Abstract
Misfolding of proteins containing abnormal expansions of polyglutamine (polyQ) repeats is associated with cytotoxicity in several neurodegenerative disorders, including Huntington's disease. Recently, the eukaryotic chaperonin TRiC hetero-oligomeric complex has been shown to play an important role in protecting cells against the accumulation of misfolded polyQ protein aggregates. It is essential to elucidate how TRiC function is regulated to better understand the pathological mechanism of polyQ aggregation. Here, we propose that vaccinia-related kinase 2 (VRK2) is a critical enzyme that negatively regulates TRiC. In mammalian cells, overexpression of wild-type VRK2 decreased endogenous TRiC protein levels by promoting TRiC ubiquitination, but a VRK2 kinase-dead mutant did not. Interestingly, VRK2-mediated downregulation of TRiC increased aggregate formation of a polyQ-expanded huntingtin fragment. This effect was ameliorated by rescue of TRiC protein levels. Notably, small interference RNA-mediated knockdown of VRK2 enhanced TRiC protein stability and decreased polyQ aggregation. The VRK2-mediated reduction of TRiC protein levels was subsequent to the recruitment of COP1 E3 ligase. Among the members of the COP1 E3 ligase complex, VRK2 interacted with RBX1 and increased E3 ligase activity on TRiC in vitro. Taken together, these results demonstrate that VRK2 is crucial to regulate the ubiquitination-proteosomal degradation of TRiC, which controls folding of polyglutamine proteins involved in Huntington's disease.
INTRODUCTION
Mutant proteins containing abnormal polyglutamine (polyQ) expansions beyond a critical threshold are involved in several late-onset neurodegenerative disorders (1). For example, the mutation responsible for Huntington's disease (HD) is >40 polyQ expansions in the N terminus of huntingtin (Htt), a ubiquitously expressed cytoplasmic protein. HD is characterized by the selective and progressive dysfunction and death of neurons, mainly in the striatum, which manifest as voluntary movement dysfunction, personality changes, and dementia (2, 3). The causative mutant Htt protein is prone to proteolysis, and the resulting N-terminal fragments containing expanded polyQ repeats transition into a toxic conformation (4, 5). This eventually leads to the accumulation of intracellular aggregates or inclusion bodies, often consisting of insoluble, heat-stable β-sheet amyloid deposits (6). The length of polyQ tracts and the age-dependent accumulation of the mutant Htt contribute to HD onset and progression (7). Aggregation-based diseases disproportionately affect postmitotic cells, such as neurons, presumably because they cannot dilute the toxic protein aggregates during cell division (6).
Molecular chaperones are the first line of defense against misfolded, aggregation-prone proteins and are the most potent suppressors of neurodegeneration (8). Heat shock protein 70 (HSP70) and HSP40 have been shown to protect cells against polyQ aggregation and toxicity in various cellular and animal models (9–12). In addition, the eukaryotic chaperonin TCP-1 ring complex (TRiC) (also known as CCT [chaperonin-containing TCP-1]), a member of the HSP60 family, was also identified to have a significant role in protecting against polyQ aggregation in Caenorhabditis elegans (13). TRiC consists of eight homologous subunits arranged in two octameric rings stacked back to back that form a central cavity in which substrate proteins are captured and properly folded in an ATP-dependent manner (14, 15). Potential TRiC substrates include skeletal proteins, Von Hippel-Lindau tumor suppressor protein (VHL), and many WD40 repeat proteins (16). Recently, several studies provided evidence that TRiC inhibits polyQ aggregation and cytotoxicity during the early stages of the aggregation process (17–19). It is therefore necessary to reveal the mode of TRiC regulation to better understand the upstream pathway of TRiC-assisted polyQ protein folding.
Vaccinia-related kinase 2 (VRK2) is a novel serine/threonine kinase similar in sequence and structure to the catalytic domain of the casein kinase 1 (CK1) family (20). There are two isoforms of human VRK2; VRK2A is primarily in the endoplasmic reticulum (ER) and mitochondria because of its C-terminal transmembrane (TM) domain, and VRK2B, which lacks this domain, is found in both the cytosol and the nucleus (21). Functionally, VRK2 suppresses the hypoxia-induced TAK1–JIP1–Jun N-terminal protein kinase cascade by interacting with TAK1, which results in the downregulation of AP-1-dependent transcription (22). VRK2 also inhibits MEK/extracellular signal-regulated kinase signal transduction by interacting with the KSR1 scaffold protein in breast cancer cell lines, which leads to a signaling imbalance (23). In addition, VRK2 enhances cell survival through interacting with BHRF1, an antiapoptotic Epstein-Barr virus protein (24), and Bcl-xL, an antiapoptotic BH (Bcl-2 homology) domain protein (25). Because VRK2 has a sequence similar to that of the catalytic domain of vaccinia-related kinase 1 (VRK1), VRK2 also phosphorylates histone H3 (26), p53 (21), and barrier to autointegration factor (BAF) (27), which supposedly compensates for the VRK1 function. Recently, VRK2 was reported to phosphorylate NFAT1, which affects cell invasion through enhanced Cox2 expression (28). However, other VRK2 functions remain largely unexplored because VRK2 substrates have rarely been identified.
In the study described in this report, we addressed whether and how VRK2 regulates polyQ aggregation. The eukaryotic chaperonin TRiC was degraded by the ubiquitin-proteasome system (29), but its molecular mechanism has remained largely unexplored. We describe the molecular mechanism of TRiC protein ubiquitination through recruitment of the mammalian constitutive photomorphogenic 1 (COP1) E3 ligase complex. These findings suggest that VRK2 plays a critical role in regulating the function of the chaperonin TRiC, which is involved in polyQ protein aggregation.
MATERIALS AND METHODS
Plasmids.
VRK2 (VRK2A; accession number NM_027260) and TRiC expression constructs were generated by amplifying mouse full-length VRK2 and TRiC subunits (CCT1 to CCT8) by PCR from a day 16 mouse embryo cDNA library (Clontech, Mountain View, CA). For mammalian expression constructs, VRK2 and its kinase-dead (KD) mutant (VRK2-KD), generated by site-directed mutagenesis (Lys61 to Ala), were subcloned into the pFlag-CMV2 (Sigma, St. Louis, MO) and the pDsRed1-N1 and pDsRed1-C1 (BD Biosciences, San Jose, CA) vectors. TRiC subunits were subcloned into the pEGFP-C1 (BD Biosciences) and pcDNA3 (Invitrogen, Carlsbad, CA) vectors modified with a human influenza virus hemagglutinin (HA) epitope. For domain mapping, VRK2-N (amino acids 1 to 300), VRK2-C (amino acids 301 to 508), CCT4-N term (amino acids 1 to 220), CCT4 apical (amino acids 221 to 392), CCT4 api-C (amino acids 221 to 539), and CCT4-C term (amino acids 393 to 539) were subcloned into a pGEX-4T-3 vector. For expression in Escherichia coli, the VRK2 and TRiC subunits were subcloned into the pProEX, pGEX-4T-1, and pGEX-4T-3 (Amersham, Piscataway, NJ) and pET-SUMO (Invitrogen) vectors. Htt-exon1-GFP-pcDNA3.1 (where the length of the polyQ tract was 25 or 103) was a gift from Judith Frydman (Stanford University, Stanford, CA). pGEX6P1-COP1 (accession number AF527539) was kindly provided by Vishva M. Dixit (Genentech, Inc., South San Francisco, CA). The RING-box 1 (RBX1; accession number NM_014248) and Cul4A (accession number AY365124) constructs were gifts from Jong-Bok Yoon (Yonsei University, Seoul, South Korea). The DDB1 (accession number NM_001923) construct was a gift from Yue Xiong (Addgene plasmid 19909).
Biochemical methods.
Extract and protein preparation, immunoprecipitation, glutathione S-transferase (GST) pulldown, and Western blotting were performed as previously described (30).
In vitro kinase assay.
Kinase assays utilizing 1 μg recombinant GST-VRK2 (or HIS-VRK2) and 1 μg recombinant proteins (HIS-CCTx [where x is subunit 1 to 8], GST-COP1, or GST-RBX1) as the substrates were performed at 30°C in a 20-μl reaction mixture containing kinase buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 0.5 mM dithiothreitol, 150 mM KCl, [γ-32P]ATP). After 30 min, reactions were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and visualized with autoradiography.
Fluorescence microscopy.
Mammalian cells were grown on coverslips until they reached 50 to 60% confluence. Transfected cells were maintained for 24 h, fixed with 4% paraformaldehyde for 15 min, and, if necessary, immunostained with appropriate primary and fluorescence-conjugated secondary antibodies. Coverslips were mounted onto a slide with Fluoromount mounting medium (Sigma). An Axioplan2 fluorescence imaging microscope (Carl Zeiss, Jena, Germany) equipped with an ApoTome device (Carl Zeiss) was used to obtain fluorescence or differential interference contrast images. We also used superresolution structured illumination microscopy (SIM; N-SIM; Nikon, Tokyo, Japan). The raw images were reconstructed to three-dimensional SIM images using NIS-E software (Nikon). Images were taken with an Eclipse Ti-E research inverted microscope with Nikon's CFI Apo TIRF ×100 oil objective lens (numerical aperture, 1.49) and a 512- by 512-pixel resolution with an iXon DU-897 electron-multiplying charge-coupled-device camera (Andor Technology, Belfast, United Kingdom). Multicolor fluorescent images were acquired using a diode laser (488 nm, 561 nm). MitoTracker staining was performed with 250 nM MitoTracker red (CMXRos; Invitrogen) for 30 min at 37°C. The cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 40 min at room temperature. Mounting was done with Cytoseal XYL mounting medium (UltraCruz hard-set mounting medium; Santa Cruz).
Yeast two-hybrid assay.
A genetic screening using the yeast interaction trap with bait plasmid pEG202 harboring a full-length mouse VRK2 gene and mouse liver cDNA library in the pJG4-5 plasmid was carried out in Saccharomyces cerevisiae strain EGY48 as previously described (31).
Cell culture and transfection.
HEK293T, SK-N-BE(2)C, A549, U2OS, SH-SY5Y, and HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml each of penicillin G and streptomycin. Transient transfection was carried out with the Metafectene reagent (Biontex, Munich, Germany) or a Microporator system (Invitrogen), as described in the manufacturers' protocols.
siRNA sequence.
The small interference RNA (siRNA) duplex pool targeting human VRK2 (siVRK2) and COP1 (siCOP1) was obtained from Dharmacon (Lafayette, CO). The siRNA sequence for the nonspecific control oligonucleotide was CCUACGCCACCAAUUUCGUdTdT.
Antibodies.
Commercially available antibodies were as follows: anti-Flag epitope (M2) from Sigma; anti-HA epitope from Roche (Mannheim, Germany); anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) from Calbiochem (San Diego, CA); anti-CCT1, anti-CCT2, anti-CCT4, anti-CCT7, anticalnexin, and anti-COP1 from Abcam (Cambridge, United Kingdom); anti-VRK2, anti-green fluorescence protein (anti-GFP), antiubiquitin, anti-HIS epitope, anti-GST, anti-Tom20, and anti-lamin B from Santa Cruz Biotechnology (Santa Cruz, CA); and all fluorescence-conjugated secondary antibodies from Invitrogen.
In vitro ubiquitination assay.
For the ubiquitination assay, 1 μg HIS-CCT4, 5 μg Flag-ubiquitin (Sigma), 1 μM UbcH5b (Boston Biochem, Cambridge, MA), 200 nM rabbit E1 (Boston Biochem), 1 μg GST-COP1 (E3), 1 μg GST-RBX1, 1 μg GST-DDB1, 1 μg Cul4A, and 1 μg SUMO-VRK2 were preincubated with 20 mM HEPES (pH 7.3) for 30 min at room temperature and then incubated in a buffer containing 20 mM HEPES (pH 7.3), 4 mM ATP, 10 mM MgCl2, and 4 mM dithiothreitol. After incubation for 2 h at 30°C with gentle agitation, the reaction solutions were boiled in PBS containing 0.5% Triton X-100 (PBST) with 1% SDS for 5 min and reduced to 0.1% SDS with PBST for immunoprecipitation with anti-CCT4. Finally, samples were subjected to SDS-PAGE, followed by immunoblotting with anti-Flag to detect polyubiquitinated CCT4.
Protein and RNA stability assay.
For cycloheximide (CHX)-chase assays, cells were treated with cycloheximide (Calbiochem) (HEK293T cells, 100 μg/ml; HeLa cells, 40 μg/ml) in isopropanol and harvested at the indicated times for Western blot analysis. For RNA stability, cells were treated with 10 μg/ml actinomycin D (Sigma) at 0 h and harvested at the indicated times. Total RNA was isolated using the TRI reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. RNA was reverse transcribed using oligo(dT) (Promega, Fitchburg, WI) and Moloney murine leukemia virus reverse transcriptase (Roche, Indianapolis, IN) in the presence of RNase inhibitor (RNasin; Promega). CCT4 mRNA levels were detected by quantitative real-time PCR using StepOnePlus real-time PCR systems (Applied Biosystems, Foster City, CA) with a SYBR green PCR mixture (Roche). We used primer pairs specific for GAPDH (forward primer, 5′-GCCATCAATGACCCCTTCATT-3′; reverse primer, 5′-GCTCCTGGAAGATGGTGATGG-3′) and CCT4 (forward primer, 5′-AAGCCTTGGACCAAAAGGAA-3′; reverse primer, 5′-AAGAGGGAGCCAGCAATGAT-3′).
Filter-trap assay.
Cells were lysed in PBST, and protein concentrations were measured. Lysates were diluted in 1% SDS in PBS and boiled for 5 min. Immediately after cooling, the indicated amounts of proteins were loaded onto nitrocellulose membranes (pore size, 0.2 μm) settled on a dot blotter (Bio-Rad, Hercules, CA). After blocking with 5% skim milk and PBST for 1 h, the membranes were incubated with mouse anti-GFP and horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibodies. HRP activity was detected by development in SUPEX solution (Neuronex, South Korea).
Statistical analysis.
All statistical analyses were performed with one-way analysis of variance and post hoc Tukey's multiple-comparison tests using GraphPad software. Differences were considered significant when P was <0.05.
RESULTS
VRK2 and chaperonin TRiC interaction.
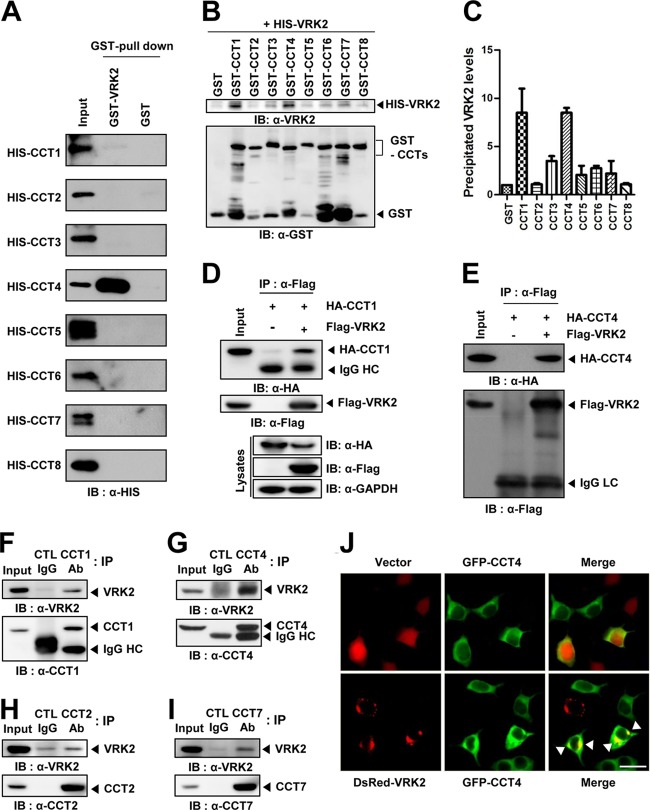
To explore the functional roles of VRK2 (which represents VRK2A), we performed yeast two-hybrid screening and found that VRK2 interacts with the CCT2 subunit of the mammalian chaperonin TRiC. In order to verify which subunit of TRiC directly binds to VRK2 in vitro, all TRiC subunits and VRK2 were purified as HIS (HIS-CCT) and GST (GST-VRK2) fusion protein forms, respectively, and GST pulldown assays were performed. HIS-CCT1 and -4 were shown to interact with GST-VRK2 (Fig. 1A), which was confirmed using GST-CCT and HIS-VRK2 constructs by inversely exchanging fusion proteins. GST-CCT1 and -4 also bound to HIS-VRK2 (Fig. 1B and C). To test whether VRK2 interacts with TRiC in vivo, immunoprecipitation experiments were carried out in HEK293T cells. When HA-CCT1 or -4 was coexpressed with Flag-VRK2, it was efficiently coimmunoprecipitated by an anti-Flag antibody (Fig. 1D and E). Even though purified CCT2 did not bind directly to VRK2 (Fig. 1A and B), a GST pulldown assay using HA tagging of all CCT subunits also demonstrated that it was efficiently pulled down by GST-VRK2 (see Fig. S1 in the supplemental material) because all eight TRiC subunits exist assembled to form a complex. These results provide evidence that VRK2 interacts with assembled TRiC complex proteins through specific binding between VRK2 and CCT1 or -4. Notably, interaction between endogenous VRK2 and CCT subunits (1, 2, 4, and 7) was also detected by immunoprecipitation with extracts of A549 cells, which are known to express high levels of VRK2 (Fig. 1F to I). Fluorescent imaging analysis has previously shown VRK2 in the ER, mitochondria, and nuclear envelope, likely because of its TM domain (21). However, our fluorescent imaging analysis with superresolution SIM (Nikon N-SIM) clearly showed that VRK2 was mainly present at the ER and the outer nuclear membrane (see Fig. S2A to C in the supplemental material); VRK2 colocalized with calnexin (ER protein marker) but not MitoTracker (mitochondria marker). We observed the images generated with Tom20 in detail and found that VRK2 slightly overlaps Tom20 at certain spots. It seems that a subpopulation of VRK2 is localized in the outer mitochondrial membrane. VRK2 was reported to regulate apoptosis through interaction with Bcl-xL in the outer mitochondrial membrane (25). Although VRK2 was observed in the nuclear membrane, it did not perfectly overlap the lamin B (an inner nuclear envelope protein marker). Scanning profiles of merged images of VRK2 and lamin B suggested that VRK2 localizes at the outer nuclear membrane (see Fig. S2D in the supplemental material). Unlike VRK2, GFP-CCT4 was diffuse throughout the cytoplasm, but it was enriched in DsRed-VRK2-positive areas (Fig. 1J). Taken together, these results indicate that VRK2 interacts with the eukaryotic chaperonin TRiC in vitro and in vivo.
FIG 1.
Interaction and colocalization of VRK2 with chaperonin TRiC. (A, B) GST pulldown assay of VRK2 and TRiC subunit proteins purified from E. coli cultures. (A) GST-VRK2 bound to HIS-CCT1 or -4. (B) HIS-VRK2 bound to GST-CCT1 or -4. (C) We calculated the amounts of bound VRK2 protein with GST-CCT proteins. (D, E) Immunoprecipitation assay was performed with the lysates of HEK293T cells cotransfected with Flag-VRK2 and HA-CCT1 (D) or HA-CCT4 (E) vectors. Encoded HA-CCT1 and HA-CCT4 were coimmunoprecipitated with an anti-Flag antibody. (F to I) The interaction between endogenous VRK2 and CCT subunit (1, 2, 4, and 7) proteins in A549 cells. Endogenous VRK2 was immunoprecipitated with anti-CCT subunit (1, 2, 4, and 7) antibody, and immunoprecipitated proteins were analyzed by Western blotting. (J) Colocalization of VRK2 and CCT4. HEK293T cells were cotransfected with Flag-VRK2 and HA-CCT4 and then stained for Flag (red), HA (green), and nuclei (Hoechst; blue). Arrowheads, concentrated colocalization of CCT4 and VRK2. Bar, 20 μm. IB, immunoblot; IP, immunoprecipitation; Ab, antibody.
Role of VRK2 in CCT4 downregulation.
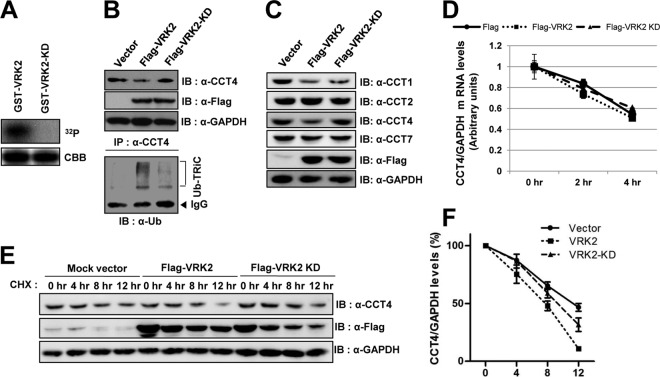
To determine whether VRK2 is a substrate for TRiC, we performed pulldown assays to observe the interactions between VRK2 and various CCT4 fragments. In the substrate discrimination of TRiC, the apical domain of TRiC is important for binding its substrates (32). However, VRK2 interacted with the C-terminal equatorial domain of TRiC, which is required for ATP binding (see Fig. S3A in the supplemental material). This suggests that VRK2 could bind to CCT1 and -4 due to the highly conserved equatorial domains of all chaperonin subunits (33). VRK2 domain mapping indicated that the C-terminal domain of VRK2 (with the exception of the kinase domain) binds to HIS-CCT4 (see Fig. S3B in the supplemental material). On the other hand, VRK2 is a serine-threonine protein kinase known to phosphorylate substrates such as p53 (21), BAF (27), and NFAT1 (28), and the ability of VRK2 to phosphorylate TRiC was of great interest. However, none of the purified HIS-CCTs were phosphorylated by GST-VRK2 (see Fig. S4A and B in the supplemental material). Although TRiC did not appear to be a VRK2 substrate, the interaction between VRK2 and TRiC led us to hypothesize that VRK2 may be an upstream regulator of TRiC. A VRK2 kinase-dead mutant construct (VRK2-KD) was generated by point mutating Lys61 into Ala, which eliminated enzymatic activity and effectively prevented autophosphorylation (Fig. 2A). VRK2 enzymatic activity was not necessary for VRK2 and CCT4 interaction (see Fig. S4C and D in the supplemental material). Surprisingly, Flag-VRK2 overexpression in mammalian cells significantly decreased endogenous CCT4, but VRK2-KD did not (Fig. 2B, top). As previously reported, the ubiquitin-proteasome system (UPS) plays an important role in determining TRiC's half-life (6 to 7 h) (29). Therefore, we examined the possibility that VRK2 enhances TRiC degradation by promoting TRiC subunit ubiquitination. We immunoprecipitated the same extracts used in the assay whose results are shown at the top of Fig. 2B with an anti-CCT4 antibody and immunoblotted with an antiubiquitin antibody. As hypothesized, polyubiquitinated CCT4 was obviously identified in the presence of VRK2 but not in the presence of VRK2-KD (Fig. 2B, bottom). We also tested whether VRK2 could downregulate other CCT subunits. Endogenous CCT1 protein levels were decreased in cells overexpressing VRK2, but endogenous CCT2 and CCT7 were not (Fig. 2C). Downregulation of CCT subunits might result from a direct interaction with VRK2, because CCT1 and -4 directly bound to VRK2 (Fig. 1A and B). To further confirm VRK2-mediated CCT4 ubiquitination, CCT4 turnover rates were measured in the presence of cycloheximide, an inhibitor of protein biosynthesis, or actinomycin D, an inhibitor of transcription. Although CCT4 mRNA stability was not affected by VRK2 (Fig. 2D), CCT4 protein turnover rates were accelerated in Flag-VRK2-overexpressing cells (Fig. 2E and F). Collectively, these results suggest that VRK2 decreases CCT4 protein levels via enzyme activity-dependent ubiquitination.
FIG 2.
VRK2 activity-dependent polyubiquitination and degradation of TRiC. (A) Construction of VRK2-KD (K61A). Purified GST-VRK2-KD did not autophosphorylate because it cannot bind ATP. CBB, Coomassie brilliant blue. (B) Polyubiquitination and degradation of CCT4 are dependent on VRK2 enzymatic activity. Lysates of cells overexpressing Flag-VRK2 or VRK2-KD were immunoblotted with anti-CCT4 antibody. TRiC levels decreased following Flag-VRK2 overexpression (top). The same lysates were immunoprecipitated with an anti-CCT4 antibody and immunoblotted with an antiubiquitin antibody. CCT4 polyubiquitination was promoted by Flag-VRK2 but not Flag-VRK2-KD (bottom). Ub, ubiquitin. (C) CCT1 was also degraded in cells overexpressing VRK2. VRK2 has no effect on degradation of CCT2 and CCT7. (D) VRK2 does not affect CCT4 mRNA stability. Cells were transfected with Flag-VRK2 or Flag-VRK2-KD and then treated with actinomycin D (10 μg/ml), a transcription inhibitor. (E) Decreased TRiC protein stability was dependent on VRK2 enzymatic activity through the proteasomal degradation pathway. Cells were harvested at the indicated times after cycloheximide (100 μg/ml) treatment to inhibit protein synthesis, and endogenous CCT4 was detected by Western blotting. The endogenous CCT4 protein turnover rate accelerated in cells overexpressing Flag-VRK2 but not in those overexpressing Flag-VRK2-KD. (F) The CCT4 protein turnover rate was quantified. The error bars represent the mean ± SEM (n = 3).
Physiological role of VRK2 in polyQ aggregation.
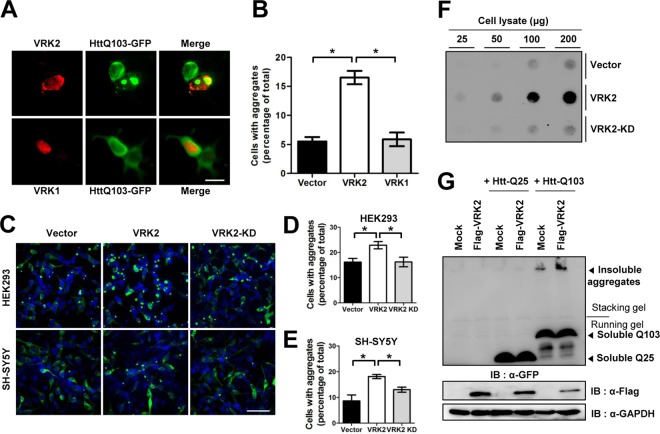
To further examine the physiological role of VRK2-mediated CCT4 downregulation, we visualized the polyQ-expanded Htt fragment fused to GFP. Htt with 103 Q residues (HttQ103) fused to GFP (HttQ103-GFP) has been used to demonstrate that functional TRiC is involved in molecular mechanisms that contribute to polyQ pathogenesis. For instance, TRiC impairment increases the extent of pathogenic HttQ103-GFP aggregate formation (17). Conversely, overexpression of either a specific TRiC subunit or all eight TRiC subunits significantly suppresses aggregate formation and neuronal cell death (18, 19). We coexpressed DsRed1-VRK2 with pathogenic HttQ103-GFP in HEK293T cells and manually counted the number of double-labeled cells containing large foci of aggregated HttQ103-GFP. Whereas VRK1 had no effect on HttQ103-GFP aggregate formation, VRK2 increased the number of cells containing foci approximately 3-fold over the number for a control vector (Fig. 3A and B). This result that VRK2 suppresses the function of TRiC in preventing polyQ aggregation suggests that VRK2 facilitates CCT4 degradation (Fig. 2B). Consistent with the notion that VRK2 enzymatic activity is necessary for modulating TRiC, VRK2-KD did not promote HttQ103-GFP aggregate formation (Fig. 3C). The pattern of VRK2-induced polyQ aggregation was also observed in neuroblastoma SH-SY5Y cells, suggesting the possibility that the levels of wild-type VRK2 in neuronal cells are critical for the onset and progression of polyQ diseases. Accordingly, it would be important to determine whether VRK2 levels are increased in HD patient brain. Consistent with the fluorescence imaging observations, VRK2 overexpression increased SDS-resistant aggregates detected by filter-trap assay (Fig. 3D). Conversely, expression of nonpathogenic HttQ25-GFP remained diffuse regardless of VRK2 because it folds well, despite TRiC disruption (Fig. 3E; see Fig. S5 in the supplemental material) (17). Taken together, these results suggest that enhanced expression of wild-type VRK2 accelerates multiple processes to trigger polyQ aggregation.
FIG 3.
VRK2-mediated degradation of TRiC increases polyQ-expanded Htt aggregation in mammalian cells. (A) HttQ103-GFP and DsRed1-C1-VRK2 (or -VRK1) were coexpressed in HEK293T cells, and fluorescent images were analyzed. (B) We calculated the percentage of cells containing foci of HttQ103-GFP aggregates in double-fluorescence-positive cells. The error bars represent the mean ± SD (n = 3). *, P < 0.05. (C) Fluorescence microscopic images of HttQ103-GFP expressed in HEK293 and neuroblastoma SH-SY5Y cells. GFP aggregates in both cell types were increased only by overexpression of wild-type VRK2 and not by overexpression of VRK2-KD. (D, E) We calculated the percentage of HEK293 (D) and SH-SY5Y (E) cells containing foci of HttQ103-GFP aggregates. The error bars represent the mean ± SEM (HEK293 cells, n = 4; SH-SY5Y cells, n = 3). *, P < 0.05. (F) Filter-trap assay of HttQ103-GFP aggregates. Cell lysates containing the indicated amount of protein were filtered on cellulose acetate membranes and immunoblotted with anti-GFP antibody to detect HttQ103-GFP aggregates. (G) Flag-VRK2 with HttQ103-GFP or HttQ25-GFP was coexpressed in HEK293T cells and then analyzed by Western blotting to detect polyQ aggregates. Bars, 10 μm (A) and 80 μm (C).
Attenuated polyQ aggregation following CCT4 rescue.
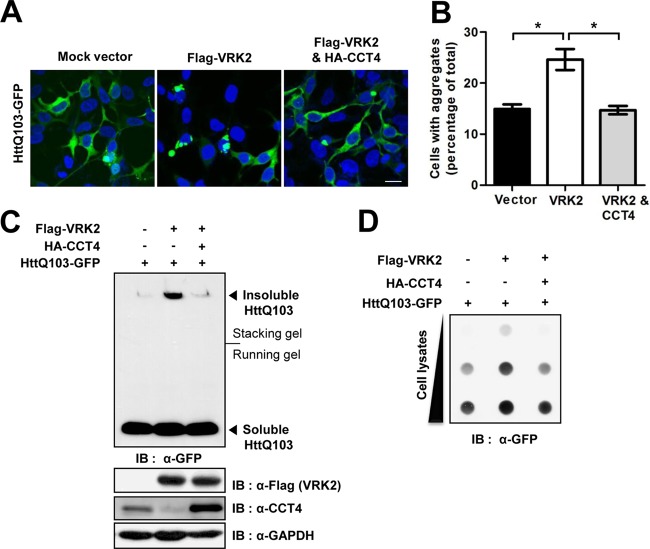
Because VRK2 manifests TRiC protein turnover and increases polyQ aggregation, we tested whether VRK2-mediated reduction of TRiC protein levels results in a greater number of polyQ aggregates. To clarify the observed association between VRK2 levels and polyQ aggregation, we carried out a CCT4 rescue experiment and measured polyQ aggregate levels with fluorescence imaging and Western blotting. Dense foci of polyQ aggregates were dramatically increased in cells overexpressing wild-type VRK2, whereas cells coexpressing CCT4 showed attenuated polyQ aggregation and a diffuse fluorescent signal similar to those of cells expressing the control vector (Fig. 4A and B). To gain more insight into the effects of CCT4 rescue on polyQ aggregation, we performed Western blotting with anti-GFP against HttQ103. Cells were lysed with radioimmunoprecipitation assay buffer to obtain soluble and insoluble polyQ aggregates. VRK2 overexpression markedly increased the levels of insoluble aggregates that were detected in the stacking gel due to their SDS insolubility and high molecular weight (Fig. 4C). Surprisingly, CCT4 cooverexpression effectively reduced the increased levels of insoluble aggregates induced by VRK2 overexpression down to basal aggregate levels, as observed in cells transfected with HttQ103 alone. In contrast, VRK2 overexpression and CCT4 rescue had no effect on soluble HttQ103 levels. Consistent with the results of fluorescence imaging and Western blotting, CCT4 rescue also decreased the level of SDS-insoluble aggregates detectable by filter-trap assay (Fig. 4D). These data suggest that VRK2-mediated TRiC protein turnover is required for the accumulation of polyQ aggregates.
FIG 4.
Rescue of TRiC alleviates polyQ-expanded huntingtin aggregation. (A) HttQ103-GFP and Flag-VRK2 with or without HA-CCT4 were coexpressed in HEK293T cells, and fluorescence microscopic images were analyzed. Bar, 20 μm. (B) The percentage of cells with HttQ103-GFP aggregate foci was calculated. The error bars represent the mean ± SEM (n = 4). *, P < 0.05. (C) The amounts of HttQ103-GFP aggregates were investigated by Western blotting using an anti-GFP antibody. The insoluble aggregates remained at the top of the gel, and soluble HttQ103-GFP entered the resolving gel. (D) Filter-trap assay for analysis of the insoluble aggregates in cells transfected with the indicated vectors.
Decreased polyQ aggregation following VRK2 knockdown.
To assess the role of endogenous VRK2 on TRiC-assisted polyQ protein folding, U2OS cells were transfected with HttQ103-GFP and small interference RNA (siRNA) against VRK2 (siVRK2) to confirm that efficient knockdown of negative regulators can change endogenous CCT4 levels. CCT4 levels were slightly increased, and SDS-resistant polyQ aggregates were also comparatively decreased in cells transfected with siRNA for VRK2 (Fig. 5A). The effect of the targeted knockdown of VRK2 on polyQ aggregation was examined by fluorescence imaging of cells expressing HttQ103-GFP for 3 days. In the case of control cells transfected with the siRNA control (siCont), HttQ103-GFP aggregates were shown in ∼20% of the GFP-positive cells at day 3. However, siVRK2-mediated knockdown decreased the proportion of cells containing aggregates to less than 5% of the GFP-positive cells at day 3. From days 1 to 3, the increasing rate of GFP foci in VRK2-depleted cells was significantly lower than that in siCont-treated cells (Fig. 5B and C). These results suggest that knockdown of VRK2 enhances the functional capacity of TRiC and eventually results in the suppression of polyQ aggregation. This provides more reliable evidence that VRK2 negatively regulates TRiC.
FIG 5.

Knockdown of VRK2 decreases formation of polyQ-expanded huntingtin aggregates. (A) HttQ103-GFP with or without siRNA against VRK2 was coexpressed in U2OS cells, and HttQ103 aggregates were investigated by immunoblotting. (B) Fluorescence microscopy of HttQ103-GFP expressed in HeLa cells treated with siVRK2 or siCont. VRK2 depletion decreased the number of cells containing HttQ103-GFP aggregates. Bar, 20 μm. (C) The percentage of aggregate-positive cells from panel B was calculated. The error bars represent the mean ± SD (n = 3). **, P < 0.01 versus siCont-treated cells at 2 and 3 days; n.s, not significant.
Involvement of COP1 E3 ligase in VRK2-mediated TRiC turnover.
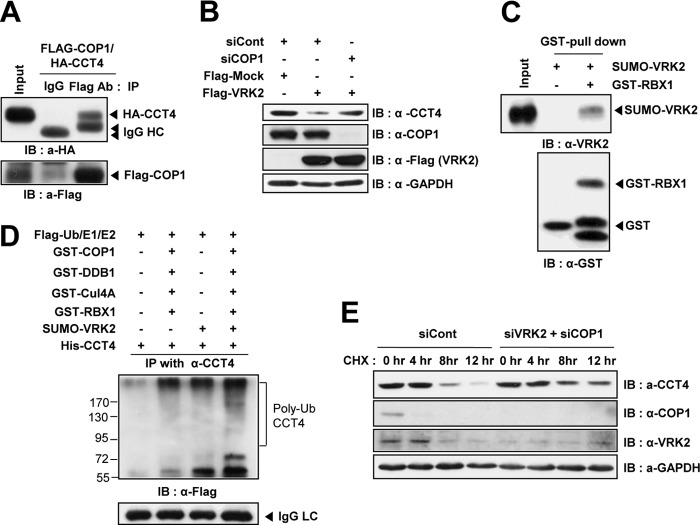
VRK2 increased CCT4 polyubiquitination of CCT4, which induced CCT4 turnover (Fig. 2). We were interested in determining which E3 ligase was activated by VRK2 and whether it could function as a ubiquitin ligase for CCT4. Because increased CCT4 ubiquitination was dependent on VRK2 enzymatic activity, it raises the possibility that phosphorylation could affect ubiquitination. Phosphorylation-dependent ubiquitination is a well-known phenomenon described for the cullin-based RING E3 ligase complex (34). To identify which E3 ligase was involved in CCT4 protein ubiquitination, we screened RING E3 ligases using siRNA and determined which one restored CCT4 protein levels in VRK2-overexpressing cells. COP1 E3 ligase was previously reported to interact with TRiC (35) and strongly bound to it (Fig. 6A). The observed VRK2-mediated downregulation of TRiC in Fig. 2B was reversed in siRNA against COP1-transfected cells (Fig. 6B). COP1 functions as an E3 ligase by forming a supercomplex that also includes heterodimeric substrate receptor DET1, adaptor DDB1, scaffold Cul4A, and RBX1 to recruit the E2 enzyme (36). To verify this observation, we first tested the functional interaction between VRK2 and COP1 with an in vitro kinase assay. Although RBX1 directly bound to VRK2 (Fig. 6C), we could not detect the phosphorylated RBX1, nor could we detect COP1 or the E2 enzyme (see Fig. S6 in the supplemental material). Next, we analyzed the action of the COP1 E3 ligase supercomplex on CCT4 ubiquitination with an in vitro ubiquitination assay. The recombinant COP1 supercomplex derived from E. coli showed an increased ability to ubiquitinate CCT4 in vitro, and COP1-mediated polyubiquitination of CCT4 was markedly enhanced in the presence of VRK2 (Fig. 6D). The results suggest that VRK2 enables the COP1 complex to efficiently attach ubiquitin on the CCT4 by providing a close interaction between CCT4 and the COP1 complex. As previously reported, the ubiquitin-proteasome system (UPS) plays an important role in determining TRiC's half-life (6 to 7 h) (29). The effect of targeted knockdown of VRK2 and COP1 on the CCT4 protein turnover rate was examined after treatment with the translation inhibitor cycloheximide (CHX). The protein stability of CCT4 was significantly enhanced in cells transfected with siRNAs for VRK2 and COP1 (Fig. 6E). Thus, the results suggest that increased CCT4 ubiquitination by VRK2 was mediated by COP1 E3 ligase, and the interaction with RBX1 was especially important.
FIG 6.
COP1 E3 ligase is involved in VRK2-mediated TRiC protein turnover. (A) Immunoprecipitation assay using HEK293T cells cotransfected with Flag-COP1 and HA-CCT4 vectors. Encoded HA-CCT4 was coimmunoprecipitated with an anti-Flag antibody. (B) VRK2-mediated downregulation of TRiC disappeared in cells transfected with siCOP1. (C) GST pulldown assay using in vitro-purified SUMO-VRK2 and GST-RBX1 recombinant proteins. (D) VRK2 enhanced CCT4 ubiquitination by the COP1 E3 ligase supercomplex in vitro. Purified HIS-CCT4 was incubated with components of the ubiquitin system in the presence or absence of the COP1 E3 ligase supercomplex with or without VRK2. Polyubiquitinated CCT4 was immunoblotted with an anti-Flag antibody to detect Flag-ubiquitin. (E) Knockdown of both VRK2 and COP1 enhanced CCT4 protein stability. Cells were harvested at the indicated times after cycloheximide (40 μg/ml) treatment, and then endogenous CCT4 was detected by Western blotting.
DISCUSSION
Although HD is a monogenic neurodegenerative disorder known to be caused by expansion of Htt's polyQ tract, there is no therapeutic treatment. Mutant Htt causes dysfunction in numerous signaling pathways and affects several molecular mechanisms, including transcription, axonal transport, and mitochondrial and synaptic functions (7). Although mutant Htt aggregation is known to be involved in HD development, the pathogenic mechanism leading to cell damage and death remains controversial. Because the formation of polyQ-containing aggregates is considered the hallmark of HD, it would be beneficial to prevent this process. Therefore, popular therapeutic strategies, including the inhibition of misfolded aggregation-prone proteins through molecular chaperone expression (17, 37–39) or clearance through the autophagic pathway (40–42), have been proposed for HD and other neurodegenerative disorders.
Our present results provide evidence for molecular chaperone-mediated alleviation of protein aggregation through TRiC protein turnover. In summary, we have shown that VRK2 specifically binds TRiC, which affects its degradation (Fig. 1 and 2). Physiologically, impairment of TRiC by wild-type VRK2 hyperactivation increases polyQ aggregation, whereas VRK2 KD has no effect (Fig. 3). VRK2-mediated downregulation of TRiC required COP1 E3 ligase activity (Fig. 6). A previous study demonstrated that overexpression of TRiC subunit CCT1 (or CCT4) reduced polyQ foci by directing newly made polyQ proteins to nonpathogenic conformations via an interaction between CCT1 (or CCT4) and polyQ proteins (17). Our system clearly showed that the reduction of polyQ aggregation occurred in the presence of HA-CCT4 overexpression, despite VRK2 overexpression (Fig. 4).
Aggregation-based diseases disproportionately affect postmitotic cells, such as neurons, presumably because toxic protein aggregates accumulate easily in undividing cells (6). In our study, neuroblastoma SH-SY5Y cell viability was reduced by pathogenic HttQ103-GFP expression, and VRK2 overexpression increased HttQ103-GFP aggregate formation in SH-SY5Y cells, suggesting that the level of active VRK2 in neuronal cells may be important for the onset and progression of polyQ diseases. Moreover, genome-wide association studies have revealed that VRK2 has emerged as one of the risk factors against neurological disorders, including schizophrenia (43, 44) and epilepsy (45, 46). VRK2 expression, however, is known to be much higher in actively proliferating cells and is maintained at a low level in the brain (47). Analysis of VRK2 mRNA levels revealed that the relative amount in neural SK-N-BE(2)C cells was 600-fold lower than that in nonneural HEK293T cells (unpublished data). These results raise the possibility that neuronal cells maintain very low levels of VRK2 because it makes cells prone to polyQ aggregation. To prove this, it would be very important to determine whether VRK2 levels are increased in HD brain tissue. In support of this concept, a progressive decrease of molecular chaperones was observed in a mouse model of HD, and this affected disease symptoms (38). According to transcriptome analysis, expression of Cul4A, a component of the COP1 E3 ligase supercomplex, is upregulated in a mouse model of HD (48), which supports the possibility of impaired TRiC function in HD pathogenesis.
Our findings demonstrate that the enzymatic activity of wild-type VRK2 protein correlates with pathogenic polyQ aggregation in cells and support the hypothesis that VRK2 mediates activity-dependent functional impairment of TRiC. TRiC is known to affect polyQ aggregation (13); growing evidence demonstrates that it is essential for trapping aggregation-prone structures to prevent expanded polyQ proteins from misfolding and aggregating. However, the upstream regulator of TRiC and the exact function of VRK2 had been unknown. Thus, our results are important in that we identified a regulator of TRiC, discovered a new function of VRK2 in the aggregation process of polyQ-containing proteins, and demonstrated that COP1 can be recruited to TRiC by interacting with VRK2 and another upstream regulator to destabilize TRiC and facilitate polyQ protein aggregation (Fig. 7). Taken together, our results identify VRK2 as a modulator that suppresses the TRiC-assisted folding pathway of polyQ proteins. Further research is necessary to identify the target substrate of VRK2 in TRiC downregulation. Our study provides clues for the genesis of polyQ diseases and for developing VRK2-targeted therapeutic inhibitors of neurodegenerative disorders caused by polyQ-containing protein aggregates that play a role in many neurodegenerative diseases.
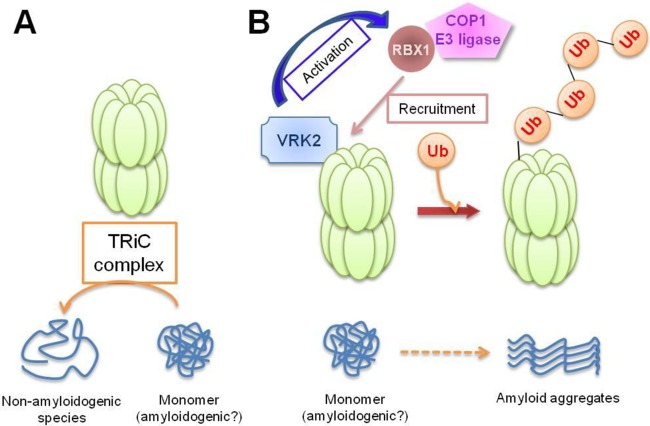
FIG 7.
Diagram of the action of VRK2 on negative regulation of TRiC. (A) The TRiC complex reduces the formation of misfolded polyQ-containing protein aggregates. (B) VRK2 facilitates TRiC protein degradation through activation of COP1 E3 ligase activity and then induces polyQ aggregation.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Frydman, V. Dixit, J. Yoon, Y. Xiong, and J. Yoo for providing plasmids.
This work was supported by grants from the National Research Foundation of Korea (NRF; no. 20110027957, 20110031234, 20120005830, and 20110031517), the Brain Korea 21 plus program funded by the South Korean Ministry of Education, Science, and Technology (MEST), and the Next-Generation BioGreen 21 Program (no. PJ00950301) funded by the South Korean Rural Development Administration.
Footnotes
Published ahead of print 2 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00756-13.
REFERENCES
- 1.Ross CA. 2002. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron 35:819–822. 10.1016/S0896-6273(02)00872-3 [DOI] [PubMed] [Google Scholar]
- 2.Young AB. 2003. Huntingtin in health and disease. J. Clin. Invest. 111:299–302. 10.1172/JCI200317742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald ME, Gines S, Gusella JF, Wheeler VC. 2003. Huntington's disease. Neuromol. Med. 4:7–20. 10.1385/NMM:4:1-2:7 [DOI] [PubMed] [Google Scholar]
- 4.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. 2007. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 14:332–340. 10.1038/nsmb1215 [DOI] [PubMed] [Google Scholar]
- 5.Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. 2001. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98:12784–12789. 10.1073/pnas.221451398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClellan AJ, Tam S, Kaganovich D, Frydman J. 2005. Protein quality control: chaperones culling corrupt conformations. Nat. Cell Biol. 7:736–741. 10.1038/ncb0805-736 [DOI] [PubMed] [Google Scholar]
- 7.Zoghbi HY, Orr HT. 2000. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23:217–247. 10.1146/annurev.neuro.23.1.217 [DOI] [PubMed] [Google Scholar]
- 8.Muchowski PJ, Wacker JL. 2005. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6:11–22. 10.1038/nrn1587 [DOI] [PubMed] [Google Scholar]
- 9.Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ. 2004. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 11:1215–1222. 10.1038/nsmb860 [DOI] [PubMed] [Google Scholar]
- 10.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23:425–428. 10.1038/70532 [DOI] [PubMed] [Google Scholar]
- 11.Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU. 2004. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15:95–105. 10.1016/j.molcel.2004.06.029 [DOI] [PubMed] [Google Scholar]
- 12.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. 2000. Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. U. S. A. 97:7841–7846. 10.1073/pnas.140202897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. U. S. A. 101:6403–6408. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwich AL, Fenton WA, Chapman E, Farr GW. 2007. Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23:115–145. 10.1146/annurev.cellbio.23.090506.123555 [DOI] [PubMed] [Google Scholar]
- 15.Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, Frydman J. 2003. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113:369–381. 10.1016/S0092-8674(03)00307-6 [DOI] [PubMed] [Google Scholar]
- 16.Spiess C, Meyer AS, Reissmann S, Frydman J. 2004. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 14:598–604. 10.1016/j.tcb.2004.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam S, Geller R, Spiess C, Frydman J. 2006. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 8:1155–1162. 10.1038/ncb1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, Hartl FU. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell 23:887–897. 10.1016/j.molcel.2006.08.017 [DOI] [PubMed] [Google Scholar]
- 19.Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. 2006. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 8:1163–1170. 10.1038/ncb1478 [DOI] [PubMed] [Google Scholar]
- 20.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 21.Blanco S, Klimcakova L, Vega FM, Lazo PA. 2006. The subcellular localization of vaccinia-related kinase-2 (VRK2) isoforms determines their different effect on p53 stability in tumour cell lines. FEBS J. 273:2487–2504. 10.1111/j.1742-4658.2006.05256.x [DOI] [PubMed] [Google Scholar]
- 22.Blanco S, Santos C, Lazo PA. 2007. Vaccinia-related kinase 2 modulates the stress response to hypoxia mediated by TAK1. Mol. Cell. Biol. 27:7273–7283. 10.1128/MCB.00025-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez IF, Blanco S, Lozano J, Lazo PA. 2010. VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol. Cell. Biol. 30:4687–4697. 10.1128/MCB.01581-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li LY, Liu MY, Shih HM, Tsai CH, Chen JY. 2006. Human cellular protein VRK2 interacts specifically with Epstein-Barr virus BHRF1, a homologue of Bcl-2, and enhances cell survival. J. Gen. Virol. 87:2869–2878. 10.1099/vir.0.81953-0 [DOI] [PubMed] [Google Scholar]
- 25.Monsalve DM, Merced T, Fernandez IF, Blanco S, Vazquez-Cedeira M, Lazo PA. 2013. Human VRK2 modulates apoptosis by interaction with Bcl-xL and regulation of BAX gene expression. Cell Death Dis. 4:e513. 10.1038/cddis.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanz-Garcia M, Lopez-Sanchez I, Lazo PA. 2008. Proteomics identification of nuclear Ran GTPase as an inhibitor of human VRK1 and VRK2 (vaccinia-related kinase) activities. Mol. Cell. Proteomics 7:2199–2214. 10.1074/mcp.M700586-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols RJ, Wiebe MS, Traktman P. 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17:2451–2464. 10.1091/mbc.E05-12-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez-Cedeira M, Lazo PA. 2012. Human VRK2 (vaccinia-related kinase 2) modulates tumor cell invasion by hyperactivation of NFAT1 and expression of cyclooxygenase-2. J. Biol. Chem. 287:42739–42750. 10.1074/jbc.M112.404285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota S, Kayano T, Ohta T, Kurimoto M, Yanagi H, Yura T, Kubota H. 2000. Proteasome-dependent degradation of cytosolic chaperonin CCT. Biochem. Biophys. Res. Commun. 279:712–717. 10.1006/bbrc.2000.4011 [DOI] [PubMed] [Google Scholar]
- 30.Kang TH, Kim KT. 2006. Negative regulation of ERK activity by VRK3-mediated activation of VHR phosphatase. Nat. Cell Biol. 8:863–869. 10.1038/ncb1447 [DOI] [PubMed] [Google Scholar]
- 31.Jung H, Kim T, Chae HZ, Kim KT, Ha H. 2001. Regulation of macrophage migration inhibitory factor and thiol-specific antioxidant protein PAG by direct interaction. J. Biol. Chem. 276:15504–15510. 10.1074/jbc.M009620200 [DOI] [PubMed] [Google Scholar]
- 32.Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. 1998. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93:125–138. 10.1016/S0092-8674(00)81152-6 [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Willison KR, Horwich AL. 1994. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 19:543–548. 10.1016/0968-0004(94)90058-2 [DOI] [PubMed] [Google Scholar]
- 34.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 35.Yi C, Li S, Wang J, Wei N, Deng XW. 2006. Affinity purification reveals the association of WD40 protein constitutive photomorphogenic 1 with the hetero-oligomeric TCP-1 chaperonin complex in mammalian cells. Int. J. Biochem. Cell Biol. 38:1076–1083. 10.1016/j.biocel.2005.12.019 [DOI] [PubMed] [Google Scholar]
- 36.Yi C, Deng XW. 2005. COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15:618–625. 10.1016/j.tcb.2005.09.007 [DOI] [PubMed] [Google Scholar]
- 37.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. 2000. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 9:2811–2820. 10.1093/hmg/9.19.2811 [DOI] [PubMed] [Google Scholar]
- 38.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. 2004. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 13:1389–1405. 10.1093/hmg/ddh144 [DOI] [PubMed] [Google Scholar]
- 39.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. 2002. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295:865–868. 10.1126/science.1067389 [DOI] [PubMed] [Google Scholar]
- 40.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, Nukina N. 2010. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 28:256–263. 10.1038/nbt.1608 [DOI] [PubMed] [Google Scholar]
- 41.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. 2004. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med. 10:148-154. 10.1038/nm985 [DOI] [PubMed] [Google Scholar]
- 42.Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, Yamamoto A, Krainc D. 2009. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell 137:60–72. 10.1016/j.cell.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Wang Y, Zheng XB, Ikeda M, Iwata N, Luo XJ, Chong SA, Lee J, Rietschel M, Zhang F, Muller-Myhsok B, Cichon S, Weinberger DR, Mattheisen M, Schulze TG, Martin NG, Mitchell PB, Schofield PR, Liu JJ, Su B, Moo DSC. 2012. Meta-analysis and brain imaging data support the involvement of VRK2 (rs2312147) in schizophrenia susceptibility. Schizophr. Res. 142:200–205. 10.1016/j.schres.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 44.Steinberg S, de Jong S, Irish Schizophrenia Genomics Consortium. Andreassen OA, Werge T, Borglum AD, Mors O, Mortensen PB, Gustafsson O, Costas J, Pietilainen OP, Demontis D, Papiol S, Huttenlocher J, Mattheisen M, Breuer R, Vassos E, Giegling I, Fraser G, Walker N, Tuulio-Henriksson A, Suvisaari J, Lonnqvist J, Paunio T, Agartz I, Melle I, Djurovic S, Strengman E, Jurgens G, Glenthoj B, Terenius L, Hougaard DM, Orntoft T, Wiuf C, Didriksen M, Hollegaard MV, Nordentoft M, van Winkel R, Kenis G, Abramova L, Kaleda V, Arrojo M, Sanjuan J, Arango C, Sperling S, Rossner M, Ribolsi M, Magni V, Siracusano A, et al. 2011. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum. Mol. Genet. 20:4076–4081. 10.1093/hmg/ddr325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helbig I, Lowenstein DH. 2013. Genetics of the epilepsies: where are we and where are we going? Curr. Opin. Neurol. 26:179–185. 10.1097/WCO.0b013e32835ee6ff [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.EPICURE Consortium, EMINet Consortium, Steffens M, Leu C, Ruppert AK, Zara F, Striano P, Robbiano A, Capovilla G, Tinuper P, Gambardella A, Bianchi A, La Neve A, Crichiutti G, de Kovel CG, Kasteleijn-Nolst Trenite D, de Haan GJ, Lindhout D, Gaus V, Schmitz B, Janz D, Weber YG, Becker F, Lerche H, Steinhoff BJ, Kleefuss-Lie AA, Kunz WS, Surges R, Elger CE, Muhle H, von Spiczak S, Ostertag P, Helbig I, Stephani U, Moller RS, Hjalgrim H, Dibbens LM, Bellows S, Oliver K, Mullen S, Scheffer IE, Berkovic SF, Everett KV, Gardiner MR, Marini C, Guerrini R, Lehesjoki AE, Siren A, Guipponi M, Malafosse A, et al. 2012. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum. Mol. Genet. 21:5359–5372. 10.1093/hmg/dds373 [DOI] [PubMed] [Google Scholar]
- 47.Nezu J, Oku A, Jones MH, Shimane M. 1997. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45:327–331. 10.1006/geno.1997.4938 [DOI] [PubMed] [Google Scholar]
- 48.Tang B, Seredenina T, Coppola G, Kuhn A, Geschwind DH, Luthi-Carter R, Thomas EA. 2011. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lengths reveals genes associated with disease onset and progression in Huntington's disease. Neurobiol. Dis. 42:459–467. 10.1016/j.nbd.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.