Abstract
The mitochondria of cancer cells are characterized by elevated oxidative stress caused by reactive oxygen species (ROS). Such an elevation in ROS levels contributes to mitochondrial reprogramming and malignant transformation. However, high levels of ROS can cause irreversible damage to proteins, leading to their misfolding, mitochondrial stress, and ultimately cell death. Therefore, mechanisms to overcome mitochondrial stress are needed. The unfolded protein response (UPR) triggered by accumulation of misfolded proteins in the mitochondria (UPRmt) has been reported recently. So far, the UPRmt has been reported to involve the activation of CHOP and estrogen receptor alpha (ERα). The current study describes a novel role of the mitochondrial deacetylase SirT3 in the UPRmt. Our data reveal that SirT3 acts to orchestrate two pathways, the antioxidant machinery and mitophagy. Inhibition of SirT3 in cells undergoing proteotoxic stress severely impairs the mitochondrial network and results in cellular death. These observations suggest that SirT3 acts to sort moderately stressed from irreversibly damaged organelles. Since SirT3 is reported to act as a tumor suppressor during transformation, our findings reveal a dual role of SirT3. This novel role of SirT3 in established tumors represents an essential mechanism of adaptation of cancer cells to proteotoxic and mitochondrial stress.
INTRODUCTION
Mitochondrial reprogramming associated with elevated reactive oxygen species (ROS) levels is a hallmark of cancers. Altered mitochondrial metabolism and ROS, both required during oncogenic transformation (1–4), are regulated by the mitochondrial deacetylase SirT3 (5–7). Reduction in SirT3 levels and the resulting elevated ROS levels have been directly linked to the switch to glycolysis, known as the Warburg effect (8). While ROS are required for glucose metabolism and metastasis in triple-negative breast cancers (9), decreased SirT3 levels concomitant with high ROS levels are frequently observed in all breast cancers (8). Based on these findings, SirT3 is considered a tumor suppressor (5, 8).
SirT3 regulates the activity of magnesium superoxide dismutase (MnSOD), which detoxifies superoxide to hydrogen peroxide (5, 7, 10–14). Moreover, SirT3 has been linked to augmented transcription of MnSOD and catalase, both known targets of the transcription factor FOXO3A (15). The SirT3-dependent deacetylation of FOXO3A promotes both its nuclear translocation and its transcriptional activity (16, 17). Therefore, mechanistically, the reduction of SirT3 levels leads to an elevation in ROS levels by compromising the mitochondrial antioxidant machinery.
Mitochondria are the main source of ROS production (18). However, they are also the main targets of oxidative stress. Excessive ROS induce oxidative damage to DNA, lipids, and proteins, leading to their misfolding and aggregation in the mitochondria. Upon accumulation of misfolded and aggregated proteins in the mitochondria, cells mount the unfolded protein response (UPRmt), a mitochondrial-to nuclear cross talk. This UPRmt aims at reducing proteotoxic stress and reestablishing protein homeostasis of the organelle by elevating the levels of mitochondrial chaperones and proteases. The UPRmt acts similarly to the UPR of the endoplasmic reticulum (19). While the UPR of the endoplasmic reticulum was the first to be identified, the activation of the UPRmt has only recently begun to be appreciated (20–24) and has yet to be elucidated. Moreover, its relation to cancer has not been established.
The UPRmt is regulated by the transcription factor CHOP, which is required to activate transcription of the mitochondrial chaperones and proteases (25–27). Furthermore, we previously reported that the UPRmt relies on estrogen receptor alpha (ERα), which confers cytoprotection against proteotoxic stress in the mitochondria (28).
In addition to the UPRmt, mitophagy plays an important role in removing irreversibly damaged mitochondria from the mitochondrial network. More specifically, mitochondrial proteotoxic stress was shown to induce mitophagy in Drosophila melanogaster (29); however, the mechanism remains unknown.
In the current study, we report a novel role of SirT3 in the UPRmt. Our results implicate SirT3 as a major coordinator of the UPRmt that orchestrates both the antioxidant machinery and mitophagy. These outcomes are required to overcome proteotoxic stress and reestablish homeostasis in cancer cells. Importantly, we found that these functions of SirT3 are independent of either CHOP or ERα. Therefore, our study establishes SirT3 as being critical to monitor the functional integrity of the organelle. Collectively, our results highlight a dual role for SirT3. During malignant transformation, a reduction in the SirT3 level is required to enhance ROS and assist mitochondrial metabolic reprogramming (8). However, in the context of the tumor phenotype, an elevation in the SirT3 level is vital to reduce proteotoxic stress and keep ROS levels below a critical threshold. Therefore, these dual roles allow SirT3 to act as a rheostat of ROS that is essential not only for malignant transformation but also for survival and maintenance of malignant cells.
MATERIALS AND METHODS
Reagents, cell culture, and Western analysis.
Mitochondrial mutant endonuclease G (Endo G) (N174A) cloned into pEGFP-N1 was kindly provided by Gregor Meiss (Justus Liebig University Giessen, Giessen, Germany). pEGFP-N1 expressing green fluorescent protein (GFP) alone was used as a control. Mitochondrion-targeted mutant SOD1-GFP plasmids were gifts from G. Sobue (Nagoya University, Japan). The mammalian expression vector pCAGGS−OTCΔ (deletion of amino acids 30 to 114) was kindly provided by N. J. Hoogenraad (La Trobe University, Australia). Ammonium chloride (NH4Cl), leupeptin, and N-acetylcysteine (NAC) were obtained from Sigma. MitoSox Red, tetramethylrhodamine ethyl ester (TMRE), 4′,6-diamidino-2-phenylindole (DAPI), and LysoTracker Red (LTR) were obtained from Invitrogen (Molecular Probes). Breast cancer cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Western blotting was performed as described previously (30), using antibodies to GFP, ornithine transcarbamylase (OTC) (Santa Cruz), phosphorylated AKT, AKT, SirT3, LC3B, CHOP, FOXO3A (Cell Signaling), Omi (BioVision), NRF-1 (Abcam), and MnSOD (Millipore).
Oxidative stress, mitochondrial potential, and fractionation.
Mitochondrial superoxide anion production was detected by using MitoSox Red fluorescent dye (Molecular Probes), while mitochondrial membrane potential was assessed by staining cells with 50 nM TMRE dye for 20 min according to the manufacturer's protocols. Subcellular fractionation was performed as described previously (30).
Cell viability.
Viability was determined by using a colorimetric 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega) according to the manufacturer's instruction.
siRNA transfection.
Transfections were performed with Lipofectamine 2000 (Invitrogen), using control small interfering RNA (siRNA) against luciferase (5′-CUUACGCUGAGUACUUCGATT-3′), siRNA against SirT3 (siRNA 1, 5′-GCCCAACGUCACUCACUACTT-3′; siRNA 2, 5′ ACUCCCAUUCUUCUUUCACTT-3′), or siRNA against ATG9 (5′-GUACAUGAAUUGCUUCUUGTT-3′) (GeneLink).
RNA isolation and qRT-PCR.
Isolation of RNA was performed with the RNeasy Minikit (Qiagen Inc.) according to the manufacturer's protocol. One-step quantitative real-time PCR (qRT-PCR) was performed by using a SYBR Premix Ex Taq TaKaRa kit on a DNA Opticon Engine system. The human forward primer sequences were as follows: GCATTCCAGACTTCAGATCGC for SirT3, GGAGTGATGTCCGCACAGAA for NRF-1, 5′-CATGAGCGAGTTGGTCAAGA-3′ for ATG8, CGCACCTTCGAACAAAGAG for LC3B, 5′-ATGTTTGGCTTTGGGGCTA-3′ for BNIP3L, 5′-CGGGTGCCGGTGATAGTAGA-3′ for GABARAP, 5′-TCAGCTGCGTGGACTATGAC-3′ for ATG9, 5′-GACCGGCACCCTTTCTTG-3′ for Omi, 5′-CGACTATGCAGTGACAGGTTGTG-3′ for FOXO3A, and 5′-GGCCTACGTGAACAACCTGAA-3′ for MnSOD (GeneLink). Expression levels were normalized to the level of beta-actin, which was used as a loading control.
LysoTracker Red staining.
LTR was added (100 nM) and incubated for 30 min at 37°C. Cells were then incubated with 10 μg/ml Hoechst for an additional 10 min. After fixation, fluorescence was measured by using a fluorescent Spectra Max MS plate reader.
Immunofluorescence and transmission electron microscopy.
LC3B assessment was performed by incubation overnight with LC3B antibody. LC3B was detected by using fluorescently labeled Alexa Fluor 599 secondary antibody. Acidic vesicles were evaluated by loading the cells with 0.5 μM LTR for 20 min. For electron microscopy, cells were fixed at 4°C overnight in 2.5% glutaraldehyde–4.0% paraformaldehyde in 0.1 M phosphate buffer and were processed by the electron microscopy facility at Mount Sinai.
Immunohistochemistry.
Tissues were processed by using the Histostain-Plus broad-spectrum (DAB) substrate kit for peroxidase (Invitrogen) according to the manufacturer's protocol. Sections were then counterstained in hematoxylin for 30 s and mounted with DePEX.
Statistical analysis.
Statistical significance was determined by using GraphPad Prism 6.0 software, and P values were calculated by a one-way analysis of variance (ANOVA) followed by a pairwise contrast (Bonferroni) analysis. Significance was considered to be a P value of <0.05 or less.
RESULTS
Accumulation of misfolded proteins in the mitochondria induces proteotoxic stress.
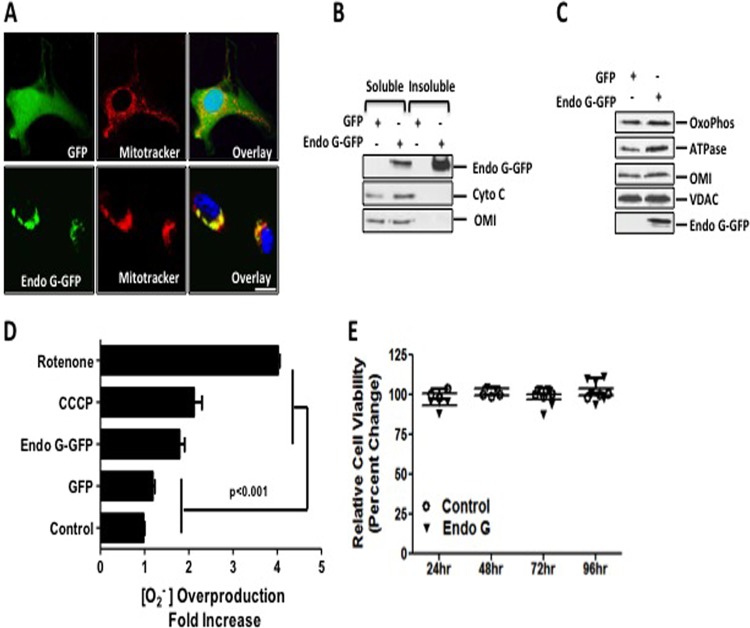
We reported the activation of the UPRmt by ERα using a mutant of the mitochondrial protein endonuclease G (Endo G-GFP). Upon its accumulation in the mitochondria, Endo G-GFP is misfolded and induces mitochondrial proteotoxic stress (30). While expanding our analysis of the effects of such stress in breast cancer cells, immunofluorescence analysis revealed that Endo G staining exhibited severe mitochondrial clustering, especially in cells lacking ERα (Fig. 1A). This finding indicated that proteotoxic stress results in a heterogeneity among the mitochondrion population, with a fraction of them being severely stressed.
FIG 1.
Accumulation of Endo G aggregates in the mitochondria induces proteotoxic stress. (A) Localization of GFP or Endo G-GFP (green) was detected by immunofluorescence in cells transfected with the indicated plasmids. Mitochondria (red) were stained with Mitotracker orange dye, whereas nuclei were counterstained with DAPI (blue). Bar, 1 μm. (B) Soluble and insoluble fractions of purified mitochondria treated with sodium carbonate were analyzed by immunoblotting for Endo G, Omi, and cytochrome c. (C) Mitochondrial lysates of cells overexpressing either Endo G-GFP or GFP were used for Western analysis of oxidative phosphorylation (OxoPhos) complex III, the FoF1 ATP synthase α subunit, Omi, VDAC, and Endo G-GFP. (D) Mitochondrial superoxide anion (O2−) production was detected by FACS analysis of live cells transfected with either control GFP or mutant Endo G-GFP as well as of untreated cells or cells treated overnight with 10 μM CCCP or 20 μM rotenone (used as positive controls). (E) Cell viability was measured by an MTS assay at 24, 48, 72, and 96 h after transfection with either the control vector GFP or mutant Endo G-GFP.
In agreement with this finding, we found that Endo G was present in the soluble fraction along with endogenous Omi and cytochrome c. However, Endo G was evident mostly in the insoluble fraction (Fig. 1B). This result indicates that upon its accumulation, Endo G forms insoluble aggregates in the mitochondria. However, these aggregates did not impact the general import of a variety of mitochondrial proteins (Fig. 1C). While the formation of these aggregates had no effect on cellular viability (Fig. 1E), they led to a significant increase in mitochondrial superoxide anion O2− levels (Fig. 1D). Fluorescence-activated cell sorter (FACS) analysis revealed that proteotoxic stress increased O2− to a level that is comparable to the level induced by the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. 1D), while rotenone induced a massive increase in the amount of ROS (Fig. 1D). We also confirmed that the downstream effectors of ERα, such as AKT phosphorylation, proteasome, and Omi, were not activated in these cells (see Fig. S1 in the supplemental material).
Taken together, these results suggest that proteotoxic stress does not compromise cellular integrity despite the elevation in ROS levels and the absence of ERα. Therefore, additional mechanisms must take place to maintain the viability of cells undergoing proteotoxic stress.
Proteotoxic stress activates mitophagy.
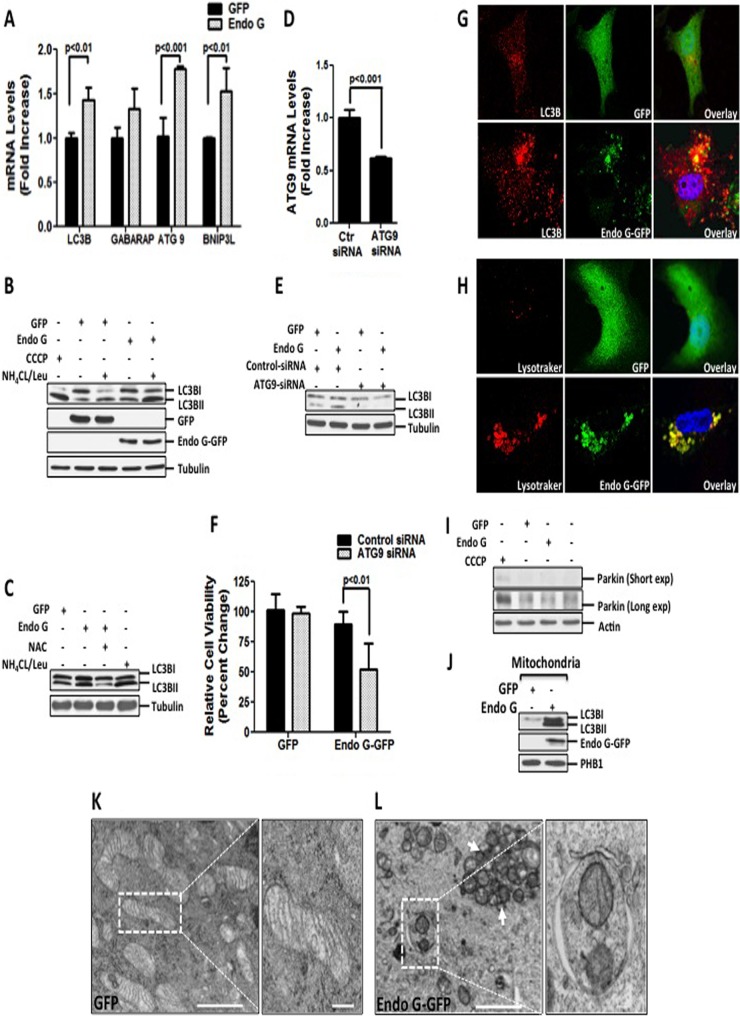
Autophagy acts as a survival mechanism by eliminating irreversibly damaged organelles. Since a fraction of the cells exhibited mitochondrial clustering and insoluble protein aggregates, we tested whether autophagy is activated by proteotoxic stress. We found significant elevations in the levels of a variety of ATG genes, such as LC3B, ATG9, and BNIP3L (Fig. 2A). GABARAP levels were also moderately elevated (Fig. 2A). In addition, proteotoxic stress stimulated an increase in phosphatidylethanolamine (PE) lipidation of endogenous LC3B (LC3BII), a marker of autophagy (Fig. 2B). Furthermore, we evaluated the lysosomal turnover of PE-lipidated LC3B to verify autophagic flux by NH4CL/Leu treatment and found an increase in the accumulation of LC3BII (Fig. 2B). Similar results were obtained by using other mutated mitochondrial proteins, such as SOD1-GFP, to generate proteotoxic stress (see Fig. S2A and B in the supplemental material) in ERα-negative breast cancer cell lines, including MDA-MB231 and MDA-MB157 (see Fig. S2A, C, and D in the supplemental material). To further verify that autophagy induction is due to the accumulation of proteins in the mitochondria, we also tested the effects of expression of ΔMLS-Endo G-His, which lacks the mitochondrial localization sequence (MLS) and thus localizes in the cytosol. Since this construct contains a His tag rather than a GFP tag, we also included in these experiments a vector expressing full-length Endo G-His as a control (see Fig. S2B in the supplemental material). In agreement with our previous findings, we found that full-length Endo G-His also induced an increase in LC3BII levels, while deletion of the MLS abolished this effect.
FIG 2.
ROS-mediated mitophagy is required to maintain the viability of cells undergoing proteotoxic stress. (A) mRNA levels of LC3B, GABARAP, ATG9, and BNIP3L were assessed by qRT-PCR in MDA-MB231 cells transfected with the indicated plasmids at 24 h. (B) MDA-MB231 cells were treated with 10 μM CCCP overnight or transfected with the indicated plasmids for 48 h, followed by treatment with a combination of 20 μM NH4CL and 0.1 mM leupeptin for 3 h prior to harvesting. Protein extracts were used for Western analysis of lipidated LC3BII. (C) Lysates from cells transfected with the indicated plasmids in the presence or absence of 5 mM NAC were used for Western analysis of LC3BII. (D) Efficiency of knockdown and ATG9 mRNA levels were evaluated by qRT-PCR of cells transfected with 20 nM siRNA against luciferase or siRNA against ATG9 followed by overexpression with Endo G-GFP for 48 h. (E) Crude extracts from MDA-MB231 cells transfected with either 20 nM siRNA against luciferase or 20 nM siRNA against ATG9, followed by transfection with the indicated plasmids, were analyzed by Western blotting for LC3B lipidation. (F) Viability of cells transfected as described below for panel H was determined by trypan blue staining at 48 h. (G) LC3B punctum formation was assessed by confocal microscopy in cells stained with anti-LC3B antibody (red) and counterstained with DAPI to visualize nuclei (blue), whereas Endo G-GFP or GFP is indicated in green. (H) Transfected cells loaded with LysoTracker Red (LTR) were analyzed by confocal microscopy to detect the overlay (yellow) of LTR-labeled acidic autolysosomal compartments (red) with GFP or Endo G-GFP (green). (I) Crude lysates from untreated cells or cells treated with 10 μM CCCP or transfected as indicated were tested for parkin by Western blotting. (J) Mitochondrial fractions were isolated from cells transfected with the indicated plasmids and subjected to Western blotting to detect levels of LC3BII. Overexpression of Endo G-GFP was confirmed by immunoblotting with Endo G antibody. PHB1 were used as a loading control for mitochondrial fractions. (K) GFP-overexpressing cells were subjected to electron microscopy (magnification, ×5,000; scale bar, 2 μm). Also shown is a higher-magnification view of mitochondria from the selected area (magnification, ×10,000; scale bar, 1 μm). (L) Electron micrographs of Endo G-GFP-overexpressing cells (magnification, ×5,000; scale bar, 2 μm) and a magnified view of the selected area indicating fragmented mitochondria engulfed in the autophagosome (magnification, ×20,000; scale bar, 0.5 μm).
Since accumulation of misfolded proteins in the mitochondria increases ROS levels, we tested whether inhibition of ROS by the antioxidant N-acetylcysteine (NAC) affects the induction of autophagy. We found that NAC abolished the increase in LC3BII levels induced by proteotoxic stress (Fig. 2C), indicating that ROS are required for autophagy. Furthermore, we found that inhibition of ATG9 by siRNA abolished the increase in LC3BII levels (Fig. 2D and E) and led to a 40% reduction in viability (Fig. 2F). Therefore, we concluded that cells undergoing proteotoxic stress activate ROS-dependent autophagy to maintain their viability.
To gain further insights into the autophagy triggered by proteotoxic stress, we assessed punctum formation that results from lipidation of endogenous LC3B. This analysis revealed that cells undergoing proteotoxic stress displayed a drastic increase in numbers of LC3B puncta, which overlaid Endo G-GFP, suggesting stimulation of mitophagy (Fig. 2G). In addition, LysoTracker Red (LTR) staining of the lysosomes showed an increase in the number of LTR-labeled punctate structures, which is indicative of autophagic activation (Fig. 2H). Moreover, a significant fraction of Endo G-GFP aggregates colocalized with the lysosomes, suggesting fusion of mitochondria with the lysosomes (Fig. 2H).
Since the ubiquitin ligase parkin has been functionally linked to mitophagy (31–33), we tested the levels of parkin following proteotoxic stress. However, we found no evidence of induction of parkin under these conditions (Fig. 2I), suggesting a parkin-independent mechanism of mitophagy. In agreement with this possibility, mitophagy induced by restricted mitochondrial protein acetylation has also been found to be independent of parkin (34).
In support of the induction of mitophagy, enhanced levels of LC3B lipidation (LC3BII) were associated with the mitochondria of cells undergoing proteotoxic stress (Fig. 2J). Mitophagy induced by proteotoxic stress was further examined by electron microscopy. This analysis revealed normal mitochondrial morphology in control cells overexpressing GFP (Fig. 2K). In contrast, the mitochondria of cells undergoing proteotoxic stress exhibited an array of alterations of mitochondrial morphology (Fig. 2L). Notably, severely fragmented mitochondria were sequestered by autophagosomes, whereas the majority of mitochondria were not (Fig. 2L, arrow). These observations indicate that only a fraction of damaged mitochondria is eliminated by mitophagy.
Proteotoxic stress activates SirT3 and the antioxidant defense mechanisms of mitochondria.
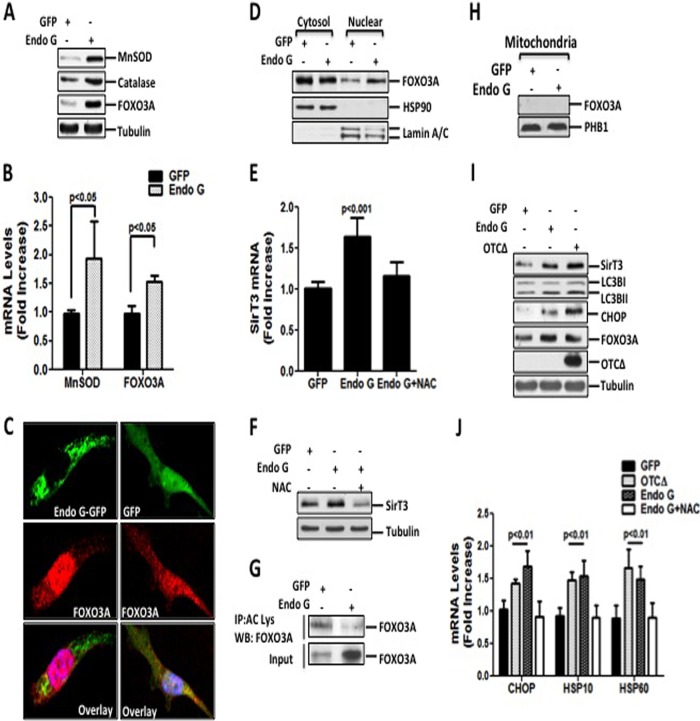
Since the majority of mitochondria are not eliminated by mitophagy, we hypothesized that activation of the antioxidant machinery may protect these mitochondria. We found drastic increases in the levels of MnSOD and catalase (Fig. 3A and B). Since FOXO3A is required for the transcription of both MnSOD and catalase, we also tested FOXO3A and found an increase in its levels (Fig. 3A and B). Furthermore, confocal microscopy and subcellular fractionation revealed an accumulation of FOXO3A in the nucleus (Fig. 3C and D).
FIG 3.
Proteotoxic stress enhances mitochondrial antioxidant defense mechanisms. (A) Crude extracts from cells transfected with either control GFP or Endo G-GFP for 48 h were analyzed by Western blotting to detect MnSOD, catalase, and FOXO3A using the respective antibodies. (B) Transcript levels of MnSOD and FOXO3A were determined by qRT-PCR of cells transfected as described above for panel A for 24 h. (C) Localization of FOXO3A (red) was detected by immunofluorescence confocal microscopy in cells transiently transfected with either GFP or Endo G-GFP (green) for 48 h. Nuclei were counterstained with DAPI (blue). (D) Cytosolic and nuclear fractions of cells transfected with either GFP or Endo G-GFP were analyzed by immunoblotting to determine FOXO3A levels. Hsp90 and lamin A/C levels were used as loading controls for cytosolic and nuclear fractions, respectively. (E) Endogenous SirT3 mRNA levels were assessed by qRT-PCR at the 24-h time point in cells transfected as described above for panel A, in the presence or absence of 5 mM NAC. (F) SirT3 protein levels were detected by Western blotting in cells treated as described above for panel E. (G) Acetylated FOXO3A levels were detected by Western blotting (WB) of the immunoprecipitated (IP) proteins using an anti-acetylated lysine antibody. Levels of FOXO3A present in the input were also detected by Western blotting. (H) Mitochondrial fractions of cells transfected with either GFP or Endo G-GFP for 24 h were analyzed by immunoblotting for the presence of FOXO3A. (I) Crude lysates of cells overexpressing the indicated constructs were immunoblotted with the indicated antibodies. Tubulin served as a loading control. OTCΔ was detected only in the insoluble fraction. (J) Transcript levels of CHOP, Hsp10, and Hsp60 were determined by qRT-PCR of cells transfected with OTC and OTCΔ or either GFP or Endo G-GFP in the presence or absence of 5 mM NAC.
The deacetylase SirT3 promotes both the activity and nuclear translocation of FOXO3A through its deacetylation (5, 10, 17, 35–38). Therefore, we evaluated SirT3 levels and found that proteotoxic stress increased SirT3 at both the transcript (Fig. 3E) and protein (Fig. 3F) levels. Treatment with the antioxidant NAC abolished the elevation in SirT3 levels (Fig. 3E and F), suggesting that the increase is dependent on ROS. Additionally, we assessed the acetylation of FOXO3A. We found a significant deacetylation of FOXO3A (Fig. 3G) despite the higher levels of FOXO3A induced by proteotoxic stress (Fig. 3A and G). This finding supports previous reports that SirT3 regulates FOXO3A. Our results suggest that the effect of SirT3 on FOXO3A must be indirect, since we found no evidence that FOXO3A localizes to the mitochondria (Fig. 3H).
Given that proteotoxic stress promotes the expression of novel markers of the UPRmt that are implicated in both mitophagy and antioxidant defense mechanisms, we next explored whether accumulation of other misfolded mitochondrial proteins also induces these markers. The mutated form of ornithine transcarbamylase (OTCΔ) has been shown to promote the formation of insoluble aggregates similar to those induced by mutant Endo G (20). We found that proteotoxic stress due to the accumulation of OTCΔ also led to elevations of SirT3, FOXO3A, as well as LC3BII levels (Fig. 3I), suggesting that these responses are not specific to misfolded Endo G.
The transcription factor CHOP, a well-characterized marker of the UPRmt, has been reported to be activated by the accumulation of OTCΔ (20). In addition, CHOP activation is required for transcription of the mitochondrial chaperones Hsp10 and Hsp60 (20). We confirmed these effects of OTC in our model (Fig. 3I and J).
To further examine the effect of proteotoxic stress caused by different misfolded mitochondrial proteins, we also tested the effect of accumulation of misfolded Endo G on CHOP, Hsp10, and Hsp60. We found that these markers were induced (Fig. 3I and J) and that their induction was dependent on ROS (Fig. 3J). Therefore, these findings indicate that proteotoxic stress is not protein specific and activates both CHOP as well as the antioxidant machinery. This finding is in agreement with our previous observation that in estrogen receptor-positive breast cancer cells, ERα can be activated using a wide array of different mitochondrial proteins (30).
CHOP is dispensable for the integrity of cells undergoing intermembrane space stress.
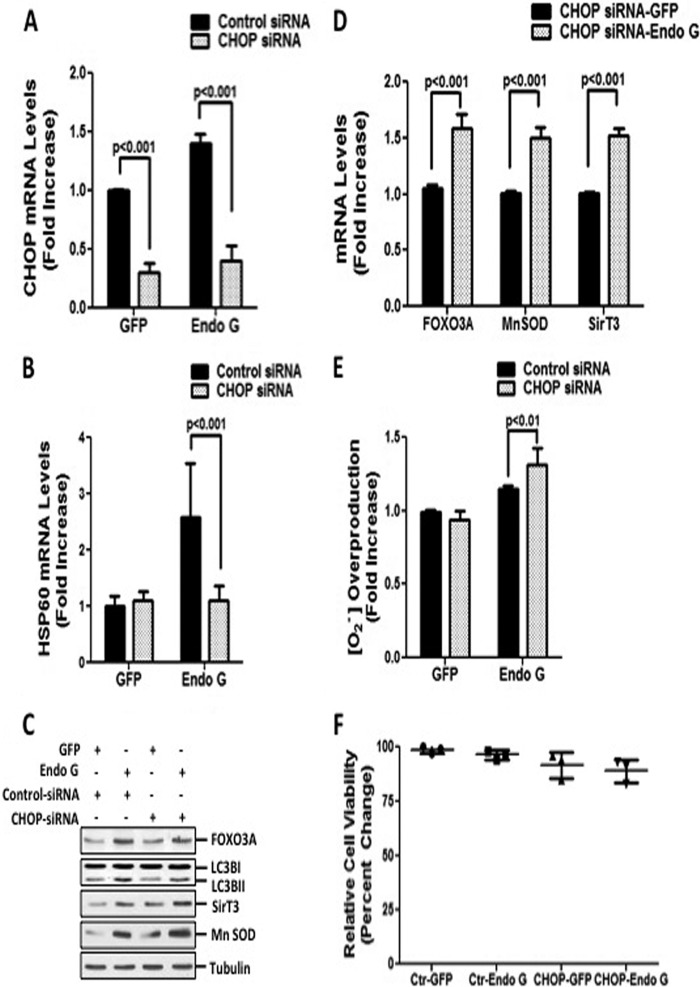
Since we found that CHOP is upregulated by proteotoxic stress in our model, we next focused on testing whether inhibition of CHOP by siRNA affects the induction of mitophagy and the antioxidant machinery. First, we confirmed that siRNA against CHOP significantly diminished the transcript levels of CHOP and its downstream target Hsp60 (Fig. 4A and B). However, inhibition of CHOP did not affect the upregulation of FOXO3A, SirT3, or MnSOD or the lipidation of LC3B by proteotoxic stress (Fig. 4C and D). Furthermore, the absence of CHOP led to a small but significant increase in mitochondrial O2− levels (Fig. 4E). However, this slight elevation in ROS levels was not sufficient to compromise cellular viability (Fig. 4F).
FIG 4.
CHOP is not essential for activation of cytoprotective responses and maintaining cellular viability. (A and B) Transcript levels of CHOP and Hsp60 were determined by qRT-PCR of cells transfected with 20 nM siRNA against CHOP or 20 nM control siRNA against luciferase, followed by transfection with either GFP or Endo G-GFP. (C) Endogenous levels of FOXO3A, lipidated LC3B, SirT3, and MnSOD were evaluated by Western blotting of the crude extract of cells treated as described above for panel A. (D) Transcript levels of endogenous FOXO3A, SirT3, and MnSOD were measured by qRT-PCR of cells treated with 20 nM siRNA against CHOP, followed by transfection with either GFP or Endo G-GFP. (E) Mitochondrial superoxide anion levels were determined by FACS analysis of cells transfected as described above for panel A and stained with MitoSox Red. (F) Viability of MDA-MB231 cells, transfected as described above for panel A, was determined by trypan blue staining at 48 h.
Taken together, these results confirm that the induction of CHOP is required to promote the transcription of the mitochondrial chaperones. However, CHOP is not essential to activate mitophagy and the antioxidant machinery.
SirT3 is required to maintain mitochondrial integrity.
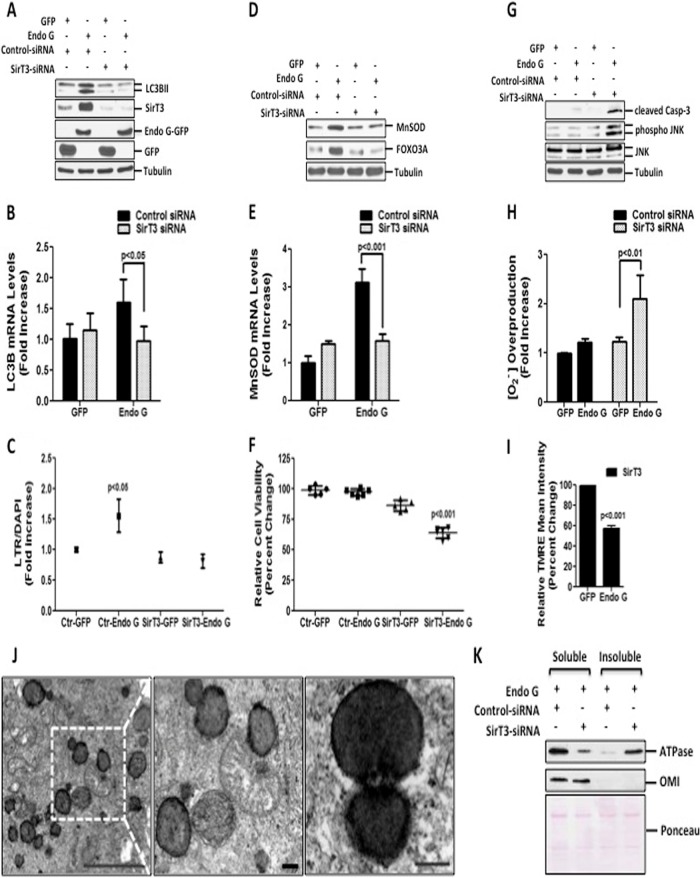
Having established that CHOP is not essential to maintain cellular viability in response to proteotoxic stress, we next assessed the role of SirT3. Thus, we tested the levels of lipidated LC3B in cells where the expression of SirT3 is inhibited by siRNA. We found that reduction of SirT3 levels inhibited LC3B lipidation (Fig. 5A) and significantly abrogated the increase in transcript levels of LC3B (Fig. 5B) and LysoTracker staining (Fig. 5C). Moreover, inhibition of SirT3 led to a failure to upregulate MnSOD (Fig. 5D and E) and FOXO3A (Fig. 5D).
FIG 5.
SirT3 is required to maintain mitochondrial integrity and cellular viability of cells undergoing proteotoxic stress. (A) Lipidated levels of LC3B were detected by immunoblotting in cells transfected with 20 nM control siRNA against luciferase or against SirT3, followed by transfection with the indicated plasmids for 48 h. Endogenous levels of SirT3 and exogenous levels of Endo G-GFP and GFP were evaluated by Western blotting. (B) Endogenous mRNA levels of LC3B were determined by qRT-PCR of cells treated as described above for panel A. (C) LTR uptake was assessed in fixed cells treated as described above for panel A. The same results were obtained by using two different siRNAs against SirT3. (D) Immunoblots of crude lysates from cells treated as described above for panel A were used to detect the levels of the indicated proteins by probing with the respective antibodies. (E) Endogenous mRNA levels of MnSOD were determined by qRT-PCR of cells treated as described above for panel A. (F) Viability of MDA-MB231 cells, transfected as described above for panel A, was determined at 48 h. (G) Crude cellular lysates, treated as described above for panel A, were used to assess cleaved caspase-3, phosphorylated JNK, and total JNK levels by Western blotting. (H) Mitochondrial superoxide anion levels were determined by FACS analysis in cells transfected as described above for panel A and stained with MitoSox Red. (I) Mitochondrial potential was evaluated by FACS analysis in cells treated with 20 nM siRNA against SirT3 followed by transfection with either GFP or Endo G and stained with TMRE dye at 48 h. (J) Representative electron micrographs of mitochondria from MDA-MB231 cells transfected with 20 nM siRNA against SirT3 followed by Endo G-GFP at 48 h (scale bar, 5 μm). Also shown is a higher-magnification view of the selected region indicating fragmented mitochondria with electron-dense aggregates (scale bar, 0.5 μm). (K) The soluble and insoluble fractions of isolated mitochondria from cells transfected as indicated for 24 h and lysed with buffer containing Triton X-100 were subjected to Western blotting to detect FoF1 ATP synthase α subunit levels. Ponceau S staining served as a loading control.
Importantly, inhibition of SirT3 significantly compromised the viability of cells undergoing proteotoxic stress (Fig. 5F), which correlated with the activation of cell death markers such as cleaved caspase-3 and Jun N-terminal protein kinase (JNK) phosphorylation (Fig. 5G). To determine the effect of SirT3 on the integrity of the mitochondria, we determined mitochondrial O2− production and membrane potential. While the significant increase in the mitochondrial O2− level induced by proteotoxic stress alone at 24 h (Fig. 1D) was no longer evident at 48 h (Fig. 5H), we found that the lack of SirT3 triggered prominent and sustained mitochondrial O2− levels following proteotoxic stress (Fig. 5H). In addition, a drastic reduction in the integrity of the mitochondrial membrane potential was also evident (Fig. 5I).
Having established that loss of SirT3 led to an inhibition of the autophagy marker LC3B, collapse of the membrane potential, and activation of apoptotic markers, we aimed at analyzing the morphology of the mitochondria under these conditions by electron microscopy. This analysis revealed the presence of extensively fragmented mitochondria (Fig. 5J). However, despite these severely stressed mitochondria, no evidence of mitophagy was observed, confirming that in the absence of SirT3, mitophagy is inhibited. Furthermore, the electron micrographs indicated electron-dense mitochondria, suggesting aggregation of mitochondrial proteins. Consistent with our evidence, identical electron-dense areas were observed in cells lacking the mitochondrial protease LonP (39).
To further test this possibility, we examined the soluble and insoluble fractions of the FoF1 ATP synthase α subunit. The FoF1 ATP synthase α subunit of complex V plays a critical role in the electron transport chain. In addition, it was reported to interact with a Hsp60 chaperone and reduce aggregate formation (40). We found that inhibition of SirT3 led to the accumulation of the FoF1 ATP synthase α subunit in the insoluble fraction, while its level decreased in the soluble fraction (Fig. 5L).
Taken together, these results indicate an essential role of SirT3 as a critical mitochondrial checkpoint required for induction of both the antioxidant machinery and mitophagy. Moreover, these results suggest that SirT3 is required to limit protein misfolding and the formation of aggregates in the mitochondria. Collectively, these observations support the notion that SirT3 plays a novel role in the UPRmt triggered by mitochondrial proteotoxic stress.
SirT3 is activated by mitochondrial stress.
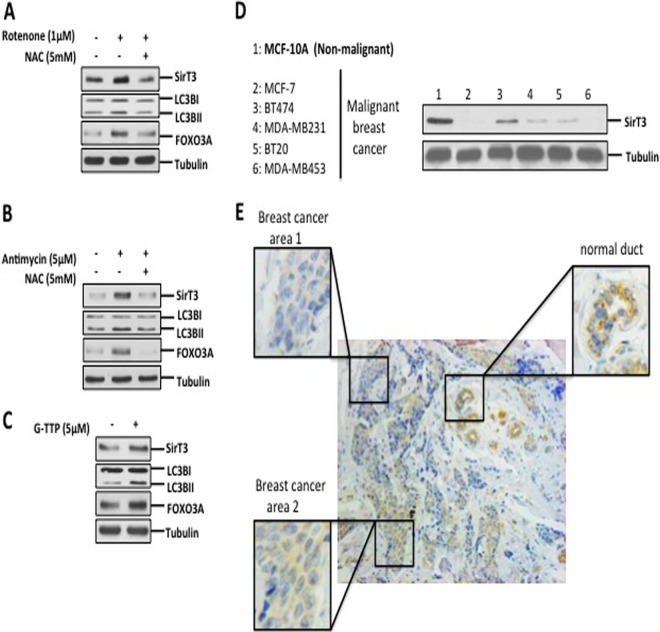
The use of specific misfolded proteins such as OTCΔ and Endo G has been useful in defining the different markers of the UPRmt. These tools have allowed the identification of CHOP (20), ERα (30), and, in the current study, SirT3 as markers of the UPRmt. Since ROS induce the formation of a broad array of protein aggregates, rather than accumulation of protein-specific aggregates used in our model, we aimed at determining whether stimulators of ROS activate SirT3 and its downstream targets. To test this possibility, cells were treated with rotenone, which inhibits complex I, leading to accumulation of mitochondrial ROS. We found that levels of SirT3, lipidation of LC3B, and FOXO3A were increased under these conditions (Fig. 6A). Furthermore, treatment with the antioxidant NAC abolished this effect of rotenone (Fig. 6A). In addition, antimycin A, an inhibitor of complex III of the electron transport chain, induced the same effect as rotenone (Fig. 6B).
FIG 6.
SirT3 induction is triggered by other mitochondrial stressors and detected in human breast adenocarcinoma. (A and B) Crude extracts from cells treated for 24 h with 1 μM rotenone or 5 μM antimycin A in the presence or absence of 5 mM NAC were subjected to Western blotting and evaluated for levels of SirT3, FOXO3A, and lipidation of LC3B. (C) Lysates of cells treated with 5 μM G-TPP for 4 h were tested to determine the levels of the indicated proteins. (D) Crude lysates from the indicated cells were evaluated to determine SirT3 levels. (E) Representative histological image of a primary breast adenocarcinoma tumor tissue stained for SirT3. Higher-magnification views of the boxed region are shown.
To further validate the activation of SirT3 by a broad array of protein aggregates, we treated cells with gamitochondrial matrix inhibitor (G-TPP), an inhibitor of mitochondrial Hsp90 chaperones (41). G-TPP has been shown to promote the aggregation of a variety of proteins (41). We found that G-TPP led to increases in the levels of SirT3, FOXO3A, and lipidation of LC3BII (Fig. 6C). Therefore, these results indicate that both ROS and accumulation of misfolded proteins in the mitochondria induce activation of SirT3 and its downstream targets.
Since SirT3 has been reported to be a tumor suppressor, our finding that mitochondrial stress stimulates SirT3 activity raises the possibility that SirT3 plays a dual role in tumorigenesis. Whereas a reduction in SirT3 levels is required to enhance ROS and assists in metabolic reprogramming, proteotoxic stress activates SirT3 to reduce ROS levels below a critical threshold. In agreement with the role of SirT3 as a tumor suppressor, we found that compared to the levels in MCF-10A cells, which are transformed but nonmalignant, the levels of SirT3 were reduced in all breast cancer cell lines tested (Fig. 6D). Furthermore, we confirmed this observation using immunohistochemistry of primary breast cancer, where normal ducts stained strongly for SirT3 but invasive breast cancer areas of the same section showed either reduced or a complete loss of SirT3 staining (Fig. 6E). However, this analysis also revealed a heterogeneity of SirT3 staining among the areas of invasive breast cancer of the same histological grade (Fig. 6E), with staining being undetectable in some areas (area 1) but clearly present in other areas (area 2). While this observation on its own does not prove the dual role of SirT3, it is nevertheless consistent with the possibility that in areas of the tumor experiencing local mitochondrial stress, SirT3 is upregulated.
DISCUSSION
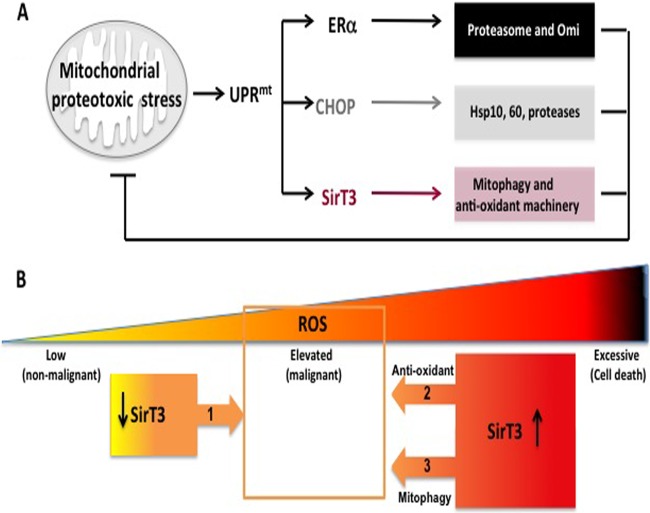
Cancer cells survive in a highly oxidative environment. High ROS levels, mutation in mitochondrial DNA, oxidative damage to lipids, and misfolded proteins characterize cancer cells. Ultimately, each of these interconnected events leads to mitochondrial stress. The UPRmt represents a powerful mechanism that allows the adaptation of the mitochondrial network to such an environment. Prior to the current study, two coordinators of this pathway had been described using OTC and Endo G: CHOP, which acts to upregulated mitochondrial chaperones and proteases, and ERα, which acts to upregulate the activity of the proteasome and the protease Omi. Using the same tools, we now report SirT3 as a third coordinator of the UPRmt, which acts to upregulate mitophagy and the antioxidant machinery. Collectively, these respective outcomes complement each other to result in an impressive array of cytoprotective responses that limit mitochondrial damage triggered by proteotoxic stress (Fig. 7A). The discovery of these coordinators of the UPRmt raises an important analogy to the endoplasmic reticulum UPR (19). PERK, ATF6, and IRE are critical coordinators of this UPR and activate different cytoprotective outcomes that also complement each other (19).
FIG 7.
(A) Diagram of the critical effectors of the UPRmt. Mitochondrial proteotoxic stress induces the UPRmt, which involves activation of CHOP, ERα, and SirT3. ERα, which is activated only in estrogen receptor-positive breast cancer cells, promotes the activity of the proteasome and of the protease Omi to limit the accumulation of misfolded proteins in the mitochondria. CHOP activates the heat shock proteins Hsp10 and Hsp60 as well as the protease LonP, which collectively also prevent the accumulation of misfolded proteins in the mitochondria. SirT3, described in the current study, confers additional but essential cytoprotective effects by activating antioxidant defense and mitophagy. SirT3 is critical for cellular viability under conditions of mitochondrial proteotoxic stress. Collectively, the combined outcome of the UPRmt is the reduction of proteotoxic stress and the maintenance of the integrity of the organelle. (B) SirT3 plays a dual role in breast cancer. On the one hand, a reduction in SirT3 levels is necessary to allow an increase in ROS levels and assist in the metabolic reprogramming of the mitochondria during transformation (arrow 1). On the other hand, by inducing the antioxidant machinery (arrow 2), SirT3 is required to repress ROS levels within a window (indicated by the orange square) that is compatible with the maintenance of cellular viability. In addition, by inducing the transcription of mitophagy genes (arrow 3), SirT3 primes the cells for the elimination of severely damaged mitochondria. However, the selective recruitment of the mitophagy machinery to these damaged organelles requires an additional step, which remains unknown. As these severely damaged organelles have excessive ROS, their elimination also contributes to reducing ROS within a window that is compatible with cellular viability (arrow 3). In the absence of SirT3, ROS increase to excessive levels, leading to cell death.
Our findings indicate that by activating the antioxidant machinery and coordinating mitophagy, SirT3 serves an essential role in the maintenance of mitochondrial network integrity. In contrast, activation of CHOP is not vital in retaining cellular viability, although it is required to enhance the mitochondrial chaperone and protease capacity. Moreover, our data also implicate SirT3 as a key regulator of a mitochondrial quality control checkpoint.
It has been reported that the expression level of SirT3 is reduced in breast cancers (8). In agreement with these observations, we also found that in both cell lines and primary breast cancers, SirT3 expression is reduced or lost. Collectively, these results support the notion that a reduction in SirT3 levels is required to elevate the levels of ROS and assist in metabolic reprogramming during malignant transformation (Fig. 7B, arrow 1). Importantly, our findings also demonstrate that the expression of SirT3 is inducible by mitochondrial proteotoxic stress. We propose that by activating the antioxidant machinery and mitophagy, SirT3 acts to lower ROS levels below a critical threshold and retain cellular viability (Fig. 7B, arrows 2 and 3). Therefore, we propose that SirT3 plays a dual role in cancer.
In support of previous reports, our data indicate that SirT3 is required for FOXO3A activity. Whether this effect is due to a direct association between FOXO3A and SirT3 in the mitochondria (16, 17) is a matter of debate. However, our data suggest that the effect of SirT3 on FOXO3A is likely to be indirect.
Our findings suggest that if proteotoxic stress cannot be resolved, SirT3-dependent activation of the mitophagy machinery primes the cells for the elimination of irreversibly damaged mitochondria. In this context, while the manuscript was in preparation, a study by Webster et al. reported that starvation induces SirT3-dependent mitophagy (42). While starvation activates a broad array of cellular stresses, which are not specific to mitochondria, our study links mitophagy and SirT3 to mitochondrial proteotoxic stress specifically. Therefore, we propose that SirT3 leads to the lipidation of LC3B and the transcription of genes involved in mitophagy. The selective recruitment of the mitophagy machinery to severely defective organelles required an additional step, which is initiated by excessive damage due to ROS. The recruitment of the ubiquitin ligase parkin to mitochondria with collapsed membrane potential (43) appears to be a possible mechanism underlying such selectivity. However, in agreement with the study by Webster et al. (42), we did not find any evidence of the involvement of parkin. Therefore, the mechanism by which SirT3 coordinates the selection of irreversibly damaged mitochondria remains to be determined.
Collectively, the discovery of these coordinators of the UPRmt illustrates the remarkable plasticity of the mitochondrial network to adapt to stress. Clearly, the complexity of the UPRmt is only beginning to be revealed. As it may represent an Achilles' heel of cancer cells, a full understanding of the UPRmt is critical to enhance our ability to target the mitochondrial network for therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dario Altieri for providing us with G-TPP. We also thank Mary Hahn for her technical assistance with the FACS analysis.
This work was funded by NIH grant RO1 CA109482 to D.G. L.P. is supported by National Institutes of Health/National Cancer Institute training grant T32 CA78207.
We declare that we do not have any conflicts of interest.
Footnotes
Published ahead of print 9 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01337-13.
REFERENCES
- 1.Ward PS, Thompson CB. 2012. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21:297–308. 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantor JR, Sabatini DM. 2012. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2:881–898. 10.1158/2159-8290.CD-12-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ralph SJ, Rodriguez-Enriquez S, Neuzil J, Saavedra E, Moreno-Sanchez R. 2010. The causes of cancer revisited: “mitochondrial malignancy” and ROS-induced oncogenic transformation—why mitochondria are targets for cancer therapy. Mol. Aspects Med. 31:145–170. 10.1016/j.mam.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 4.Hamanaka RB, Chandel NS. 2010. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35:505–513. 10.1016/j.tibs.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. 2010. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17:41–52. 10.1016/j.ccr.2009.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombard DB, Tishkoff DX, Bao J. 2011. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb. Exp. Pharmacol. 206:163–188. 10.1007/978-3-642-21631-2_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. 2010. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40:893–904. 10.1016/j.molcel.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. 2011. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19:416–428. 10.1016/j.ccr.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, Lorkiewicz P, St. Clair D, Hung MC, Evers BM, Zhou BP. 2013. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23:316–331. 10.1016/j.ccr.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. 2008. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 28:6384–6401. 10.1128/MCB.00426-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SH, Lu HF, Alano CC. 2011. Neuronal Sirt3 protects against excitotoxic injury in mouse cortical neuron culture. PLoS One 6:e14731. 10.1371/journal.pone.0014731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. 2011. SIRT3 and cancer: tumor promoter or suppressor? Biochim. Biophys. Acta 1816:80–88. 10.1016/j.bbcan.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. 2010. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12:662–667. 10.1016/j.cmet.2010.11.015 [DOI] [PubMed] [Google Scholar]
- 14.Finley LW, Haigis MC. 2012. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol. Med. 18:516–523. 10.1016/j.molmed.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemoto S, Finkel T. 2002. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295:2450–2452. 10.1126/science.1069004 [DOI] [PubMed] [Google Scholar]
- 16.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. 2008. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int. J. Biol. Sci. 4:291–299. 10.7150/ijbs.4.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. 2009. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 119:2758–2771. 10.1172/JCI39162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sena LA, Chandel NS. 2012. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48:158–167. 10.1016/j.molcel.2012.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutkowski DT, Kaufman RJ. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20–28. 10.1016/j.tcb.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21:4411–4419. 10.1093/emboj/cdf445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. 1999. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 18:522–533. 10.1093/emboj/18.3.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldridge JE, Horibe T, Hoogenraad NJ. 2007. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One 2:e874. 10.1371/journal.pone.0000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan MT, Hoogenraad NJ. 2007. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76:701–722. 10.1146/annurev.biochem.76.052305.091720 [DOI] [PubMed] [Google Scholar]
- 24.Pellegrino MW, Nargund AM, Haynes CM. 2013. Signaling the mitochondrial unfolded protein response. Biochim. Biophys. Acta 1833:410–416. 10.1016/j.bbamcr.2012.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker MJ, Tatsuta T, Langer T. 2011. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3:a007559. 10.1101/cshperspect.a007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horibe T, Hoogenraad NJ. 2007. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One 2:e835. 10.1371/journal.pone.0000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13:467–480. 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 28.Radke S, Chander H, Schafer P, Meiss G, Kruger R, Schulz JB, Germain D. 2008. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J. Biol. Chem. 283:12681–12685. 10.1074/jbc.C800036200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SH, Martins LM. 2012. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 19:1308–1316. 10.1038/cdd.2012.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papa L, Germain D. 2011. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J. Cell Sci. 124:1396–1402. 10.1242/jcs.078220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. 2010. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6:1090–1106. 10.4161/auto.6.8.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380. 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, Daniels MP, McCoy JP, Jr, Combs CA, Gucek M, Sack MN. 2013. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 126:4843–4849. 10.1242/jcs.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calnan DR, Brunet A. 2008. The FoxO code. Oncogene 27:2276–2288. 10.1038/onc.2008.21 [DOI] [PubMed] [Google Scholar]
- 36.van der Horst A, Burgering BM. 2007. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 8:440–450. 10.1038/nrm2190 [DOI] [PubMed] [Google Scholar]
- 37.Vogt PK, Jiang H, Aoki M. 2005. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 4:908–913. 10.4161/cc.4.7.1796 [DOI] [PubMed] [Google Scholar]
- 38.Peserico A, Chiacchiera F, Grossi V, Matrone A, Latorre D, Simonatto M, Fusella A, Ryall JG, Finley LW, Haigis MC, Villani G, Puri PL, Sartorelli V, Simone C. 2013. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell. Mol. Life Sci. 70:2015–2029. 10.1007/s00018-012-1244-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein SH, Venkatesh S, Li M, Lee J, Lu B, Hilchey SP, Morse KM, Metcalfe HM, Skalska J, Andreeff M, Brookes PS, Suzuki CK. 2012. The mitochondrial ATP-dependent Lon protease: a novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood 119:3321–3329. 10.1182/blood-2011-02-340075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray RE, Grasso DG, Maxwell RJ, Finnegan PM, Nagley P, Devenish RJ. 1990. Identification of a 66 KDa protein associated with yeast mitochondrial ATP synthase as heat shock protein hsp60. FEBS Lett. 268:265–268. 10.1016/0014-5793(90)81024-I [DOI] [PubMed] [Google Scholar]
- 41.Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, Ross AH, Plescia J, Altieri DC. 2011. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Invest. 121:1349–1360. 10.1172/JCI44855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, Daniels MP, McCoy JP, Jr, Combs CA, Gucek M, Sack MN. 2013. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 126:4843–4849. 10.1242/jcs.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazarou M, Jin SM, Kane LA, Youle RJ. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22:320–333. 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.