Abstract
Atopobium species are Gram-positive, anaerobic, catalase-negative, fastidious bacteria belonging to the family Coriobacteriaceae. We report the isolation of an Atopobium-like species in a patient with Fournier's gangrene and highlight the role of 16S rRNA gene sequencing in the identification of fastidious organisms in the clinical laboratory.
CASE REPORT
A 43-year-old mentally retarded, obese woman, without relevant medical history, was admitted to the hospital with fever and an extensive perianal abscess with spontaneous drainage during 1 week. Clinical examination showed an erythematous, swollen, warm perianal region with ulcers and regional lymphadenopathy. Hematological investigations revealed a white blood cell (WBC) count of 34.73 × 103 cells/μl (reference range of 3.90 × 103/μl to 11.10 × 103/μl) with 91% neutrophils (reference range of 46 to 64%), a hemoglobin level of 11.5 g/dl (reference range of 11.8 to 14.8 g/dl), and a platelet count of 418 × 103/μl (reference range of 150 × 103/μl to 450 × 103/μl). C-reactive protein was increased up to 448.0 mg/liter (reference range of 0 to 7.0 mg/liter). Biochemical testing revealed increased glucose (154 mg/dl [reference range of 65 to 110 mg/dl]), decreased serum iron (13 μg/dl [reference range of 50 to 150 μg/dl]), and decreased serum transferrin (194 mg/dl [reference range of 200 to 360 mg/dl]). Liver function tests showed increased alkaline phosphatase (178 U/liter [reference range of 42 to 98 U/liter]), gamma-glutamyltransferase (65 U/liter [reference range of 7 to 32 U/liter]), aspartate transaminase (AST) (51 U/liter [reference range of 10 to 35 U/liter]), and alanine aminotransferase (ALT) (56 U/liter [reference range of 10 to 35 U/liter]) enzymes. There were signs of prerenal kidney failure (urea, 93 mg/dl [reference range of <50 mg/dl]; creatinine, 1.70 mg/dl [reference range of 0.51 to 0.95 mg/dl]).
Before empirical antibiotic treatment with amoxicillin-clavulanic acid (1 g every 6 hours [q6h] intravenously [i.v.]) was initiated, two aerobic and two anaerobic blood culture bottles (Bactec FX; Becton, Dickinson, Sparks, MD) were drawn, and the patient was transferred to the operation room for incision of an ischiorectal abscess. During this procedure, a diagnosis of Fournier's gangrene (FG) was made, followed by an extended surgical debridement of all necrotic and infectious tissue. Purulent fluid was obtained and sent to the laboratory for microbiological examination. Gram staining of the pus showed predominantly Gram-positive rods and cocci. Specimens were plated onto homemade Columbia sheep blood agar and commercial chocolate agar (bioMérieux, Marnes-la-Coquette, France), both incubated in anaerobic (80% N2, 10% H2, 10% CO2) and aerobic conditions at 37°C. After 48 h of incubation, catalase-negative, anaerobic, Gram-positive rods and two types of Gram-positive cocci were isolated. The cocci were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Microflex; Brüker, Bremen, Germany) as vancomycin-susceptible Enterococcus faecalis and Staphylococcus epidermidis. After a mean incubation time of 82.7 h at 35°C, both anaerobic blood culture bottles yielded short, rod-shaped, Gram-positive, catalase-negative bacteria. At this time, antibiotic therapy was switched to meropenem (2 g q8h i.v.). The strain was tested using the commercially available API RAPID 32A test (bioMérieux), which produced a presumptive identification of Bacillus species with poor probability (53.1%) of correct identification. MALDI-TOF MS identification of the Gram-positive rods revealed no clear match with any of the species in the database, with the best identification score being 1.304 for Bacillus cereus.
Because conventional identification was inconclusive, 16S rRNA gene sequencing was performed. Bacterial DNA extraction, PCR amplification, and 16S rRNA gene sequencing were performed as described in previous studies (1) using abNOT (forward primer) (5′-GGTATCGCTGCTGCTCTGC-3′) and omegaMB (reverse primer) (5′-AGCCAACGTACCCAGGTC-3′) as the PCR and sequencing primers (2). The obtained sequence was compared with the publically available bacterial 16S rRNA gene sequences deposited in GenBank using the Basic Local Alignment Search Tool (BLAST) software (www.ncbi.nlm.nih.gov/BLAST/). There was only 95% nucleotide identity between the 16S rRNA gene sequence of the isolate and that of Atopobium minutum. Because the closest relatives (95%) were found to be Atopobium minutum strains, all available complete 16S rRNA gene sequences of Atopobium minutum, Atopobium fossor, Atopobium parvulum, Atopobium rimae, and Atopobium vaginae were aligned with the obtained sequence and with the 16S rRNA sequence of Coriobacterium glomerans, the closest relative of the genus Atopobium (3), as an outgroup using the online Clustal Omega Software (http://www.ebi.ac.uk/Tools/msa/clustalo) with default parameters. After alignment, the gaps at the 5′ and 3′ ends of the alignment were omitted from further analysis. The alignment was subsequently used to calculate evolutionary distances by Kimura's two-parameter model (4) using the MEGA 5.2.2 software package (http://www.megasoftware.net) (5). Phylogenetic trees were constructed using the neighbor-joining (6), maximum parsimony (7), and maximum likelihood (8) methods, utilizing Coriobacterium glomerans PW2T (9) as the outgroup.
Antimicrobial susceptibility of this Atopobium species was determined according to the protocol described by De Backer et al. (10). Briefly, an inoculum taken from Columbia agar was suspended in 0.5 ml of physiological water and adjusted to a turbidity of 1 McFarland standard. Before the plates were inoculated and testing was done for the described antibiotics, plates were reduced in an anaerobic environment for 24 h prior to the Etests being set up. EUCAST clinical breakpoints for Gram-positive anaerobic bacteria were applied (http://www.eucast.org). The following MIC results were obtained: penicillin, 0.016 μg/ml, susceptible (clinical breakpoint, 0.25 μg/ml); meropenem, 0.012, susceptible (clinical breakpoint, 2 μg/ml); metronidazole, 24 μg/ml, resistant (clinical breakpoint, 4 μg/ml); clindamycin, <0.016 μg/ml, susceptible (clinical breakpoint, 4 μg/ml); vancomycin, 0.5 μg/ml, susceptible (clinical breakpoint, 2 μg/ml); cefotaxime, 0.016 μg/ml (no clinical breakpoint available).
The patient responded very well after surgery with meropenem and topical application of povidone iodine. Clinical and biochemical signs of infection and fever disappeared gradually, and the patient was discharged in good health 9 weeks after admission. A checkup after 25 weeks revealed the patient to be symptom free.
FG is a fulminant form of infective necrotizing fasciitis of the perineal, genital, or perianal regions. Men are more frequently affected than women and children (11). Early surgical debridement of necrotic tissues and administration of antibiotics are fundamental in the treatment of FG. Mostly, a polymicrobial infection of genitourinary or perianal source is found in these patients, but the portal of entry is often difficult to establish. Microbial invasion usually occurs either through direct injury or through a direct spread from the urogenital tract or from a perforated viscus like the colon, rectum, or anal orifice. In a meta-analysis, the portal of entry was found to be dermatological in 24%, colorectal in 21%, and urological in 19% of cases, whereas in 36% of the cases no definite portal of entry was established (12). The portal of entry in our case was thought to be perirectal.
In practice, three types of necrotizing soft tissue infections are seen. Type I infections, such as FG, are polymicrobial in origin, and a combination of Gram-positive and Gram-negative bacteria along with anaerobes can be cultured. Type II infections are monomicrobial and are mostly caused by Streptococcus pyogenes (group A streptococci) but may be associated with Staphylococcus aureus. Type II is less common than type I and is usually seen in healthy, immunocompetent patients. Type III infections are usually caused by Vibrio vulnificus (13).
Predisposing factors for FG include diabetes mellitus, which is reported to be present in 20 to 70% of patients with FG, and chronic alcoholism, which is found in 25 to 50% of the patients (14). In our patient, neither of these conditions was present. Isolation of Atopobium species has only rarely been recognized in patients with necrotizing fasciitis (15). The lack of hygiene and social neglect together with morbid obesity and mental retardation are possible causes that have given rise to this case of FG. Our patient was undoubtedly septic, with the probable cause of the sepsis being the infected necrotic perianal ulcers. No single bacterial species was exclusively responsible for the sepsis. Interestingly, the Atopobium species was the only bacterial pathogen identified in both blood cultures and perioperatively obtained pus. Therefore, its virulence and pathogenic role in the clinical sepsis are strongly suggested.
Phylogenetic analysis based on the 16S rRNA sequences of Atopobium strain HHRM1715 and the 12 available Atopobium sequences indicate that Atopobium strain HHRM1715 forms a distinct phylogenetic lineage within the genus Atopobium (Fig. 1), most closely related to the lineage comprising Atopobium fossor and Atopobium minutum strains. Similar tree topologies were generated using the maximum likelihood and the maximum parsimony algorithms (trees not shown).
FIG 1.
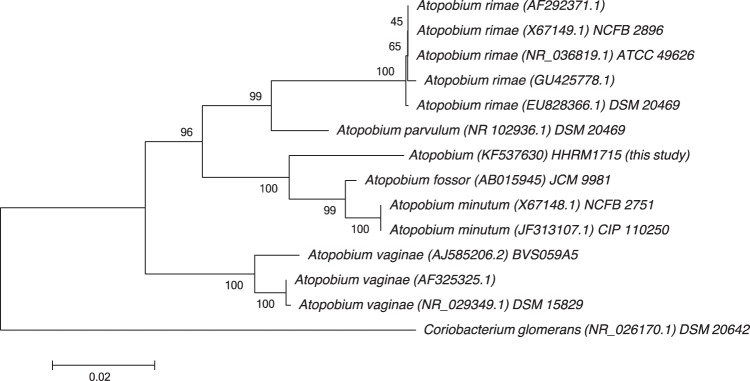
Phylogenetic tree based on 16S rRNA gene sequences of Atopobium species strain HHRM1715 and 16S rRNA gene sequences of the genus Atopobium. Coriobacterium glomerans, the closest relative of the genus, was used as an outgroup. Evolutionary distances were computed using Kimura's two-parameter model. The tree was constructed using the neighbor-joining method. Bootstrap values are expressed as percentages of 1,000 replications. Scale bar represents 0.02 substitutions per nucleotide position. Accession numbers are shown between brackets, followed by strain name if applicable.
The genus Atopobium was described as a new bacterial genus in 1992 with reclassification of the species Lactobacillus minutum, Lactobacillus rimae, and Streptococcus parvulus based on their phylogenetic relatedness determined by 16S rRNA gene sequencing (16). Atopobium species are found in human gingival crevices and rarely have been described in various human infections, including dental abscesses, abdominal wound infections, pelvic abscesses, and bacteremia. Mostly, these bacteria were found associated with other microorganisms (3, 17, 18). Currently, the genus consists of five species: A. rimae, A. parvulum, A. minutum, A. fossor, and A. vaginae (3, 16, 19). A. vaginae was first isolated from the vaginal flora in a healthy woman in 1999 (3). Subsequently, the bacterium has also been found in patients with bacterial vaginosis, usually cooccuring with other anaerobic bacteria like Gardnerella vaginalis (20, 21). As in our experience, identification and differentiation of Atopobium species from other non-spore-forming Gram-positive coccobacilli by the use of conventional phenotypic and biochemical tests, including the commercially available test kits, are often laborious and may carry the risk of misidentification (22–25). The identification process is especially difficult in the presence of coexisting colonizing organisms (25). Newer identification techniques, particularly 16S rRNA gene sequencing, have allowed the accurate and rapid identification of Atopobium species, like A. vaginae (23, 24, 26). MALDI-TOF MS has also been used with success in one previously reported case (24) but was inaccurate in our case, which was due to the fact that our species was not included in the databases.
In conclusion, this case demonstrated that Atopobium species must be considered a potential pathogenic microorganism that can lead to sepsis in patients with Fournier's gangrene. Commercial identification as well as more advance identification systems may fail to identify or misidentify Atopobium species. In such situations, 16S rRNA gene sequencing may be considered a useful alternative method for rapid and accurate identification of the organism.
Nucleotide sequence accession number.
The GenBank accession number for the partial 16S rRNA gene sequence of Atopobium species strain HHRM1715 is KF537630.
ACKNOWLEDGMENT
There are no conflicts of interests to declare.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Cools P, Haelters J, Santiago GLS, Claeys G, Boelens J, Leroux-Roels I, Vaneechoutte M, Deschaght J. 2013. Edwardsiella tarda sepsis in a live-stranded sperm whale (Physeter macrocephalus). Vet. Microbiol. 166:311–315. 10.1016/j.vetmic.2013.05.020 [DOI] [PubMed] [Google Scholar]
- 2.Vaneechoutte M, Claeys G, Steyaert S, De Baere T, Peleman R, Verschraegen G. 2000. Isolation of Moraxella canis from an ulcerated metastatic lymph node. J. Clin. Microbiol. 38:3870–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez Jovita M, Collins MD, Sjöden B, Falsen E. 1999. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. Int. J. Syst. Bacteriol. 49:1573–1576. 10.1099/00207713-49-4-1573 [DOI] [PubMed] [Google Scholar]
- 4.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 5.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 7.Fitch Walter M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Biol. 20:406–416. 10.1093/sysbio/20.4.406 [DOI] [Google Scholar]
- 8.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368–376. 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- 9.Haas F, Konig H. 1988. Coriobacterium glomerans gen. nov. spp. nov. from the intestinal tract of the red soldier bug. Int. J. Syst. Bacteriol. 38:382–384. 10.1099/00207713-38-4-382 [DOI] [Google Scholar]
- 10.De Backer E, Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Temmerman M, Vaneechoutte M. 2006. Antibiotic susceptibility of Atopobium vaginae. BMC Infect. Dis. 6:51. 10.1186/1471-2334-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thwaini A, Khan A, Malik A, Cherian J, Barua J, Shergill I, Mammen K. 2006. Fournier's gangrene and its emergency management. Postgrad. Med. J. 82:516–519. 10.1136/pgmj.2005.042069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eke N. 2000. Fournier's gangrene: a review of 1726 cases. Br. J. Surg. 87:718–728. 10.1046/j.1365-2168.2000.01497.x [DOI] [PubMed] [Google Scholar]
- 13.Sarani B, Strong M, Pascual J, Schwab CW. 2009. Necrotizing fasciitis: current concepts and review of the literature. J. Am. Coll. Surg. 208:279–288. 10.1016/j.jamcollsurg.2008.10.032 [DOI] [PubMed] [Google Scholar]
- 14.Vick R, Carson CC. 1999. Fournier's disease. Urol. Clin. North. Am. 26:841–849. 10.1016/S0094-0143(05)70224-X [DOI] [PubMed] [Google Scholar]
- 15.Marvaud JC, Mory F, Lambert T. 2011. Clostridium clostridioforme and Atopobium minutum clinical isolates with VanB-type resistance in France. J. Clin. Microbiol. 49:343–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins MD, Wallbanks S. 1992. Comparative sequence analyses of the 16S rRNA genes of Lactobacillus minutus, Lactobacillus rimae and Streptococcus parvulus: proposal for the creation of a new genus Atopobium. FEMS Microbiol. Lett. 74:235–240 [DOI] [PubMed] [Google Scholar]
- 17.Olson I, Johnson JL, Moore LVH, Moore WEC. 1991. Lactobacillus uli sp. nov. and Lactobacillus rimae sp. nov. from the human gingival crevice and emended descriptions of Lactobacillus minutus and Streptococcus parvulus. Int. J. Syst. Bacteriol. 41:261–266. 10.1099/00207713-41-2-261 [DOI] [PubMed] [Google Scholar]
- 18.Cools P, Verstraelen H, Vaneechoutte M, Verhelst R. 2011. Atopobium, p 31–43 In Liu D, Molecular detection of human bacterial pathogens, 1st ed. CRC Press, Boca Raton, FL [Google Scholar]
- 19.Kageyama A, Benno Y, Nakase T. 1999. Phylogenic and phenotypic evidence for the transfer of Eubacterium fossor to the genus Atopobium as Atopobium fossor comb. nov. Microbiol. Immunol. 43:389–395 [DOI] [PubMed] [Google Scholar]
- 20.Bradshaw CS, Tabrizi SN, Fairly CK, Morton AN, Rudland E, Garland SM. 2006. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J. Infect. Dis. 194:828–836. 10.1086/506621 [DOI] [PubMed] [Google Scholar]
- 21.Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L, De Ganck C, Temmerman M, Vaneechoutte M. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae with bacterial vaginosis. Am. J. Obstet. Gynecol. 191:1130–1132. 10.1016/j.ajog.2004.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferris MJ, Masztal A, Aldridge KE, Fortenbert JD, Fidel PL, Martin DH. 2004. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect. Dis. 4:5. 10.1186/1471-2334-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissdörfer W, Böhmer C, Pelz K, Schoerner C, Frobenius W, Bogdan C. 2003. Tuboovarian abscess caused by Atopobium vaginae following transvaginal oocyte recovery. J. Clin. Microbiol. 41:2788–2790. 10.1128/JCM.41.6.2788-2790.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoester M, Lashley LE, Wessels E, Oepkes D, Kuijper EJ. 2011. First report of Atopobium vaginae bacteremia with fetal loss after chorionic villus sampling. J. Clin. Microbiol. 49:1684–1686. 10.1128/JCM.01655-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan J, Lau S, Curreem S, To K, Leung S, Cheng V, Yuen K, Woo P. 2012. First report of spontaneous intrapartum Atopobium vaginae bacteremia. J. Clin. Microbiol. 50:2525–2528. 10.1128/JCM.00212-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagishi Y, Mikamo H, Tanaka K, Watanabe K. 2011. A case of uterine endometritis caused by Atopobium vaginae. J. Infect. Chemother. 17:119–121. 10.1007/s10156-010-0100-6 [DOI] [PubMed] [Google Scholar]


