Abstract
Transforming growth factor β (TGF-β)-activated kinase 1 (TAK1) is a key regulator in the signals transduced by proinflammatory cytokines and Toll-like receptors (TLRs). The regulatory mechanism of TAK1 in response to various tissue types and stimuli remains incompletely understood. Here, we show that ribosomal S6 kinase 1 (S6K1) negatively regulates TLR-mediated signals by inhibiting TAK1 activity. S6K1 overexpression causes a marked reduction in NF-κB and AP-1 activity induced by stimulation of TLR2 or TLR4. In contrast, S6K1−/− and S6K1 knockdown cells display enhanced production of inflammatory cytokines. Moreover, S6K1−/− mice exhibit decreased survival in response to challenge with lipopolysaccharide (LPS). We found that S6K1 inhibits TAK1 kinase activity by interfering with the interaction between TAK1 and TAB1, which is a key regulator protein for TAK1 catalytic function. Upon stimulation with TLR ligands, S6K1 deficiency causes a marked increase in TAK1 kinase activity that in turn induces a substantial enhancement of NF-κB-dependent gene expression, indicating that S6K1 is negatively involved in the TLR signaling pathway by the inhibition of TAK1 activity. Our findings contribute to understanding the molecular pathogenesis of the impaired immune responses seen in type 2 diabetes, where S6K1 plays a key role both in driving insulin resistance and modulating TLR signaling.
INTRODUCTION
Transforming growth factor β-activated kinase 1 (TAK1) is a member of the mitogen-activated protein kinase (MAPK) kinase kinase (MAP3K) family (1). TAK1 is essential for the production of tumor necrosis factor (TNF-α) and other inflammatory mediators by activating several MAPKs, such as p38α MAPK, Jun N-terminal protein kinases 1 and 2 (JNK1 and JNK2), and extracellular signal-regulated kinases 1 and 2 (ERK1/2). TAK1 also plays a key regulatory role in several cytokine-mediated innate immunity signal transduction cascades, including interleukin-1 (IL-1) and the downstream signaling of Toll-like receptors (TLRs) and NOD1/2 (2, 3). In these pathways, various proinflammatory cytokines and TLR agonists trigger TAK1 activity, leading to its autophosphorylation and subsequent recruitment to the IκB kinase (IKK) complex, ultimately resulting in the activation of the transcription factor NF-κB and the upregulation of genes encoding proinflammatory cytokines, chemokines, adhesion molecules, and proteolytic enzymes.
Several binding partners of TAK1, including TAK1-binding protein 1 (TAB1), TAB2, and TAB3, have been implicated in the regulation of TAK1 activity in response to various stimuli (1, 4). Previous reports demonstrated that the native forms of TAK1 comprise a catalytic kinase subunit in complex with the regulatory subunit TAB1 and either of two homologous proteins, TAB2 and TAB3 (2, 5, 6). Importantly, it has been reported that TAB1 might play a key role in the regulation of the TAK1 complex (7, 8). Studies with TAB1-deficient mouse embryonic fibroblasts (TAB1−/− MEFs) demonstrated that TAB1 is able to recruit p38α MAPK to the TAK1 complex for TAB3 phosphorylation, resulting in the induction of TAK1 catalytic activity (8). In addition, TAB1−/− MEFs do not activate TAK1 in response to IL-1 and TNF-α, strongly suggesting a pivotal role of TAB1 in TAK1 signaling. Moreover, other studies have demonstrated that TAB1 specifically regulates TAK1 activity induced by proinflammatory responses by disrupting a MEKK3-TAK1 complex under unstimulated conditions, which may be important in preventing basal NF-κB signaling (9).
Recently, the biological significance of heteromeric complex formation between different MAP3Ks has been proposed as a critical mechanism for cells to fine-tune cellular responses when faced with a wide range of stimuli. A previous report showed that apoptosis signal-regulating kinase 1 (ASK1), a member of the MAP3K family, inhibits the activation of NF-κB induced by IL-1 through disruption of TRAF6-TAK1 interaction (10). Moreover, it has recently been proposed that TAK1 inhibits S6 kinase1 (S6K1) phosphorylation by interfering with the interaction between raptor and S6K1, inducing autophagy (11). Nevertheless, the mechanisms that allow TAK1 to be regulated by so many different stimuli and tissue types and the kinetics of the regulatory response are not completely understood. Here, we demonstrate that S6K1 is negatively involved in the TLR2- or TLR4-mediated signaling pathway. In line with our in vitro results, we found that S6K1−/− mice exhibit increased lethality after challenge with lipopolysaccharide (LPS) in vivo.
MATERIALS AND METHODS
Animals.
Mice lacking S6K1 were kindly provided by George Thomas (University of Cincinnati). Age- and sex-matched S6K1−/− mice were originally generated in a mixed 129/SveJ×C57BL/6 line. Male S6K-dko and wild-type (wt) mice were housed in individual cages under a 12-h/12-h light/dark cycle (7 a.m. lights on) with ad libitum access to standard chow (6% kcal from fat; Harlan Teklad Laboratory Diets, Madison, WI), fat-free diet (0% kcal from fat; Research Diets, Inc., New Brunswick, NJ), or high-fat diet (60% kcal from fat [formulation D12492] and 45% kcal from fat [formulation D12451]) (Research Diets, Inc.) and tap water. The Sungkyunkwan University School of Medicine Institutional Animal Care and Use Committee approved all animal protocols.
Cells.
HEK293 cells (human embryonic kidney cells) were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Corp., Carlsbad, CA) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 unit/ml penicillin, 100 μg/ml streptomycin, and 5 × 10−5 M β-mercaptoethanol. THP-1 cells (human monocytic cells) were purchased from ATCC and maintained in RPMI medium (Invitrogen) containing 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 × 10−5 M β-mercaptoethanol. HEK293 cells expressing human TLR2 (catalog number hkb-htlr2) and human TLR4 (catalog number hkb-htlr4) were purchased from InvivoGen (San Diego, CA) and maintained according to the manufacturer's protocol.
Production of S6K1 gene-containing lentivirus particles.
The S6K1 gene was inserted into the pLVX-IRES-Puro (catalog number 632183; Clontech) lentiviral vector according to the manufacturer's protocol. pLVX-Mock-Puro and pLVX-S6K1-Puro were transfected into HEK293T packaging cells along with vesicular stomatitis virus (VSV) G protein and Δ8.2 vector by using Lipofectamine (Invitrogen). The supernatants were harvested at ∼40 h posttransfection, and then lentivirus particles were collected using ultracentrifugation. The collected lentivirus particles were filtered with a 45-μm filter and transferred to a sterile storage tube, and the viral particles were frozen at −80°C for long-term storage.
Control THP-1 and S6K1 knockdown THP-1 cells.
Lentiviral particles containing small hairpin RNA (shRNA) targeting human S6K1 (sc-36165-V) or control shRNA lentiviral particles (sc-108080) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). THP-1 cells were cultured in a 24-well plate (2 × 105 cells/well) and infected with each lentiviral particle according to the manufacturer's protocol. Control THP-1 cells generated by control shRNA lentiviral particles and S6K1 knockdown (KD) THP-1 cells generated by S6K1 shRNA lentiviral particles (S6K1KD THP-1 cells) were cultured in puromycin-containing (4 to 8 μg/ml) medium for 2 weeks to select stable clones (12, 13). To generate S6K1-overexpressing Raw 264.7 cells, Raw 264.7 cells (2 × 104) were infected with a mock (empty) vector (pLVX-Mock-Puro) or the lentivirus containing the Flag-tagged S6K1 expression construct (pLVX-S6K1-Puro) and incubated for 18 to 20 h at 37°C in a humidified CO2 incubator, and then the culture medium was removed from the wells and replaced with fresh medium containing puromycin (4 to 8 μg/ml). The fresh medium containing puromycin was replaced every 3 to 4 days until resistant colonies were obtained. Puromycin-resistant colonies were picked, and then the expression of Flag-tagged S6K1 was examined with anti-Flag antibody by using Western blot analysis.
Luciferase reporter assay.
HEK293, HEK293-TLR2, HEK293-TLR4, control THP-1, or S6K1KD THP-1 cells grown on 12-well plates were transiently transfected using the Neon transfection system (Invitrogen) with either the NF-κB-dependent reporter construct pBIIx-luc or the AP-1-dependent reporter construct AP-1-luc with Renilla luciferase vector (Promega Corp., Madison, WI). Cells were treated with or without FSL-1 (synthetic diacylated lipoprotein, a TLR2 agonist; 10 μg/ml) or LPS (TLR4 agonist; 100 ng/ml) for 6 h, and luciferase activity was measured using the dual-luciferase assay kit (Promega) (14). Mock-Raw 264.7 and Flag-tagged S6K1-expressing Raw 264.7 cells grown on 12-well plates were transiently transfected using the Neon transfection system (Invitrogen, Carlsbad, CA) with either the NF-κB-dependent reporter construct pBIIx-luc or the AP-1-dependent reporter construct AP-1-luc with Renilla luciferase vector (Promega Corporation, Madison, WI) according to the respective manufacturer's instructions. The total DNA concentration in each experiment was maintained by adding the appropriate empty vector to the DNA mixture. Typically at 24 h after transfection, cells were lysed and luciferase activity was measured using the dual-luciferase assay kit (Promega). HEK293-TLR4 cells were cotransfected with wt S6K1, dominant-negative S6K1 (DN-S6K1), or constitutively active S6K1 (CA-S6K1) vector, together with pBIIx-luc reporter and Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with or without LPS for 6 h and then analyzed for luciferase activity. DN-S6K1 and CA-S6K1 constructs, originally provided by G. Tomas, were supplied by J. W. Han (Sungkyunkwan University, Suwon, Republic of Korea).
Measurement of proinflammatory cytokines.
Splenocytes were isolated from wt or S6K1−/− mice, and 5 × 105 cells were seeded into U-bottom 96-well plates. The cells were treated with or without the TLR agonists to TLR2 (FSL-1) or TLR4 (LPS) (InvivoGen). After 9 h, the levels of mouse TNF-α, IL-1β, and IL-6 were measured in the supernatant according to the manufacturer's protocol (enzyme-linked immunosorbent assay [ELISA] kit; R&D Systems, Minneapolis, MN). Wild-type THP-1 or S6K1KD THP-1 cells were treated with or without TLR agonists for 9 h, and then the supernatants were harvested. The levels of human TNF-α, IL-1β, and IL-6 were measured in the supernatants according to the manufacturer's protocol (R&D Systems). Mock-Raw 264.7 and S6K1-Raw 264.7 cells were treated with or without TLR2 or TLR4 agonist (FSL-1 and LPS, respectively) for 9 h, and then supernatants were harvested. The levels of mouse IL-6 and mouse IL-1β were measured in the supernatants according to the manufacturer's protocol (R&D Systems, Minneapolis, MN).
Survival analysis of S6K1−/− mice.
Six- to 8-week-old S6K1−/− mice (n = 12) and wild-type controls (n = 12) were intraperitoneally injected with a lethal dose of LPS (40 mg/kg) and monitored for mortality. The survival of the mice was analyzed using Kaplan-Meier survival curves, and the difference in median survival times of S6K1−/− and wild-type mice was assessed by log rank tests.
Truncated mutant plasmids.
G. Thomas kindly provided the wt S6K1 vector. The Flag-tagged S6K1 mutants Flag-S6K1 (66-333) (comprising amino acids 66 to 333 of S6K1; the internal catalytic domain) and Flag-S6K1 (333-502) (comprising amino acids 333 to 502 of S6K1; the regulator domain) were generated by PCR, using Flag-S6K1 wt as a template, and inserted into pcDNA3. Myc-tagged TAK1 mutants Myc-TAK1 (1-500), Myc-TAK1 (1-400), Myc-TAK1 (1-300), Myc-TAK1 (1-200), and Myc-TAK1 (1-100) (each comprising the indicated N-terminal amino acids of TAK1) were generated by PCR using wt Myc-TAK1 as the template and inserted into pcDNA3 (13, 14).
Immunoprecipitation and Western blotting.
HEK293 cells transfected with appropriate expression vectors were lysed in a lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% Triton X-100, 1% deoxycholate, 1.5% aprotinin, and 1 mM phenylmethylsulfonyl fluoride. Cellular debris was removed by centrifugation. For immunoprecipitation (IP) and Western blotting, we used anti-Myc and anti-Flag antibodies (Sigma-Aldrich). The proteins were detected using the enhanced chemiluminescence system (Amersham Pharmacia Biotech, Amersham, United Kingdom). For endogenous immunoprecipitation, THP-1 cells were treated with or without LPS (100 ng/ml) for 45 min. Cells were extracted and immunoprecipitated with anti-S6K1 antibody. The interaction was detected by Western blotting with anti-TAK1 antibody. The same lysates were verified with anti-S6K1 antibody. For Western blotting, wt and S6K1KD THP-1 cells were treated with or without TLR4 and TLR2 agonists (LPS and FSL-1, respectively) for different times. The lysates were examined by Western blot analysis with anti-phosphorylated (anti-pho)-TAK1, anti-TAK1, anti-pho-IKKαβ, anti-IKKαβ, anti-pho-p38, and anti-p38 antibodies. Immunoblotting with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was performed to generate a control for gel loading. For the domain mapping of TAK1, HEK293 cells were transfected with vectors for the expression of Flag-S6K1, Myc-TAK1 wt, Myc-TAK1 (1-500), Myc-TAK1 (1-400), Myc-TAK1 (1-300), Myc-TAK1 (1-200), Myc-TAK1 (1-100), Flag-S6K1 and Myc-TAK1 wt, Flag-S6K1 and Myc-TAK1 (1-500), Flag-S6K1 and Myc-TAK1 (1-400), Flag-S6K1 and Myc-TAK1 (1-300), Flag-S6K1 and Myc-TAK1 (1-200), or Flag-S6K1 and Myc-TAK1 (1-100), as indicated below. At 36 h after transfection, transfected cells were extracted and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of Flag-S6K1, Myc-TAK1 wt, or Myc-TAK1 truncated mutants in the pre-IP lysates was verified by Western blotting. For the domain mapping of S6K1, HEK293 cells were transfected with vectors for the expression of Myc-TAK1, Flag-S6K1 wt, Flag-S6K1 (66-333), Flag-S6K1 (333-502), Myc-TAK1 and Flag-S6K1 wt, Myc-TAK1 and Flag-S6K1 (66-333), or Myc-TAK1 and Flag-S6K1 (333-502), as indicated below. At 36 h after transfection, transfected cells were extracted and immunoprecipitated with anti-Flag antibody. The interaction was detected by Western blotting with anti-Myc antibody. The presence of Myc-TAK1, Flag-S6K1, or Flag-S6K1 truncated mutants in the pre-IP lysates was verified by Western blotting (12–14).
In vitro kinase assay for TAK1.
Control (wt) THP-1 and S6K1KD THP-1 cells were treated with or without LPS (100 ng/ml) or FSL-1 (10 μg/ml) for different times. The kinase assay for TAK1 was performed using the c-TAK1 kinase assay kit (U-TRF number 17; PerkinElmer, Branchburg, NJ) in accordance with the manufacturer's protocol.
Microarray analysis.
For microarray analysis, control THP-1 and S6K1KD THP-1 cells were treated with or without LPS (100 ng/ml) and FSL-1 (10 μg/ml) for 3 or 9 h. Total RNA was extracted using TRIzol (Invitrogen) and purified using RNeasy columns (Qiagen, Valencia, CA) according to the manufacturer's protocol. After DNase digestion processing and cleanup procedures, RNA samples were quantified, aliquoted, and stored at −80°C until use. For quality control, RNA purity and integrity were evaluated by denaturing gel electrophoresis, using a 260-nm/280-nm optical density ratio, and analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Total RNA was amplified and purified using the Ambion Illumina RNA amplification kit (Ambion, Austin, TX) to yield biotinylated cRNA according to the manufacturer's instructions. Briefly, 550 ng of total RNA was reverse transcribed to cDNA using a T7 oligo(dT) primer. Second-strand cDNA was synthesized, in vitro transcribed, and labeled with biotin-nucleoside triphosphate. After purification, the cRNA was quantified using an ND-1000 spectrophotometer (NanoDrop, Wilmington, NC). Typically, 750-ng amounts of labeled cRNA samples were hybridized to each human HT-12 expression bead array, version 4, for 16 to 18 h at 58°C, according to the manufacturer's instructions (Illumina, Inc., San Diego, CA). Detection of the array signal was carried out using Amersham FluoroLink streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, United Kingdom) following the directions in the bead array manual. Arrays were scanned using an Illumina bead array reader confocal scanner according to the manufacturer's instructions.
For raw data preparation and statistical analysis, the quality of hybridization and overall chip performance were monitored by visual inspection of both internal quality control checks and the raw scanned data. Raw data were extracted using the software provided by the manufacturer (Illumina GenomeStudio version 2009.2 [Gene Expression Module version 1.5.4]). Array data were filtered by detection of a P value of <0.05 (similar to the signal-to-noise ratio) in at least 50% of samples (we applied a filtering criterion for data analysis; a higher signal value was required to obtain a detection P value of <0.05). Selected gene signal values were transformed by logarithm and normalized by the quantile method. A comparative analysis between the test and control samples was carried out using fold change as the measure. Hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. GO ontology analysis for the significant probe list was performed with PANTHER (http://www.pantherdb.org/panther/ontologies.jsp), using text files containing the Gene ID list and the accession number of the Illumina probe ID. Gene set enrichment analysis (GSEA) was performed to check whether the a priori defined set of genes showed a differential pattern in both the biological process and the molecular function state. A one-tail Fisher's exact test was adopted to measure the gene enrichment in annotation terms. All the data analysis and visualization of differentially expressed genes was conducted using R 2.4.1 (www.r-project.org). For analysis of NF-κB-dependent upregulated and downregulated genes, genes that contained a κB-binding site were sorted (see Table S1 in the supplemental material) and analyzed in different combinations as follows: 1, control THP-1 cells treated with LPS or FSL-1 for 3 h versus control THP-1 cells; 2, control THP-1 cells treated with LPS or FSL-1 for 9 h versus control THP-1 cells; 3, S6K1KD THP-1 cells treated with LPS or FSL-1 for 3 h versus S6K1KD THP-1 cells; 4, S6K1KD THP-1 cells treated with LPS or FSL-1 for 9 h versus S6K1KD THP-1 cells; 5, S6K1KD THP-1 cells versus control THP-1 cells; 6, S6K1KD THP-1 cells treated with LPS or FSL-1 for 3 h versus control THP-1 cells treated with LPS or FSL-1 for 3 h; and 7, S6K1KD THP-1 cells treated with LPS or FSL-1 for 9 h versus control THP-1 cells treated with LPS or FSL-1 for 9 h.
Gene expression analysis by qRT-PCR.
Control THP-1 and S6K1KD THP-1 cells were treated with or without LPS (100 ng/ml) and FSL-1 (10 μg/ml) for different times. Isolation of total RNA and cDNA synthesis were performed following the protocols provided along with the kit (Qiagen). For quantitative real-time PCR (qRT-PCR) analysis, the following qPCR primers were purchased from Qiagen: IL-8 (PPH 00568A), IRF7 (PPH02014E), CD44 (PPH 00114A), NF-κB2 (PPH 00782E), IER3 (PPH 10008E), BCL3 (PPH 02009C), and IL-1B (PPH 00171C). The qRT-PCR analysis was performed using Rotor-Gene Q (Qiagen) according to the manufacturer's protocol (13).
Statistical analysis.
Data are presented as the mean value ± standard deviation (SD) as indicated below and analyzed using Student's two-tailed t tests. P values of <0.05, <0.01, and <0.001 were considered statistically significant.
RESULTS
S6K1 is negatively involved in the TLR2- or TLR4-mediated signaling pathway.
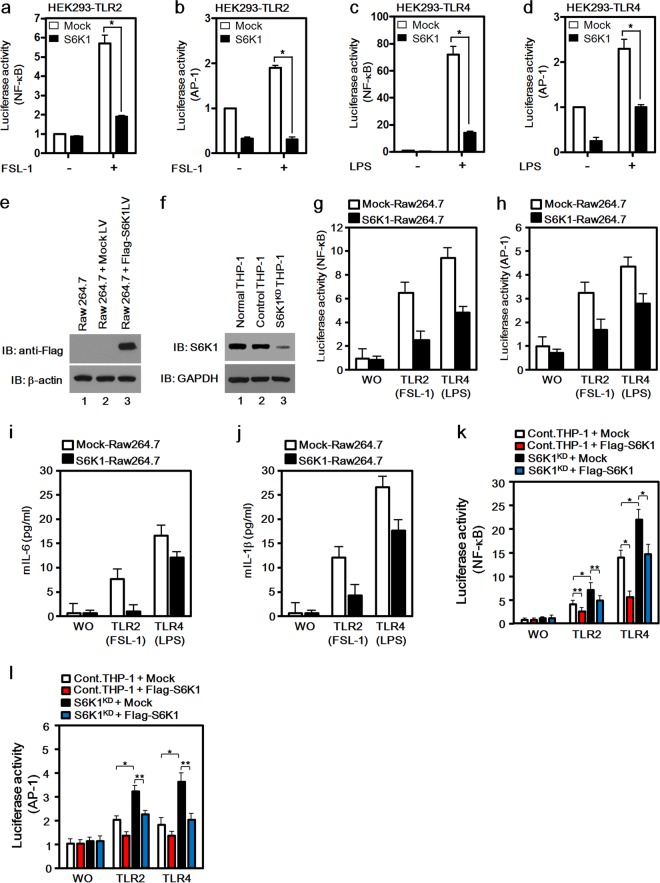
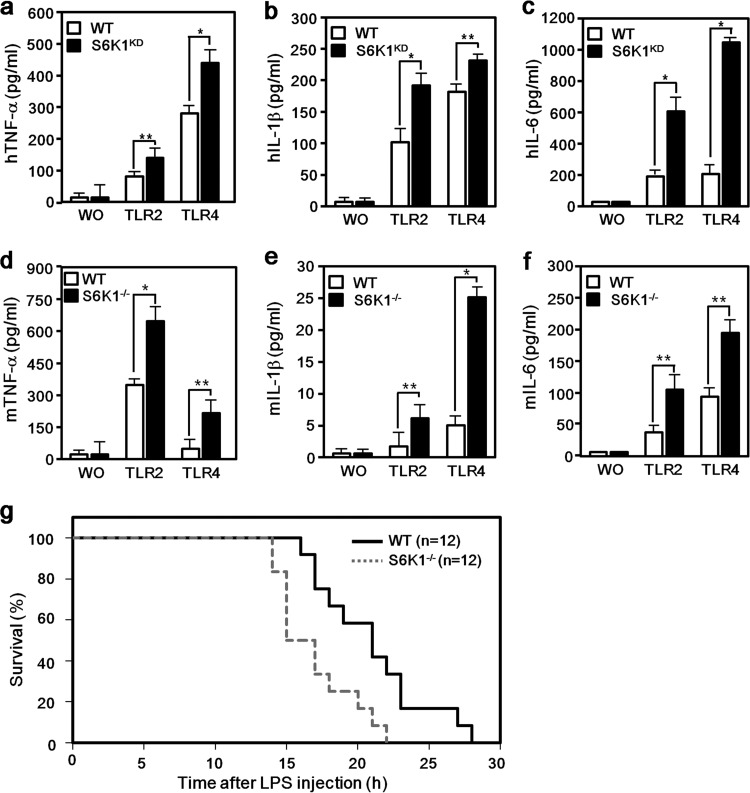
To examine whether S6K1 is involved in TLR-mediated signaling, TLR2- or TLR4-expressing HEK293 cells were utilized. The Flag-tagged S6K1 vector was transiently cotransfected into TLR2-expressing or TLR4-expressing HEK293 cells along with a NF-κB or AP-1 reporter vector, and their activities in the presence or absence of a synthetic diacylated lipoprotein (FSL-1, a TLR2 agonist) (15) or lipopolysaccharide (LPS, a TLR4 agonist) were measured. Both NF-κB and AP-1 activities were significantly elevated in mock-transfected TLR2-expressing HEK293 cells (Fig. 1a and b, open bars), whereas S6K1 expression induced marked suppression of NF-κB and AP-1 activities in TLR2-expressing HEK293 cells (Fig. 1a and b, closed bars). Similarly, overexpression of S6K1 suppressed TLR4-induced NF-κB and AP-1 activities (Fig. 1c and d, closed bars) compared with those seen with mock transfection (Fig. 1c and d, open bars). To further verify these results, S6K1-overexpressing Raw 264.7 cells were generated by transduction with lentivirus containing the Flag-S6K1 expression construct (Fig. 1e) and S6K1 knockdown THP-1 cells were generated by transduction with lentivirus containing shRNA targeting S6K1 (Fig. 1f). Accordingly, both TLR4 and TLR2 agonist-induced NF-κB and AP-1 activities were significantly lower in the S6K1-expressing Raw 264.7 cells than in mock-transfected Raw 264.7 cells (Fig. 1g and h). Moreover, the production of mouse IL-6 (mIL-6) or mIL-1β was significantly attenuated in S6K1-expressing Raw 264.7 cells (Fig. 1i and j). In contrast, TLR2 and TLR4 agonist-induced NF-κB and AP-1 activities were significantly higher in S6K1KD THP-1 cells than in control THP-1 cells, whereas the activities were significantly reduced by the reexpression of Flag-S6K1 in S6K1KD THP-1 cells, up to the levels in control THP-1 cells treated with FSL-1 or LPS (Fig. 1k and l). To further investigate whether the negative regulation of NF-κB activity by S6K1 is able to affect the production of proinflammatory cytokines like TNF-α, IL-1β, and IL-6, the levels were measured in S6K1KD THP-1 cells and S6K1−/− splenocytes treated with or without LPS or FSL-1. Interestingly, marked increases in TNF-α, IL-1β, and IL-6 levels could be detected in both S6K1KD cells (Fig. 2a, b, and c) and S6K1−/− splenocytes (Fig. 2d, e, and f) compared with the levels in control THP-1 cells and splenocytes. Interestingly, according to in vivo challenge with a lethal dose of LPS, S6K1−/− mice showed a statistically significant (P = 0.0064; hazard ratio 2.55; 95% confidence interval [95% CI] of ratio, 1.05 to 6.22) decrease of survival, with a median survival time of 16.0 h (95% CI, 15.0 to 18.0) compared to the wild-type mouse median survival time of 21.0 h (95% CI, 18.0 to 23.0) (Fig. 2g). Together, these results suggest that S6K1 is negatively involved in the NF-κB and AP-1 activation induced by TLR stimulation and thereby attenuates the production of proinflammatory cytokines induced by TLR stimulation and affects survival in response to LPS challenge.
FIG 1.
S6K1 is negatively involved in the activation of NF-κB and AP-1 induced by TLR2 and TLR4. (a to d) HEK293-TLR2 cells (a and b) and HEK293-TLR4 cells (c and d) were cotransfected with a mock or Flag-S6K1 vector together with pBIIx-luc (a and c) or with AP-1-luc together with the Renilla luciferase vector (b and d). Twenty-four hours after transfection, cells were treated with or without FSL-1 (a and b) or LPS (c and d) for 6 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that in untreated cells. (e) The mock or Flag-S6K1 lentivirus vector was used to infect Raw 264.7 cells as described in Materials and Methods. The expression of Flag-S6K1 was examined with anti-Flag antibody by using Western blot assay. Immunoblotting (IB) with anti-β-actin antibody was performed to generate a control for gel loading. (f) A total of 2 × 105 THP-1 cells were cultured in a 24-well plate and infected with lentiviral particles containing shRNA targeting human S6K1 (S6K1KD THP-1) or control shRNA lentiviral particles (Control THP-1). Cells were cultured in medium containing puromycin (4 to 8 μg/ml) for 2 weeks to select stable clones. The knockdown efficacy was examined with anti-S6K1 antibody using Western blotting. Immunoblotting with anti-GAPDH antibody was performed to generate a control for gel loading. (g and h) Mock-Raw 264.7 and S6K1-Raw 264.7 cells were cotransfected with pBIIx-luc (g) or AP1-luc (h) reporter together with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with or without (WO) FSL-1 or LPS for 6 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that in untreated cells. The data shown are the averages of a minimum of three independent experiments, with error bars denoting standard deviations (±SD). (i and j) Mock-Raw 264.7 and S6K1-Raw 264.7 cells were treated with or without TLR2 or TLR4 agonist (FSL-1 and LPS, respectively) for 9 h, and then supernatants were harvested. Levels of mouse IL-6 and mouse IL-1β were measured in the supernatants according to the assay manufacturer's protocol. (k and l) Control THP-1 and S6K1KD THP-1 cells were transfected with the mock or Flag-S6K1 vector with pBIIx-luc (k) or AP1-luc (l) together with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with or without FSL-1 or LPS for 6 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that in untreated cells. The data shown are the averages of a minimum of three independent experiments, with error bars denoting ±SD. *P < 0.01, **P < 0.05.
FIG 2.
S6K1 deficiency increases proinflammatory cytokine production induced by TLR2 and TLR4. (a to c) Wild-type and S6K1KD THP-1 cells were treated with or without (WO) FLS-1 or LPS for 9 h. Human TNF-α (hTNF-α) (a), hIL-1β (b), and hIL-6 (c) production was analyzed by enzyme-linked immunosorbent assay (ELISA). The data shown are the averages of a minimum of three independent experiments, with error bars denoting ±SD. *, P < 0.01; **, P < 0.05. (d to f) Splenocytes were isolated from wt or S6K1−/− mice (n = 3 each). Cells were plated into 96-well plates (5 × 105 cells/well) and treated with or without TLR2 and TLR4 agonists (FSL-1 and LPS, respectively) for 24 h. Mouse TNF-α (mTNF-α) (d), mIL-1β (e), and mIL-6 (f) production was analyzed by ELISA. Data are shown as the mean ± SD. *, P < 0.01; **, P < 0.05. (g) Six- to 8-week-old S6K1−/− mice and wild-type controls were intraperitoneally injected with a lethal dose of LPS (40 mg/kg of body weight) and monitored for mortality. Survival of mice was analyzed using Kaplan-Meier survival curves, and the difference in median survival times of S6K1−/− and wild-type mice was assessed by log rank tests (P = 0.0064; hazard ratio, 2.55; 95% CI of ratio, 1.05 to 6.22).
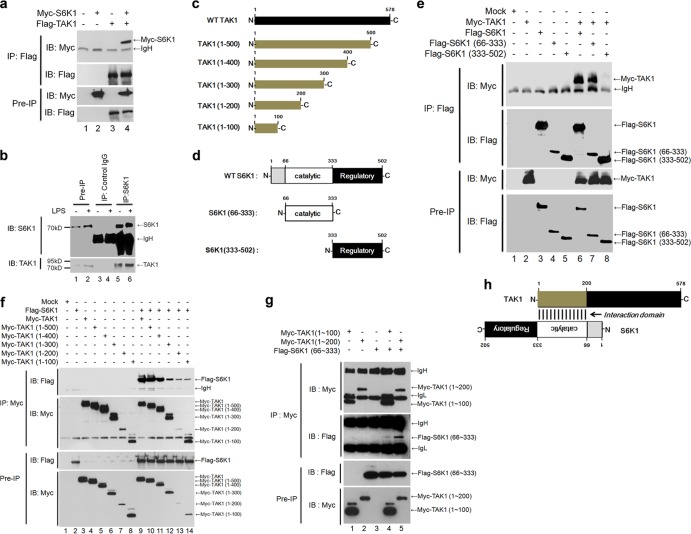
The N terminus of TAK1 interacts with the internal catalytic domain of S6K1.
Having shown that S6K1 has an inhibitory effect on TLR2- or TLR4-mediated signaling and the production of proinflammatory cytokines, we next explored the molecular mechanism of this phenomenon. It has recently been shown that TAK1 interacts with S6K1 and that this interaction inhibits S6K1 activation (11). We therefore hypothesized that S6K1 is able to negatively regulate TAK1 activity. Consistent with previous results, S6K1 coprecipitated with TAK1 (Fig. 3a, lanes 4). In addition, the interaction between TAK1 and S6K1 was confirmed by an endogenous immunoprecipitation assay (Fig. 3b). Based on these results, we further tried to identify the domains involved in the interaction. For this purpose, five different truncation mutants were generated for TAK1 and two different truncated mutants were generated for S6K1 (Fig. 3c and d). As shown in Fig. 3e, Flag-tagged wild-type S6K1 and the Flag-S6K1 (66–333) mutant coprecipitated with Myc-tagged TAK1, whereas no significant interaction of Flag-S6K1 (333–502) with Myc-TAK1 could be seen, indicating that TAK1 interacts with the internal catalytic domain of S6K1. In the case of TAK1 truncation mutants, Flag-S6K1 significantly coprecipitated with all truncation mutants of TAK1, but two N-terminal TAK1 truncation mutants, Myc-TAK1 (1–100) and TAK1 (1–200), interacted weakly with TAK1 (Fig. 3f). To verify the interaction between TAK1 and S6K1, two Myc-tagged N-terminal mutants of TAK1, Myc-TAK1 (1–100) and Myc-TAK1 (1–200), and a Flag-S6K1 (66–333) mutant were expressed and an IP assay was performed with anti-Myc antibodies. Interestingly, the Flag-S6K1 (66–333) mutant coprecipitated strongly with Myc-TAK1 (1–200) but weakly with Myc-TAK1 (1–100) (Fig. 3g), indicating that the N terminus of TAK1, presumably through the region comprising amino acids 1 to 200, interacts with the internal catalytic domain of S6K1, as depicted in Fig. 3h.
FIG 3.
The TAK1 N terminus interacts with the catalytic domain of S6K1. (a) HEK293 cells were transfected with Myc-S6K1, Flag-TAK1, or Myc-S6K1 and Flag-TAK1 vector. At 36 h after transfection, cells were extracted and immunoprecipitated (IP) with anti-Flag antibody. The interaction was detected by Western blotting with anti-Myc antibody. The presence of Myc-S6K1 and Flag-TAK1 in the pre-IP lysates was verified by Western blotting. (b) HEK293 cells were treated with or without LPS (100 ng/ml) for 45 min. Cells were extracted and immunoprecipitated with anti-S6K1 or control IgG antibody. The endogenous interaction was detected by Western blotting with anti-TAK1 antibody. (c) Five different Myc-tagged truncated mutants of TAK1 were generated from the control TAK1 vector as described in Materials and Methods. (d) Two different Flag-tagged truncated mutants of S6K1 were generated from the control S6K1 vector as described in Materials and Methods. (e) HEK293 cells were transfected with combinations of proteins as indicated above the lanes. At 36 h after transfection, transfected cells were extracted and immunoprecipitated with anti-Flag antibody. The interaction was detected by Western blotting with anti-Myc antibody. The presence of proteins in the pre-IP lysates was verified by Western blotting as indicated to the right. (f) HEK293 cells were transfected with combinations of proteins as indicated above the lanes. At 36 h after transfection, transfected cells were extracted and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of proteins in the pre-IP lysates was verified by Western blotting as indicated to the right. (g) HEK293 cells were transfected with combinations of proteins as indicated above the lanes. At 36 h after transfection, transfected cells were extracted and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of the proteins in the pre-IP lysates was verified by Western blotting. (h) A model of the interaction between the N terminus of TAK1 and catalytic domain of S6K1.
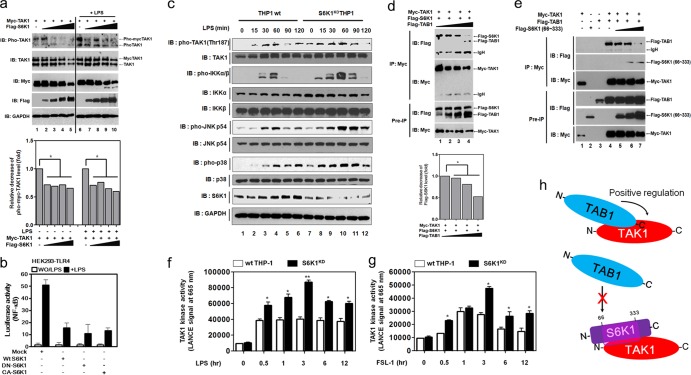
S6K1 interacts competitively with TAB1, inhibiting TAK1 kinase activity.
We next examined whether the interaction of S6K1 with TAK1 alters the activation of TAK1 induced by TLR stimulation. Increases in Flag-S6K1 significantly decreased TAK1 phosphorylation in a dose-dependent manner (Fig. 4a). Moreover, no significant changes in the NF-κB activation induced by TLR4 were observed with the overexpression of wt S6K1, dominant-negative S6K1 (DN-S6K1), or constitutively active S6K1 (CA-S6K1) protein (Fig. 4b), indicating a kinase-independent inhibition. In contrast, upon stimulation of TLR4, S6K1KD THP-1 cells exhibited a marked increase in TAK1 phosphorylation, with subsequent activation of molecules downstream from TAK1, such as IKKs, p38, and JNK (Fig. 4c). This indicates that S6K1 functions to inhibit the TAK1 activation induced by stimulation of TLR4. Previous reports showed that the N terminus of TAK1 interacts with the C terminus of TAB1 (16, 17) and that this interaction plays a crucial role in TAK1 signaling. As shown by the results in Fig. 2, S6K1 interacts with the N terminus of TAK1. Based on these results, we thought it possible that the interaction of S6K1 and TAK1 affected the interaction between TAB1 and TAK1. As expected, increased TAB1 expression resulted in a decrease in the interaction between TAK1 and S6K1 (Fig. 4d). Inversely, an increase in the expression of Flag-S6K1 (66-333) decreased the interaction between TAK1 and TAB1 (Fig. 4e), strongly suggesting that S6K1 might be capable of inhibiting the interaction between TAK1 and TAB1. We further asked whether S6K1 deficiency could enhance the induction of TAK1 kinase activity by TLR stimulation. TAK1 kinase activity was significantly enhanced in control (wt) THP-1 cells (Fig. 4f and g, open bars). Interestingly, the kinase activities were markedly increased in S6K1KD THP-1 cells treated with LPS and FSL-1 compared with those in control (wt) THP-1 cells (Fig. 4f and g, closed bars). These results suggest that S6K1 negatively regulates the TAK1 kinase activity induced by TLR2 or TLR4 stimulation by interrupting the molecular interaction between TAK1 and TAB1, as depicted in Fig. 4h.
FIG 4.
S6K1 competitively interferes with the binding of TAB1 to TAK1, which inhibits TAK1 kinase activity. (a) HEK293 cells were transfected with Myc-TAK1 and different concentrations of Flag-S6K1. At 36 h after transfection, cells were treated with or without LPS (100 ng/ml) for 45 min, extracted, and Western blotting was performed with the antibodies indicated to the left. The band intensity of pho-TAK1 was analyzed with Image J (bottom). Data shown are the averages from a minimum of three independent experiments (±SD). *, P < 0.05. (b) HEK293-TLR4 cells were cotransfected with wt S6K1, DN-S6K1, or CA-S6K1 vector, together with pBIIx-luc reporter and Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with or without (WO) LPS for 6 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that in untreated cells. The data shown are the averages of a minimum of three independent experiments, with error bars denoting ±SD. (c) Wild-type and S6K1KD THP-1 cells were treated with or without LPS for different times, and then Western blotting was performed as described in Materials and Methods. (d) HEK293 cells were transfected with Myc-TAK1, Flag-S6K1, and different concentrations of Flag-TAB1 as indicated. At 36 h after transfection, cells were extracted and immunoprecipitated with anti-Myc antibody. Interaction was detected by Western blotting with anti-Flag antibody. The presence of Myc-TAK1, Flag-S6K1, and Flag-TAB1 in the pre-IP lysates was verified by Western blotting. The band intensity of Flag-S6K1 was analyzed with Image J (bottom). The data shown are the averages of a minimum of three independent experiments (±SD). *, P < 0.05. (e) HEK293 cells were transfected with Myc-TAK1, Flag-TAB1, and different concentrations of Flag-S6K1 (66-333) as indicated. At 36 h after transfection, cells were extracted and immunoprecipitated with anti-Myc antibody. The interaction was detected by Western blotting with anti-Flag antibody. The presence of Myc-TAK1, Flag-TAB1, and Flag-S6K1 (66-333) in the pre-IP lysates was verified by Western blotting. (f and g) Wild type and S6K1KD THP-1 cells were treated with or without LPS (f) or FSL-1 (g) for different times as indicated. The kinase assay for TAK1 was performed using a c-TAK1 kinase assay kit in accordance with the manufacturer's protocol. The data shown are the averages of a minimum of three independent experiments (±SD). *, P < 0.05; **, P < 0.01. (h) A model for the negative regulation of TAK1. The C terminus of TAB1 interacts with the N terminus of TAK1, and the TAB1 association to TAK1 positively regulates TAK1 activity via the recruitment of p38 and induction of catalytic activity (top). In contrast, the interaction between the internal catalytic domain of S6K1 and the N terminus of TAK1 inhibits the TAB1 interaction with TAK1, which results in the inhibition of TAK1 catalytic activity.
S6K1 knockdown THP-1 cells exhibit enhanced NF-κB-dependent gene expression induced by TLR2 and TLR4.
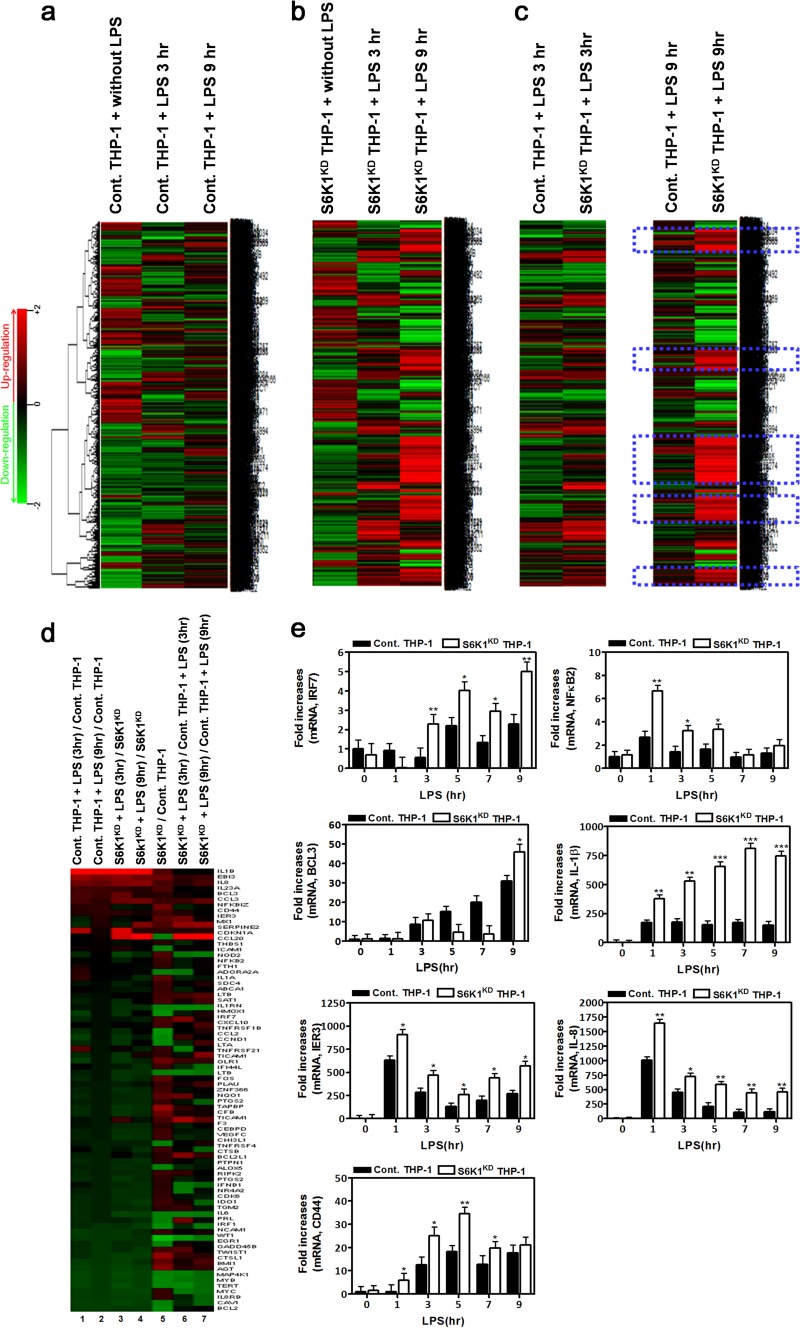
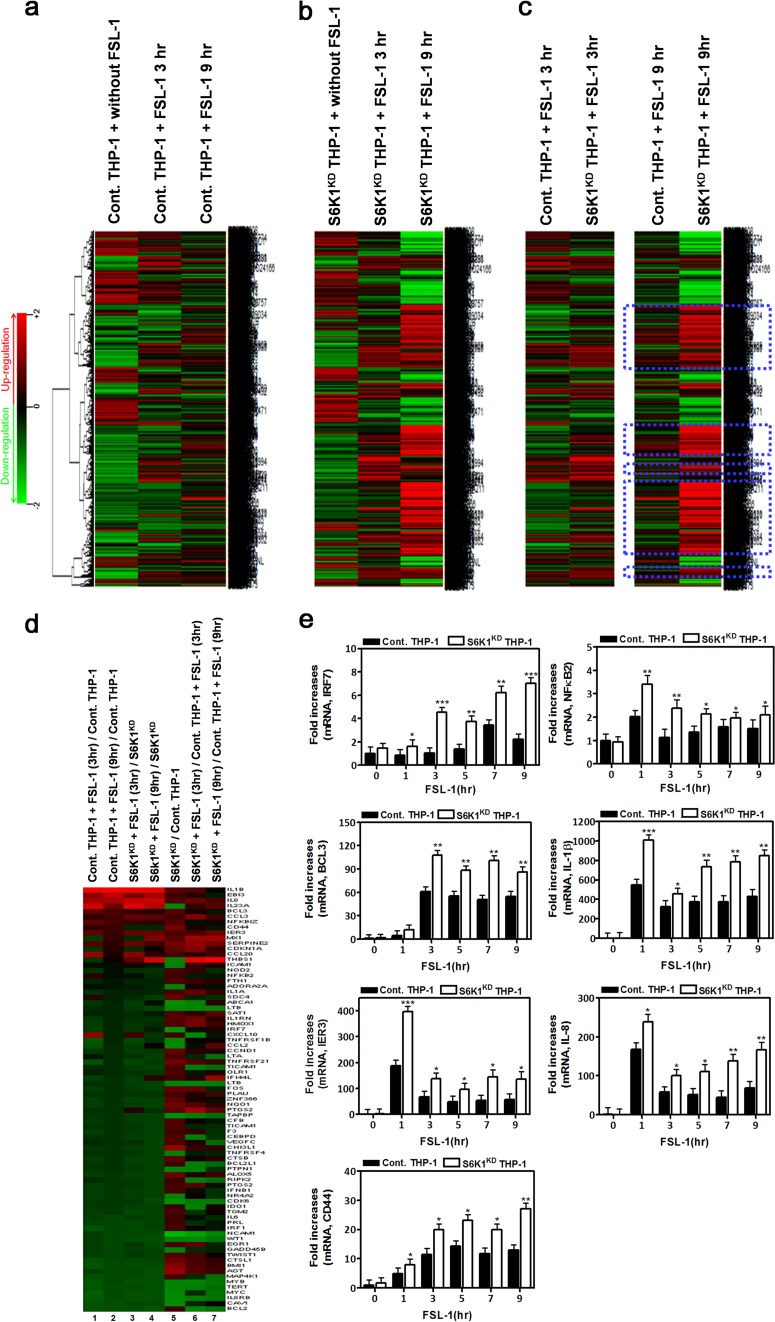
Since it is well known that TAK1 activation results in the expression of NF-κB-dependent genes through the activation of downstream signaling molecules like IKKs and NF-κB (4, 18), we examined the effects of S6K1 on the expression of NF-κB-dependent genes induced by stimulation of TLR2 or TLR4. Control and S6K1KD THP-1 cells were treated with LPS or FSL-1 for different exposure times, and then gene expression profiles were analyzed by microarray assay. As expected, stimulation of TLR4 or TLR2 induced marked changes in the total gene expression in both control and S6K1KD THP-1 cells (Fig. 5a and b and 6a and b). Compared to control THP-1 cells treated with LPS or FSL-1 for 3 h, many genes were highly upregulated in S6K1KD THP-1 cells under the same treatment conditions (Fig. 5c and 6c). Furthermore, the upregulation of these genes was greatly magnified after treatment with LPS for 9 h (Fig. 5c and 6c, blue dashed boxes). To further assess NF-κB-dependent gene expression, the expression of genes containing specific NF-κB-binding DNA sequences was further analyzed (see Tables S1 and S2 in the supplemental material). Upon stimulation with LPS, NF-κB-dependent gene expression was markedly enhanced in both control and S6K1KD THP-1 cells in a time-dependent manner (Fig. 5d, columns 1 to 4). Their expression was significantly higher in S6K1KD than in control THP-1 cells (Fig. 5d, columns 5 to 7). To verify the microarray data, we selected 7 different genes, IRF7, BCL3, IER3, CD44, NF-κB2, IL-1β, and IL-8, which were upregulated in S6K1KD THP-1 cells treated with LPS (see Table S2), and their expression was confirmed by quantitative real-time PCR (qRT-PCR) analysis with specific primers targeted to each gene. As expected, these genes were upregulated in LPS-treated S6K1KD THP-1 cells in a time-dependent manner (Fig. 5e). Stimulation of TLR2 similarly resulted in enhanced NF-κB-dependent gene expression in S6K1KD THP-1 cells (Fig. 6d, columns 1 to 4). The expression of these genes was significantly higher in S6K1KD THP-1 cells (Fig. 6d, column 5), and that difference was magnified with FSL-1 stimulation (Fig. 6d, columns 6 and 7). These microarray results were confirmed by qRT-PCR analysis with specific primers targeted to IRF7, BCL3, IER3, CD44, NF-κB2, IL-1β, and IL-8 (Fig. 6e). These results suggest that S6K1 negatively regulates the expression of NF-κB-dependent genes induced by stimulation of TLR2 or TLR4.
FIG 5.
S6K1 knockdown THP-1 cells exhibit a marked induction in NF-κB-dependent gene expression in response to stimulation of TLR4. (a) Control THP-1 cells were treated with or without TLR4 agonist LPS (100 ng/ml) for 3 h or 9 h. Microarray analysis was performed as described in Materials and Methods. (b) S6K1KD THP-1 cells were treated with or without LPS for 3 h or 9 h. Microarray analysis was performed as described in Materials and Methods. (c) Comparison of microarray data between control THP-1 and S6K1KD THP-1 cells treated with LPS for 3 h or 9 h. Highly upregulated genes in S6K1KD THP-1 cells treated with LPS are indicated with dashed blue boxes. (d) Microarray analysis of the NF-κB-dependent upregulated and downregulated genes is shown. The experimental conditions are indicated above the columns. (e) Control THP-1 and S6K1KD THP-1 cells were treated with or without LPS (100 ng/ml) for different times as indicated. Total RNAs were isolated from each sample, and quantitative RT-PCR analysis was performed with specific primers targeted to the genes indicated on the y axes. Data represent the averages of data from two independent experiments, each done with triplicates. Error bars represent the means ± SD based on these six samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 6.
S6K1 knockdown THP-1 cells exhibit a marked induction in NF-κB-dependent gene expression in response to stimulation of TLR2. (a) Control THP-1 cells were treated with or without FSL-1 (10 μg/ml) for 3 h or 9 h. Microarray analysis was performed as described in Materials and Methods. (b) S6K1KD THP-1 cells were treated with or without FSL-1 for 3 h or 9 h. Microarray analysis was performed as described in Materials and Methods. (c) Comparison of microarray data between control THP-1 and S6K1KD THP-1 cells treated with FSL-1 for 3 h or 9 h. The highly upregulated genes in S6K1KD THP-1 treated with FSL-1 are indicated with dashed blue boxes. (d) Microarray analysis of the NF-κB-dependent upregulated and downregulated genes. The experimental conditions are indicated above the columns. (e) Control THP-1 and S6K1KD THP-1 cells were treated with or without FSL-1 (100 ng/ml) for different times as indicated. Total RNAs were isolated from each sample, and quantitative RT-PCR analysis was performed with specific primers targeted to the genes indicated on the y axes. Data represent the averages of data from two independent experiments, each done with triplicates. Error bars represent the means ± SD based on these six samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
TLR signaling pathways are initiated when cytoplasmic Toll/IL-1 receptor (TIR) domains of TLRs recruit binding of TIR-containing adaptor proteins, such as MyD88, TIR domain-containing adaptor protein (TIRAP)/MyD88-adaptor-like (Mal), and TIR domain-containing adaptor-inducing IFN-β (TRIF)/TIR domain-containing adaptor molecule-1 (TICAM-1) (19–21). TLR signaling pathways can be classified as MyD88-dependent pathways that are common to all TLRs or as MyD88-independent pathways that are particular to signaling by TLR3 and TLR4 (19). In MyD88-dependent pathways, MyD88 further recruits IL-1 receptor-associated kinase (IRAK) to the TLRs, and then activated IRAK associates with TRAF6, eventually leading to the activation of two distinct transcriptional factors, NF-κB and AP-1. TRAF6 plays a key role in TLR signaling pathways through the formation of a signaling complex containing TAK1, TAB1, and TAB2, which induces the activation of IKKs, leading to the activation of NF-κB, MAPK pathways, and AP-1 (4, 22–24).
The data presented here show that S6K1 negatively regulates TLR2- or TLR4-mediated signaling. Through biochemical and molecular studies, we found that S6K1 overexpression suppressed NF-κB activity and the production of proinflammatory cytokines, such as TNF-α, IL-6, and IL-1β. By introducing shRNA targeting S6K1 by lentivirus transfection, we were able to confirm that S6K1 is a negative regulator of TLR2- and TLR4-mediated signaling. S6K1 deficiency resulted in increased TAK1 kinase activity and increased activation of TAK1-downstream molecules, including IKKs and p38. This results in increased production of proinflammatory cytokines, such as TNF-α, IL-6, and IL-1β. S6K1 interacts with the N terminus of TAK1, which has been defined as a TAB1 binding region. Interestingly, we found that the TAK1-TAB1 interaction or TAK1-S6K1 interaction was markedly attenuated in the presence of S6K1 or TAB1, respectively. These results strongly suggest that S6K1 interacts with the N terminus of TAK1 and that this interaction negatively regulates the activation of TAK1 through competition with the TAK1-TAB1 interaction. Microarray analysis revealed that S6K1 deficiency enhanced the expression of NF-κB-dependent genes in response to stimulation of TLR2 or TLR4. Furthermore, upon LPS challenge, the survival of S6K1−/− mice was significantly decreased, and this decrease in survival correlated with increases in the expression of proinflammatory cytokines.
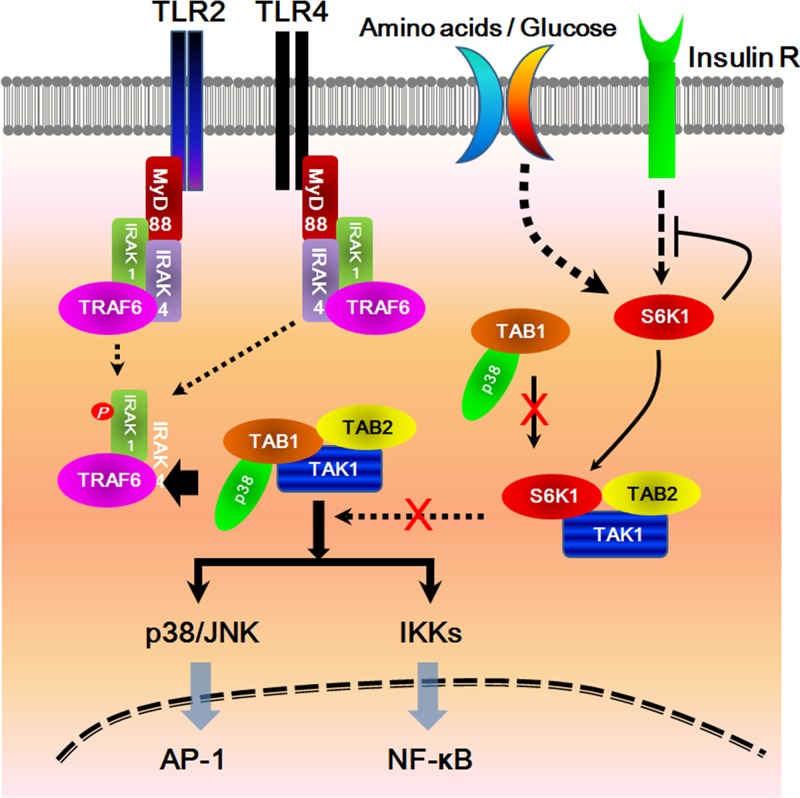
Taken together, the results of our study demonstrate that S6K1 acts as a negative regulator of TLR signaling by interrupting the interaction between TAK1 and TAB1 (Fig. 7). Interruption of the TAK1-TAB1 interaction may also attenuate the MAPK pathway that activates AP-1, as TAB1 is responsible for recruiting the p38 molecule into the TAK1 complex (17, 25). Therefore, the interaction between S6K1 and TAK1 might be critical for TAK1 signaling for both NF-κB activation via the activation of IKKs and AP-1 activation via the p38/JNK pathway. Based on our current results, we speculate that this mechanism may contribute to the bacterial and viral infection susceptibility seen in patients with type 2 diabetes. Clinical reports have demonstrated that type 2 diabetic patients have a high risk for infection (26–28). Recent reports have shown that type 2 insulin resistance is tightly linked with S6K1 activity via a negative feedback loop involving S6K1 that suppresses insulin signaling under conditions of nutrient overload (29). Moreover, increasing evidence has shown that cross talk between inflammatory macrophages and adipocytes is involved in insulin resistance (30), suggesting a pivotal role for macrophages in the pathogenesis of type 2 diabetes. In line with these results, previous evidence suggested an intimate association of reduced monocyte insulin receptor activity with systemic insulin resistance in humans (31). Since macrophages are pivotal effector cells capable of mounting an innate immune response against viral and bacterial infections (32) and also are able to express insulin signaling molecules capable of inducing the insulin receptor/IRS2/PI3K/AKT signaling cascade (30, 33), these results provide a framework for understanding how S6K1 might be involved in both insulin receptor signaling and TLR signaling in macrophages in the setting of type 2 diabetes. As shown in Fig. 7, we propose that in macrophages derived from type 2 diabetics, S6K1 leads to an insulin resistance response through a negative feedback loop under conditions of nutrient overload. Simultaneously, S6K1 is able to interact with TAB1, leading to the suppression of TLR signaling through inhibition of TAK1 activity, which might eventually be implicated in the impairment of innate immune responses against infections. We believe that this model provides a novel mechanism by which type 2 diabetic patients are sensitive to microbial and viral infections.
FIG 7.
A schematic model for how S6K1 inhibits TLR2- or TLR4-mediated signaling pathways. Upon stimulation of TLR2 or TLR4 receptors, MyD88 binds to the cytoplasmic portion of the TLRs through interactions between individual TIR domains. IRAKs, such as IRAK-4 and IRAK-1, and TRAF6 are recruited to the receptor, and then phosphorylated IRAK-1 (by IRAK-4) dissociates from the receptor together with TRAF6. TRAF6 further interacts with TAK1, TAB1, and TAB2. TAB1 also interacts with p38α through its p38α-binding domain. TAB1 plays several roles in the regulation of the TAK1 complex, such as the recruitment of p38α MAPK to the TAK1 complex for the phosphorylation of TAB1 and induction of TAK1 catalytic activity. The activated TAK1 eventually induces the activation of NF-κB and AP-1 transcription factors through activation of the IKK complex and MAP kinases, respectively. In this study, we speculate on the inhibitory role of S6K1 in type 2 diabetes, inducing insulin resistance under conditions of nutrient overload. Under high-nutrient conditions, S6K1 acts as a negative regulator in insulin signaling. Simultaneously, S6K1 interacts with TAK1, which results in the inhibition of the association of TAB1 and p38α to TAK1. The inhibitory effect eventually induces the inhibition of NF-κB and AP-1 activation by the suppression of IKKs and p38/JNK activation, respectively.
Supplementary Material
ACKNOWLEDGMENTS
The Mid-Career Researcher Program supported this work through an NRF grant (NRF-2012R1A2A2A01005659).
We thank Sara Kozma and George Thomas for kindly providing S6K1−/− mice.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 25 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01225-13.
REFERENCES
- 1.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. 1999. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398:252–256. 10.1038/18465 [DOI] [PubMed] [Google Scholar]
- 2.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. 1996. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science 272:1179–1182. 10.1126/science.272.5265.1179 [DOI] [PubMed] [Google Scholar]
- 3.Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Núñez G, Inohara N. 2008. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 27:373–383. 10.1038/sj.emboj.7601962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. 2001. TAK1 is an ubiquitin-dependent kinase of MKK and IKK. Nature 412:346–351. 10.1038/35085597 [DOI] [PubMed] [Google Scholar]
- 5.Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. 2003. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 22:6277–6288. 10.1093/emboj/cdg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung PC, Nebreda AR, Cohen P. 2004. TAB3, a new binding partner of the protein kinase TAK1. Biochem. J. 378:27–34. 10.1042/BJ20031794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conner SH, Kular G, Peggie M, Shepherd S, Schüttelkopf AW, Cohen P, Van Aalten DM. 2006. TAK1-binding protein 1 is a pseudophosphatase. Biochem. J. 399:427–434. 10.1042/BJ20061077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendoza H, Campbell DG, Burness K, Hastie J, Ronkina N, Shim JH, Arthur JS, Davis RJ, Gaestel M, Johnson GL, Ghosh S, Cohen P. 2008. Roles for TAB1 in regulating the IL-1-dependent phosphorylation of the TAB3 regulatory subunit and activity of the TAK1 complex. Biochem. J. 409:711–722. 10.1042/BJ20071149 [DOI] [PubMed] [Google Scholar]
- 9.Di Y, Li S, Wang L, Zhang Y, Dorf ME. 2008. Homeostatic interactions between MEKK3 and TAK1 involved in NF-kappaB signaling. Cell. Signal. 20:705–713. 10.1016/j.cellsig.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mochida Y, Takeda K, Saitoh M, Nishitoh H, Amagasa T, Ninomiya-Tsuji J, Matsumoto K, Ichijo H. 2000. ASK1 inhibits interleukin-1-induced NF-kappa B activity through disruption of TRAF6-TAK1 interaction. J. Biol. Chem. 275:32747–32752. 10.1074/jbc.M003042200 [DOI] [PubMed] [Google Scholar]
- 11.Shin JH, Min SH, Kim SJ, Kim YI, Park J, Lee HK, Yoo OJ. 2013. TAK1 regulates autophagic cell death by suppressing the phosphorylation of p70 S6 kinase 1. Sci. Rep. 3:1561. 10.1038/srep01561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SY, Chun E, Lee KY. 2011. Phospholipase A(2) of peroxiredoxin 6 has a critical role in tumor necrosis factor-induced apoptosis. Cell Death Differ. 18:1573–1583. 10.1038/cdd.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Jeong S, Jung E, Baik KH, Chang MH, Kim SA, Shim JH, Chun E, Lee KY. 2012. AMP-activated protein kinase-α1 as an activating kinase of TGF-β-activated kinase 1 has a key role in inflammatory signals. Cell Death Dis. 3:e357. 10.1038/cddis.2012.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, Shim JH, Chun E, Lee KY. 2012. Reciprocal inhibition between the transforming growth factor-β-activated kinase 1 (TAK1) and apoptosis signal-regulating kinase 1 (ASK1) mitogen-activated protein kinase kinase kinases and its suppression by TAK1-binding protein 2 (TAB2), an adapter protein for TAK1. J. Biol. Chem. 287:3381–3391. 10.1074/jbc.M111.317875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiura K, Kataoka H, Yasuda M, Inoue N, Shibata K. 2006. The diacylated lipopeptide FSL-1 induces TLR2-mediated Th2 responses. FEMS Immunol. Med. Microbiol. 48:44–55. 10.1111/j.1574-695X.2006.00119.x [DOI] [PubMed] [Google Scholar]
- 16.Brown K, Vial SC, Dedi N, Long JM, Dunster NJ, Cheetham GM. 2005. Structural basis for the interaction of TAK1 kinase with its activating protein TAB1. J. Mol. Biol. 354:1013–1020. 10.1016/j.jmb.2005.09.098 [DOI] [PubMed] [Google Scholar]
- 17.Pathak S, Borodkin VS, Albarbarawi O, Campbell DG, Ibrahim A, Van Aalten DM. 2012. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 31:1394–1404. 10.1038/emboj.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19:2668–2681. 10.1101/gad.1360605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akira S, Takeda K, Kaisho T. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675–680. 10.1038/90609 [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, McMurray D, Smith DE, Sims JE, Bird TA, O'Neill LA. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413:78–83. 10.1038/35092578 [DOI] [PubMed] [Google Scholar]
- 21.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161–167. 10.1038/ni886 [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Akira S. 2004. TLR signaling pathways. Semin. Immunol. 16:3–9. 10.1016/j.smim.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 23.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361. 10.1016/S0092-8674(00)00126-4 [DOI] [PubMed] [Google Scholar]
- 24.Sakurai H, Miyoshi H, Mizukami J, Sugita T. 2000. Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS Lett. 474:141–145. 10.1016/S0014-5793(00)01588-X [DOI] [PubMed] [Google Scholar]
- 25.Lu G, Kang YJ, Han J, Herschman HR, Stefani E, Wang Y. 2006. TAB-1 modulates intracellular localization of p38 MAP kinase and downstream signaling. J. Biol. Chem. 281:6087–6095. 10.1074/jbc.M507610200 [DOI] [PubMed] [Google Scholar]
- 26.Sangiorgio L, Attardo T, Gangemi R, Rubino C, Barone M, Lunetta M. 2000. Increased frequency of HCV and HBV infection in type 2 diabetic patients. Diabetes Res. Clin. Pract. 48:147–151. 10.1016/S0168-8227(99)00135-7 [DOI] [PubMed] [Google Scholar]
- 27.Danquah I, Bedu-Addo G, Mockenhaupt FP. 2010. Type 2 diabetes mellitus and increased risk for malaria infection. Emerg. Infect. Dis. 16:1601–1604. 10.3201/eid1610.100399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller LM, Gorter KJ, Hak E, Goudzwaard WL, Schellevis FG, Hoepelman AI, Rutten GE. 2005. Increased risk of common infections in patients with type 1 and type 2 diabetes mellitus. Clin. Infect. Dis. 41:281–288. 10.1086/431587 [DOI] [PubMed] [Google Scholar]
- 29.Um SH, D'Alessio D, Thomas G. 2006. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 3:393–402. 10.1016/j.cmet.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 30.Liang CP, Han S, Okamoto H, Carnemolla R, Tabas I, Accili D, Tall AR. 2004. Increased CD36 protein as a response to defective insulin signaling in macrophages. J. Clin. Invest. 113:764–773. 10.1172/JCI200419528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang CP, Han S, Senokuchi T, Tall AR. 2007. The macrophage at the crossroads of insulin resistance and atherosclerosis. Circ. Res. 100:1546–1555. 10.1161/CIRCRESAHA.107.152165 [DOI] [PubMed] [Google Scholar]
- 32.West AP, Koblansky AA, Ghosh S. 2006. Recognition and signaling by toll-like receptors. Annu. Rev. Cell Dev. Biol. 22:409–437. 10.1146/annurev.cellbio.21.122303.115827 [DOI] [PubMed] [Google Scholar]
- 33.Welham MJ, Bone H, Levings M, Learmonth L, Wang LM, Leslie KB, Pierce JH, Schrader JW. 1997. Insulin receptor substrate-2 is the major 170-kDa protein phosphorylated on tyrosine in response to cytokines in murine lymphohemopoietic cells. J. Biol. Chem. 272:1377–1381. 10.1074/jbc.272.2.1377 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.