Abstract
Long-term antiviral therapy of chronic hepatitis B virus (HBV) infection can lead to the selection of drug-resistant HBV variants and treatment failure. Moreover, these HBV strains are possibly present in treatment-naive patients. Currently available assays for the detection of HBV drug resistance can identify mutants that constitute ≥5% of the viral population. Furthermore, drug-resistant HBV variants can be detected when a viral load is >104 copies/ml (1,718 IU/ml). The aim of this study was to compare matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and multitemperature single-strand conformation polymorphism (MSSCP) with commercially available assays for the detection of drug-resistant HBV strains. HBV DNA was extracted from 87 serum samples acquired from 45 chronic hepatitis B (CHB) patients. The 37 selected HBV variants were analyzed in 4 separate primer extension reactions on the MALDI-TOF MS. Moreover, MSSCP for identifying drug-resistant HBV YMDD variants was developed and turned out to be more sensitive than INNOLiPA HBV DR and direct sequencing. MALDI-TOF MS had the capability to detect mutant strains within a mixed viral population occurring with an allelic frequency of approximately 1% (with a specific value of ≥102 copies/ml, also expressed as ≥17.18 IU/ml). In our study, MSSCP detected 98% of the HBV YMDD variants among strains detected by the MALDI-TOF MS assay. The routine tests revealed results of 40% and 11%, respectively, for INNOLiPA and direct sequencing. The commonly available HBV tests are less sensitive than MALDI-TOF MS in the detection of HBV-resistant variants, including quasispecies.
INTRODUCTION
Hepatitis B virus (HBV) infection remains a global health problem affecting about 2 billion people. It is estimated that there are 240 to 350 million chronic HBV carriers who developed chronic hepatitis B (CHB) with high risk of liver cirrhosis and hepatocellular carcinoma (HCC) (1–3). The main goals of antiviral therapy are the prevention of liver disease progression and the prolongation of patient survival. Since the HBV viral load has been shown to be a crucial determinant of the progression of liver damage, these goals can be achieved as long as HBV replication is sustained (4, 5). However, the existence of covalently closed circular DNA (cccDNA) in infected hepatocytes (1 to 50 copies/hepatocyte) make the complete eradication of HBV infection currently impossible (6, 7). In recent years, treatment of CHB has been improved by nucleoside/nucleotide analogues (NAs), such as lamivudine, adefovir, tenofovir, and entecavir, that inhibit the reverse transcriptase activity of HBV polymerase and suppress virus replication (8, 9).
Unfortunately, because of the high HBV replication rate and the lack of proofreading activity of the viral polymerase enzyme, long-term therapy can lead to the emergence or selection of drug-resistant mutants and treatment failure. Moreover, there is a possible preexistence of drug-resistant HBV variants in treatment-naive patients (10–12). This phenomenon of HBV replication is related to the coexistence of numerous virus variants and quasispecies. New variants appearing during the natural course of HBV infection are more viable, spread rapidly in the liver, and accumulate in infected hepatocytes. After suppression of the dominant strain, the drug-resistant minor strain emerges (8, 13).
Nowadays, an increasing number of patients experience multiple NA treatment failures, especially patients who are sequentially treated with low genetic barrier drugs (e.g., lamivudine, telbivudine, and adefovir) (14, 15). Sequential therapy leads to the accumulation of resistance mutations on the same viral genome and increases the risk of the emergence of multidrug resistance (13, 16–18). If initial monotherapy fails, a second drug with a different resistance profile should be added or a patient's therapy should be switched to a more potent combination of drugs (19). There are few data to demonstrate the clinical relevance of detecting resistance mutations that are present before treatment.
The NA treatment strategies should be based on the detection of drug-resistant HBV variants as early as possible, before virological and clinical breakthrough. When the drug-resistant variant appears during the treatment, it could predict therapy failure and viral repopulation in hepatocytes. The ideal assay should be sensitive, specific, reproducible, and easy to perform (20). Direct sequencing of PCR products (Sanger sequencing), restriction fragment length polymorphisms (RFLPs), mutation-specific real-time PCR, and reverse hybridization (Line Probe Assay) are the most common assays for the detection of drug-resistant HBV variants. However, most of these methods detect viral mutants that constitute 5% or more (20% in direct sequencing) of the viral population. Furthermore, drug-resistant mutants can be identified in patients with a viral load of more than 104 copies/ml (1,718 IU/ml) (21, 22).
The multitemperature single-strand conformation polymorphism (MSSCP) assay is a new approach based on the electrophoretic mobility differences of single-stranded DNA. The electrophoretic separation is performed under sequentially changed gel temperatures. Data in the literature show that results obtained by MSSCP are much more sensitive (0.1% of minor strains) than those obtained by three classical SSCP analyses of the same samples at different but constant gel temperatures (23).
Recently, the MassARRAY system (Sequenom Inc., San Diego, CA), based on nucleic acid analysis by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), has become a powerful and widespread analytical tool in all fields of life science. High-throughput MALDI-TOF MS provides an alternative approach to HBV genotyping. This assay has the capability to detect mutant strains within a mixed viral population occurring with an allelic frequency of 1% (24).
The aim of this study was an assessment of the prevalence of HBV drug-resistant variants in NA treatment-naive and experienced CHB patients with the use of MALDI-TOF MS and MSSCP assays. Additionally, the study results were compared with commonly used assays in HBV drug-resistant YMDD variant detection assays.
MATERIALS AND METHODS
Sample handling and DNA extraction.
Eighty-seven blood samples from 45 CHB patients referred to Department of Infectious Diseases, Medical University of Gdansk, and the outpatients' clinic at the Pomeranian Centre for Infectious Diseases and Tuberculosis in Gdansk were enrolled for this study in 2011 to 2013. The study group consisted of NA treatment-naive patients (17/45) and treatment-experienced patients (28/45). The blood samples were stored after weeks 0, 12, and 24 of NA therapy. The Local Ethics Committee accepted the protocol of this study and all of the patients provided a written informed consent to participate in this study.
Whole-blood samples were collected into Vacutainer tubes without anticoagulant and then incubated in an upright position for 30 to 45 min (no longer than 60 min) to allow clotting. The clot was then removed by centrifuging at 3,500 × g for 15 min in a refrigerated centrifuge. The resulting supernatant (serum) was immediately transferred into a clean Eppendorf tube and stored at −20°C until further analysis. Viral DNA was extracted from 200 μl of serum samples using a High Pure viral nucleic acid kit (Roche Diagnostics, Germany) with a slightly modified manufacturer's protocol. The incubation time with proteinase K was 1 h instead of 10 min and the final elution volume was 30 μl. Quality and quantity of the DNA were measured using a spectrophotometer (NanoDrop ND-1000, Thermo Scientific).
Multitemperature single-strand conformation polymorphism.
In order to conduct the MSSCP assay the DNA Pointer System (BioVectis, Poland) was used. To increase the sensitivity of the detection of HBV YMDD (rt204) variants, nested PCR was performed. The reaction mixture for the first step (25 μl) contained 2 μl of HBV DNA, 1× chelating buffer, 1.2 mM Mg(OAc)2, 0.2 mM deoxynucleoside triphosphate (dNTP), 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Germany), 0.1 mg/ml of casein, 0.01% (vol/vol) formamide, and 0.125 μM primers 1F and 1R (1F, 5′-GACTCGTGGTGGACTTCTCTC-3′; 1R, 5′-TGATCCTGTGGCAAAGTTCC-3′). Amplification conditions for the first PCR were 95°C for 10 min, and then 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s, and finally 72°C for 10 min. The reaction mixture for the second step (25 μl) contained 1× chelating buffer, 1.2 mM MgCl2, 0.5 mM deoxynucleoside triphosphate (dNTP), 1 U of AmpliTaq Gold polymerase, 0.25% (vol/vol) glycerol, 0.4% (vol/vol) bovine serum albumin (BSA), 0.125 μM primers 2F (25) and 2R (5′-TAACAGCGGTATAAAGGGCCT-3′), and 2 μl of the first amplification step product. Amplification conditions were 95°C for 10 min, and then 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 30 s, and finally 72°C for 10 min. Positive and negative controls were used at each step (HBV DNA External Quality Control, PeliSpy PRO; AcroMetrix). To determine the detection limit, serial dilutions of quantification standard (QS) HBV Real Star (Altona Diagnostics, Germany) were used: 104 copies/μl, 103 copies/μl, 102 copies/μl, 10 copies/μl, 5 copies/μl, 2 copies/μl, and 1 copy/μl.
One microliter of PCR products (159 bp) containing HBV mutant and wild type (WT) sequences at the codons of interest was mixed with 2 μl of denaturing buffer A (BioVectis, Poland), heated for 5 min at 95°C, and then immediately cooled on ice. After the addition of 1 μl of denaturing buffer B, 3 μl of denatured sample was loaded onto a polyacrylamide gel (29:1 acrylamide/bisacrylamide). The conditions for electrophoretic separation of conformers of the HBV YMDD region were determined in respect to percentage of the gel (9%, 10%, or 11%), addition of glycerol (5% or none), and different sequentially changed sets of gel temperature. The electrophoresis was run at a constant voltage of 40 V in 0.5× Tris-borate-EDTA (TBE), and after silver staining, specific single-stranded DNA (ssDNA) band patterns appeared that were characteristic for each HBV variant. All band patterns were cut out from the gel, incubated overnight at −20°C, purified using the QIAquick gel extraction kit (Qiagen, Germany), and then sequenced.
MALDI-TOF MS sample preparation.
The first step of the MALDI-TOF MS-based HBV genotyping assay was amplification of a 754-bp HBV DNA fragment containing the polymorphic site of our interest (codons 80, 169, 173, 180, 181, 184,194, 202, 204, 233, 236, and 250). PCR conditions were the same as those described previously (26). In order to remove the remaining, nonincorporated dNTPs from the amplification products, 5 μl of each PCR product was treated with 2 μl of shrimp alkaline phosphatase (SAP) solution. This procedure was performed for 40 min at 37°C and stopped by inactivation at 85°C for 5 min.
MALDI-TOF MS—iPLEX Gold reaction.
The iPLEX Gold reaction was done on the Mass Array genotyping platform (Sequenom, Inc., San Diego, CA). The 37 selected HBV variants, C/T/A/G at position 367 (27), T/C at position 635 (28), G/C/T at position 646 (29), C/T/A at position 667 (30), G/A/T at position 670 (30), C/T at position 671 (30), A/G at position 679 (28), C/G at position 680 (28), G/A at position 709 (31), G/T at position 734 (28), A/G at position 739 (32), T/G at position 740 (33), G/C/T at position 741 (32), A/G at position 826 (34), A/C at position 836 (16), and A/G at position 877 (28), were analyzed in separate primer extension reactions (9-plex in assay 1, 2,6-plex in assay 3, and 13-plex in assay 4). During the assay, oligonucleotide primers annealed at positions directly adjacent to the single-nucleotide polymorphisms (SNP) of interest. The primer was extended, dependent upon the template sequence, resulting in an allele-specific difference in mass between extension products. This mass difference allowed the data analysis software to differentiate between SNP alleles. A specific extension product was generated for each allele. In the case of heterozygosity, both products were generated simultaneously. The two SNP alleles appeared as two distinct mass signals.
Each reaction mixture contained 7 μl of PCR/SAP product and 2 μl of iPLEX mix. and the procedure followed the iPLEX kit standard protocol (Sequenom, Inc., San Diego, CA). Allele-specific products resulting from the iPLEX reaction were desalted through the addition of deionized water and 6 mg of an anion-exchange resin and after centrifugation at 360 × g for 5 min and analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). All data analyses were carried out with the Typer Analyzer Application, version 4 (Sequenom, Inc., San Diego, CA).
INNOLiPA assay and direct sequencing.
Using INNOLiPA HBV DR, wild-type and mutations or polymorphisms, at codons 80, 173, 180, 181, 204, and 236 of the HBV polymerase gene were detected simultaneously. HBV DNA was amplified in one PCR round with INNOLiPA HBV DR primers according to the manufacturer's instructions (Innogenetics, Belgium). The 867-bp amplified product was denatured and hybridized to specific oligonucleotide probes coated onto the INNOLiPA HBV DR strip. After hybridization, in order to remove remaining unhybridized DNA from the strip, wash steps were performed. Streptavidin labeled with alkaline phosphatase was added and bound to any biotinylated hybrid previously formed. Incubation with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) chromogen resulted in a purple/brown precipitate. The INNOLiPA HBV DR strip contained 2 control lines, the conjugate control (control for the color development reaction) and the amplification control line (to check for the presence of amplified HBV genomic material).
In order to analyze the accuracy of the assay, sequencing reference results were required for each sample. The sequencing was performed by the Genomed (Warsaw, Poland) with sense and antisense primers as previously described (35) and analyzed by our group.
RESULTS
Within the analyzed serum samples, 0.02 and >170,000 kIU/ml viremia were detected. All samples were easily amplified by the PCR steps preceding the MSSCP, MALDI-TOF MS, and direct sequencing. Five of carrier patients had undetectable viral DNA after the PCR step of the HBV drug resistance line probe assay (INNOLiPA HBV DR). These patients had HBV levels of <104 copies/ml (1,718 IU/ml). The remaining 82 samples were analyzed by direct sequencing, INNOLiPA assay, and MALDI-TOF MS and compared. Moreover, the YMDD motif was also assessed by the MSSCP method.
Detection of drug-resistant HBV variants.
MALDI-TOF MS was the most sensitive method, allowing the detection of drug- resistant HBV variants in 98% of all samples. Almost all (except rtA181S and rtM204S) of the HBV mutants were found in the study group with different allelic frequencies. Of 87 serum samples taken consecutively from 45 patients, the MALDI-TOF MS detected mutant virus with at least one of the rt80, rt180, rt204, rt233, and rt250 codons in 83% of the sera (72/87). The mutant type of rt233 was the most common and could be found in 71% (62/87) of samples. Mutant viruses in the remaining codons were detected as follows: rt80 (55%), rt169 (4%), rt173 (15%), rt180 (38%), rt181 (18%), rt184 (5%), rt194 (2%), rt202 (2%), rt204 (51%), rt236 (11%), and rt250 (61%) (Table 1). MALDI-TOF MS had the capability to detect mutant strains within a mixed viral population occurring with an allelic frequency of 1% (Fig. 1b). Minor amounts of virus in mixed population were found at each codon. The analysis was possible for plasma viral loads of more than 102 copies/ml (17.18 IU/ml). In one patient, wild-type virus was observed with respect to each codon.
TABLE 1.
Prevalence of the HBV reverse transcriptase polymorphisms in MALDI-TOF MS assay in selected patients
| Patient no. | Analyzed HBV reverse transcriptase polymorphisms (allele %) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L80I(V) | I169T | V173L | L180M | A181T(V) | T184G | A194T | S202I | M204V(I) | I233V | N236T | M250V | |
| 23 | V (4) | WTa | L (3) | WT | T (2) | WT | WT | WT | I (30) | V (10) | WT | V (4) |
| 28 | I (2) | WT | L (4) | M (13) | WT | WT | WT | WT | V (16) | V (11) | WT | V (3) |
| 69III | WT | WT | L (1) | WT | WT | WT | WT | WT | WT | WT | T (7) | WT |
| 95 | WT | T (11) | WT | M (29) | WT | G (5) | WT | WT | V (6) | V (4) | WT | V (5) |
| 97 | I (3) | WT | L (4) | WT | V (3) | WT | WT | WT | I (5) | WT | V (4) | (8) |
| 115 | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | T (3) | WT |
| 118IV | I (13) | WT | L (3) | M (12) | T (2) | WT | G (17) | WT | I (21) | V (2) | WT | V (6) |
| 123 | I (1) | WT | L (2) | M (48) | WT | WT | WT | WT | V (31) | V (3) | WT | V (4) |
| 139 | I (2) | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| 144 | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
WT, wild type of HBV.
FIG 1.
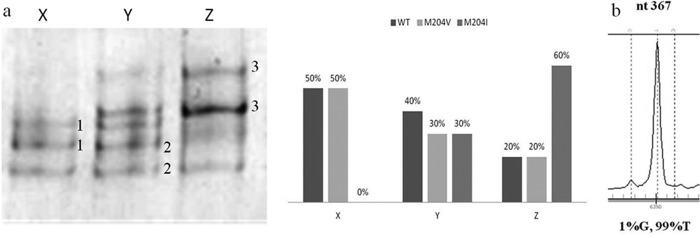
(a) YMDD variants with their frequencies identified by MSSCP and MALDI-TOF MS for 3 patients, 1-YMDD, 2-YVDD, and 3-YIDD. (b) MALDI-TOF MS capability to detect mutant strains with an allelic frequency of 1%. nt, nucleotide.
The detection limit of MSSCP after nested PCR was 1 copy/μl (Fig. 2a). The best results were achieved on 11% polyacrylamide gel with addition of 5% glycerol and three temperatures of 15°C, 10°C, and 5°C for 900 V × h each. Different band patterns were obtained for different variants of viral YMDD (rt204), including mixtures of variants (Fig. 2b). Band patterns after sequencing and reamplification were used as a standards for further analysis. Using the MSSCP technique, we found 98% of the YMDD mutants detected by MALDI-TOF MS (Fig. 1a). The mutant type of rt204 was detected in up to 2% of the total virus population (Table 2).
FIG 2.
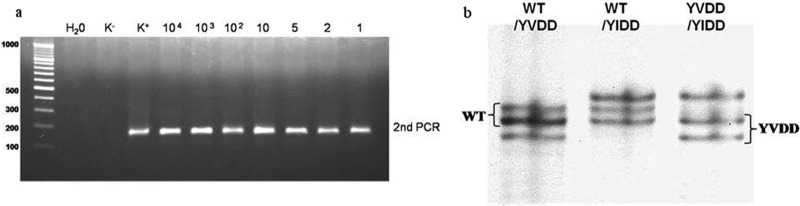
(a) Detection limit after second round of PCR (K−, negative control; K+, positive control; HBV DNA, external quality control [PeliSpy PRO; AcroMetrix]) and (b) band patterns for mixtures of YMDD variants after optimized MSSCP (WT, YMDD; YVDD, M204V; YIDD, M204I).
TABLE 2.
Assessment of efficacies of MALDI-TOF MS, MSSCP, INNOLiPA HBV DR, and direct sequencing assays in detection of the HBV polymorphism YMDD motif
| YMDD motif (amino acid)a | No. of motifs with the indicated method of YMDD polymorphism detection (n = 82) |
|||
|---|---|---|---|---|
| MALDI-TOF MS | MSSCP | INNOLiPA HBV DR | Direct sequencing | |
| M | 40 | 40 | 65 | 77 |
| V | 14 | 14 | 8 | 3 |
| I | 10 | 10 | 4 | 2 |
| M/V | 12 | 12 | 3 | 0 |
| M/I | 2 | 2 | 1 | 0 |
| I/V | 3 | 4 | 1 | 0 |
| M/I/V | 1 | 0 | 0 | 0 |
HBV wild type, amino acid M.
Eighty-seven samples were tested with INNOLiPA HBV DR. Six out of 87 samples gave no band on 1.5% agarose gel. For these samples, a repeat extraction was performed. After amplification only one sample was amplified and resulted in an interpretable hybridization test. With the use of the INNOLiPA assay we were able to detect 40% of the YMDD motifs and 58% of all HBV variants in the study population (Table 2). We were able to detect combinations of wild and mutant variants of HBV strains if variants were present at a concentration higher than 5% of the total virus population. With the INNOLiPA assay, it was possible to detect only 28% of all minor HBV variants that were detectable with the MassARRAY system.
Direct sequencing did not identify specific genotypes with a double mutation in one codon. Using direct sequencing, we detected only 11% of the YMDD variants and 34% of all HBV variants (Table 2). Such results show the inability of direct sequencing to correctly determine the variants present in the sample.
The HBV drug-resistant variants in treatment-naive patients were detected at codons rt80, rt173, rt180, rt181, rt204, rt233, rt236, and rt250 (Table 3). The most common HBV variants were L80I (62%), I233V (81%), M250V (76%), and M204V and -I (68%), of which M204V was the more frequent (60%). Additional HBV variants occurring at one codon were present with an allelic frequency of <5%. Only one patient had a single wild-type HBV variant at all codons (Table 1). In the group of experienced patients, drug resistance mutations were detected additionally at codons rt169, rt194, and rt184. The most common mutations were L80I (50%), L180M (54%), I233V (75%), and M204V and -I (92%), of which M204I appeared more often (68%). All treated patients possessed mutations causing drug resistance, with an allelic frequency at least 2%. Based on the results obtained from MALDI-TOF MS analysis, a retrospective study of hepatitis B virus (HBV) resistance mutations in the study groups was done (Table 3).
TABLE 3.
Comparison of MALDI-TOF MS, INNOLiPA DR, and direct sequencing in detection of HBV drug-resistant variants in selected treatment-naive and experienced patients
| Patient no. | Therapy experience | Drug-resistant variant(s) found with the indicated method of HBV reverse transcriptase polymorphism detection |
||
|---|---|---|---|---|
| MALDI-TOF MS | INNOLiPA HBV DR | Direct sequencing | ||
| 21 | Experienced | L180M, T184S, M204I, I233V | L180M, M204V | WTa |
| 23 | Experienced | L80I, V173L, A 181 T, M204I, I233V, M250V | M204I | WT |
| 28 | Experienced | L80V, V173L, L180M, M204V, I233V, M250V | L180M, M204V | WT |
| 30 | Naive | L80I, M204V, I233V | WT | WT |
| 32 | Experienced | L80I, L180M, A181T, M204V | WT | WT |
| 101 | Naive | L80I, N236T | WT | WT |
| 115 | Naive | N236T | WT | WT |
| 119 | Naive | I233V, M250V | WT | WT |
| 124 | Experienced | L80I, N236T | WT | WT |
| 125 | Naive | I233V, I250V | WT | WT |
| 126 | Experienced | L180M, A181T, M204V, I233V, M250V | L80 M, M204V | WT |
| 127 | Naive | L80I, V173L, I233V, M250V | WT | WT |
| 133 | Naive | L80I, M204I | WT | WT |
| 134 | Experienced | I169P, L180M, T184A, M204V | M204V, L180M | WT |
| 143 | Experienced | L80I, L180M, A181T, M204I, I233V | L80I, M204I | WT |
WT, wild type of HBV.
DISCUSSION
The high replication rate of HBV together with the absence of an effective proofreading mechanism in HBV polymerase contributes significantly to the development of drug resistance mutations and treatment failure (36). Despite the constant searching for new antiviral drugs, CHB remains a clinical challenge. Although emergence of drug-resistant strains is closely related to the duration of the antiviral treatment, such variants have also been shown to exist in treatment-naive CHB patients. Therefore, faithful analysis of all mutations associated with antiviral resistance is essential for optimal management of CHB (37).
Currently, the identification of resistant HBV strains in hepatitis B patients is usually performed by direct sequencing and reverse hybridization (INNOLiPA) (9). In our study we compared these two methods with a new, non-gel-based MALDI-TOF MS technique for the detection of drug resistance mutations in CHB patients. Moreover, we designed a new method based on multitemperature single-strand conformation polymorphism for identifying YMDD variants of HBV. The results obtained by MSSCP were compared with those from direct sequencing, INNOLiPA assay, and MALDI-TOF MS.
The ability of MALDI-TOF MS to detect minor mutations was superior to that of direct sequencing and reverse hybridization. Literature data show that direct sequencing of PCR products does not detect minor virus populations that comprise <20% of the total viral population. In case of the INNOLiPA assay, it is possible to detect drug-resistant variants at frequencies as low as 5% in mixed viral populations (21, 22). In our study, direct sequencing did not identify specific genotypes with a double mutation in one codon, whereas INNOLiPA HBV DR detected 58% of all variants that were present in all 82 samples compared with a mass spectrometry method (100%). What is more, 5 of 87 samples could not be analyzed because of the low viral load (<104 copies/ml, <1,718 IU/ml) and a minor mutation was identified only in 28% of cases.
The HBV variants of the YMDD motif detected by MSSCP were concordant with 98% of those detected by MALDI-TOF MS. There was only one case for which MSSCP detected a mixture of YVDD and YIDD, whereas during mass spectrometry analysis a wild-type (YMDD) motif was identified as well. The percentage of wild-type virus in this sample was 1%, which is below the detection limit of MSSCP. These results suggest that MSSCP can be a sensitive and high-accuracy technique for detecting drug-resistant HBV strains. However, it requires time-consuming primer design and experimental optimization for each codon and is still laborious and far from being an automated method.
The most probable cause of such huge differences between routine tests and MALDI-TOF MS in detecting minor HBV mutations is that almost half of the samples were taken from treatment-naive patients. Recently, it has been shown that there is a natural prevalence of HBV reverse transcriptase amino acid substitutions in treatment-naive patients with CHB, which may decrease susceptibility to available oral antiviral drugs (38, 39). This study shows that the presence of minor mutations in treatment -naive patients is common. However, the frequency of these mutations in the total HBV population is often below the detection limit of routine detection methods. Similar situations appear in experienced patients. On the basis of the findings of this study, new genotypic tests of high sensitivity may be recommended before the start of therapy in some treatment-naive patients, especially in treatment regimens for low genetic barrier NAs. The monitoring of drug resistance during treatment could bring benefits in the treatment of difficult subjects (such as patients who are immunocompromised or have had previous therapy failure).
MALDI-TOF mass spectrometry seems to be an ideal assay for the discrimination of single point mutations within the HBV genome. Compared with other techniques, it has high accuracy, sensitivity and the capability to detect mutant strains within a mixed viral population occurring with an allelic frequency on the level of 1% (24). The newly available ultradeep pyrosequencing (UDPS) technique, currently the most competitive for mass spectrometry, detects HBV drug-resistant strains in plasma samples if they show HBV DNA levels ≥103 IU/ml, which is 10 times higher than those detected using MALDI-TOF MS. Moreover, several drug resistance mutations were detected at frequencies of >1% (40, 41). Thus, high-throughput MALDI-TOF MS provides an alternative approach to HBV genotyping. Using such a sensitive method for monitoring drug resistance in CHB patients during antiviral therapy allows the change from an applied drug to a new one that is effective before an outbreak of viral replication. When choosing new treatment strategies, antiviral-resistant mutants should be identified by means of sensitive HBV DNA monitoring as early as possible before clinical breakthrough of the disease.
ACKNOWLEDGMENTS
This work was supported by grant numbers N N302 183639 from the Ministry of Science and Higher Education and ST79 from the Medical University of Gdansk.
Magda Rybicka is the recipient of financial support from the European Social Fund in the framework of the project “InnoDoktorant”—Scholarships for PhD students, 5th edition.
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Seeger C, Mason WS. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51–68. 10.1128/MMBR.64.1.51-68.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielawski KP, Dybikowska A, Lisowska-Charmuszko U, Stalke P, Gregorowicz K, Trocha H, Podhajska A. 2004. Distribution of HBV geno-types and mutants among hepatitis B infected patients from northern Poland. Int. J. Mol. Med. 14:301–304 [PubMed] [Google Scholar]
- 3.World Health Organization 2013. WHO HBV fact sheet no. 204, revised July 2012. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- 4.Bielawski KP, Stalke P. 2005. Molecular epidemiology of chronic hepatitis B virus infection in northern Poland. J. Clin. Virol. 34(Suppl 1):S63–S69. 10.1016/S1386-6532(05)80012-5 [DOI] [PubMed] [Google Scholar]
- 5.Chen CJ, Yang HI. 2011. Natural history of chronic hepatitis B REVEALed. J. Gastroenterol. Hepatol. 26:628–638. 10.1111/j.1440-1746.2011.06695.x [DOI] [PubMed] [Google Scholar]
- 6.Cai D, Mills C, Yu W, Yan R, Aldrich CE, Saputelli JR, Mason WS, Xu X, Guo JT, Block TM, Cuconati A, Guo H. 2012. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob. Agents Chemother. 56:4277–4288. 10.1128/AAC.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locarnini S, Mason WS. 2006. Cellular and virological mechanisms of HBV drug resistance. J. Hepatol. 44:422–431. 10.1016/j.jhep.2005.11.036 [DOI] [PubMed] [Google Scholar]
- 8.Samal J, Kandpal M, Vivekanandan P. 2012. Molecular mechanisms underlying occult hepatitis B virus infection. Clin. Microbiol. Rev. 25:142–163. 10.1128/CMR.00018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoulim F. 2011. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 31(Suppl 1):111–116. 10.1111/j.1478-3231.2010.02399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielawski KP, Charmuszko U, Dybikowska A, Stalke P, Podhajska AJ. 2006. Genetic variability of hepatitis B virus isolates in Poland. Virus Genes 33:77–86. 10.1007/s11262-005-0040-x [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi S, Ide T, Sata M. 2001. Detection of YMDD motif mutations in some lamivudine-untreated asymptomatic hepatitis B virus carriers. J. Hepatol. 34:584–586. 10.1016/S0168-8278(00)00023-4 [DOI] [PubMed] [Google Scholar]
- 12.Rybicka M, Stalke P, Charmuszko U, Bielawski KP. 2011. The influence of hepatitis B virus polymorphism on the progression of chronic liver disease. Postepy. Hig. Med. Dosw. 65:244–254 (In Polish.) 10.5604/17322693.939665 [DOI] [PubMed] [Google Scholar]
- 13.Zoulim F. 2004. Resistance to antiviral drugs, p 43–52 In Honore S. (ed), Virology of hepatitis B, 1st ed, vol 1 Elsevier SAS, Paris, France [Google Scholar]
- 14.Kim SS, Cho SW, Kim SO, Hong SP, Cheong JY. 2013. Multidrug-resistant hepatitis B virus resulting from sequential monotherapy with lamivudine, adefovir, and entecavir: clonal evolution during lamivudine plus adefovir therapy. J. Med. Virol. 85:55–64. 10.1002/jmv.23440 [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Fan R, Sun J, Hou J. 2012. Prevention and management of drug resistant hepatitis B virus infections. J. Gastroenterol. Hepatol. 27:1432–1440. 10.1111/j.1440-1746.2012.07198.x [DOI] [PubMed] [Google Scholar]
- 16.Angus P, Vaughan R, Xiong S, Yang H, Delaney W, Gibbs C. 2003. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology 125:292–297. 10.1016/S0016-5085(03)00939-9 [DOI] [PubMed] [Google Scholar]
- 17.Marcellin P, Jacobson I, Habersetzer F, Senturk H, Andreone P, Moyes C, Horban A, Teuber G, Sorbel J, Mandou J, Quinn J, Rousseau F. 2008. Tenofovir disoproxil fumarate (TDF) for the treatment of HBeAg-negative chronic hepatitis B: week 72 TDF data and week 24 adefovir dipivoxil switch data (study 102). J. Hepatol. 48(Suppl 2):S26. 10.1016/S0168-8278(08)60059-8 [DOI] [Google Scholar]
- 18.Zoulim F. 2005. Combination of nucleoside analogues in the treatment of hepatitis B chronic infection: lesson from experimental models. J. Antimicrob. Chemother. 55:608–611. 10.1093/jac/dki095 [DOI] [PubMed] [Google Scholar]
- 19.Zoulim F, Locarnini S. 2009. Hepatitis B virus resistance to nucleos (t) ide analogues. Gastroenterology 137:1593–1608. e1-2. 10.1053/j.gastro.2009.08.063 [DOI] [PubMed] [Google Scholar]
- 20.Zoulim F, Perrillo R. 2008. Hepatitis B: reflections on the current approach to antiviral therapy. J. Hepatol. 48(Suppl 1):S2–S19. 10.1016/j.jhep.2008.01.011 [DOI] [PubMed] [Google Scholar]
- 21.Keeffe EB, Dieterich DT, Pawlotsky JM, Benhamou Y. 2008. Chronic hepatitis B: preventing, detecting, and managing viral resistance. Clin. Gastroenterol. Hepatol. 6:268–274. 10.1016/j.cgh.2007.12.043 [DOI] [PubMed] [Google Scholar]
- 22.Paik YH, Han KH, Hong SP. 2006. The clinical impact of early detection of YMDD mutants on the outcomes of long-term lamivudine therapy in patients with chronic hepatitis B. Antivir. Ther. 11:447–455 [PubMed] [Google Scholar]
- 23.Kaczanowski R, Trzeciak L, Kucharczyk K. 2001. Multitemperature single-strand conformation polymorphism. Electrophoresis 22:3539–3545. [DOI] [PubMed] [Google Scholar]
- 24.Jurinke C, Oeth P, van den Boom D. 2004. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol. Biotechnol. 26:147–164. 10.1385/MB:26:2:147 [DOI] [PubMed] [Google Scholar]
- 25.Bielawski KP, Al-Soud WA, Stalke P, Charmuszko U, Wadstrom T. 2008. Determination of lamivudine-resistant variants of hepatitis B virus by denaturing gradient gel electrophoresis: a novel approach to monitoring drug resistance. Med. Sci. Monit. 14:CR281–CR285 [PubMed] [Google Scholar]
- 26.Luan J, Yuan J, Li X, Jin S, Yu L, Liao M. 2009. Multiplex detection of 60 hepatitis B virus variants by MALDI-TOF mass spectrometry. Clin. Chem. 55:1503–1509. 10.1373/clinchem.2009.124859 [DOI] [PubMed] [Google Scholar]
- 27.Warner N, Locarnini S, Kuiper M, Bartholomeusz A, Ayres A, Yuen L, Shaw T. 2007. The L80I substitution in the reverse transcriptase domain of the hepatitis B virus polymerase is associated with lamivudine resistance and enhanced viral replication in vitro. Antimicrob. Agents Chemother. 51:2285–2292. 10.1128/AAC.01499-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L. 2004. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to lamivudine. Antimicrob. Agents Chemother. 48:3498–3507. 10.1128/AAC.48.9.3498-3507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney WE, Yang H, Westland CE, Das K, Arnold E, Gibbs CS. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77:11833–11841. 10.1128/JVI.77.21.11833-11841.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Invest. 107:449–455. 10.1172/JCI11100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheldon J, Camino N, Rodes B, Bartholomeusz A, Kuiper M, Tacke F. 2005. Selection of hepatitis B virus polymerase mutations in HIV-coinfected patients treated with tenofovir. Antivir. Ther. 10:727–734 [PubMed] [Google Scholar]
- 32.Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology 27:1670–1677. 10.1002/hep.510270628 [DOI] [PubMed] [Google Scholar]
- 33.Bozdayi AM, Uzunalimoglu O, Turkyilmaz AR, Aslan N, Sezgin O, Sahin T. 2003. YSDD: a novel mutation in HBV DNA polymerase confers clinical resistance to lamivudine. J. Viral Hepat. 10:256–265. 10.1046/j.1365-2893.2003.00435.x [DOI] [PubMed] [Google Scholar]
- 34.Schildgen O, Sirma H, Funk A, Olotu C, Wend UC, Hartmann H. 2006. Variant of hepatitis B virus with primary resistance to adefovir. N. Engl. J. Med. 354:1807–1812. 10.1056/NEJMoa051214 [DOI] [PubMed] [Google Scholar]
- 35.Niesters HG, Zoulim F, Pichoud C, Buti M, Shapiro F, D'Heuvaert N, Celis L, Doutreloigne J, Sablon E. 2010. Validation of the INNO-LiPA HBV DR assay (version 2) in monitoring hepatitis B virus-infected patients receiving nucleoside analog treatment. Antimicrob. Agents Chemother. 54:1283–1289. 10.1128/AAC.00970-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung J, Lai CL, Seto WK, Yuen MF. 2011. Nucleoside/nucleotide analogues in the treatment of chronic hepatitis B. J. Antimicrob. Chemother. 66:2715–2725. 10.1093/jac/dkr388 [DOI] [PubMed] [Google Scholar]
- 37.Han KH, Sun Hong P, Choi SH, Shin SK, Cho SW, Ahn SH, Hahn JS, Kim SO. 2011. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir. Ther. 16:77–87. 10.3851/IMP1702 [DOI] [PubMed] [Google Scholar]
- 38.Singla B, Chakraborti A, Sharma BK, Kapil S, Chawla Arora YKSK, Das A, Dhiman RK, Duseja A. 2013. Hepatitis B virus reverse transcriptase mutations in treatment Naïve chronic hepatitis B patients. J. Med. Virol. 85:1155–1162. 10.1002/jmv.23608 [DOI] [PubMed] [Google Scholar]
- 39.Tan YW, Ge GH, Zhao W, Gan JH, Zhao Y, Niu ZL, Zhang DJ, Chen L, Yu XJ, Yang LJ. 2012. YMDD motif mutations in chronic hepatitis B antiviral treatment naïve patients: a multi-center study. Braz. J. Infect. Dis. 16:250–255. 10.1590/S1413-86702012000300006 [DOI] [PubMed] [Google Scholar]
- 40.Ko SY, Oh HB, Park CW, Lee HC, Lee JE. 2012. Analysis of hepatitis B virus drug-resistant mutant haplotypes by ultra-deep pyrosequencing. Clin. Microbiol. Infect. 18:404–411. 10.1111/j.1469-0691.2012.03951.x [DOI] [PubMed] [Google Scholar]
- 41.Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, Holmes SP, Babrzadeh F, Gharizadeh B, Hanczaruk B, Simen BB, Egholm M, Shafer RW. 2009. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naïve patients. J. Infect. Dis. 199:1275–1285. 10.1086/597808 [DOI] [PMC free article] [PubMed] [Google Scholar]


