Abstract
The rapidly growing mycobacterium M. abscessus sensu lato is the causative agent of emerging pulmonary and skin diseases and of infections following cosmetic surgery and postsurgical procedures. M. abscessus sensu lato can be divided into at least three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii. Clinical isolates of rapidly growing mycobacteria were previously identified as M. abscessus by DNA-DNA hybridization. More than 30% of these 117 clinical isolates were differentiated as M. abscessus subsp. massiliense using combinations of multilocus genotyping analyses. A much more cost-effective technique to distinguish M. abscessus subsp. massiliense from M. abscessus subsp. abscessus, a multiplex PCR assay, was developed using the whole-genome sequence of M. abscessus subsp. massiliense JCM15300 as a reference. Several primer sets were designed for single PCR to discriminate between the strains based on amplicons of different sizes. Two of these single-PCR target sites were chosen for development of the multiplex PCR assay. Multiplex PCR was successful in distinguishing clinical isolates of M. abscessus subsp. massiliense from samples previously identified as M. abscessus. This approach, which spans whole-genome sequencing and clinical diagnosis, will facilitate the acquisition of more-precise information about bacterial genomes, aid in the choice of more relevant therapies, and promote the advancement of novel discrimination and differential diagnostic assays.
INTRODUCTION
Members of the Mycobacterium chelonae-M. abscessus group of rapidly growing mycobacteria (RGM), M. abscessus sensu lato, have been identified not only as sources of pulmonary infections but also as emerging pathogens of nosocomial infections following cosmetic surgery and postsurgical procedures (1–4). The taxonomic status of M. abscessus sensu lato has not been resolved; however, M. massiliense and M. bolletii were characterized as new species distinct from M. abscessus (5, 6). Although the clinical significance of M. massiliense has been emphasized (7, 8), it was proposed by Leao et al. in 2011 that M. massiliense and M. bolletii should be reclassified as a united subspecies of M. abscessus, M. abscessus subsp. bolletii, and that a new subspecies, M. abscessus subsp. abscessus, should be designated (9). However, a recent whole-genome study strongly supported the hypothesis that the species can be divided into at least three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii (10). M. abscessus subsp. massiliense was initially isolated from the sputum of a patient with pneumonia in France in 2004 (5). In 2005, an outbreak of M. abscessus subsp. massiliense infection was linked to intramuscular injections of an antimicrobial agent in South Korea (11). This bacterium was also the source of a lethal case of sepsis in Italy (12) and has been found in cystic fibrosis patients in France (13). Several cases of bacteremia and cutaneous pulmonary infections have also been reported in Japan (14–17).
A novel approach is required to differentiate M. abscessus subsp. massiliense from M. abscessus subsp. abscessus and M. abscessus subsp. bolletii because conventional methods such as biochemical assays and 16S rRNA genotyping cannot make the discrimination. Moreover, the clinical profile of M. abscessus subsp. massiliense is different from those of M. abscessus subsp. abscessus and M. abscessus subsp. bolletii. In particular, antibiotic treatment with clarithromycin is more effective against M. abscessus subsp. massiliense lung infections, with resistance developing more readily in cases of M. abscessus subsp. abscessus lung disease. Therefore, differentiating M. abscessus subsp. massiliense from M. abscessus subsp. abscessus is critical in the clinical environment (7). A significant difference between M. abscessus subsp. massiliense and M. abscessus subsp. abscessus-M. abscessus subsp. bolletii in susceptibility to various antimycobacterial drugs has also been observed in our laboratory (17). A significant difference between M. abscessus subsp. abscessus and M. abscessus subsp. massiliense in treatment response was also noted (18). However, the incidence of M. abscessus subsp. bolletii infection is very rare, making it difficult to separate its clinical profile from that of an M. abscessus subsp. abscessus infection.
In Japanese hospitals, a commercially available DNA-DNA hybridization (DDH) assay is frequently used for the clinical identification of mycobacterial strains. However, the reference panel for the DDH Mycobacteria kit consists of only the 18 most common strains of mycobacteria. Although the DDH test is able to clearly differentiate M. chelonae from the rest of the M. chelonae-M. abscessus group, M. abscessus subsp. massiliense and M. abscessus subsp. bolletii are not included in the panel (19). In fact, isolates from different subpopulations of patients were all identified as M. abscessus by the DDH assay. However, these isolates appeared to have different responses to several antimycobacterial drugs (20). These observations led us to develop a simple genotyping test to discriminate M. abscessus subsp. massiliense from M. abscessus using the whole-genome data of M. abscessus subsp. massiliense as a reference sequence.
MATERIALS AND METHODS
Bacterial strains.
An environmental isolate (strain LRC AbsB-1) and 117 clinical isolates were obtained for differential diagnosis from hospitals in Japan (see the Appendix for the list of the hospitals). Of these, 109 strains were isolated from sputum samples and 8 were obtained from skin lesions (see Table S1 in the supplemental material). All of the isolates had been classified as M. abscessus based on the results of DDH analysis (DDH Mycobacteria kit; Kyokuto Pharmaceutical Industrial, Tokyo, Japan). This kit contains 18 strains of mycobacteria on the reference panel, which includes M. abscessus but not M. abscessus subsp. massiliense or M. abscessus subsp. bolletii (19).
Reference strains of the rapidly growing mycobacteria M. abscessus subsp. massiliense JCM 15300T, M. chelonae JCM 6388T, M. abscessus subsp. bolletii JCM 15297T, and M. abscessus subsp. abscessus JCM 13569T (ATCC 19977) were obtained from the Japan Collection of Microorganisms of the Riken Bio-Resource Center (BRC-JCM; Ibaraki, Japan). All bacterial strains were subcultured on 2% Ogawa egg slants or 7H11 agar plates.
Following development of the multiplex PCR assay, several laboratory and clinical isolates that had been classified using DDH assays and/or sequencing were applied to this assay. They included isolates of M. avium complex, M. fortuitum, M. gordonae, M. kansasii, M. lentiflavum, M. peregrinum, M. shimoidei, M. szulgai, M. triplex, M. tuberculosis, and M. xenopi.
DNA extraction.
DNA extraction was performed as described previously (17). In brief, a loopful of bacilli was suspended in 400 μl sterilized phosphate-buffered saline supplemented with 0.05% Tween 80 and stored at −80°C until the extraction was performed. A frozen sample was crushed with zirconia beads (2 mm in diameter) in a bead-beating instrument. Total genomic DNA was purified from the crushed suspension using a High Pure PCR template preparation kit according to the manufacturer's instructions (Roche Diagnostics) and stored at −20°C.
Sequence analysis.
The sequences of the clinical and environmental isolates, which had been preliminarily identified as M. abscessus with the DDH kit, were compared to those of the M. abscessus subsp. massiliense JCM 15300T, M. abscessus subsp. bolletii JCM 15297T, M. chelonae JCM 6388T, and M. abscessus subsp. abscessus JCM 13569T reference strains. The sequences of the majority of the 16S rRNA gene, partial hsp65 and rpoB genes, and the internal transcribed spacer (ITS) region between the 16S and 23S rRNA genes were amplified using AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) with the primers listed in Table 1. Both strands were sequenced with BigDye Terminator cycle sequencing kit ver. 3.1 (Applied Biosystems) and run on an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). Analyses were performed after removing the primers from the sequences (24).
TABLE 1.
Primers used in this study
| Primer | Sequence | Target and/or purpose (amplified fragment size) | Reference |
|---|---|---|---|
| 8F16S | 5′-AGAGTTTGATCCTGGCTCAG-3′ (positions 8 to 27)a | Mycobacterial 16S rRNA gene, PCR (ca. 1,500 bp), sequencing | 21 |
| 1047R16S | 5′-TGCACACAGGCCACAAGGGA-3′ (positions 1047 to 1028)a | ||
| 830F16S | 5′-GTGTGGGTTTCCTTCCTTGG-3′ (positions 830 to 849)a | ||
| 1542R16S | 5′-AAGGAGGTGATCCAGCCGCA-3′ (positions 1542 to 1523)a | ||
| TB11 | 5′-ACCAACGATGGTGTGTCCAT-3′ | Mycobacterial hsp65 gene, PCR (441 bp), sequencing | 22 |
| TB12 | 5′-CTTGTCGAACCGCATACCCT-3′ | ||
| MabrpoF | 5′-GAGGGTCAGACCACGATGAC-3′ (positions 2112–2131)b | Mycobacterial rpoB gene, PCR (449 bp), sequencing | 17 |
| MabrpoR | 5′-AGCCGATCAGACCGATGTT-3′ (positions 2559–2541)b | ||
| ITSF | 5′-TTGTACACACCGCCCGTC-3′ | Mycobacterial 16S-23S ITS region, PCR (ca. 340 bp), sequencing | 23 |
| ITSR | 5′-TCTCGATGCCAAGGCATCCACC-3′ |
Nucleotide positions were assigned using the Escherichia coli 16S rRNA gene sequence as a reference.
Primer design and nucleotide positions were based on the M. tuberculosis rpoB gene sequence (GenBank/EMBL/DDBJ accession no. L27989).
Short-read DNA sequencing using Illumina Genome Analyzer II.
A cDNA library of M. abscessus subsp. massiliense JCM 15300T, containing fragments of approximately 500 bp in length, was prepared using a genomic DNA Sample Prep kit (Illumina, San Diego, CA). DNA clusters were generated on a slide using a Cluster Generation kit (ver. 2) on an Illumina Cluster Station (Illumina), according to the manufacturer's instructions. All sequencing runs for 83-mers were performed using Illumina Genome Analyzer II (GA II) and an Illumina sequencing kit (ver. 3). Fluorescent images were analyzed using Illumina base-calling pipeline 1.4.0 to obtain FASTQ-formatted sequence data.
De novo assembly of short DNA reads.
Prior to de novo assembly, reads were divided into 40-, 50-, 60-, or 70-mers from the 5′ end of 83-mer reads followed by nucleotide trimming based on the phred quality value (cutoff of 14) using the Euler-SR qualitytrimmer command (25). These trimmed sequences were then assembled using Euler-SR v1.0 (25) with the default parameters (vertex size, 25).
Genome scaffold analysis using reference sequences.
Reference sequence-assisted gap closing was performed with OSLay v1.0 software (26) using the Mycobacterium abscessus ATCC 19977 chromosome DNA sequence as a reference genome (GenBank accession no. NC_010397). Homologous regions between the de novo assembly of short reads and ATCC 19977 chromosome DNA were identified by BLASTN searches with 1E-10 as a cutoff value (setting parameters, -m 8 -e 1E-10). Predicted supercontigs (an ordered and oriented set of contigs that contained gaps) were visualized by OSLay (26). Tentative scaffolds of M. abscessus subsp. massiliense chromosomal DNA sequence were obtained in the same manner as the supercontigs. Pairwise alignment between those genome sequences was performed using a BLASTN homology search (27) followed by visualization of the aligned images with the Artemis Comparison Tool (ACT) (28).
PCR assays.
Single-PCR and multiplex PCR assays differentiating M. abscessus subsp. abscessus and M. abscessus subsp. massiliense were conducted with the listed primers (see Table 4). In brief, 50 μl of a mixture containing 50% AmpliTaq Gold 360 Master Mix (Applied Biosystems), 2% GC enhancer, 0.5 μM (each) primer, and 0.1 μg template DNA was used for PCR with a single set of primers. Amplification was performed in the Mastercycler gradient (Eppendorf) using 95°C for 10 min; 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min. The PCR products were separated by 2% agarose gel electrophoresis and stained with ethidium bromide.
TABLE 4.
Primers to discriminate reference strains of M. abscessus subsp. abscessus and M. abscessus subsp. massiliense
| Primer | Sequence (locations)a | Amplified fragment size (ca. bp) of M. abscessus subsp. abscessus/M. abscessus subsp. massilienseb |
|---|---|---|
| MAB 0022cF | 5′-TTCGATTCTCCTAGGCTCCA-3′ (22165–22184) | 220/270 |
| MAB 0022cR | 5′-GGCGTACATGACCGCATACT-3′ (22386–22367) | |
| MAB 0104cF | 5′-GGTGACTGAACTCCACGAAGA-3′ (105362–105382) | 590/160 |
| MAB 0104cR | 5′-CGATATCGTGAGCCATCCTC-3′ (105948–105929) | |
| MAB 0357cF | 5′-GGTAGCTCTTCCAGCCGAAT-3′ (354468–354487) | 900/300 |
| MAB 0357cR | 5′-CAGCACGCA AAGGTACGAC-3′ (355376–355358) | |
| MAB 1112cF | 5′-CCAAAACCGTTGAGCGTTAT-3′ (1125571–1125590) | 200/1,020 |
| MAB 1112cR | 5′-ATCATGAACCGCAAGTACCG-3′ (1125776–1125757) | |
| MAB 1176cF | 5′-CACACCACGTGTCCTACAGC-3′ (1193378–1193397) | 210/860 |
| MAB 1176cR | 5′-AGTCCATCGAACGAACTTGG-3′ (1193594–1193575) | |
| MAB 2847cF | 5′-CCCACAAATTTCGTAAGACCA-3′ (2896496–2896516) | 130/200 |
| MAB 2847cR | 5′-ACCCAGGTGGAACTCTTCAC-3′ (2896628–2896609) | |
| MAB 3644F | 5′-GTCACCGCAGAAATCGAGTC-3′ (3694129–3694148) | 200/700 |
| MAB 3644R | 5′-GGGGTGGTTGACGTGTTC-3′ (3694310–3694293) | |
| MAB 3644 (Rev2R)c | 5′-CGAGGTCAAGTGGCTTCTTT-3′ (3694262–3694243) | 150/650 |
| MAB 4614F | 5′-CCTTCACCCTTCCGTTCAT-3′ (4698970–4698988) | 215/535 |
| MAB 4614R | 5′-GTTGGGACTGCAGGTATTGC-3′ (4699184–4699165) |
Nucleotide positions were based on the complete sequence of the M. abscessus subsp. abscessus chromosome (accession no. CU458896) as a reference.
Fragment sizes were predicted from the results of sequencing of the draft genome of M. abscessus subsp. massiliense (accession no. BAOM01000001 to BAOM01000060).
MAB 3644 (Rev2R) was used only in the multiplex PCR.
Nucleotide sequence accession numbers.
The DNA sequences of the 16S rRNA (1,468-bp), hsp65 (401-bp), rpoB (409-bp), and ITS (298-bp) fragments from the reference strains (type strains of M. abscessus subsp. massiliense JCM 15300T, M. chelonae JCM 6388T, M. abscessus subsp. bolletii JCM 15297T, M. abscessus subsp. abscessus JCM 13569T, and M. abscessus subsp. massiliense cutaneous isolate strain A1) were deposited into the International Nucleotide Sequence Databases (INSD) through the DNA Databank of Japan (DDBJ) under accession numbers AB548592 to AB548611 (see Table 2). The draft genome sequences of M. abscessus subsp. massiliense were deposited under accession numbers BAOM01000001 to BAOM01000060.
TABLE 2.
List of DNA sequence accession numbers (AB548592 to AB548611)
| Strain | 16S rRNA (1,468 bp) | hsp65 (401 bp) | rpoB (409 bp) | ITS (298 bp) |
|---|---|---|---|---|
| M. abscessus subsp. massiliense JCM 15300T | AB548602 | AB548601 | AB548600 | AB548603 |
| M. abscessus subsp. bolletii JCM 15297T | AB548606 | AB548605 | AB548604 | AB548607 |
| M. abscessus subsp. abscessus JCM 13569T | AB548599 | AB548598 | AB548597 | AB548596 |
| M. abscessus subsp. massiliense strain A1 | AB548592 | AB548593 | AB548594 | AB548595 |
| M. chelonae JCM 6388T | AB548610 | AB548609 | AB548608 | AB548611 |
RESULTS
Multilocus sequence analysis.
Nucleotide sequence analysis was performed with the isolates and reference strains (M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, M. abscessus subsp. bolletii, and M. chelonae). The sequences of the 1,468-bp fragment of the 16S rRNA genes of the 118 isolates and the reference strains were almost identical, with only 1-bp mismatches or no mismatches with M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii found at nucleotide position 1007 or 1008 or 1407 or 1408 and additional 3-bp mismatches with M. chelonae at nucleotide positions 999, 1039, and 1265 (17). However, differences were observed with the sequences of hsp65, rpoB, and the ITS region (Table 3). There were two distinct groups. The strains in the first group either had the same hsp65/rpoB/ITS sequence as the M. abscessus subsp. abscessus type strain (type 1) or had a 1- or 2-bp difference in hsp65 and/or the ITS region (type 1a or type 1b). In this group, 58.5% of the strains were classified as M. abscessus subsp. abscessus, in accordance with the DDH results. The strains in the second group had the same hsp65/rpoB/ITS sequence as the M. abscessus subsp. massiliense type strain, with the exception of one base in the ITS region (type 2) or one or two or no base pair differences in the ITS region and/or rpoB (type 2a, type 2b, type 2c, and type 2d). In this group, 36.4% of the strains classified as M. abscessus by DDH were actually M. abscessus subsp. massiliense. The strains in the third group, with only 3 isolates included, had the same hsp65/rpoB/ITS sequence as the M. abscessus subsp. bolletii type strain, with the exception of 1-bp differences in rpoB (type 3a) and/or the ITS region and/or hsp65 (type 3b and type 3c). In this group, 2.5% of the strains classified as M. abscessus by DDH were actually M. abscessus subsp. bolletii. The remaining three clinical isolates could not be identified by the sequences, because they had discordant sequencing results. The sequences of the hsp65 genes and ITS regions of the two isolates were identical to those of M. abscessus subsp. abscessus; however, they carried the rpoB sequence of M. abscessus subsp. bolletii with the 1-bp mismatch (DS type 4). The third isolate had the M. abscessus subsp. massiliense hsp65 and ITS region sequences and the rpoB sequence of M. abscessus subsp. abscessus (DS type 5). An examination of the data from combinations of multilocus sequence analyses can be used to clearly discriminate M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii with 97.5% accuracy.
TABLE 3.
The sequential differences in clinical isolates in M. abscessus sensu lato
| Group | No. of tested strains | Predicted DDH result | Nucleotide sequence positiona |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
hsp65b |
rpoBc |
ITS region |
|||||||||||||||||||||||||||||
| 115 | 118 | 127 | 187 | 190 | 229 | 340 | 10 | 31 | 79 | 88 | 124 | 127 | 136 | 193 | 202 | 277 | 283 | 316 | 343 | 376 | 379 | 25 | 45 | 48 | 60 | 101–103d | 180 | 276 | |||
| M. abscessus subsp. abscessusT | 1 | M. abscessus | T | T | C | C | C | G | C | T | T | C | T | G | C | T | C | C | C | C | T | C | C | C | T | A | C | A | C-G | C | G |
| Type 1 | 61 | M. abscessus | T | T | C | C | C | G | C | T | T | C | T | G | C | T | C | C | C | C | T | C | C | C | T | A | C | A | C-G | C | G |
| Type 1a | 6 | M. abscessus | T | T | C | C | C | G | C | T | T | C | T | G | C | T | C | C | C | C | T | C | C | C | T | A | T | A | C-G | C | G |
| Type 1b | 2 | M. abscessus | T | T | C | C | C | G | T | T | T | C | T | G | C | T | C | C | C | C | T | C | C | C | T | A | T | A | C-G | C | G |
| M. abscessus subsp. massilienseT | 1 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | C | T | T | T | T | A | C | G | CCG | C | A |
| Type 2 | 29 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | C | T | T | T | T | A | C | G | CCG | T | A |
| Type 2a | 10 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | C | T | T | T | C | A | C | G | CCG | T | A |
| Type 2b | 2 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | T | T | T | T | T | A | C | G | CCG | C | A |
| Type 2c | 1 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | T | T | T | T | C | A | C | G | CCG | C | A |
| Type 2d | 1 | M. abscessus | G | C | T | C | T | G | T | C | C | C | C | A | T | C | C | G | T | C | C | T | T | T | T | A | C | G | CCG | C | A |
| M. abscessus subsp. bolletiiT | 1 | M. abscessus | G | C | T | T | T | C | C | G | T | T | C | A | T | C | T | T | T | T | T | T | T | T | T | A | C | A | C-G | C | G |
| Type 3a | 1 | M. abscessus | G | C | T | T | T | C | C | G | T | T | C | A | T | C | T | C | T | T | T | T | T | T | T | A | C | A | C-G | C | G |
| Type 3b | 1 | M. abscessus | G | C | T | T | T | C | C | G | T | T | C | A | T | C | T | C | T | T | T | T | T | T | T | G | C | A | C-G | C | G |
| Type 3c | 1 | M. abscessus | G | C | T | C | T | C | C | G | T | T | C | A | T | C | T | T | T | T | T | T | T | T | T | G | C | A | C-G | C | G |
| DSe type 4 | 2 | M. abscessus | T | T | C | C | C | G | C | G | T | T | C | A | T | C | T | C | T | T | T | T | T | T | T | A | C | A | C-G | C | G |
| DS type 5 | 1 | M. abscessus | G | C | T | C | T | G | T | T | T | C | T | G | C | T | C | C | C | C | T | C | C | C | T | A | C | G | CCG | C | A |
Bold letters indicate nucleotides different from those of the type strains.
Nucleotide positions were based on the M. abscessus subsp. massiliense sequence of the partial hsp65 gene (accession no. AB548601).
Nucleotide positions were based on the M. abscessus subsp. massiliense sequence of the partial rpoB gene (accession no. AB548600).
Base deficient.
Isolate showing discordant sequencing results.
Whole-genome sequence analysis and primer design.
The whole-genome sequence of the M. abscessus subsp. massiliense type strain was compared with that of the M. abscessus subsp. abscessus type strain (GenBank accession no. NC_010397) using the ACT to visualize pairwise alignments between the sequences. At least eight notable regions, containing 50- to 800-bp differences, were identified as candidates for PCR targets (i.e., 50- to 800-bp insertions or deletions in M. abscessus subsp. massiliense compared to M. abscessus subsp. abscessus). Figure S1 in the supplemental material shows a representative region, which had a 494-bp insertion in M. abscessus subsp. massiliense at position 3694233 of M. abscessus subsp. abscessus. The eight regions were labeled MAB_0022c, MAB_0104c, MAB_0357c, MAB_1112c, MAB_1176c, MAB_2847c, MAB_3644, and MAB_4614 with reference to the locations of the open reading frames (ORFs). Regions around MAB_0022c, MAB_0104c, MAB_3644, and MAB_4614 were associated with coding sequences, whereas MAB_0357, MAB_1112c, MAB_1176, and MAB_2847 were associated with noncoding sequences. Eight primer sets were designed using locations in the borders of regions that would sharply differentiate M. abscessus subsp. abscessus and M. abscessus subsp. massiliense based on the sizes of their PCR amplicons (Table 4). Single-PCR tests were performed to cull the two best primer sets. The following check points were used as selection criteria. (i) Were the isolates amplified? (ii) Were the amplicons clear single bands? (iii) Were the amplicons the correct size? (iv) Were the PCR results discriminating M. abscessus subsp. abscessus and M. abscessus subsp. massiliense consistent with the multilocus sequence analysis? All clinical isolates were used in the single-PCR tests; however, tests were terminated when check point 1, 2, or 3 was negative, with the first 20 strains selected. Figure S2 in the supplemental material shows single-PCR results targeting MAB_1176c as representative. As shown in that figure, PCR of all M. abscessus subsp. abscessus strains produced an amplicon of the expected size, ca. 210 bp. Four clinical M. abscessus subsp. massiliense strains and the M. abscessus subsp. massiliense type strain produced an amplicon of ca. 860 bp. However, 8 clinical strains produced an unexpected size of ca. 400 bp. These results suggested that the MAB 1176c primer sets were not suitable for PCR, since the insertion sequence in the M. abscessus subsp. massiliense clinical strains was not conserved as a uniformly sized sequence of ca. 860 bp. Single-PCR results are shown in Table S2 in the supplemental material. Amplification using MAB 3644, MAB 0022c, and MAB 1112c primer sets produced amplicons of the proper size, although some of the clinical strains displayed contrasting results. We concluded that the combined targeting of MAB_0357c and MAB_3644 would produce the best results (Fig. 1 and 2). These regions generated amplicons of the correct size, delivered results that were consistent with those of multilocus sequence analysis, and provided an easy visual means to distinguish between M. abscessus subsp. abscessus and M. abscessus subsp. massiliense.
FIG 1.
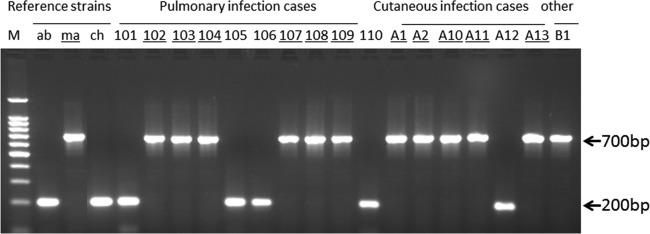
Representative single-PCR results for the reference strains and clinical isolates amplified with primer pair MAb 3644F and MAb 3644R. The numbers are the strain numbers of the clinical isolates. Underlining indicates that M. abscessus subsp. massiliense (ma) was classified after the multilocus genotyping assay shown in Table 3. The M. abscessus subsp. massiliense PCR product was fully amplified to 700 bp. However, the M. abscessus subsp. abscessus (ab) and M. chelonae (ch) amplicons amplified to 200 bp in size.
FIG 2.
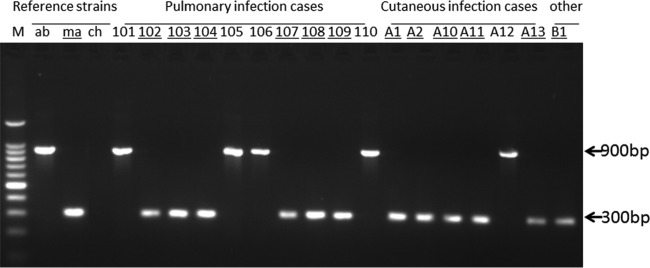
Representative single-PCR results for the reference strains and clinical isolates amplified with primer pair MAb 0357cF and MAb 0357cR. The numbers are the strain numbers of the clinical isolates. Underlining indicates that M. abscessus subsp. massiliense (ma) was classified after the multilocus genotyping assay shown in Table 3. The M. abscessus subsp. abscessus (ab) PCR product was fully amplified to 900 bp in size. However, the M. abscessus subsp. massiliense amplicon was 300 bp in size, while M. chelonae (ch) did not amplify.
Discriminatory multiplex PCR assay.
Based on the results of single PCR, we performed amplifications using the clinical strains. All of the clinical isolates that were initially identified as M. abscessus by DDH could be resolved as M. abscessus subsp. abscessus or M. abscessus subsp. massiliense using both sets of primer pairs MAB 0357c and MAB 3644. However, the amplification results were not always clear enough: it has been speculated that Taq polymerase is heavily utilized to amplify shorter bands, leaving many weak longer bands (data not shown). For more clearly balanced multiplex amplification, MAB 3644R was replaced with MAB 3644 (Rev2R) (Fig. 3). Using MAB 0357c and MAB 3644 (Rev2R) allowed a clear separation of M. abscessus subsp. abscessus isolates from M. abscessus subsp. massiliense isolates (Table 5; see also Table S1 in the supplemental material). Multiplex PCR results from the three type strains and 118 clinical isolates are shown in Table S1. A total of 70 M. abscessus subsp. abscessus isolates showed the same multiplex PCR pattern (ca. 900 bp and ca. 150 bp), and 44 M. abscessus subsp. massiliense isolates showed ca. 300 bp and ca. 650 bp. However, the results for 4 isolates of M. abscessus subsp. bolletii were not converged. One of the clinical isolates (type 3a) and the type strain showed an amplification pattern that was identical to that of M. abscessus subsp. abscessus (ca. 900 bp and ca. 150 bp). The two other clinical isolates (types 3b and 3c) showed different patterns, ca. 150 bp and ca. 300 bp and a weak pattern of ca. 300 bp and ca. 650 bp, respectively. The discordant results from two type 4 isolates were identical to those obtained for M. abscessus subsp. bolletii type 3b, showing a multiplex PCR pattern of ca. 150 bp and ca. 300 bp. The pattern determined for the remaining type 5 discordant isolate was identical to that of M. abscessus subsp. massiliense, ca. 300 bp and ca. 650 bp. All other mycobacterial isolates prepared as negative controls were negative in this multiplex PCR assay. The only exception was the M. chelonae type strain, which showed a single weak band of ca. 200 bp (Fig. 3).
FIG 3.
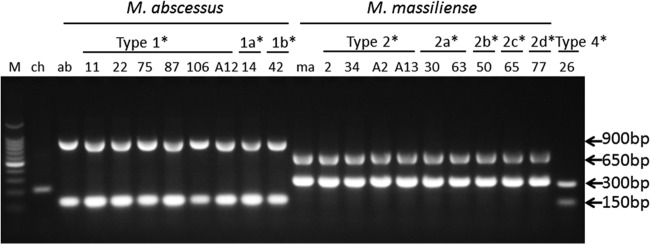
Representative multiplex PCR results for the reference strains and clinical isolates amplified with primer pair MAB 3644F and MAB 3644 (Rev2R) and primer pair MAB 0357cF and MAB 0357cR. *, types 1, 1a, and 1b, types 2, 2a, 2b, 2c, and 2d, and type 4 are the groupings of the clinical isolates based on their sequences (Table 3). The numerals below the groupings are the strain numbers of the clinical isolates.
TABLE 5.
PCR results compared with other genetic classifications
| Group | No. of tested strains | Predicted DDH result | Classification by hsp65/rpoB/ITS sequencesa | PCR result(s) (ca. bp) with primer pair(s): |
||
|---|---|---|---|---|---|---|
| MAB 3644F + MAB 3644R | MAB 0357cF + MAB 0357cR | MAB 3644F + MAB 3644 (Rev2R), MAB 0357cF + MAB 0357cR | ||||
| M. abscessus subsp. abscessusT | 1 | M. abscessus | M. abscessus subsp. abscessus | 200 | 900 | 900, 150 |
| Type 1 | 61 | M. abscessus | M. abscessus subsp. abscessus | 200 | 900 | 900, 150 |
| Type 1a | 6 | M. abscessus | M. abscessus subsp. abscessus | 200 | 900 | 900, 150 |
| Type 1b | 2 | M. abscessus | M. abscessus subsp. abscessus | 200 | 900 | 900, 150 |
| M. abscessus subsp. massilienseT | 1 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| Type 2 | 29 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| Type 2a | 10 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| Type 2b | 2 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| Type 2c | 1 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| Type 2d | 1 | M. abscessus | M. abscessus subsp. massiliense | 700 | 300 | 300, 650 |
| M. abscessus subsp. bolletiiT | 1 | M. abscessus | M. abscessus subsp. bolletii | 200 | 900 | 900, 150 |
| Type 3a | 1 | M. abscessus | M. abscessus subsp. bolletii | 200 | 900 | 900, 150 |
| Type 3b | 1 | M. abscessus | M. abscessus subsp. bolletii | 200 | 300 | 300, 150 |
| Type 3c | 1 | M. abscessus | M. abscessus subsp. bolletii | 700 | 300b | 300b, 650 |
| DSc type 4 | 2 | M. abscessus | M. abscessus subsp. abscessus-M. abscessus subsp. bolletiid | 200 | 300 | 300, 150 |
| DS type 5 | 1 | M. abscessus | M. abscessus subsp. massiliense-M. abscessus subsp. abscessuse | 700 | 300 | 300, 650 |
Classifications were determined on the basis of the overall results of sequencing of hsp65, rpoB, and the ITS region (Table 3).
Weak band.
Isolate showing discordant sequencing results.
Isolate showing M. abscessus subsp. abscessus hsp65 and M. abscessus subsp. bolletii rpoB sequences.
Isolate showing M. abscessus subsp. massiliense hsp65 and ITS region and M. abscessus subsp. abscessus rpoB sequences.
DISCUSSION
We have developed a simple, cost-effective discriminative multiplex PCR to differentiate M. abscessus subsp. massiliense from M. abscessus subsp. abscessus and from other RGM. The multiplex PCR expanded upon the results of two sets of discriminative single PCRs to concurrently amplify M. abscessus subsp. abscessus and M. abscessus subsp. massiliense and distinguish them based upon differences in amplicon size. In order to achieve the different amplifications in length, clear insertion or deletion regions between M. abscessus subsp. massiliense and M. abscessus subsp. abscessus whole-genome sequences were selected for the single-PCR targets using the Artemis Comparison Tool. The combination of MAB 0357c and MAB 3644 (Rev2R) for targeted PCR mainly leads to two distinct visual patterns that are easily read by novice PCR technologists. Previously, clinicians identified all isolates as M. abscessus because the use of the DDH assay was very common in Japan. The isolates were also grouped together because of their colony morphologies, growth profiles, and biochemical characteristics. Even the majority of the sequences of their 16S rRNA genes are the same. However, clinicians began to suspect that different strains were present because there were significantly different clinical outcomes and drug susceptibility groups among these isolates. For example, M. abscessus subsp. massiliense is more susceptible to azithromycin than M. abscessus subsp. abscessus but not to clofazimine, meropenem, and panipenem (17). Thus, correct and rapid species identification could facilitate the clinical treatment of mycobacterial infections (7). Two distinct genotypes were eventually observed in isolates identified as M. abscessus by the DDH mycobacterial assays (17). These two groups can be separated by a combinational genotypic analysis of the sequences of the ITS region and hsp65 and rpoB genes. Similarly, in other reports of multilocus sequencing analysis performed with hsp65, rpoB, and secA1 (4) or with eight housekeeping genes (29), M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii are clearly differentiated. But this methodology is not practical due to the cost and effort involved. There have been other PCR-based methods, such as erythromycin ribosome methyltransferase (erm) PCR (8) and variable-number tandem-repeat (VNTR) analysis (30). erm PCR is also a very simple and accurate method but, having only one target, can easily lead to false-negative results. The VNTR method is not easy for nonexperts.
A study in South Korea found that 51% of the M. chelonae-M. abscessus group is comprised of M. abscessus subsp. abscessus and 47% is M. abscessus subsp. massiliense (11). A typing study of M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii performed in the United States showed that 64% of the isolates were M. abscessus subsp. abscessus and 28% were M. abscessus subsp. massiliense (4). In France, 60% of the isolates belonged to M. abscessus subsp. abscessus and 22% to M. abscessus subsp. massiliense (13), while in Japan, 71% belonged to M. abscessus subsp. abscessus and 26% to M. abscessus subsp. massiliense (20). In pulmonary patients in the Netherlands diagnosed with infections by strains from the M. chelonae-M. abscessus group, 50% of the isolates were identified as M. abscessus subsp. abscessus and 29% as M. abscessus subsp. massiliense (31). In accordance with the recent study in Japan mentioned above (20), we can estimate that more than 30% of patients diagnosed with M. abscessus subsp. abscessus should differentiate as M. abscessus subsp. massiliense patients. Some isolates might be colonizers whereas others might have appeared after prolonged treatment of infections by other nontuberculous mycobacteria. Therefore, we are currently collecting patient treatment data from the hospitals that participated in this study and are analyzing the relationship between clinical isolates and treatment history.
Three clinical isolates were not identified to the species level by multilocus sequence analysis. They are discordant isolates that were previously reported by Zelazny et al. (4) and Macheras et al. (29). The sequences of the hsp65 genes and ITS regions of the two isolates were identical to those of M. abscessus subsp. abscessus; however, they carried the rpoB sequence of M. abscessus subsp. bolletii with the 1-bp mismatch (DS type 4). Both strains produced multiplex PCR amplicons of ca. 300 bp and ca. 150 bp, like those of M. abscessus subsp. bolletii type 3b (see Table S1 in the supplemental material). The third isolate which had the M. abscessus subsp. massiliense hsp65 and ITS regions and the rpoB sequence of M. abscessus subsp. abscessus (DS type 5) had the multiplex PCR pattern of M. abscessus subsp. massiliense (ca. 300 bp and ca. 650 bp). Although those strains are very rare, they are interesting in that they suggest the occurrence of horizontal gene transfer. Such discordant isolates would produce different amplicons (ca. 300 bp and ca. 150 bp) from M. abscessus subsp. abscessus and M. abscessus subsp. massiliense (Table 5).
In the case of RGM infection, we propose the use of this multiplex PCR assay as a first step, because it can be used by inexperienced technicians to identify M. abscessus subsp. abscessus and M. abscessus subsp. massiliense quickly and accurately. In addition, there is no need to prepare purified DNA; the supernatant from a boiled bacterial suspension can be used (data not shown). The resulting band patterns of 900 bp and 150 bp and of 300 bp and 650 bp imply the identification of M. abscessus subsp. abscessus and M. abscessus subsp. massiliense with 97.2% and 95.7% accuracy. Another rare pattern, 300 bp and 150 bp, implies identification of a discordant strain with 66.7% accuracy. Although this multiplex PCR method conduces to reasonably accurate discrimination, it cannot be used to differentiate M. abscessus subsp. bolletii from M. abscessus subsp. abscessus or M. abscessus subsp. massiliense, because all three clinical isolates of M. abscessus subsp. bolletii showed different multiplex PCR patterns in this study (Table 5).
As a result of this study, we have developed a simple, rapid methodology to distinguish between M. abscessus subsp. abscessus and M. abscessus subsp. massiliense. This approach, which spans whole-genome sequencing and clinical diagnosis, will facilitate the acquisition of more precise information about bacterial genomes, aid in the choice of more-relevant therapies, and promote the advancement of novel discrimination and differential diagnostic assays.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate Ms Katsue Ishii (Fukujuji Hospital) for preparing negative control of clinical isolates.
This work was supported in part by a Grant-in-Aid for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare of Japan for Y.H., M.M., and N.I. and by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan for Y.H. and by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science for K.N.
APPENDIX
The clinical isolates used in this study were sent from the hospitals and universities described below. Specimens were originally collected for disease diagnosis. The portion remaining after diagnosis was used for this study. We appreciate the work of all of the clinicians in the following institutions who took care of patients infected with these mycobacteria: Hokkaido Social Insurance Hospital, Japan Anti-Tuberculosis Association (JATA) Fukujuji Hospital, Saitama Medical University, National Hospital Organization (NHO) Tokyo National Hospital, Showa University Fujigaoka Hospital, National Defense Medical College Hospital, Kyorin University Hospital, NHO Minami-Kyoto Hospital, Kyoto Prefectural University of Medicine, JATA Osaka Hospital, NHO Kinki-Chuo Chest Medical Center, NHO Matsue Medical Center, NHO Higashihiroshima Medical Center, Kawasaki Medical School, Kyosai-Yoshijima Hospital, and NHO Omuta Hospital.
Footnotes
Published ahead of print 6 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01327-13.
REFERENCES
- 1.Viana-Niero C, Lima KV, Lopes ML, Rabello MC, Marsola LR, Brilhante VC, Durham AM, Leão SC. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J. Clin. Microbiol. 46:850–855. 10.1128/JCM.02052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmon KE, Pounder JI, Greene JN, Walsh F, Anderson CM, Cohen S, Petti CA. 2007. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J. Clin. Microbiol. 45:1978–1980. 10.1128/JCM.00563-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso AM, Martins de Sousa E, Viana-Niero C, Bonfim de Bortoli F, Pereira das Neves ZC, Leão SC, Junqueira-Kipnis AP, Kipnis A. 2008. Emergence of nosocomial Mycobacterium massiliense infection in Goiás, Brazil. Microbes Infect. 10:1552–1557. 10.1016/j.micinf.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 4.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacteriumabscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J. Clin. Microbiol. 47:1985–1995. 10.1128/JCM.01688-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, Raoult D, Drancourt M. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493–5501. 10.1128/JCM.42.12.5493-5501.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adékambi T, Berger P, Raoult D, Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56(Pt 1):133–143. 10.1099/ijs.0.63969-0 [DOI] [PubMed] [Google Scholar]
- 7.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am. J. Respir. Crit. Care Med. 183:405–410. 10.1164/rccm.201003-0395OC [DOI] [PubMed] [Google Scholar]
- 8.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, Kook YH. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol. Immunol. 54:347–353. 10.1111/j.1348-0421.2010.00221.x [DOI] [PubMed] [Google Scholar]
- 9.Leao SC, Tortoli E, Euzéby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int. J. Syst. Evol. Microbiol. 61(Pt 9):2311–2313. 10.1099/ijs.0.023770-0 [DOI] [PubMed] [Google Scholar]
- 10.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. 10.1016/S0140-6736(13)60632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HY, Yun YJ, Park CG, Lee DH, Cho YK, Park BJ, Joo SI, Kim EC, Hur YJ, Kim BJ, Kook YH. 2007. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J. Clin. Microbiol. 45:3127–3130. 10.1128/JCM.00608-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tortoli E, Gabini R, Galanti I, Mariottini A. 2008. Lethal Mycobacterium massiliense sepsis, Italy. Emerg. Infect. Dis. 14:984–985. 10.3201/eid1406.080194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux AL, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet MA, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard JL; Jean-Louis Herrmann for the Group OMA 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J. Clin. Microbiol. 47:4124–4128. 10.1128/JCM.01257-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobashi Y, Mouri K, Obase Y, Miyashita N, Nakanaga K, Oka M. 2011. Pulmonary Mycobacterium massiliense disease with septicemia during immunosuppressive treatment. Intern. Med. 50:1069–1073. 10.2169/internalmedicine.50.4733 [DOI] [PubMed] [Google Scholar]
- 15.Hamamoto T, Yuki A, Naoi K, Kawakami S, Banba Y, Yamamura T, Hikota R, Watanabe J, Kimura F, Nakanaga K, Hoshino Y, Ishii N, Shimazaki H, Nakanishi K, Tamai S. 2012. Bacteremia due to Mycobacterium massilinese in a patient with chronic myelogenous leukemia: case report. Diagn. Microbiol. Infect. Dis. 74:183–185. 10.1016/j.diagmicrobio.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 16.Otsuki T, Izaki S, Nakanaga K, Hoshino Y, Ishii N, Osamura K. 2012. Cutaneous Mycobacterium massiliense infection: a sporadic case in Japan. J. Dermatol. 39:569–572. 10.1111/j.1346-8138.2011.01339.x [DOI] [PubMed] [Google Scholar]
- 17.Nakanaga K, Hoshino Y, Era Y, Matsumoto K, Kanazawa Y, Tomita A, Furuta M, Washizu M, Makino M, Ishii N. 2011. Multiple cases of cutaneous Mycobacterium massiliense infection in a “hot spa” in Japan. J. Clin. Microbiol. 49:613–617. 10.1128/JCM.00817-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi GE, Shin SJ, Won CJ, Min KN, Oh T, Hahn MY, Lee K, Lee SH, Daley CL, Kim S, Jeong BH, Jeon K, Koh WJ. 2012. Macrolide treatment for Mycobacterium abscessus and Mycobacterium massiliense infection and inducible resistance. Am. J. Respir. Crit. Care Med. 186:917–925. 10.1164/rccm.201111-2005OC [DOI] [PubMed] [Google Scholar]
- 19.Kusunoki S, Ezaki T, Tamesada M, Hatanaka Y, Asano K, Hashimoto Y, Yabuuchi E. 1991. Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 Mycobacterium species. J. Clin. Microbiol. 29:1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada T, Akiyama Y, Kurashima A, Nagai H, Tsuyuguchi K, Fujii T, Yano S, Shigeto E, Kuraoka T, Kajiki A, Kobashi Y, Kokubu F, Sato A, Yoshida S, Iwamoto T, Saito H. 2012. Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J. Clin. Microbiol. 50:3556–3561. 10.1128/JCM.01175-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer B, Wu WK, Bodmer T, Haase G, Pfyffer GE, Kroppenstedt RM, Schroder KH, Emler S, Kilburn JO, Kirschner P, Telenti A, Coyle MB, Böttger EC. 1996. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J. Clin. Microbiol. 34:1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth A, Fischer M, Hamid ME, Michalke S, Ludwig W, Mauch H. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanaga K, Ishii N, Suzuki K, Tanigawa K, Goto M, Okabe T, Imada H, Kodama A, Iwamoto T, Takahashi H, Saito H. 2007. “Mycobacterium ulcerans subsp. shinshuense” isolated from a skin ulcer lesion: identification based on 16S rRNA gene sequencing. J. Clin. Microbiol. 45:3840–3843. 10.1128/JCM.01041-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaisson MJ, Pevzner PA. 2008. Short read fragment assembly of bacterial genomes. Genome Res. 18:324–330. 10.1101/gr.7088808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter DC, Schuster SC, Huson DH. 2007. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics 23:1573–1579. 10.1093/bioinformatics/btm153 [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- 29.Macheras E, Roux AL, Ripoll F, Sivadon-Tardy V, Gutierrez C, Gaillard JL, Heym B. 2009. Inaccuracy of single-target sequencing for discriminating species of the Mycobacterium abscessus group. J. Clin. Microbiol. 47:2596–2600. 10.1128/JCM.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong YL, Ong CS, Ngeow YF. 2012. Molecular typing of Mycobacterium abscessus based on tandem-repeat polymorphism. J. Clin. Microbiol. 50:3084–3088. 10.1128/JCM.00753-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Ingen J, de Zwaan R, Dekhuijzen RP, Boeree MJ, van Soolingen D. 2009. Clinical relevance of Mycobacterium chelonae-abscessus group isolation in 95 patients. J. Infect. 59:324–331. 10.1016/j.jinf.2009.08.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


