclonal antibodies and later confirmed using SBT.
TABLE 1.
Outbreaks with matching clinical and environmental isolates investigated by the CDC between 1982 and 2012
| Yr | State or districta | No. of cases | Community, travel, or health care associated | MAb patternb | MAb pattern name | SBT profilec | Associated ST |
|---|---|---|---|---|---|---|---|
| 1982 | St. Croix, Virgin Islands | 12 | Travel | 1,2,3 | Knoxville | 3-4-1-1-1-9-1 | ST35 |
| 1993 | Rhode Island | 17 | Community | 1,2,5,6 | Philadelphia | 3-4-1-1-14-9-11 | ST37 |
| 1994 | Connecticut | 22 | Health care | 1,2,5,6 | Philadelphia | 11-14-16-1-15-13-1 | ST150 |
| 1994 | Delaware | 21 | Community | 1,2,5,6 | Philadelphia | 3-4-1-1-14-9-1 | ST36 |
| 1995 | Pennsylvania | 22 | Health care | 1,2,5,6 | Philadelphia | 3-4-1-1-14-9-3 | ST771 |
| 1996 | Virginia | 23 | Community | 1,2,5,6 | Philadelphia | 8-24-3-15-21-12-20 | ST775 |
| 1996 | Missouri | 3 | Community | 1,2,3 | Knoxville | 6-10-15-10-9-14-9 | ST555 |
| 1997 | California | 8 | Community | 1,2,5,6 | Philadelphia | 3-4-1-1-14-9-11 | ST37 |
| 2001 | Nevada* | 21 | Travel | 1,2,5,6 | Philadelphia | 3-4-1-1-1-9-1 | ST35 |
| 2002 | Vermont | 13 | Community | 1,2,3 | Knoxville | 2-19-5-10-18-1-10 | ST222 |
| 2002 | Pennsylvania | 12 | Community | 1,2,5,6 | Philadelphia | 21-27-28-2-15-29-6 | ST259 |
| 2003 | St. Croix, Virgin Islands | 3 | Travel | 1,2,3 | Knoxville | 11-14-16-16-15-13-2 | ST154 |
| 2005 | North Carolina | 4 | Travel | 1,2,5d | N/Ag | 3-4-1-1-14-9-1 | ST36 |
| 2005 | South Dakota | 18 | Community | 1,2,5,7 | Benidorm | 4-7-11-3-11-12-9 | ST42 |
| 2005 | New Mexico | 2 | Travel | 1,2,5,7d | Benidorm | 2-10-3-5-19-4-9 | ST275 |
| 2006 | Texas | 6 | Health care | 1,2,5,ve | N/A | 3-4-1-1-14-9-1 | ST36 |
| 2009 | Georgia | 5 | Health care | 1,2,5d | N/A | 3-4-1-1-14-9-1 | ST36 |
| 2009 | Maryland | 10 | Community | 1,2,3 | Knoxville | 2-19-5-10-18-1-10 | ST222 |
| 2010 | Mississippi* | 8 | Travel | 1,2,5d | N/A | 3-4-1-1-1-9-1 | ST35 |
| 2010 | Michigan | 29 | Community | 1,2,3 | Knoxville | 2-19-5-10-18-1-10 | ST222 |
| 2010 | Utah | 2 | Travel | 1,2,5d | N/A | 5-1-22-15-6-10-6 | ST109 |
| 2011 | Washington* | 3 | Health care | 1f | N/A | 7-6-17-3-13-11-11 | ST59 |
| 2012 | Nevada | 3 | Travel | 1f | N/A | 1-4-3-1-1-1-1 | ST1 |
| 2012 | Illinois | 11 | Travel | 1,2f | N/A | 3-4-1-1-14-9-1 | ST36 |
| 2012 | Pennsylvania | 21 | Health care | 1,2f | N/A | 14-18-8-10-28-19-2 | ST1395 |
| 2012 | Wisconsin | 3 | Community | 1,2f | N/A | 4-8-11-16-42-12-2 | ST224 |
Outbreaks that appeared to be recurrent are indicated by an asterisk.
The MAb reactivity pattern is shown (e.g., 1,2,3 indicates that the isolate was reactive with MAb1, MAb2, and MAb3).
The SBT profile is shown in the order flaA, pilE, asd, mip, mompS, proA, and neuA (e.g., 1-4-3-1-1-1-1).
MAb6 reactivity was not tested.
MAb6 reactivity is variable (v).
Only reactivity with MAb1, MAb75, and MAb2 was tested.
N/A, not available. The MAb pattern name cannot be established due to the incomplete information of MAb reactivities.
Sporadic isolates.
States are not obligated to send Legionella isolates to the CDC lab; when they choose to do so, it is typically to verify identification done by the local lab or for further identification to the species level and/or typing. Fifteen representative sporadic LD isolates from different states, when possible, were selected from the CDC Legionella reference collection for each year between 1982 and 2005. For 2006 to 2012, all Lp1 sporadic isolates sent to the CDC reference collection underwent SBT analysis and were included in this study (see Table S1 in the supplemental material).
Environmental isolates.
Fifty environmental isolates were randomly selected from PathCon Laboratories' (Norcross, GA) culture collection of isolates obtained from water samples collected from buildings that underwent routine surveillance sampling (7) (see Table S2 in the supplemental material). Five environmental isolates were selected from monochloramine remediation studies conducted in California in 2002 to 2003 and 2008 from hospital buildings (17). Two environmental isolates were selected from a cooling tower surveillance study conducted in collaboration with Pinellas County Utilities Laboratory in Florida (C. E. Lucas, unpublished data). Five environmental isolates were collected from water cisterns in households located on the Navajo Nation territory (Lucas, unpublished). The Navajo Nation is a sovereign American Indian Nation located in the southwestern region of the United States within the borders of Arizona, New Mexico, and Utah. In addition, 87 environmental isolates were sent to the CDC between 2010 and 2012 from EMSL Analytical Laboratories' (Cinnaminson, NJ) culture collection of isolates obtained during routine sampling for its clients. There was no known association with cases of LD for the environmental isolates.
Legionella MAb test.
Between 1989 and 2006, all Lp1 isolates associated with LD outbreaks were typed by a full “Joly” panel using MAb1 to MAb7 (5). Subsequent to that time, MAb3 to MAb7 became unavailable. Hence, since 2006, all Lp1 isolates have been typed by only MAb1 (5), MAb75 (18), and MAb2 (5) using an immunodot method as previously described (19). Briefly, a suspension of L. pneumophila cultures was prepared in 0.6% formalin–phosphate-buffered saline (PBS), immobilized on a nitrocellulose membrane, and washed with PBS. Then, the antigen was overlaid with the MAbs. A mix of MAb1 and MAb75 was used to confirm that the L. pneumophila isolates tested were serogroup 1; MAb2 was used to identify MAb2-positive strains. After the excess MAbs were washed away, a goat anti-mouse antibody labeled with horseradish peroxidase (Bio-Rad, Hercules, CA) was added. The membrane was washed again and placed in peroxidase substrate (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) to allow colorimetric detection of the antigen/antibody reaction.
DNA extraction, PCR amplification, and sequencing.
Genomic DNA was extracted using the InviMag Bacteria DNA kit/KFmL (Invitek, Berlin, Germany) on the KingFisher mL platform (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's guidelines. PCR was performed on a DNA Engine Dyad thermal cycler system (Bio-Rad) using the European Society of Clinical Microbiology and Infectious Diseases Study Group for Legionella Infections (ESGLI) SBT protocol for epidemiological typing of L. pneumophila (version 5.0) with M13-tagged primers (20). In cases when the neuA gene fragment failed to amplify following the standard protocol, an alternative ESGLI protocol “Analysis of the N-Acylneuraminate Cytidyltransferase homologue (neuAh) found in some non-serogroup 1 strains of Legionella pneumophila” (version 1) was used (21). PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA). Nucleotide sequences were determined using M13 primers together with the BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, Grand Island, NY), and the products were analyzed on a model 3130xl or 3730xl genetic analyzer (Life Technologies).
SBT analysis.
Genotyping was performed using the seven-gene protocol from the ESGLI SBT scheme as described previously (12, 14). The online Legionella SBT Quality Tool (www.hpa-bioinformatics.org.uk/cgi-bin/legionella/sbt/seq_assemble_legionella1.cgi) was used to assign individual allele numbers. For each isolate, the combination of seven alleles was defined as a seven-digit allelic profile by using the predetermined order flaA, pilE, asd, mip, mompS, proA, and neuA (e.g., 1-4-3-1-1-1-1) and a sequence type (ST) represented by a number (e.g., ST1). Novel alleles and STs identified for the first time in this study were submitted to the ESGLI SBT database (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php).
Phylogenetic analysis.
Examination of the relationships between STs and within clonal complexes was conducted using the eBURST v3 website (http://eburst.mlst.net). We used the stringent group definition; according to this definition, a clonal complex consists of STs that share six of seven evaluated alleles with at least one other member of the group and are all believed to be descended from the same founding genotype (the primary founder) (22). Comparative eBURST analysis was employed to relate STs of sporadic and environmental Lp1 isolates characterized in this study.
Statistical analysis.
Diversity was estimated by calculating Hunter and Gaston's modification of Simpson's index of diversity (23) as previously described (24). Indexes of diversity (IODs) were calculated for outbreak-associated, sporadic, and environmental isolates and, separately, for potable and nonpotable environmental isolates. The respective samples were expanded by bootstrap methods to estimate variance across each group, e.g., environmental nonpotable. Simulated populations were compared using a t test after testing the simulated results for normality. Simulation and statistical tests were performed using SAS version 9.3 (SAS Institute Inc., Cary, NC, USA) executing on the X64_7PRO platform.
RESULTS
Outbreaks with matching clinical and environmental isolates.
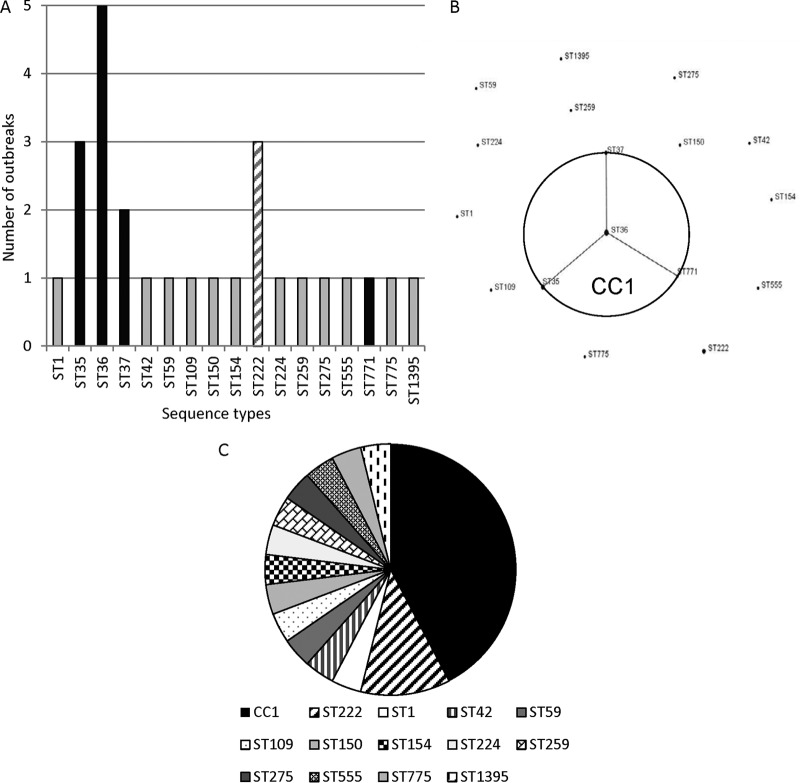
The 26 outbreaks took place in 20 U.S. states and territories (two outbreaks each in Nevada and St. Croix, U.S. Virgin Islands; three outbreaks in Pennsylvania) (Table 1). All outbreaks were caused by Lp1 strains, 24/26 (92%) of which were MAb2 positive. The outbreak-associated strains belonged to 17 different STs (IOD, 0.948), with the most frequent STs being ST36 (five outbreaks) and ST35 and ST222 (three outbreaks each) (Fig. 1A). eBURST analysis was used to determine the phylogenetic relationship between outbreak-associated STs and indicated that ST35, ST36, ST37, and ST771 formed a clonal complex (clonal complex 1 [CC1]), whereas the remaining 13 STs did not relate to each other (Fig. 1B). If one considers STs that form CC1 as a single group of closely related strains, then over half (53.8%) of characterized outbreaks were associated with Lp1 from CC1 (42.3%) and ST222 (11.5%) (Fig. 1C). Three outbreaks, two associated with ST35 and one associated with ST59, appeared to be recurrent. The Lp1 strains associated with each of the recurrent outbreaks were counted only once in this study (Table 1). In Nevada, the outbreak strain (ST35) was initially isolated in 2001 and later isolated from the same facility in 2002 and 2008 (25). For two more recent outbreaks that took place in Mississippi (ST35) and Washington (ST59), the outbreak strains continued to be detected in environmental samples and cause disease over a time period of at least a year despite remediation efforts. Although unlikely, the possibility does exist that each of these “recurrent” outbreaks may indeed represent several independent outbreaks occurring in the same facility.
FIG 1.
Lp1 associated with outbreaks investigated by CDC between 1982 and 2012. (A) Frequency of the 17 outbreak-associated sequence types (STs). STs composing clonal complex 1 (black bars), ST222 (hatched bar), and the other 12 STs that do not form a clonal complex and are associated with only a single outbreak (gray bars) are indicated. (B) Phylogenetic relationships between outbreak-associated STs determined by eBURST analysis. A population snapshot of outbreak-associated STs is shown: four STs grouped into a clonal complex (CC1), and 13 STs were singletons. The area of each dot represents the prevalence of the ST in the input data. (C) Frequencies of STs among outbreaks. STs composing clonal complex 1, ST35, ST36, ST37, and ST771, are grouped in a single sector.
Clinical isolates from sporadic cases.
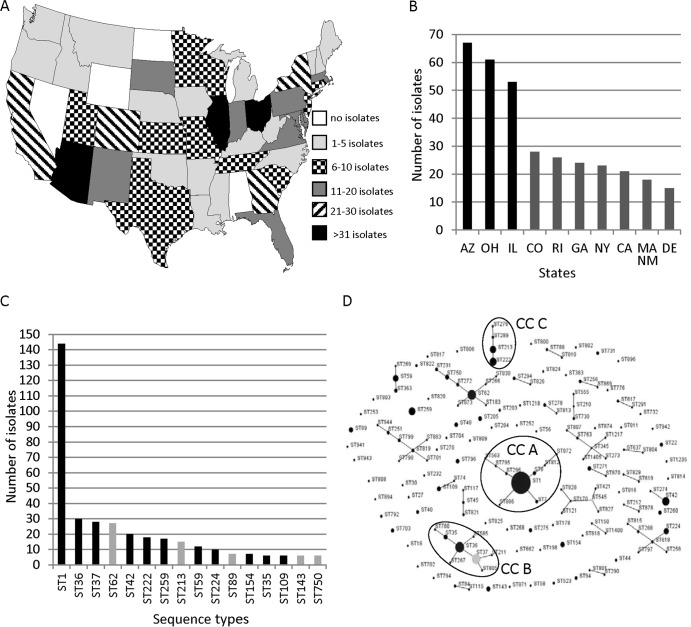
A total of 571 clinical Lp1 isolates from sporadic cases of LD originated in 46 states and the District of Columbia (Fig. 2A and Table S1 in the supplemental material). There were no isolates from Alabama, North Dakota, Nevada, or Wyoming. The three states that submitted the majority of sporadic isolates were Arizona (12%), Ohio (11%), and Illinois (9%) (Fig. 2B).
FIG 2.
Lp1 isolates from sporadic cases of legionellosis sent to the CDC Legionella laboratory between 1982 and 2012. (A) Geographical distribution of 571 sporadic clinical isolates. Alaska (1 isolate), District of Columbia (2 isolates), and Hawaii (13 isolates) are not shown on the map. (B) States with the most Lp1 clinical sporadic isolates. The three states with the most isolates (black bars) are indicated. Massachusetts and New Mexico sent the same number of sporadic isolates (18 isolates). (C) Most frequent STs of Lp1 sporadic isolates. STs that were also the outbreak-associated STs are indicated by black bars. (D) Phylogenetic relationship between sporadic STs identified by eBURST analysis shown as a population snapshot of 153 STs. The light gray dot represents a subgroup founder of clonal complex (an ST that has at least two descendant single-locus variants). Clonal complexes of sporadic STs discussed in the text are circled and named CCA, CCB, and CCC.
The results of SBT analysis indicated that 571 sporadic isolates belonged to 153 STs (IOD, 0.924). Two of these STs, ST1400 and ST1405, carried the N-acylneuraminate cytidyltransferase homolog (neuAh) typically found in non-Lp1 strains of L. pneumophila and not amplified with the standard neuA primers (26). Both ST1400 and ST1405 contained the same neuAh allele, 207, and this appears to be the first instance of the neuAh allele (in contrast to a typical neuA allele) in Lp1 isolates. The most prevalent ST among the sporadic isolates was ST1, accounting for 25% (144/571) of all sporadic isolates in this study (Fig. 2C). ST35, ST36, ST37, ST42, ST59, ST109, ST154, ST222, ST224, and ST259 were also among the most frequent STs of sporadic isolates. These same 10 STs were also responsible for 73% (19/26) of CDC-investigated outbreaks (Fig. 2C and Table 1). eBURST analysis of the 153 sporadic STs showed that 91 of these STs formed 25 clonal complexes, while 62 STs did not relate to each other and existed as singletons (Fig. 2D). The largest clonal complex, clonal complex A (CCA), included nine STs which comprised 28% (158/571) of sporadic isolates. ST1, the most prevalent ST in the world, was the primary founder of CCA. ST36 was the primary founder of the second largest clonal complex, CCB, that was formed by eight STs and included 43 sporadic isolates (Fig. 2D). ST35 and ST37 were single-locus variants of ST36, and all three STs were among the most frequent outbreak-associated and sporadic STs. Clonal complex C was comprised of four STs, ST213, ST222, ST276, and ST289, and included 36 isolates (Fig. 2D). All but one of 36 sporadic isolates that belonged to this clonal complex were from the northeastern region of the United States. Only one isolate was from Colorado, which is in the mountain region. Both ST222 and ST213 were among the top 10 most frequent sporadic STs, and ST222 was also one of the most frequent outbreak-associated STs (Fig. 2C and 1A). According to the ESGLI SBT database (accessed 24 October 2013), 20/42 Lp1 isolates that belonged to ST222 and that were deposited into the database were from Canada. Only three recently isolated ST222 Lp1 isolates were from outside North America: Austria (isolated in 2010), Germany (isolated in 2010), and Great Britain (isolated in 2012). All 18 ST213 isolates listed in the database were from either Canada or the United States. Both ST276 and ST289 appeared to be present only among isolates submitted from the United States, with the ST276 strain isolated from Connecticut and ST289 strains isolated from New York and Indiana.
Environmental isolates with no known association with LD.
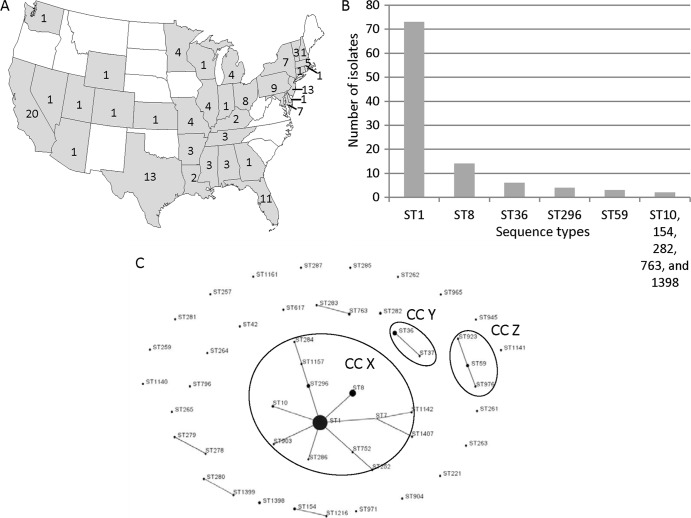
A total of 149 Lp1 isolates collected from facilities with no known association with LD were obtained from 35 states (142 isolates), the Navajo Nation territory (five isolates), and Puerto Rico (one isolate). The origin of one isolate was unknown. The majority of isolates were from California (14%), New Jersey (9%), Texas (9%), and Florida (7%) (Fig. 3A). For 99 environmental isolates, the information on the water system type from which the isolates were obtained was available and indicated that 45 were from plumbing systems, 40 from cooling towers, eight from nonspecified nonpotable sources, five from cisterns, and one from a decorative fountain (see Table S2 in the supplemental material). The environmental isolates analyzed in this study belonged to 49 STs (IOD, 0.751). The most prevalent environmental ST was ST1, with 49% (73/149) of isolates belonging to this sequence type (Fig. 3B). The other STs that appeared more than once among environmental isolates were ST8 (9%), ST36 (4%), ST296 (3%), ST59 (2%), and STs 10, 154, 282, 763, and 1398 (1% each) (Fig. 3B). Based on the analysis of the 99 isolates for which the water system type was known, ST1 was the predominant ST for both potable and nonpotable water systems (data not shown). The IODs for potable and nonpotable water systems were 0.615 and 0.787, respectively. eBURST analysis grouped 26 environmental STs into seven clonal complexes, whereas 23 STs were singletons. The largest clonal complex, CCX, consisted of 13 STs, and 68.5% (102/149) of environmental isolates belonged to this complex (Fig. 3C). The primary founder of CCX was ST1, which is similar to the sporadic isolates, but not outbreak-associated isolates. The two second largest environmental clonal complexes were CCY (ST36 and ST37; 5% isolates) and CCZ (ST59, ST923, and ST976; 3% isolates) (Fig. 3C).
FIG 3.
Lp1 environmental isolates with no known association with legionellosis. (A) Geographical distribution of 142 environmental isolates. The states depicted in white had no environmental isolates analyzed in this study. The numbers inside the states represents the number of environmental isolates that originated from each state. Alaska (no isolates), Hawaii (one isolate), the Navajo Nation territory (five isolates), and Puerto Rico (one isolate) are not shown on the map. (B) Frequency of STs that appeared more than once among environmental isolates. ST10, ST154, ST282, ST763, and ST1398 were represented twice each among environmental isolates. (C) Phylogenetic relationship between 49 environmental STs determined by eBURST analysis depicted as a population snapshot. Clonal complexes of environmental STs discussed in the text are circled and named CCX, CCY, and CCZ.
Comparison of STs from different source types.
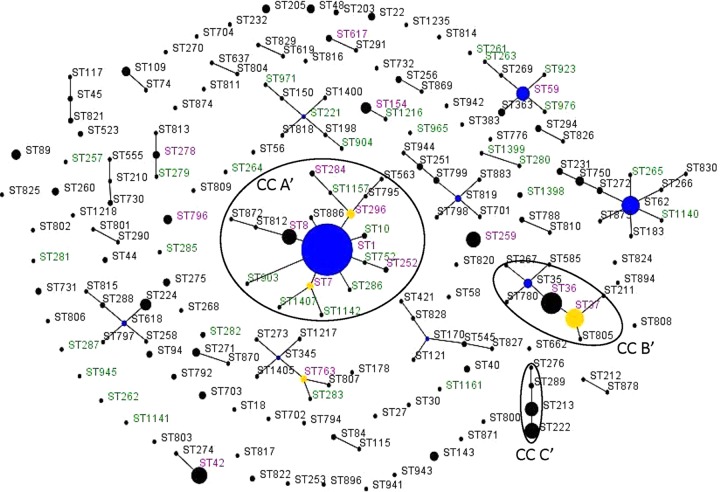
Comparison of outbreak-associated STs with STs of isolates from sporadic cases showed that 82% (14/17) of outbreak-associated STs were also found among sporadic STs, with 11 outbreak-associated STs present among the most frequent sporadic STs (Fig. 2C and 4). Three unique outbreak-associated STs, ST771, ST775, and ST1395, which were not isolated from sporadic cases of disease, were associated with only a single outbreak. The overall diversity of outbreak-associated and sporadic isolates calculated based on IODs was not significantly different (IODs of 0.948 and 0.924, respectively; P = 0.14). Comparison of outbreak-associated STs with environmental STs with no known association with LD revealed that there were seven STs in common between these groups: ST1, ST36, ST37, ST42, ST59, ST154, and ST259 (Fig. 4). Notably, ST1, ST36, ST59, and ST154 were among the most frequent environmental STs (Fig. 3B). The IODs of the outbreak-associated (0.948) and environmental (0.751) isolates differed significantly (P < 0.001). eBURST comparative analysis of the 153 sporadic STs and 49 environmental STs indicated that these populations shared only 16 STs (Fig. 4 and 5). However, 7 of the 16 shared STs (ST1, ST36, ST37, ST42, ST59, ST154, and ST259) also belonged to the group of 16 most prevalent sporadic STs (Fig. 2C). In addition, the seven shared STs (ST1, ST8, ST36, ST59, ST154, ST296, and ST763) were among the 10 most prevalent environmental STs (Fig. 3B). There were 33 unique environmental STs (67% of all environmental STs) and 137 unique sporadic STs (89.5% of all sporadic STs). ST1 was the primary founder of the largest clonal complex, CCA′, of combined environmental and sporadic isolates (Fig. 5). This complex contained more unique environmental STs than shared or unique sporadic STs. The clonal complex CCB′, with the primary founder ST36, contained only STs unique to sporadic isolates and two shared STs (ST36 and ST37). The clonal complex CCC′, which contained STs primarily from the northeastern region of North America, was composed of only sporadic STs (Fig. 5). The IODs of sporadic (0.924) and environmental (0.751) isolates differed significantly (P < 0.001).
FIG 4.
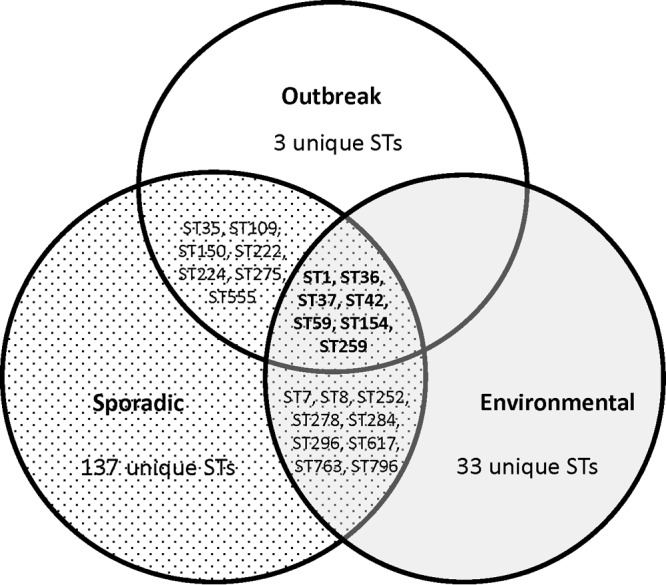
STs shared by outbreak-associated, sporadic, and environmental Lp1 isolates.
FIG 5.
Comparative analysis between the environmental and clinical sporadic STs analyzed in this study done by eBURST analysis. The population snapshot contained 27 clonal complexes and 70 singletons. STs in green font were unique to environmental STs, STs in black font were unique to sporadic STs. STs in magenta font were found among both the environmental and sporadic STs. Blue circles indicate the predicted primary founder of the clonal complex. Yellow dots and circles represent a subgroup founder. The clonal complexes of interest are circled and named CCA′, CCB′, and CCC′.
DISCUSSION
In the last 6 years, SBT analysis has become the gold standard method for typing L. pneumophila isolates. This method is used not only for comparing clinical strains to environmental isolates from a suspect water source during outbreak investigations but also for the study of genetic diversity and clonal expansion of L. pneumophila populations. In this study, we expanded our 2009 analysis of clinical and environmental Lp1 isolates (7) by typing a larger and more comprehensive number of isolates from different source types, geographical areas, and temporal ranges. We were interested to see whether the trends of ST distribution observed on a small sample of Lp1 isolates discussed in the previous study remained reliable and consistent for the larger Lp1 population and whether strains identified as native in 2009 persisted within previously observed geographical borders or expanded to other regions.
As in our previous study, the most prevalent ST for both clinical sporadic and environmental isolates was ST1, the most widespread ST in the world. According to the studies of the distribution of Lp1 STs in different regions of the world, it appears that the most prevalent ST among clinical and environmental populations of Lp1 is either ST1 (8, 27–32) or an ST that is confined to a specific geographic region (33–35). For example, in Belgium and the United Kingdom, the most frequently detected ST among clinical isolates was ST47 (9, 35). It is most likely that ST47, the Lorraine strain, is also the most prevalent genotype among clinical Lp1 isolates in France and the Netherlands (9, 36, 37). Interestingly, ST47, which is widespread among clinical Lp1 isolates in Western Europe, has yet to be detected in the United States or any Asian countries. A Canadian study mentioned that ST47 was recovered three times in Ontario (31), and a single ST47 clinical isolate from British Columbia was deposited into the ESGLI SBT database in 2012 (accessed 24 October 2013).
In contrast to the sporadic and environmental Lp1 groups where ST1 was the most frequently isolated ST, we did not observe ST1 associated with an LD outbreak until the spring of 2012 (Table 1). Even though ST1 was the cause of multiple outbreaks in Canada and Europe (31, 35), it has always been an enigma why ST1 strains were never the cause of an outbreak in the United States. Moreover, ST1 or a related strain of Lp1 was frequently isolated from water samples collected as a part of outbreak investigations conducted by the CDC (data not shown), but these strains have never been matched to an Lp1 clinical isolate and thus were deemed not to be the cause of the outbreak. Consequently, it was assumed that ST1 isolates were less virulent than other Lp1 strains that are frequently associated with an outbreak, such as ST36, ST35, ST37, or ST222. However, the travel-associated outbreak in Nevada that occurred in 2012 (NV12 outbreak) contradicted this assumption. Not only was the NV12 outbreak-associated Lp1 an ST1 strain, but it was also MAb2 negative, which occurred only one other time in the history of LD outbreaks investigated by the CDC (Table 1).
There are several possible explanations for this phenomenon. There could be an emergence of an ST1 strain with properties that make it capable of causing infection more readily than ST1 strains previously recovered in the United States. The acquisition of additional virulent traits could be accomplished by horizontal gene transfer from more-virulent strains that coexisted in the same environment as the NV12 outbreak strain. Whole-genome analysis of this strain may help to test this hypothesis. Another possibility is that ST1-associated outbreaks that took place in the past were not identified due to the unavailability of clinical isolates that could be compared with environmental isolates obtained during outbreak investigations. Regardless of the reason for the occurrence of the NV12 outbreak, ST1 should no longer be dismissed as a common strain that is prevalent among environmental isolates but almost never causes disease. Even though a decreased incidence of ST1 among clinical isolates was observed in Belgium, Canada, and Japan (27, 31, 35), we have not observed this trend in the United States. The number of sporadic ST1 isolates deposited into the CDC reference collection fluctuates between 0 (2004) and 10 (1983 and 1987) isolates per year, with 7 isolates deposited in 2012, the final year of this study (data not shown).
ST35 is an interesting outbreak-associated ST. This ST is closely related to ST36 and ST37, which caused numerous outbreaks in the United States and Europe and were suggested by Harrison et al. to belong to a select group of STs that have an enhanced ability to cause legionellosis in humans (38). In contrast to ST36 and ST37 strains, which typically cause outbreaks that are relatively easily eradicated, ST35 appears to have a tendency to cause a recurrent problem in a facility once it has colonized the water system. One of the outbreaks included in this study was associated with an ST35 strain that appeared to resurface in the same facility in Nevada in 2001, 2002, and 2008 despite periods after remediation when bacterial growth was below the level of detection (25). In another, more recent ST35-associated outbreak that took place in Mississippi in 2010, the failed remediation efforts led to more cases in 2011. The CDC was also involved in investigating two ST35-associated international outbreaks, both of which demonstrated a recurrent pattern (data not shown). All of these outbreaks differed in the type of the contaminated water source, age of the affected facilities, and the type of material used in the construction of the water system. Two common features that they shared were the ST of the outbreak-associated Lp1 strain and its ability to cause new LD cases despite remediation efforts. ST35 may represent a special strain that, in addition to having enhanced virulent properties, might be highly resistant to remediation and thus display persistence. The resistance may be attributed to changes in the physical properties of the bacterial cell surface, preference for infection of protozoa that provide better protection from adverse environmental factors, and the overall ability of this Lp1 strain to form biofilms that are more resistant to chlorine and high water temperature.
In this study, we identified for the first time two sporadic STs, ST1400 and ST1405, that had the neuAh allele previously only found in non-Lp1 strains of L. pneumophila (26). The ESGLI SBT database showed that ST1400 closely relates to ST12, ST150, ST159, ST221, and 1433, all of which are Lp1 STs that differ from ST1400 at only a single allele, the neuA or neuAh. Similarly, STs related to ST1405 include ST2, ST273, ST322, ST345, and ST502, and with the exception of ST322, they are Lp1 isolates. ST322 is an L. pneumophila serogroup 6 isolate, yet it carries the “typical” neuA allele 3 that could be amplified with the standard neuA primers. Interestingly, the ST273 isolate, which is identical to ST1405 except for carrying the neuA allele 2, originated from the same state, Arizona, as the ST1405 isolate. It is likely that Lp1 isolates that belonged to ST1400 and ST1405 acquired atypical neuAh alleles during genetic information exchange with non-Lp1 L. pneumophila strains that coresided in the same biofilm, and we may expect submission of more Lp1 isolates with atypical neuAh alleles to the ESGLI SBT database in the future.
The clonal complex C identified during eBURST analysis of 153 sporadic STs contained ST222, ST213, ST289, and ST276 (Fig. 2D). This is very similar to clonal complex A, which was identified in the 2009 study, with the exception that CCA from 2009 also contained ST227, which was detected in Canada (7). In 2009, the Lp1 isolates of CCA were identified only in the northeastern states of the United States and Ontario, Canada. In their study published in 2010, Tijet et al. indicated that strains of CCA have been detected in Ontario, Canada, since 1992, with ST222 first identified in 1999 (31). They proposed that CCA represented a recently emerged group of strains that were replacing widespread strains such as ST1. The first ST222 sporadic strain was sent to the CDC reference collection in 1998 from Pennsylvania, whereas the ST213 strain, which is a single-locus variant of ST222, was sent to CDC in 1992 from Ohio. In recent years, we have not observed a substantial increase in representation among sporadic isolates of either ST from this clonal complex. However, both ST222 and ST213 were among the most prevalent sporadic STs (Fig. 2C), with ST222 being responsible for 3/26 outbreaks analyzed in this study (Fig. 1A and C). Importantly, ST222 was also the cause of one of the biggest LD outbreaks in North America that took place in Ontario, Canada, in the fall of 2005 and caused 112 illnesses and 23 deaths (39). This suggests that the strains within clonal complex C may indeed have higher virulence potential. The geographic distribution of CCC strains in the United States was still confined to the northeastern region of the United States, with the exception of one isolate from Colorado. With epidemiological data from this case undetermined, it is possible this could be a travel-associated case that originated somewhere in the northeastern states. On the other hand, the relatively recent submission to the ESGLI SBT database of three ST222 isolates from Europe indicates that ST222 may be expanding to other continents. This is reminiscent of ST47, which only recently expanded from Europe to Canada.
Previous studies of environmental Lp1 populations showed differences in ST distribution between various water system types. In South Korea, ST1 was the predominant ST in isolates from cooling towers, whereas ST-K1, unique to South Korea, was the dominant ST in hot water samples (29). In Japan, ST1 was more prevalent in cooling tower water (74%) than in bathwater (12%) and was not isolated from any soil samples (28). In our study, the analysis of STs for 99 environmental isolates for which the water system type was known indicated that ST1 was the predominant ST among both cooling tower isolates and potable water isolates. The differences in the environmental ST distribution between the United States and South Korea could be due to the predominance of the unique South Korean strain in hot water samples. U.S. soil samples may also have different ST diversity in comparison to the potable and nonpotable water system types.
The comparison of ST distribution among populations of outbreak-associated, sporadic, and environmental Lp1 isolates showed that the STs from outbreaks and sporadic cases were more similar to each other than either group was to environmental STs. The STs associated with multiple outbreaks, such as ST35, ST36, ST37, and ST222, were also among the most prevalent sporadic STs (Fig. 1A and 2C). The closely related ST35, ST36, and ST37 are widely distributed in the world and were responsible for multiple sporadic cases and outbreaks in the past, including the first described outbreak in Philadelphia, Pennsylvania, in 1976 due to ST36. In contrast, ST222 is an emerging strain identified only 14 years ago which now may be expanding from North America to other continents. As was hypothesized previously (38), STs such as these which are common to clinical isolates may have an enhanced ability to cause legionellosis.
In contrast to our previous study, in which populations of clinical and environmental Lp1 isolates shared only three STs, this study showed a larger overlap of the environmental population with both outbreak-associated and clinical Lp1 isolates (Fig. 4). Environmental and outbreak-associated isolates shared seven STs, three of which, ST1, ST36, and ST154, were also shared by clinical and environmental isolates in the 2009 study. Together with ST59, these STs were among the most frequent environmental STs identified in the current study. With the exception of ST36 and ST37, the other STs shared between outbreak-associated and environmental isolates were responsible for a single outbreak each (Table 1 and Fig. 1A). It would be of interest to see whether these STs will be associated with more LD outbreaks in the future. It seems this may be likely for ST42, since it was associated with a recent international LD outbreak.
There were 16 STs shared between environmental and sporadic Lp1 isolates. Among the shared STs are the same seven STs that were shared between environmental and outbreak-associated STs. In addition, ST1, ST36, ST59, and ST154 were the most frequent STs for both clinical sporadic and environmental Lp1 isolates. Thus, the STs common to all three populations are ST1, ST36, ST37, ST42, ST59, ST154, and ST259. With the exception of ST259, which has been reported only in the United States and France according to the ESGLI SBT database, the six other STs are well represented throughout the world. In contrast, the majority of STs that are unique to each of the sample type populations were isolated infrequently and found only in the United States. Hence, similar to other studies, the Lp1 population of the United States is represented by a combination of STs that are frequent and widespread throughout the world and those that are found only in the United States and rarely isolated.
A limitation of this study is that the majority of Lp1 sporadic isolates from the CDC reference collection analyzed were submitted only from those requesting additional typing or confirmation of the species identification and thus may not accurately reflect the general population of sporadic Lp1 strains in the country. Similarly, not every SBT-analyzed Lp1 strain is submitted to the EGLI SBT database unless the strain belongs to a novel ST that requires a unique identifier. Hence, while providing a good representation of the diversity of Lp1 STs, the database may not provide a complete picture of an ST's frequency or geographical distribution. In addition, the CDC lab does not routinely receive environmental Lp1 isolates with no known association with cases of legionellosis, with the exception of special studies and recently established collaborations with commercial labs. Therefore, in this study, a bias exists between Lp1s from different source types. Specifically, the isolation time of outbreak and sporadic isolates was more evenly distributed between 1982 and 2012, whereas the majority of environmental isolates were collected in 2006 and 2010 to 2012.
Despite multiple guidelines and official recommendations for preventing the transmission of LD, the prevalence of legionellosis remains high in the United States. Whereas many facilities opt for routine sampling for Legionella as a means of assessing risk from Legionella growth in their water system, their action plans are often based on the concentration of legionellae or the number of sites that are positive for the bacteria. However, it appears that an action plan should also take into account the type of Legionella isolated from a facility. The most prevalent STs shared by both clinical sporadic and outbreak-associated Lp1 isolates may have an increased ability to cause disease and thus may require special attention when detected.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the CDC NCEH/NCZVED Safe Water Foundation.
We thank Norman Fry and Massimo Mentasti for help and insightful discussions of sequence-based typing methodology. We also thank Maja Kodani and Tatiana Travis for processing environmental samples from outbreaks and environmental studies and Patrick Yang for SBT analysis of a number of clinical isolates.
Footnotes
Published ahead of print 6 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01973-13.
REFERENCES
- 1.Hicks LA, Garrison LE, Nelson GE, Hampton LM. 2012. Legionellosis—United States, 2000–2009. Am. J. Transplant. 12:250–253. 10.1111/j.1600-6143.2011.03938.x [DOI] [PubMed] [Google Scholar]
- 2.Brunkard JM, Ailes E, Roberts VA, Hill V, Hilborn ED, Craun GF, Rajasingham A, Kahler A, Garrison L, Hicks L, Carpenter J, Wade TJ, Beach MJ, Yoder JS, CDC 2011. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2007–2008. MMWR Surveill. Summ. 60:38–68 http://www.ncbi.nlm.nih.gov/pubmed/21937977 [PubMed] [Google Scholar]
- 3.Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506–526. 10.1128/CMR.15.3.506-526.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127–128. 10.1086/341087 [DOI] [PubMed] [Google Scholar]
- 5.Joly JR, McKinney RM, Tobin JO, Bibb WF, Watkins ID, Ramsay D. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23:768–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luck PC, Helbig JH, Ehret W, Marre R, Witzleb W. 1992. Subtyping of Legionella pneumophila serogroup 1 strains isolated in Germany using monoclonal antibodies. Zentralbl. Bakteriol. 277:179–187. 10.1016/S0934-8840(11)80611-0 [DOI] [PubMed] [Google Scholar]
- 7.Kozak NA, Benson RF, Brown E, Alexander NT, Taylor TH, Jr, Shelton BG, Fields BS. 2009. Distribution of lag-1 alleles and sequence-based types among Legionella pneumophila serogroup 1 clinical and environmental isolates in the United States. J. Clin. Microbiol. 47:2525–2535. 10.1128/JCM.02410-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borchardt J, Helbig JH, Luck PC. 2008. Occurrence and distribution of sequence types among Legionella pneumophila strains isolated from patients in Germany: common features and differences to other regions of the world. Eur. J. Clin. Microbiol. Infect. Dis. 27:29–36. 10.1007/s10096-007-0392-3 [DOI] [PubMed] [Google Scholar]
- 9.Harrison TG, Afshar B, Doshi N, Fry NK, Lee JV. 2009. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000–2008). Eur. J. Clin. Microbiol. Infect. Dis. 28:781–791. 10.1007/s10096-009-0705-9 [DOI] [PubMed] [Google Scholar]
- 10.Dournon E, Bibb WF, Rajagopalan P, Desplaces N, McKinney RM. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157:496–501. 10.1093/infdis/157.3.496 [DOI] [PubMed] [Google Scholar]
- 11.Helbig JH, Bernander S, Castellani Pastoris M, Etienne J, Gaia V, Lauwers S, Lindsay D, Luck PC, Marques T, Mentula S, Peeters MF, Pelaz C, Struelens M, Uldum SA, Wewalka G, Harrison TG. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. 21:710–716. 10.1007/s10096-002-0820-3 [DOI] [PubMed] [Google Scholar]
- 12.Gaia V, Fry NK, Afshar B, Luck PC, Meugnier H, Etienne J, Peduzzi R, Harrison TG. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43:2047–2052. 10.1128/JCM.43.5.2047-2052.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaia V, Fry NK, Harrison TG, Peduzzi R. 2003. Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J. Clin. Microbiol. 41:2932–2939. 10.1128/JCM.41.7.2932-2939.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratzow S, Gaia V, Helbig JH, Fry NK, Luck PC. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45:1965–1968. 10.1128/JCM.00261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luck C, Fry NK, Helbig JH, Jarraud S, Harrison TG. 2013. Typing methods for Legionella. Methods Mol. Biol. 954:119–148. 10.1007/978-1-62703-161-5_6 [DOI] [PubMed] [Google Scholar]
- 16.Tablan OC, Anderson LJ, Besser R, Bridges C, Hajjeh R, CDC, Healthcare Infection Control Practices Advisory Committee 2004. Guidelines for preventing health-care–associated pneumonia, 2003: recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee. MMWR Recomm. Rep. 53(RR-3):1–36 http://www.ncbi.nlm.nih.gov/pubmed/15048056 [PubMed] [Google Scholar]
- 17.Flannery B, Gelling LB, Vugia DJ, Weintraub JM, Salerno JJ, Conroy MJ, Stevens VA, Rose CE, Moore MR, Fields BS, Besser RE. 2006. Reducing Legionella colonization in water systems with monochloramine. Emerg. Infect. Dis. 12:588–596. 10.3201/eid1204.051101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Para MF, Plouffe JF. 1983. Production of monoclonal antibodies to Legionella pneumophila serogroups 1 and 6. J. Clin. Microbiol. 18:895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanden GN, Cassiday PK, Barbaree JM. 1993. Rapid immunodot technique for identifying Bordetella pertussis. J. Clin. Microbiol. 31:170–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mentasti M, Fry NK. 8 October 2012. Sequence-based typing protocol for epidemiological typing of Legionella pneumophila, version 5.0. ESCMID Study Group for Legionella Infections, European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland: http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/protocols/ESGLI%20SBT%20GUIDELINE%20v5.0.pdf [DOI] [PubMed] [Google Scholar]
- 21.Mentasti M, Fry NK. 8 October 2012. Analysis of the N-Acylneuraminate Cytidyltransferase homologue (neuAh) found in some non-serogroup 1 strains of Legionella pneumophila, version 1.0. ESCMID Study Group for Legionella Infections, European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland: http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/protocols/ESGLI%20neuAh%20GUIDELINE%20v1.0.pdf [Google Scholar]
- 22.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison TG, Doshi N, Fry NK, Joseph CA. 2007. Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin. Microbiol. Infect. 13:78–85. 10.1111/j.1469-0691.2006.01558.x [DOI] [PubMed] [Google Scholar]
- 25.Silk BJ, Moore MR, Bergtholdt M, Gorwitz RJ, Kozak NA, Tha MM, Brown EW, Winchester JL, Labus BJ, Rowley P, Middaugh JP, Fields BS, Hicks LA. 2012. Eight years of Legionnaires' disease transmission in travellers to a condominium complex in Las Vegas, Nevada. Epidemiol. Infect. 140:1993–2002. 10.1017/S0950268811002779 [DOI] [PubMed] [Google Scholar]
- 26.Farhat C, Mentasti M, Jacobs E, Fry NK, Luck C. 2011. The N-acylneuraminate cytidyltransferase gene, neuA, is heterogenous in Legionella pneumophila strains but can be used as a marker for epidemiological typing in the consensus sequence-based typing scheme. J. Clin. Microbiol. 49:4052–4058. 10.1128/JCM.00687-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amemura-Maekawa J, Kura F, Helbig JH, Chang B, Kaneko A, Watanabe Y, Isobe J, Nukina M, Nakajima H, Kawano K, Tada Y, Watanabe H, Working Group for Legionella in Japan 2010. Characterization of Legionella pneumophila isolates from patients in Japan according to serogroups, monoclonal antibody subgroups and sequence types. J. Med. Microbiol. 59:653–659. 10.1099/jmm.0.017509-0 [DOI] [PubMed] [Google Scholar]
- 28.Amemura-Maekawa J, Kikukawa K, Helbig JH, Kaneko S, Suzuki-Hashimoto A, Furuhata K, Chang B, Murai M, Ichinose M, Ohnishi M, Kura F, Working Group for Legionella in Japan 2012. Distribution of monoclonal antibody subgroups and sequence-based types among Legionella pneumophila serogroup 1 isolates derived from cooling tower water, bathwater, and soil in Japan. Appl. Environ. Microbiol. 78:4263–4270. 10.1128/AEM.06869-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HK, Shim JI, Kim HE, Yu JY, Kang YH. 2010. Distribution of Legionella species from environmental water sources of public facilities and genetic diversity of L. pneumophila serogroup 1 in South Korea. Appl. Environ. Microbiol. 76:6547–6554. 10.1128/AEM.00422-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reimer AR, Au S, Schindle S, Bernard KA. 2010. Legionella pneumophila monoclonal antibody subgroups and DNA sequence types isolated in Canada between 1981 and 2009: laboratory component of national surveillance. Eur. J. Clin. Microbiol. Infect. Dis. 29:191–205. 10.1007/s10096-009-0840-3 [DOI] [PubMed] [Google Scholar]
- 31.Tijet N, Tang P, Romilowych M, Duncan C, Ng V, Fisman DN, Jamieson F, Low DE, Guyard C. 2010. New endemic Legionella pneumophila serogroup I clones, Ontario, Canada. Emerging Infect. Dis. 16:447–454. 10.3201/eid1603.081689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu BQ, Ren HY, Zhou HJ, Qin T, Shao ZJ. 2011. Sequence-based typing of 82 strains of serotype I Legionella pneumophila isolated from 9 provinces in China. Zhonghua Yu Fang Yi Xue Za Zhi 45:890–894 (In Chinese.) http://www.ncbi.nlm.nih.gov/pubmed/22321587 [PubMed] [Google Scholar]
- 33.Chasqueira MJ, Rodrigues L, Nascimento M, Marques T. 2009. Sequence-based and monoclonal antibody typing of Legionella pneumophila isolated from patients in Portugal during 1987–2008. Euro Surveill. 14(28):pii=19271 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19271 [DOI] [PubMed] [Google Scholar]
- 34.Kanatani JI, Isobe J, Kimata K, Shima T, Shimizu M, Kura F, Sata T, Watahiki M. 2013. Molecular epidemiology of Legionella pneumophila serogroup 1 isolates identify a prevalent sequence type, ST505, and a distinct clonal group of clinical isolates in Toyama Prefecture, Japan. J. Infect. Chemother. 19:644–652. 10.1007/s10156-012-0537-x [DOI] [PubMed] [Google Scholar]
- 35.Vekens E, Soetens O, De Mendonca R, Echahidi F, Roisin S, Deplano A, Eeckhout L, Achtergael W, Pierard D, Denis O, Wybo I. 2012. Sequence-based typing of Legionella pneumophila serogroup 1 clinical isolates from Belgium between 2000 and 2010. Euro Surveill. 17(43):pii=20302 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20302 [PubMed] [Google Scholar]
- 36.Den Boer JW, Bruin JP, Verhoef LP, Van der Zwaluw K, Jansen R, Yzerman EP. 2008. Genotypic comparison of clinical Legionella isolates and patient-related environmental isolates in the Netherlands, 2002–2006. Clin. Microbiol. Infect. 14:459–466. 10.1111/j.1469-0691.2008.01973.x [DOI] [PubMed] [Google Scholar]
- 37.Ginevra C, Forey F, Campese C, Reyrolle M, Che D, Etienne J, Jarraud S. 2008. Lorraine strain of Legionella pneumophila serogroup 1, France. Emerg. Infect. Dis. 14:673–675. 10.3201/eid1404.070961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison TG, Fry NK, Afshar B, Bellamy W, Doshi N, Underwood AP. 2005. Typing of Legionella pneumophila and its role in elucidating the epidemiology of Legionnaire's disease, p 94–99 In Cianciotto NP, Edelstein PH, Fields BS, Geary TF, Harrison TG, Joseph CA, Ratcliff RM, Stout JE, Swanson MS. (ed), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC [Google Scholar]
- 39.Gilmour MW, Bernard K, Tracz DM, Olson AB, Corbett CR, Burdz T, Ng B, Wiebe D, Broukhanski G, Boleszczuk P, Tang P, Jamieson F, Van Domselaar G, Plummer FA, Berry JD. 2007. Molecular typing of a Legionella pneumophila outbreak in Ontario, Canada. J. Med. Microbiol. 56:336–341. 10.1099/jmm.0.46738-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.