Abstract
The reversible nature of protein phosphorylation dictates that any protein kinase activity must be counteracted by protein phosphatase activity. How phosphatases target specific phosphoprotein substrates and reverse the action of kinases, however, is poorly understood in a biological context. We address this question by elucidating a novel function of the conserved PP4 family phosphatase Pph3-Psy2, the yeast counterpart of the mammalian PP4c-R3 complex, in the glucose-signaling pathway. Our studies show that Pph3-Psy2 specifically targets the glucose signal transducer protein Mth1 via direct binding of the EVH1 domain of the Psy2 regulatory subunit to the polyproline motif of Mth1. This activity is required for the timely dephosphorylation of the downstream transcriptional repressor Rgt1 upon glucose withdrawal, a critical event in the repression of HXT genes, which encode glucose transporters. Pph3-Psy2 dephosphorylates Mth1, an Rgt1 associated corepressor, but does not dephosphorylate Rgt1 at sites associated with inactivation, in vitro. We show that Pph3-Psy2 phosphatase antagonizes Mth1 phosphorylation by protein kinase A (PKA), the major protein kinase activated in response to glucose, in vitro and regulates Mth1 function via putative PKA phosphorylation sites in vivo. We conclude that the Pph3-Psy2 phosphatase modulates Mth1 activity to facilitate precise regulation of HXT gene expression by glucose.
INTRODUCTION
The ability to sense and utilize glucose is a fundamental process critical for the optimal survival of eukaryotic cells. The budding yeast Saccharomyces cerevisiae has evolved multiple interconnected signaling pathways to cope with the rapidly fluctuating glucose levels in its environment. The induction of glucose transporter (HXT) genes is precisely controlled by glucose signals through two membrane-bound glucose sensors Rgt2 and Snf3, via a pathway that leads to the phosphorylation and inhibition of a sequence-specific transcriptional repressor Rgt1 (1). In the absence of glucose, dephosphorylated Rgt1 binds to the promoters of HXT genes and recruits the Ssn6/Tup1 corepressor to repress their expression (1). That repression requires the activity of two homologous proteins, Mth1 and Std1 (2–4). Glucose induces the phosphorylation and dissociation of Rgt1 from the HXT gene promoters by promoting degradation of Mth1 and Std1, thereby leading to the derepression of HXT genes (2, 5). Another major glucose signaling pathway is initiated by the Gpr1 G protein-coupled receptor and Ras proteins that results in an increase of intracellular cyclic AMP (cAMP) level and consequently activation of cAMP-dependent kinase protein kinase A (PKA), which globally regulates cell growth (6). Therefore, the cAMP/PKA pathway needs to be coordinated with the induction of HXT genes based upon glucose availability.
Previous studies show that glucose-induced phosphorylation of Mth1/Std1 and Rgt1 plays a critical role in regulating the HXT gene expression and involves at least two protein kinases, CK1 casein kinase 1 (CK1) and PKA. The membrane-associated CK1 family protein kinases Yck1 and Yck2 (Yck1/2) are thought to phosphorylate Mth1 and Std1 in response to glucose signals (7, 8), targeting them for recognition by Grr1, the F-box protein specificity subunit of the SCFGrr1 ubiquitin protein ligase, and, thereby, ubiquitination and proteasomal degradation (7). The yeast PKA homologs, Tpk1, Tpk2, and Tpk3, have been implicated in phosphorylation of Rgt1 upon glucose stimulation (9). Rgt1 phosphorylation is thought to promote a conformational change that masks its DNA-binding domain (10). Although reversal of these phosphorylation events is required to reestablish repression of HXT genes in the absence of glucose, little is known about the role or regulation of the protein phosphatases involved in that process.
Protein phosphorylation/dephosphorylation orchestrated by protein kinases and protein phosphatases is a universal regulatory mechanism implicated in diverse cellular responses. It is assumed that for an organism to react precisely toward changes in environment stimuli, the action of phosphatases must be coordinated with that of kinases to regulate critical effector substrates. How a protein phosphatase targets a specific substrate and, by doing so, reverses a process initiated by a given kinase, however, is poorly understood. We address this issue here by focusing on the role of the yeast Pph3-Psy2 complex, a member of the PP4 phosphatase family (11), in glucose signaling. Pph3 and Psy2 are yeast orthologs of the mammalian PP4c and R3 regulatory subunit, respectively (11). The eukaryotic PP4 protein phosphatases are members of the highly conserved family of phosphoprotein phosphatases (PPPs) that are most closely related to the PP2A serine/threonine phosphatases. Similar to PP2As, PP4 phosphatases have three types of subunits: the catalytic subunit PP4c, a structural subunit (such as R1 and R2), and a variety of regulatory subunits, including R3, R4, and Gemin4 (12, 13), which presumably confer specificity to the phosphatase complex. Recent studies show that PP4 enzymes regulate a variety of biological processes, including centrosome maturation, spliceosome assembly, signaling through NF-κB, JNK, IRS4, and Wnt, histone deacetylation, and DNA damage responses (12, 14–16). It remains unclear, however, how PP4 targets phosphorylated proteins to oppose the relevant kinase activities in these processes.
The highly conserved R3/Psy2 regulatory subunit has been implicated in nutrient starvation-induced chemotaxis in Dictyostelium, where it acts as a suppressor for the mitogen-activated protein kinase kinase MEK1 and hence is known as SMEK (17). In the present study, we uncovered a novel function of Psy2 in the context of nutrient sensing in Saccharomyces cerevisiae. We found that the N-terminal EVH1 domain of Psy2, a polyproline-binding domain conserved among eukaryotes (18), exclusively interacts with two glucose signal transducers, Mth1 and Std1, in a yeast two-hybrid screen. We established a role for yeast PP4 phosphatase Pph3-Psy2 in glucose signaling mediated by the specific targeting of the Psy2 regulatory subunit via its EVH1 domain to a polyproline motif in Mth1. The PP4 activity specified by Pph3-Psy2 appears to counteract the major glucose-responsive kinase PKA by dephosphorylating the putative PKA sites in Mth1. We conclude that the opposing actions of Pph3-Psy2 phosphatase and the kinase that phosphorylates Mth1 (most likely PKA) are necessary for the precise regulation of glucose transporter (HXT) genes in response to glucose availability. This places the Pph3-Psy2-Mth1 complex at a signaling nexus that integrates two major glucose signaling inputs (the Rgt2/Snf3-Mth1-Rgt1 pathway and cAMP/PKA pathway) to coordinate optimal expression of glucose transporter genes with the cell growth.
MATERIALS AND METHODS
Yeast strains.
Yeast strains used in the present study are listed in Table 1. All strains were constructed using standard methods and were grown in standard YP medium containing 2% Bacto peptone, 1% yeast extract, and 2% glucose-galactose or in synthetic yeast nitrogen base medium with 2% glucose-galactose supplemented with the appropriate amino acids necessary for plasmid maintenance. psy2Δ pph3Δ strains and strains expressing integrated RGT1-hemagglutinin (HA), MTH1-HA, MTH1PA-HA, MTH1-4SA-HA, and Myc-tagged PPH3 were generated using standard yeast genetic methods. The strain expressing Psy2-6Myc from the endogenous locus was constructed using a knockin plasmid containing the PSY2-6Myc C-terminal region (pMAY101). PSY2-TAP strains were provided by F. Romesberg's laboratory at The Scripps Research Institute (19).
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| K699 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 | 39 |
| CWY1112 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::KanMX2 | 2 |
| CWY1919 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1PA-3×HA::URA3 | This study |
| CWY1314 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::URA3 MTH1-6×Myc::KanMX2 | This study |
| CWY1320 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 grr1-AAA-Myc::TRP1 | 7 |
| CWY1758 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::URA3 MTH1-6×Myc::KanMX2 psy2::LEU2 | This study |
| CWY1794 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::KanMX2 psy2::URA3 | This study |
| CWY1818 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::KanMX2 pph3::TRP1 | This study |
| CWY1822 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::KanMX2 psy2::URA3 pph3::TRP1 | This study |
| CWY1823 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 RGT1-3×HA::KanMX2 MTH1PA-HA::URA3 | This study |
| CWY1646 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 PSY2-6×Myc::KanMX2 | This study |
| CWY1311 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1-3×HA::KanMX2 | This study |
| CWY1895 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1-3×HA::KanMX2r PPH3-13×Myc::URA3 | This study |
| CWY1918 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1PA-3×HA::URA3 PPH3-13×Myc::KanMX2 | This study |
| CWY1916 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 PPH3-1 3×Myc::KanMX2 | This study |
| CWY1917 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1-3×HA::KanMX2 PPH3-13×Myc::URA3 psy2::LEU2 | This study |
| CWY2271 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 mth1::HIS3 pIMT52-MTH1-3×HA::URA3 | This study |
| CWY2275 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 mth1::HIS3 psy2::LEU2 pIMT52-MTH1-3×HA::URA3 | This study |
| CWY2288 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 mth1::HIS3 pIMT52-mth1-4SA-3×HA::URA3 | This study |
| CWY2290 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 mth1::HIS3 psy2::LEU2 pIMT52-mth1-4SA-3×HA::URA3 | This study |
| CWY1943 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1-3×HA::KanMX2 psy2::LEU2 | This study |
| CWY1940 | MATa ura3-1 his3-11,15 leu2-3,111 trp1-1 ade2 can1-100 MTH1-3×HA::KanMX2 pph3::TRP1 | This study |
| MAY205 | W1588-4C MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 PPH3-TAP::Kanr psy2::LEU2 | This study |
| FR796 | W1588-4C MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 PSY2-TAP::Kanr | 19 |
| FR1044 | W1588-4C MATa ura3-1 his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 PSY2-TAP::Kanr pph3::his5+ | 19 |
All strains are isogenic with W303-b (K699) unless otherwise indicated. Kanr, kanamycin resistance.
Plasmids.
The two-hybrid bait construct for the EVH1 domain of Psy2 (amino acids 1 to 151) was made by cloning the corresponding PCR fragment downstream from the Gal4 DNA-binding domain (DBD) sequence in frame on pGBT9 vector (Clontech). Detailed information related to the plasmids used in the two-hybrid screen and two-hybrid assays will be provided upon request. psy2::LEU2 plasmid (pMC178) and plasmid containing the PSY2 genomic region (pJO78) were kindly provided by Hans Ronne at the Uppsala Genetic Center. The 2μ plasmid expressing MTH1-3HA driven by its endogenous promoter (pMT52) was provided by Martin Schmidt at University of Pittsburgh. Mth1PA and Sth1PA mutants were created using QuikChange site-directed mutagenesis, in which prolines 348 to 350 of Mth1 and prolines 359 to 361 of Std1 were replaced by alanines.
The reconstituted plasmids expressing Psy2-6Myc and Psy2ΔEVH1-6Myc were created by ligating the PSY2 genomic fragment containing the PSY2 promoter and N-terminal region from pJO78 with the 6Myc-tagged PSY2 C-terminal region, including a terminator sequence from pMAY101, using pRS414 as a backbone. ΔEVH1 was generated by deleting amino acid residues 1 to 129 according to the QuikChange mutagenesis protocol.
In order to integrate hemagglutinin (HA)-tagged MTH1-4SA mutant into the yeast genome, pMT52 plasmid containing the wild-type (WT) MTH1 and its promoter sequences was digested with BsaAI and HpaI to remove the 2μ sequences and religated to generate the integration plasmid pIMT52. pIMT52-mth1-4SA was created by site-directed mutagenesis, in which serines 111, 112, 161, and 358 of Mth1 were replaced by alanines. To incorporate MTH1-4SA into the yeast genome, pIMT52-mth1-4SA was digested with EcoRV and integrated at the URA3 locus in an mth1Δ strain background. The same approach was used to integrate the wild-type control pIMT52.
Two-hybrid screen and assays.
Yeast two-hybrid screen was carried out by introducing a yeast genomic two-hybrid library (genomic bank I-1651 from the Institut Pasteur) (20) into the AH109 host strain pretransformed with the EVH1 bait construct according to the Matchmaker two-hybrid system protocol (Clontech). After library transformation, ca. 3 × 106 clones were screened for growth on selective synthetic media without His, Trp, and Leu. Subsequently, 80 clones were tested for growth on synthetic media without Ade, His, Trp, and Leu, as well as tested for β-galactosidase activity. A total of 27 confirmed positive clones were further analyzed (see Table 3). Among them, 15 were MTH1 clones and 4 were STD1 clones. The prey plasmids harbored by the positive clones were recovered from yeast cells and subjected to sequence analysis, followed by verification by cotransformation with the bait plasmid. For yeast two-hybrid interaction assays, appropriate bait and prey plasmids were cotransformed into AH109 and evaluated by their ability to confer growth on synthetic media lacking Ade, His, Trp, and Leu.
TABLE 3.
Summary of positive interacting clones identified from Psy2-EVH1 domain yeast two-hybrid screen
| Gene | No. of clones |
||
|---|---|---|---|
| Total | Containing MPPP motif | Independent | |
| MTH1 | 15 | 13 | 11 |
| STD1 | 4 | 4 | NDa |
| Ribosomal S23 protein gene | 1 | 0 | 0 |
| No insert or not sequenceable | 7 | 0 | 0 |
| Total | 27 | 0 | 0 |
ND, not determined.
Protein and RNA analysis.
For immunoblot analysis protein extracts were prepared using the trichloroacetic acid-urea extraction method as previously described (7). For reverse transcription-PCR (RT-PCR) analysis, the purification of RNA, the synthesis of cDNA, reverse transcriptase reactions, and PCRs were carried out as previously described (21).
Coimmunoprecipitation analysis.
Coimmunoprecipitation analysis was performed as previously described (7). Briefly, protein extracts were obtained using glass beads in lysis buffer (50 mM Tris-HCl [pH 7.5], 1% Triton X-100, 250 mM NaCl) containing protease and phosphatase inhibitors. Mouse 12C5 anti-HA monoclonal antibodies (kindly provided by Ian Wilson, The Scripps Research Institute, La Jolla, CA) covalently conjugated to protein A-Sepharose were used to perform immunoprecipitations. The coimmunoprecipitations were carried out with 1 mg of total protein extract incubated with the antibody described above for 1 h at 4°C and then washed three times with the same buffer before being prepared for SDS-PAGE. Rat anti-HA monoclonal antibodies (Roche) and goat anti-rat secondary antibodies were used on immunoblots to detect Mth1-3HA protein. Mouse anti-myc monoclonal antibodies (Santa Cruz) and rabbit anti-mouse secondary antibodies were used to detect Psy2-6Myc and Psy2-13Myc proteins.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation assays were performed as described previously (2). Briefly, formaldehyde (final concentration, 1%) was added to yeast culture to cross-link DNA and protein in vivo, followed by a 5-min incubation in the presence of 125 mM glycine at room temperature. The cells were frozen after being washed three times with ice-cold Tris-buffered saline. Glass beads were used to break up the yeast cells after the frozen cell pellet was resuspended in lysis buffer 2 (50 mM HEPES [pH 7.5], 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with protease inhibitors. After centrifugation (14,000 × g) the chromatin fraction (the pellet) was resuspended in the original volume of lysis buffer and processed for sonication to generate DNA fragments in the range of 500 to 1,000 bp. Immunoprecipitation of Rgt1-HA was carried out by incubating the clarified chromatin fraction with monoclonal anti-HA antibody in the presence of protein A-beads. The immune complexes were washed extensively and DNA-protein cross-linking was reversed in 1% SDS–Tris-EDTA at 65°C overnight. DNA was subsequently purified for PCR analysis, and the resulting products were visualized on 2.5% agarose gels or by quantitative PCR (qPCR) in the Chromo4 qPCR (Bio-Rad). Error bars for qPCR data represent the 95% confidence interval for triplicate samples. The sequences of PCR primers for HXT3 and actin used in this study are presented in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Analysis and gene | Oligonucleotide |
|
|---|---|---|
| Orientation | Sequence (5′–3′) | |
| HXT3 RNA | ||
| ACT1 | Forward | TGATGGTGTTACTCACGTCGTTCC |
| ACT1 | Reverse | GCAGTGGTGGAGAAAGAGTAACCA |
| HXT3 | Forward | TGAGGCCGACCAAGTACTTACCAA |
| HXT3 | Reverse | TTTGGGCGACGAAACCAGAAATGG |
| HXT3 chromatin immunoprecipitation | ||
| ACT1 | Forward | CAACCTGAAGGGACAGAGTTT |
| ACT1 | Reverse | TGTTCTCGCGTGTGTCTTATT |
| HXT3 | Forward | CGCGGATTCGGTTAAACT |
| HXT3 | Reverse | CCTCCGGAAATACCAACATC |
Quantitative RT-PCR.
RNA samples were isolated from yeast cells using the RNeasy Plus minikit [Qiagen]. All quantitative RT-PCRs were performed in triplicate using the iScript One-Step RT-PCR kit (Bio-Rad) in the Chromo4 qPCR. Error bars for qPCR data represent the 95% confidence interval for triplicate samples. The sequences of PCR primers used in this study are presented in Table 2. All qPCR data represent the averages of samples analyzed in triplicate and normalized to ACT1 RNA. HXT3 RNA is plotted relative to the wild-type time zero sample.
Purification of Pph3-Psy2 protein phosphatase complex.
WT and pph3Δ yeast strains expressing integrated TAP-tagged Psy2 were grown till late log phase in 2.5 liters of YPD, harvested by centrifugation, and washed with ice-cold water. The cell pellets were resuspended in equal volume of lysis buffer 3 (22) (20 mM Tris-HCl [pH 7.6], 10% glycerol, 200 mM potassium acetate, 1 mM EDTA, protease inhibitors) and frozen dropwise in liquid nitrogen. The frozen droplets of cells were ground for several rounds in a precooled mortar in liquid nitrogen and were then allowed to thaw completely at 4°C. The resulting lysates, after clarification at 15,000 rpm for 10 min, were then incubated with IgG-Sepharose beads overnight at 4°C. The immune complexes were washed five times in lysis buffer 3 without protease inhibitors, washed once in TEV reaction buffer (23) (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, and 1 mM dithiothreitol [DTT]), resuspended in TEV reaction buffer, and incubated with TEV protease for 3 h at room temperature. The supernatant was collected after centrifugation, dialyzed against phosphatase reaction buffer (19) (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MnCl2, 0.5 mM DTT) supplemented with glycerol at 5% final concentration for 1.5 h at 4°C, and stored in aliquots at −80°C. The phosphatase activity of the purified Pph3-Psy2 complex was assessed by dephosphorylation of a phosphopeptide K-R-pT-I-R-R in a malachite green-based assay (Upstate).
Protein phosphatase assays.
Flag-tagged Rgt1 expressed in a protease-deficient yeast strain and HA-tagged Mth1 expressed in grr1-AAA strain were immunoprecipitated using anti-Flag M2 agarose (Sigma) and anti-HA antibody coupled to protein A-beads, respectively, after cells from 200-ml late-log-phase culture were disrupted using glass beads in lysis buffer 1 supplemented with protease and phosphatase inhibitors. The immune complexes were washed four times in immunoprecipitation wash buffer (0.5% NP-40, 25 mM Tris-HCl [pH 8.0], 0.5 M NaCl), washed once in phosphatase reaction buffer, and then incubated with 10 μl of purified Psy2-Pph3 phosphatase complex in phosphatase reaction buffer at 30°C for 60 min or various times as needed, using 0.5 μl (200 U) of λ phosphatase (NEB) and the phosphopeptide substrate K-R-pT-I-R-R as positive controls.
Protein kinase assays.
Purified recombinant glutathione S-transferase (GST) fusions of the WT and mutant Mth1 or immunoprecipitated HA-tagged WT and mutant Mth1 were incubated with purified rat PKA catalytic subunit in 40 μl of kinase buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2) in the presence of 10 μCi of [γ-32P]ATP (final concentration, 82.5 nM; Perkin-Elmer) at 30°C for 15 min. The reactions were terminated by the addition of 4× sample buffer and subjected to SDS-PAGE analysis. For mass spectrometry (MS) analysis of in vitro PKA-phosphorylated Mth1, 1 mM nonradioactive ATP was used in the kinase reaction with two additions of both ATP and kinase during the reaction. The sample was denatured in 8 M urea and processed as described below.
Phosphopeptide analysis by mass spectrometry.
Log-phase grr1-AAA psy2Δ cells expressing HA-tagged Mth1 grown in 6 liters of YPD medium were harvested and homogenized in lysis buffer 1 after passing through the EmulsiFlex-C160 (Avestin) two times. The resultant lysate was centrifuged at 13,000 rpm for 20 min at 4°C, and the supernatant was incubated with anti-HA antibody coupled to protein A-agarose beads at 4°C for 2 to 4 h. The samples were washed four times in lysis buffer 1, denatured in 8 M urea, and then reduced and alkylated prior to overnight digestion with trypsin (Promega). The protein digests were loaded onto a MudPIT column that was then placed in-line with a 1110 quaternary HPLC pump (Agilent Technologies), and the eluted peptides were electrosprayed directly into an LTQ Orbi-Trap mass spectrometer (Thermo Scientific) using a 12-step MudPIT method. Tandem MS (MS/MS) spectra were extracted using RawXtract (v1.9.9) (24). MS/MS spectra were searched with the Sequest algorithm (25) against a Saccharomyces cerevisiae database concatenated to a decoy database in which the sequence for each entry in the original database was reversed (26). For analyzing in vitro PKA phosphorylated Mth1, 6 liters of wild-type cells expressing HA-tagged Mth1 were harvested from galactose-containing medium and processed for immunoprecipitation as described above followed by a “cold” PKA kinase reaction using HA-tagged Mth1 as the substrate. For the in vitro kinase reaction, a combined rat IPI database and Saccharomyces cerevisiae database was used. Sequest search results were assembled and filtered using the DTASelect (version 2.0) algorithm (27).
RESULTS
Psy2 interacts with Mth1 through a polyproline motif.
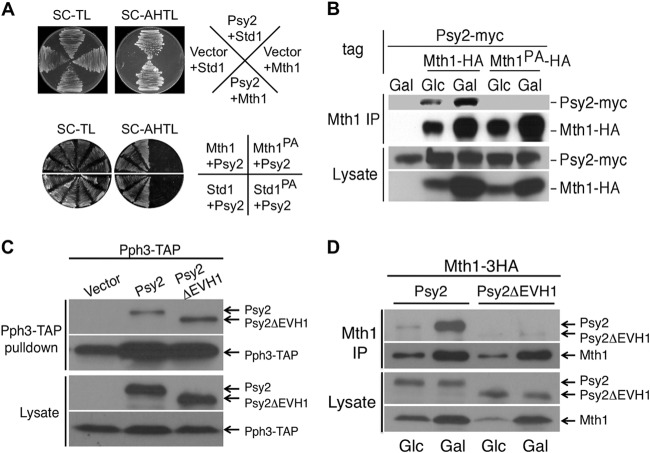
Psy2 shares a conserved EVH1 domain with other PP4R3 (SMEK) family proteins, a domain that is known to mediate binding to polyproline motifs (18). In a yeast two-hybrid screen with the Psy2 EVH1 domain, we identified both Mth1 and Std1 as the predominant Psy2-interacting proteins (Table 3 and Fig. 1A, upper). Further analysis of positive clones revealed that the majority of them expressed the C-terminal region of Mth1 and Std1, including a conserved MPPP motif (Table 3). To confirm that this proline-rich motif is required for the two-hybrid interactions, we created proline to alanine substitutions by site-directed mutagenesis and reintroduced the mutants into yeast cells. As expected, the MAAA mutation in both Mth1 and Std1 completely abolished their strong interaction with Psy2 in two-hybrid assays (Fig. 1A, lower panel). Since Mth1 is thought to play a predominant role in promoting the downstream signaling events and glucose triggers its degradation, whereas Std1 protein level remains constant regardless of glucose levels (1, 2), we focused on the interaction between Psy2 and Mth1 to further verify the initial observation. We generated a yeast strain expressing a Myc-tagged Psy2 from its genomic locus and harboring a multicopy plasmid expressing HA-tagged Mth1 from its endogenous promoter (pMT52). We performed coimmunoprecipitation experiments using cells grown in glucose (Glc)-containing medium or in medium with glucose replaced by galactose (22) as a carbon source. Immunoprecipitation with anti-HA antibody, followed by immunoblotting with anti-Myc antibody, showed that Psy2 indeed associated with Mth1 in vivo and that the association depends upon the presence of the polyproline motif (Fig. 1B). The fact that we observed more Psy2 protein associated with Mth1 in the absence of glucose (Fig. 1B, lane 3) could be accounted for by the increase in Mth1 protein, which occurs due to the enhanced stability of Mth1 under that condition. Conversely, analysis of the Psy2 mutant lacking the EVH1 domain (Psy2ΔEVH1) revealed that Psy2ΔEVH1 maintained its ability to bind to Pph3 (Fig. 1C) but no longer interacted with Mth1 (Fig. 1D).
FIG 1.
Psy2 interacts with Mth1 through a polyproline motif. (A) In the upper panel, the initial results from yeast two-hybrid screening using the EVH1 domain of Psy2 were confirmed with full-length Psy2 and Mth1/Std1 constructs. Yeast strains expressing Mth1/Std1 fused to the Gal4 DNA-binding domain (DBD) and Psy2 fused to Gal4 transcriptional activation domain or empty vector were plated on synthetic medium lacking either (i) Trp and Leu for growth or (ii) Ade, His, Trp, and Leu for selecting interactions. The lower panel shows that proline-to-alanine mutations of the MPPP motif of Mth1 (Mth1PA) and Std1 (Std1PA) abolished the interaction with Psy2. (B) Yeast cells expressing endogenous Myc-tagged Psy2 and HA-tagged wild-type Mth1 (WT) or Mth1PA were grown in glucose (Glc)- or galactose (Gal)-containing medium till log phase before being processed for coimmunoprecipitation analysis. (C) Psy2ΔEVH1 maintained the interaction with Pph3. psy2Δ strains harboring a genomic PPH3-TAP allele (MAY205) and expressing Psy2-6Myc or Psy2 ΔEVH1-6Myc from the endogenous promoter were subjected to Pph3-TAP pulldown, followed by immunoblot analysis for both Myc-tagged Psy2 variants and Pph3-TAP. (D) Psy2ΔEVH1 abolished interaction with Mth1. psy2Δ strains were transformed with the WT Psy2 and Psy2 ΔEVH1 constructs as described in panel C, together with pMT52. Cell extracts were made from cells grown in Glc-containing medium and 1 h after a shift to Gal medium. Immunoprecipitation was performed with anti-HA antibody coupled to protein A-Sepharose, followed by immunoblot analysis for both Myc-tagged Psy2 variants and Mth1-3HA.
Psy2 is required for the efficient dephosphorylation of Rgt1 and repression of HXT genes upon glucose depletion.
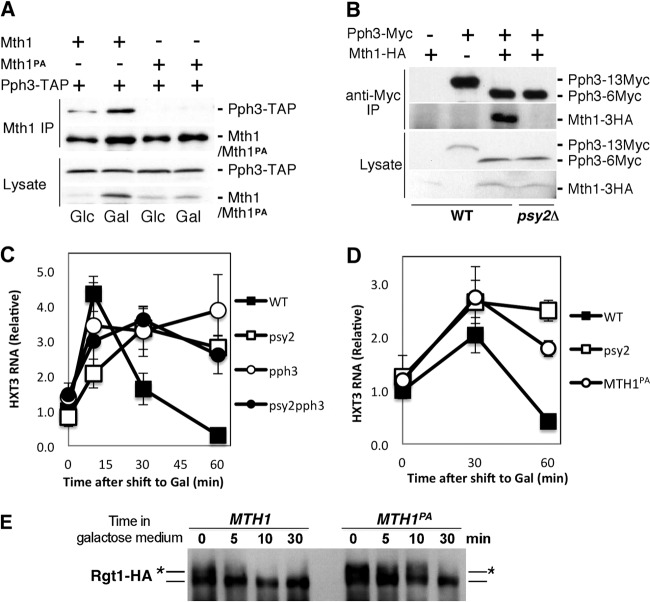
Since the main physiological function associated with Mth1 is the regulation of Rgt1 phosphorylation and HXT gene repression in response to glucose depletion (2, 5) and because Psy2 physically associates with Mth1, we hypothesized that Psy2 also plays a role in those processes. To test this hypothesis, we examined the propagation of the signal through the pathway by monitoring the phosphorylation of Rgt1 in WT and psy2Δ cells as reflected by the reduced electrophoretic mobility of the Rgt1 protein (Fig. 2A) (see also references 2 and 7). The phosphorylation of Rgt1 was analyzed by adding glucose to cells grown in galactose-containing medium. Based upon the persistence of Rgt1 protein species with decreased electrophoretic mobility, we conclude that Rgt1 is efficiently phosphorylated in the psy2Δ mutant cells (Fig. 2A, lanes 5 and 6). Conversely, when the dephosphorylation state of Rgt1 was monitored after a shift from glucose- to galactose-containing medium, we found that the extent of dephosphorylation of Rgt1 occurring over the time course was substantially reduced in the psy2Δ strain relative to wild-type cells (Fig. 2B).
FIG 2.
Psy2 promotes HXT gene repression in the absence of glucose. (A) The phosphorylation state of Rgt1 was analyzed by immunoblotting using wild-type (WT) and psy2Δ mutant cells grown in Gal-containing medium and shifted to Glc-containing medium. The position of unphosphorylated Rgt1 is indicated by a line, whereas the lines with asterisks denote the most highly phosphorylated species of Rgt1. The membrane was stained for protein prior to development for the loading control. (B) Similar analysis was performed with WT and psy2Δ mutant cells shifted from Glc- to Gal-containing medium for the time points indicated. An immunoblot with anti-PSTAIRE antibody was used for the loading control. (C) Quantitative RT-PCR analysis of HXT3 RNA with samples collected as described in panel B at the indicated time points. (D) Chromatin immunoprecipitation assay was performed using wild-type (WT) and psy2Δ mutant cells treated as in panel B, after cross-linking and immunoprecipitation of the Rgt1-HA-DNA complex using anti-HA monoclonal antibodies. The qPCR data represent the average of HXT3 RNA samples analyzed in triplicate and normalized to ACT1 RNA. HXT3 RNA is plotted relative to the wild-type time zero sample.
As Rgt1 phosphorylation correlates inversely with its function as a repressor for the HXT genes (2), we reasoned that the changes in the extent of Rgt1 phosphorylation in psy2Δ cells should cause corresponding changes in HXT gene expression. We examined the expression of the HXT3 gene, a well-established Rgt1 target, by RT-PCR following a shift in carbon source. When WT cells were shifted from glucose- to galactose-containing medium, HXT3 gene expression was repressed following a transient, but thus far unexplained, increase that is characteristic of these experiments (Fig. 2C; see also Fig. 3 and 6). In contrast, persistent expression of the HXT3 RNA was observed after depletion of glucose from psy2Δ cells (Fig. 2C). It should be noted that HXT3 gene expression returns to its basal level in both wild-type and psy2Δ mutant cells after an extended period in galactose. We conclude that Psy2 is required for the efficient dephosphorylation of Rgt1 and reestablishment of repression of HXT genes upon glucose depletion.
FIG 3.
The integrity of the Pph3-Psy2-Mth1 complex is critical for maintaining timely repression of HXT3 in response to glucose depletion. (A) Yeast cells expressing TAP-tagged Pph3 and HA-tagged Mth1/Mth1PA were subjected to coimmunoprecipitation analysis with anti-HA antibodies coupled to protein A-beads under the indicated conditions. The samples were then analyzed by immunoblotting with anti-HA to detect Mth1/Mth1PA-3HA and rabbit anti-mouse antibodies conjugated to horseradish peroxidase to detect Pph3-TAP, respectively. (B) Wild-type (WT) and psy2Δ mutant strains expressing Myc-tagged Pph3 and HA-tagged Mth1 from the endogenous loci were analyzed by coimmunoprecipitation. Pph3 expressed in the WT control is tagged with 13×Myc tag, whereas the other strains express Pph3 with a 6×Myc tag, which has more rapid gel mobility. (C) The repression of HXT3 gene upon glucose depletion in pph3Δ, psy2Δ, and pph3Δ psy2Δ cells was analyzed by quantitative RT-PCR using samples collected as described in Fig. 2B. (D and E) Yeast cells expressing the mth1PA mutant from the endogenous locus were analyzed by quantitative RT-PCR for HXT3 expression (D) and immunoblotting for Rgt1-HA (E) in comparison to WT cells at the times indicated.
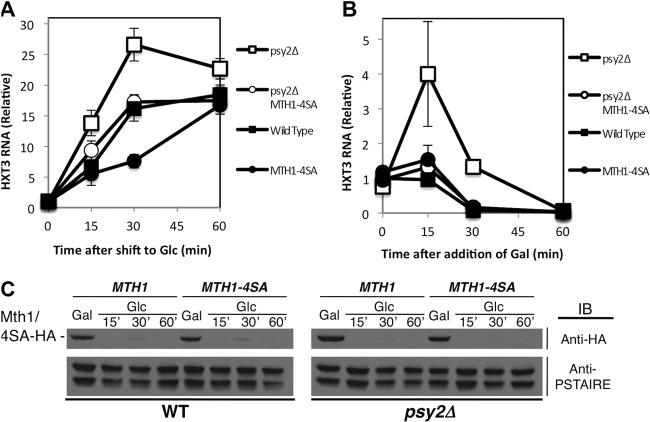
FIG 6.
Mth1-4SA mutant counteracts the effect of psy2 deletion on HXT3 expression. (A) Mth1-4SA mutant restores the induction of HXT3 gene in psy2Δ cells to wild-type level, as determined by quantitative RT-PCR. (B) Mth1-4SA mutant rescues the delayed repression of HXT3 gene in psy2Δ cells in the absence of glucose, as determined by quantitative RT-PCR. (C) Mth1-4SA mutation does not lead to significant changes in the Mth1 degradation pattern. An anti-PSTAIRE antibody immunoblot was used as a loading control.
Psy2, like Mth1, promotes chromatin binding of Rgt1 in response to glucose withdrawal.
Because the phosphorylation of Rgt1 induced by glucose is known to regulate its DNA-binding activity and because Mth1 is required for maintaining the dephosphorylated and chromatin-bound state of Rgt1 (2, 5), we sought to determine whether the delay in Rgt1 dephosphorylation is associated with the changes in the chromatin-binding activity of Rgt1 observed in psy2Δ cells, as determined by chromatin immunoprecipitation. Indeed, when glucose was withdrawn from the growth medium, the binding of Rgt1 to the HXT3 promoter was diminished in psy2Δ mutants relative to that observed in wild-type cells (Fig. 2D). That response is correlated with a decrease in the efficiency of Rgt1 dephosphorylation and the repression of HXT3 gene expression in psy2Δ mutants (Fig. 2B and C). These results suggest that Psy2 may directly or indirectly regulate Rgt1 repressor function, similar to Mth1.
Mth1 forms a complex with the Pph3-Psy2 phosphatase in vivo.
Psy2 has been shown to form a complex with Pph3, the yeast homolog of conserved PP4c (11). Therefore, we tested whether Pph3 also interacts with Mth1. Using a strain carrying an integrated TAP-tagged PPH3 gene and harboring pMT52 plasmid expressing HA-tagged Mth1, we confirmed that Pph3, like Psy2, interacts with Mth1 in vivo by coimmunoprecipitation analysis (Fig. 3A). The increase in the coimmunoprecipitation of Pph3 observed in galactose medium is likely due to an increase in the Mth1 level in the absence of glucose (Fig. 3A, lane 2). In contrast, Pph3 failed to coimmunoprecipitate with Mth1PA, suggesting that their interaction is dependent upon the polyproline motif of Mth1 (Fig. 3A, lanes 3 and 4). To further show that Mth1 interaction with Pph3 occurs at endogenous levels and requires Psy2, we generated yeast cells expressing Myc-tagged Pph3 and HA-tagged Mth1 from the endogenous loci. Immunoprecipitation with anti-Myc antibody, followed by immunoblotting with anti-Myc and anti-HA antibodies, showed that, indeed, Mth1 interacts with Pph3 at endogenous levels and that this interaction is lost in extracts from psy2Δ mutant cells (Fig. 3B). These results confirm that the Mth1 protein, stabilized by the depletion of glucose, associates with the Pph3-Psy2 phosphatase complex via an interaction between the polyproline motif of Mth1 and Psy2.
The integrity of Pph3-Psy2-Mth1 complex is critical for efficient repression of the HXT3 gene expression in response to glucose withdrawal.
The specific interaction between Pph3-Psy2 and Mth1 suggested that this complex might be important for the repression of HXT genes. We therefore predicted that conditions that disrupt integrity of that complex would result in a defect in repression of HXT3 gene expression. To evaluate that possibility, we performed quantitative RT-PCR analysis of HXT3 transcripts in psy2Δ, pph3Δ, and psy2Δ pph3Δ mutants shifted from glucose to galactose. The results show that, although HXT3 expression in both WT and mutant strains initially exhibits a transient increase, that increase persists in the mutant strains after glucose depletion, whereas in WT cells it drops quickly, resulting in HXT3 repression (Fig. 3C).
Having established a role for the complex in HXT3 regulation by glucose, we examined the consequence of loss of Mth1 binding to Pph3-Psy2 by replacing the endogenous wild-type MTH1 gene with the MTH1PA mutant allele, which is defective in Pph3-Psy2 binding. That mutation also abrogated HXT3 repression and Rgt1 dephosphorylation (Fig. 3D and E), similar to that observed in psy2Δ mutant cells (Fig. 2B and C). Together, these data demonstrate that the intact Pph3-Psy2-Mth1 complex is required for the efficient repression of the HXT3 gene following glucose withdrawal.
Mth1 is a target of the Pph3-Psy2 phosphatase complex.
We considered two hypotheses to explain the regulation of HXT gene expression by the Pph3-Psy2-Mth1 complex upon glucose depletion. First, the Pph3-Psy2 complex might be targeted to Rgt1 via the interaction of Psy2 with Mth1, promoting the dephosphorylation and DNA binding of Rgt1 and, thereby, the repression of HXT genes. Alternatively, Pph3-Psy2 might directly dephosphorylate Mth1 in response to glucose withdrawal, enhancing its activity as a corepressor with Rgt1 to repress HXT genes. To distinguish between these two possibilities, we purified the Pph3-Psy2 complex from a yeast strain expressing TAP-tagged Psy2 from its endogenous locus (Fig. 4A) and performed in vitro phosphatase assays using phosphorylated Rgt1 or Mth1 purified from yeast cells as the substrates. We evaluated dephosphorylation by monitoring the change in gel mobility using λ phosphatase as a control. Surprisingly, although the purified Pph3-Psy2 complex exhibited robust activity toward a phosphopeptide substrate (Fig. 4B), it did not alter the mobility of Rgt1 in vitro (Fig. 4C). To rule out the possibility that Mth1 is needed for the dephosphorylation of Rgt1 by Pph3-Psy2 in vitro, we carried out the same phosphatase assay in the presence of purified Mth1. However, as in its absence, we saw no change in the mobility of Rgt1 (Fig. 4D). Although we cannot exclude the possibility that Pph3-Psy2 dephosphorylates Rgt1 at residues that do not affect its electrophoretic mobility, it is the phosphorylation that retards its mobility that is associated with its activity as a transcriptional repressor (2, 7).
FIG 4.
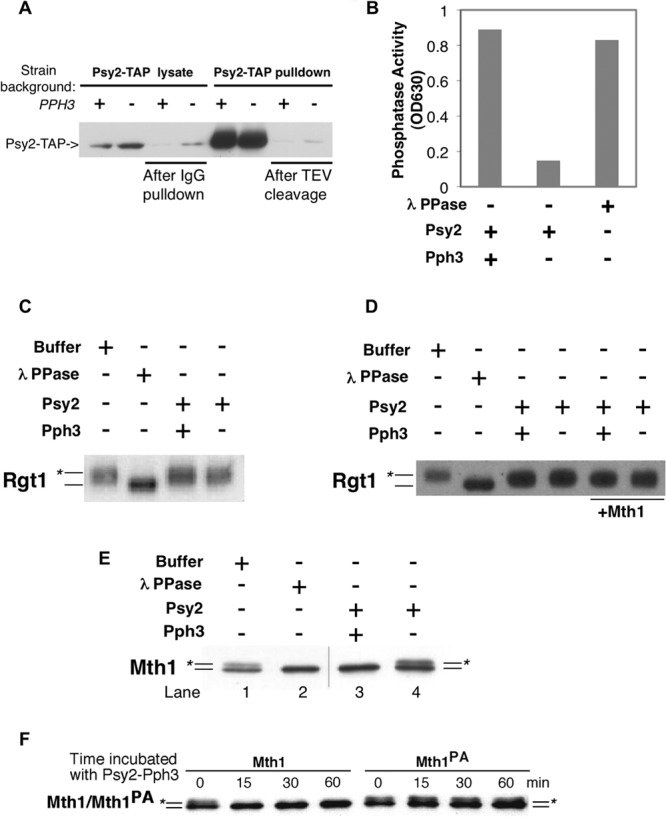
Mth1, but not Rgt1, is targeted by the Pph3-Psy2 phosphatase in vitro. (A) Psy2-TAP complex was purified from wild-type or pph3Δ strains by incubating the lysate with IgG-Sepharose, followed by TEV protease cleavage. The level of Psy2-TAP was detected by using rabbit anti-mouse antibody conjugated to horseradish peroxidase. The presence of Psy2-TAP before/after IgG pulldown and before and after TEV cleavage are shown sequentially from left to right. (B) Purified Psy2-Pph3 complex exhibited robust phosphatase activity. The results from an in vitro phosphate release assay using malachite green and a phosphopeptide substrate are shown. (C) FLAG-tagged Rgt1 purified from yeast was incubated with Psy2-TAP complex purified from wild-type (WT) or pph3Δ cells. Lambda phosphatase was used as a positive control. (D) The same experiment presented in panel C was repeated with the addition of the Mth1 protein. (E) Mth1 purified from a grr1-AAA yeast strain, which sequesters phosphorylated form of Mth1, was incubated with Psy2-TAP complex purified from WT or pph3Δ mutant cells as described in panel A. The position of the unphosphorylated Mth1 protein is indicated by a line, and the line marked with the asterisk denotes phosphorylated Mth1. The hairline marks the position of a splice between lanes from same gel. (F) Wild-type Mth1 and Mth1PA purified from grr1-AAA mutant cells were incubated with Pph3-Psy2 complex and samples were collected at the indicated time points. The positions of Mth1 and Mth1PA are labeled as in panel E.
In contrast to Rgt1, when purified Mth1 was tested as a substrate for Pph3-Psy2 in vitro, efficient dephosphorylation was observed (Fig. 4E, lane 3). The lack of dephosphorylation activity of Mth1 by the Psy2 complex purified from pph3Δ cells indicates that the Psy2-associated Mth1 phosphatase activity requires Pph3 (Fig. 4E, lane 4). To determine whether the dephosphorylation of Mth1 is dependent on the interaction between Mth1 and Psy2, we compared the ability of Pph3-Psy2 to dephosphorylate wild-type Mth1 and mutant Mth1PA proteins. The results clearly show that the Mth1PA, which fails to bind to Pph3-Psy2, was resistant to dephosphorylation by Pph3-Psy2, whereas wild-type Mth1 is efficiently dephosphorylated (Fig. 4F). This demonstrates that Mth1 is a bona fide in vitro substrate for the Pph3-Psy2 phosphatase complex and that this reaction requires the interaction between that complex and Mth1, a finding consistent with the effect of MTH1PA on glucose signaling in vivo.
Mth1 is a bona fide substrate for PKA.
We next sought to determine the kinase(s) that phosphorylates Mth1 and that is antagonized by Pph3-Psy2 phosphatase. Since Pph3-Psy2 deficiency does not have significant effects on Mth1 stability, which is thought to be regulated by Yck1/2 (Fig. 5A) (7, 8), we reasoned that there must be another kinase that acts upon Mth1 in response to glucose signal. We considered PKA a likely candidate as its activity is elevated in the presence of glucose and a survey of the Mth1 protein sequence reveals four putative PKA phosphorylation sites, including serines 111, 112, 161, and 358. Among these, Scansite predicts Ser 112 to be a high-affinity PKA site (28). To test this possibility, we first performed an in vitro PKA kinase assay using bacterially expressed GST-Mth1 fusion protein as a substrate. Indeed, GST-Mth1 is robustly phosphorylated by PKA in vitro, whereas the Mth1-4SA protein with the four serines changed to alanines completely abolished the observed phosphorylation (Fig. 5B). Furthermore, PKA phosphorylated HA-tagged Mth1 but not Mth1-4SA purified from yeast cells by immunoprecipitation (Fig. 5C).
FIG 5.
Mth1 is a bona fide substrate for PKA. (A) Pph3-Psy2 deficiency does not change the stability of Mth1. Yeast extracts from WT, psy2Δ, and pph3Δ strains expressing Mth1-HA from the endogenous promoter were collected under the conditions indicated and analyzed by immunoblotting using anti-HA antibody. The anti-PSTAIRE antibody immunoblots were used as loading controls. (B) The top panel is an autoradiograph showing that Mth1-4SA, with mutation of four serines (Ser 111, 112, 161, and 358) to alanines abolishes the GST-Mth1 phosphorylation by PKA in vitro. The bottom panel depicts Coomassie staining showing the equal amount of GST-Mth1 and Ser-to-Ala mutant substrates. (C) At the top is an autoradiograph showing that the mutation of four serines also abolishes the phosphorylation of immunoprecipitated Mth1 by PKA. At the bottom is an immunoblot using anti-HA antibody showing the protein level of WT Mth1 and Mth1-4SA. (D) AT the top left, an autoradiograph shows that PKA-phosphorylated Mth1 is dephosphorylated by Pph3-Psy2 complex in vitro. At the bottom left, an immunoblot shows the Mth1 protein levels used in the assay. Quantification of the phosphorylation levels of Mth1 (shown at the left) is depicted on the right side of the panel. (E) MS analysis identified phosphopeptides containing a single phosphoserine at either S111 or S112 in both immunoprecipitated Mth1 and in vitro PKA-phosphorylated Mth1. (F) Immunoblot with anti-HA antibody showing the comparison of the phosphorylation levels of Mth1, Mth1PA, and Mth1-4SA mutant proteins expressed from pMT52 in a grr1-AAA strain after shifting cells from galactose (Gal)- to glucose (Glc)-containing medium. Immunoblotting with anti-PSTAIRE antibody was used as a loading control.
To determine whether Pph3-Psy2 phosphatase could specifically target the PKA phosphorylation sites of Mth1, we incubated the purified Pph3-Psy2 complex with PKA-phosphorylated endogenous Mth1. Pph3-Psy2 promoted efficient dephosphorylation of Mth1 in vitro but did not remove the phosphorylation that occurs on a nonspecifically immunoprecipitated protein in the same reaction (Fig. 5D). In contrast, λ phosphatase nonselectively dephosphorylated all of the proteins targeted by PKA in that experiment (Fig. 5D).
To determine whether the phosphorylation of Mth1 on putative PKA sites does occur in vivo, we analyzed the phosphopeptides derived from immunoprecipitated Mth1 from the strain expressing Grr1-AAA, which binds and traps Mth1 in its phosphorylated form (7), using MudPIT (multidimensional protein identification technology). That analysis identified two spectra from a trypsin digestion of in vivo-phosphorylated Mth1 representing putative peptides phosphorylated at the S111/S112 site (Fig. 5E). The same spectra were identified in a similar analysis of in vitro PKA-phosphorylated Mth1. Although we did not identify peptides representative of the other putative PKA phosphorylation sites in vivo, we did find that mutation of those sites was necessary to abrogate the phosphorylation of Mth1 in vitro (Fig. 5B). Finally, whereas the phosphorylated slower-mobility wild-type Mth1 protein could be observed in strains expressing Grr1AAA, that same species was not observed in the same strain expressing the Mth1-4SA protein lacking the four putative PKA phosphorylation sites (Fig. 5F). We conclude that Mth1 is a bona fide substrate for PKA in vitro and that the same protein kinase is likely to phosphorylate Mth1 at several of those phosphorylation sites in response to glucose stimulation in vivo. Furthermore, mutation of the polyproline motif of Mth1 (Mth1PA) that abrogates dephosphorylation by Pph3-Psy2 phosphatase in vitro enhanced the accumulation of phosphorylated Mth1 in vivo, an observation consistent with a role for that phosphatase in the dephosphorylation of those sites (Fig. 5F).
Pph3-Psy2 counteracts Mth1 phosphorylation to promote Mth1 function.
Having shown that the Pph3-Psy2 phosphatase dephosphorylates Mth1 at putative PKA phosphorylation sites, we sought to determine whether these dephosphorylation events are relevant for its regulation of Mth1. To address this question, we introduced an MTH1 mutant lacking PKA phosphorylation sites (MTH1-4SA) into a psy2Δ mutant and analyzed the effect on HXT3 gene expression. We predicted that a defect in phosphatase activity would be suppressed by mutant Mth1 that lacks the capacity to be phosphorylated. Consistent with that hypothesis, we found that the elevated induction of HXT3 that occurs in the psy2Δ mutant in the presence of glucose and the delayed repression of HXT3 observed upon withdrawal of glucose were both suppressed by the MTH1-4SA phosphorylation site mutant (Fig. 6A and B, respectively). Furthermore, the regulation of HXT3 expression in cells carrying only the MTH1-4SA mutation was modestly deviated from that of wild-type cells (Fig. 6A and B) despite the level and pattern of degradation of the Mth1 protein being unaffected (Fig. 6C). We conclude that the Pph3-Psy2 phosphatase counteracts phosphorylation of Mth1, most likely by PKA, and that, together, the phosphatase and kinase are required to maintain Mth1 in a state optimal for the expression of HXT genes under a broad range of glucose concentrations.
DISCUSSION
Elucidating how Ser/Thr protein phosphatases target individual substrates to oppose the action of kinases in a biological context is of central importance to understand the signaling networks underlying diverse cellular responses as a whole. The sophisticated glucose-sensing pathways in yeast offer a unique setting for addressing this specificity issue, which provides the basis for the dynamic balance between kinases and phosphatases. We demonstrate that Pph3-Psy2 complex, the yeast homolog of mammalian PP4c-R3 phosphatase, specifically targets Mth1 in an interaction-dependent manner to antagonize a glucose-regulated kinase, most likely PKA, thereby allowing the precise regulation of glucose transporter genes according to glucose availability.
Pph3-Psy2 phosphatase complex as a novel regulator of glucose signaling.
We present here evidence that Psy2, homolog of the conserved SMEK(R3) subunit of PP4 family phosphatase, functions as a targeting subunit for Pph3 (homolog of the mammalian PP4c) in the glucose signaling pathway in budding yeast Several lines of evidence support our conclusion: first, Psy2 physically interacts with Mth1 and Std1, critical corepressors for Rgt1-mediated repression of HXT genes in response to glucose deprivation. A conserved EVH1 domain in Psy2 mediates an interaction with Mth1 via a polyproline sequence to form the Pph3-Psy2-Mth1 complex. Second, psy2Δ and pph3Δ mutants exhibit lower efficiency in dephosphorylation of the transcriptional repressor Rgt1 and, thereby, a delay in the repression of HXT gene expression. The inefficient dephosphorylation of Rgt1 in psy2Δ mutant cells correlates with a delay in binding of Rgt1 to the HXT gene promoters. Third, purified Pph3-Psy2 phosphatase complex directly dephosphorylates Mth1 in vitro. Importantly, both the Psy2-dependent dephosphorylation of Mth1 in vitro and the effect of Psy2 on glucose signaling in vivo are dependent upon the interaction between Mth1 and the Pph3-Psy2 complex, strongly suggesting a specific enzyme-substrate relationship. Based on those observations, we propose that the Pph3-Psy2 phosphatase complex promotes Rgt1-mediated repression of HXT genes in the absence of glucose by directly binding and dephosphorylating Mth1 (see Fig. 7 for a model).
FIG 7.
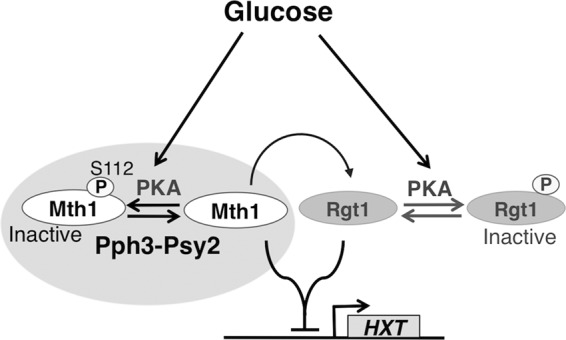
Model showing how the coordinated action of Pph3-Psy2 and PKA determines the precise expression of HXT genes in response to glucose availability. PKA phosphorylation of Mth1 regulates the repressive activity of Mth1 without affecting its abundance, which is instead a consequence of its glucose-induced phosphorylation by Yck1/2 and resulting ubiquitylation. PKA phosphorylation of Mth1 is counteracted by the Pph3-Psy2 phosphatase. The dynamic interplay between Pph3-Psy2 and PKA determines the functional state of Mth1 (possibly through spatiotemporal regulation), leading to the precise regulation of HXT gene expression in response to changes in glucose availability.
The interplay between Pph3-Psy2 phosphatase and PKA in coregulating Mth1.
Mth1 is known to be phosphorylated by Yck1/2. Our findings, however, are most consistent with Pph3-Psy2 antagonizing PKA but not Yck1/2 activity in regulating Mth1, suggesting yet another level of control for Mth1 corepressor activity independent of the Yck1/2-induced degradation. This is consistent with the recent report that PKA is not involved in the degradation of Mth1 (29). Previous genome-wide expression analysis has established PKA as the predominant glucose signaling kinase that is responsible for nearly 90% of the transcriptional changes in response to glucose (30). It has been reported to phosphorylate Rgt1, leading to the inhibition of its chromatin-binding activity (1, 29). The identification of the corepressor Mth1 as a new target for PKA indicates that the two receptor-mediated glucose-sensing pathways (the Rgt2/Snf3-Mth1-Rgt1 pathway and cAMP/PKA pathway) maybe tightly coupled. This regulatory coupling might be critical for yeast cells to coordinate growth rate with the status of available nutrients.
Because Mth1 is rapidly degraded in the presence of glucose, we speculate that the Pph3-Psy2-mediated dephosphorylation of Mth1 is most prominent during the initial phase of glucose depletion, consistent with the timing of Pph3-Psy2 action. Although the PKA activity is expected to decline from its peak under this condition, it is not immediately repressed and may suffice to limit the corepressor function of Mth1. Consequently, Pph3-Psy2 phosphatase ensures the efficient reestablishment of HXT gene repression in the absence of glucose. The tight association of Pp3-Psy2 with Mth1 via its polyproline motif may protect Mth1 from unwanted phosphorylation.
In addition to Mth1, we have also found that the Mth1 paralog Std1, which shares 61% identity with Mth1, interacts with Psy2 (Table 3; Fig. 1A). Although Std1 does not play an essential role in regulating the phosphorylation and DNA-binding activity of Rgt1, both Mth1 and Std1 are needed to establish full repression of HXT genes by Rgt1 (2, 5). We expect that, like Mth1, Std1 will prove to be a substrate for the Pph3-Psy2 phosphatase. The Ser112 residue of Mth1, however, is not conserved in Std1. Nevertheless, based on Scansite prediction, Std1 contains a medium strength PKA phosphorylation site at Ser20, which resides among a cluster of basic amino acid residues reminiscent of a nuclear localization signal (KRKS20KRHDENP-). It is possible that phosphorylation of Mth1-S112 and Std1-S20 by PKA jointly contributes to HXT gene regulation. Std1 may also integrate different signaling inputs via its connection to the Snf1 kinase, homolog for the mammalian AMP kinase that orchestrates the transcriptional response during growth in low glucose or in alternative carbon sources (31).
Consequences of Mth1 dephosphorylation.
The fact that Mth1 mutant lacking the PKA phosphorylation sites effectively reverses the HXT3 gene expression defect in psy2Δ cells underscores the importance of dephosphorylation in promoting the functional state of Mth1. Given that the stability of the Mth1-4SA mutant does not differ markedly from that of wild-type Mth1 (Fig. 6C), dephosphorylation may cause changes in the subcellular localization of Mth1 and/or its association with other proteins that, in turn, promote Rgt1 repressor function. Previous reports suggest that Mth1 and Std1 interact with the C-terminal tails of the glucose sensors Rgt2 and Snf3 at the cell surface, as well as with Rgt1 resident in the nucleus (5, 10, 32). The expected spatiotemporal regulation of those interactions requires that Mth1 actively shuttle between the plasma membrane and the nucleus to perform its role as a glucose signal transducer. Likewise, Psy2 has been shown to localize mostly in the nucleus and partly at the cell membrane (33). Thus, it is feasible that Psy2 may recruit Mth1 and retain it within the nucleus via dephosphorylation by Pph3 in the absence of glucose. Consistent with that proposal, the Dictyostelium Psy2 homology, SMEK(R3), has been reported to translocate from the cell cortex to the nucleus upon starvation (17). Furthermore, recent studies have demonstrated that the Pph3-Psy2 complex dephosphorylates and promotes the nuclear localization of yeast Maf1, the conserved “master” repressor for RNA polymerase III transcription that integrates multiple nutrient and stress signals (33). In that context, PKA is one of the kinases that phosphorylates Maf1 and maintains its cytoplasmic localization. Similarly, PKA phosphorylation of protein kinase Rim15 and transcription factor Msn2/4 has been shown to promote their export to the cytoplasm, thereby inactivating the transcription of the growth-inhibitory kinase Yak1 (34–36). By analogy, PKA and Pph3-Psy2 may well play a role in mediating the spatiotemporal dynamics of Mth1 localization.
How Mth1 dephosphorylation influences Rgt1 dephosphorylation remains unclear. Although the simple hypothesis that Mth1 targets Pph3-Psy2 to dephosphorylate Rgt1 seems economical, it appears unlikely based upon our finding that Pph3-Psy2 does not dephosphorylate Rgt1 in vitro (Fig. 4C). Furthermore, Mth1 does not facilitate dephosphorylation of Rgt1 by Pph3-Psy2 (Fig. 4D). Nevertheless, it remains possible that conditions in vivo facilitate that reaction or that Mth1 interaction promotes dephosphorylation of Rgt1 by another phosphatase.
Multiple mechanisms determine the specificity of Pph3-Psy2.
The Pph3-Psy2 phosphatase complex is well conserved and is involved in diverse cellular pathways, mostly related to stress and nutrient responses. Therefore, it must target multiple substrates to counter the actions of multiple kinases in order to achieve its diversity and specificity. Psy2 has been reported to interact with the kinase domain of the checkpoint kinase Rad53, resulting in the dephosphorylation of Rad53 due to associated Pph3 phosphatase activity (19). We found that Psy2 interacts with the polyproline motif in Mth1 via its N-terminal EVH1 domain. Since the Rad53 kinase domain does not include a proline-rich motif, it seems likely that the domains of Psy2 other than the EVH1 domain might be involved in binding to Rad53. Maf1 is also a substrate of Pph3 but it is not clear whether this physical association is direct or via Psy2 (33). It is reasonable to assume that variations in binding strategy are important in determining the differences in the affinity of Psy2-Pph3 for various substrates adapting it for the specific physiological context. Indeed, the Psy2-Rad53 interaction is considered to be weak and transient (19), whereas the Psy2-Mth1 association is strong and stable in the absence of glucose. We propose that the EVH1 domain, by specifically targeting Mth1, dictates the specific role of Pph3-Psy2 in the glucose-signaling pathway.
To our knowledge only a few cases of precise substrate targeting have been established for Ser/Thr phosphatases in which the respective binding sites are identified. These include calcineurin, which recognizes NFAT, and the Bα subunit of PP2A, which recognizes Tau, via specific docking sites (37, 38). However, in both cases the enzyme-substrate interaction is weak and transient. The highly stringent and stable interaction between Mth1 and Psy2 is uncommon. Our analysis of the interplay between Pph3-Psy2 and PKA suggests that this high-affinity association might be necessary for phosphatases to “shelter” critical substrates from unwanted kinase activity. Although obvious homologs for Mth1 have yet to be identified in mammalian cells, the EVH1 domain is highly conserved among members of the SMEK(R3) protein family, suggesting that targeting of PP4c to specific substrates containing a suitable polyproline motif involves similar mechanisms in human cells. Whether such targets might play a functionally analogous role to Mth1 in nutrient signaling or whether they have evolved to acquire new functions in multicellular organisms remains an important area for investigation.
ACKNOWLEDGMENTS
This research was funded by U.S. Public Health Service grants DK60367-02 to H.M., GM043487 and GM059441 to C.W., P41 RR011823 to J.R.Y., and CA14195, CA80100, and CA82683 to T.H. T.H. is a Frank and Else Schilling American Cancer Society Professor and holds the Renato Dulbecco Chair in Cancer Research.
We thank Brian O'Neill and Floyd Romesberg for sharing information and yeast strains related to Psy2 and Pph3 prior to publication. We also thank Tatyana Kalashnikova, Evan Hsia, and members of Vicki Lundblad's laboratory for technical support and Nathalie Spielewoy for insights early in the development of this project. We thank Jill Meisenhelder and other members of the Hunter and Eckhart laboratories at the Salk Institute for helpful discussions and Thomas Sternsdorf for critical reading of the manuscript.
Footnotes
Published ahead of print 25 November 2013
REFERENCES
- 1.Johnston M, Kim JH. 2005. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33:247–252. 10.1042/BST0330247 [DOI] [PubMed] [Google Scholar]
- 2.Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14:3230–3241. 10.1091/mbc.E03-03-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC. 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulte F, Wieczorke R, Hollenberg CP, Boles E. 2000. The HTR1 gene is a dominant-negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J. Bacteriol. 182:540–542. 10.1128/JB.182.2.540-542.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakshmanan J, Mosley AL, Ozcan S. 2003. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 44:19–25. 10.1007/s00294-003-0423-2 [DOI] [PubMed] [Google Scholar]
- 6.Tamaki H. 2007. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng. 104:245–250. 10.1263/jbb.104.245 [DOI] [PubMed] [Google Scholar]
- 7.Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994–9005. 10.1128/MCB.24.20.8994-9005.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriya H, Johnston M. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. U. S. A. 101:1572–1577. 10.1073/pnas.0305901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Johnston M. 2006. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 281:26144–26149. 10.1074/jbc.M603636200 [DOI] [PubMed] [Google Scholar]
- 10.Polish JA, Kim JH, Johnston M. 2005. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics 169:583–594. 10.1534/genetics.104.034512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. 2005. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell. Proteomics 4:1725–1740. 10.1074/mcp.M500231-MCP200 [DOI] [PubMed] [Google Scholar]
- 12.Cohen PT, Philp A, Vazquez-Martin C. 2005. Protein phosphatase 4: from obscurity to vital functions. FEBS Lett. 579:3278–3286. 10.1016/j.febslet.2005.04.070 [DOI] [PubMed] [Google Scholar]
- 13.Chen GI, Tisayakorn S, Jorgensen C, D'Ambrosio LM, Goudreault M, Gingras AC. 2008. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J. Biol. Chem. 283:29273–29284. 10.1074/jbc.M803443200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu J, Jho EH, Lu W. 2011. Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells. Cell Res. 21:911–921. 10.1038/cr.2011.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. 2005. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 19:827–839. 10.1101/gad.1286005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee DH, Chowdhury D. 2011. What goes on must come off: phosphatases gate-crash the DNA damage response. Trends Biochem. Sci. 36:569–577. 10.1016/j.tibs.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendoza MC, Du F, Iranfar N, Tang N, Ma H, Loomis WF, Firtel RA. 2005. Loss of SMEK, a novel, conserved protein, suppresses MEK1-null cell polarity, chemotaxis, and gene expression defects. Mol. Cell. Biol. 25:7839–7853. 10.1128/MCB.25.17.7839-7853.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prehoda KE, Lee DJ, Lim WA. 1999. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell 97:471–480. 10.1016/S0092-8674(00)80757-6 [DOI] [PubMed] [Google Scholar]
- 19.O'Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates YR, III, Aparicio OM, Romesberg FE. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:9290–9295. 10.1073/pnas.0703252104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fromont-Racine M, Rain JC, Legrain P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277–282. 10.1038/ng0797-277 [DOI] [PubMed] [Google Scholar]
- 21.Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21:2506–2520. 10.1128/MCB.21.7.2506-2520.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, Lieberman J, Shen X, Buratowski S, Haber JE, Durocher D, Greenblatt JF, Krogan NJ. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439:497–501. 10.1038/nature04384 [DOI] [PubMed] [Google Scholar]
- 23.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- 24.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., III 2004. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18:2162–2168. 10.1002/rcm.1603 [DOI] [PubMed] [Google Scholar]
- 25.Yates JR, III, Eng JK, McCormack AL, Schieltz D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426–1436. 10.1021/ac00104a020 [DOI] [PubMed] [Google Scholar]
- 26.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43–50. 10.1021/pr025556v [DOI] [PubMed] [Google Scholar]
- 27.Tabb DL, McDonald WH, Yates JR., III 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26. 10.1021/pr015504q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obenauer JC, Cantley LC, Yaffe MB. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635–3641. 10.1093/nar/gkg584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jouandot D, II, Roy A, Kim JH. 2011. Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J. Cell. Biochem. 112:3268–3275. 10.1002/jcb.23253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 5:245. 10.1038/msb.2009.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuchin S, Vyas VK, Kanter E, Hong SP, Carlson M. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafuente MJ, Gancedo C, Jauniaux JC, Gancedo JM. 2000. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35:161–172. 10.1046/j.1365-2958.2000.01688.x [DOI] [PubMed] [Google Scholar]
- 33.Oler AJ, Cairns BR. 2012. PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J. 31:1440–1452. 10.1038/emboj.2011.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinders A, Burckert N, Boller T, Wiemken A, De Virgilio C. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12:2943–2955. 10.1101/gad.12.18.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith A, Ward MP, Garrett S. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556–3564. 10.1093/emboj/17.13.3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597. 10.1101/gad.12.4.586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez A, Roy J, Martinez-Martinez S, Lopez-Maderuelo MD, Nino-Moreno P, Orti L, Pantoja-Uceda D, Pineda-Lucena A, Cyert MS, Redondo JM. 2009. A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol. Cell 33:616–626. 10.1016/j.molcel.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139:468–484. 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 39.Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453–463. 10.1016/S0092-8674(00)80118-X [DOI] [PubMed] [Google Scholar]