Abstract
Interferon regulatory factor 8 (IRF8), a member of the IRF transcription factor family, was recently implicated in vascular diseases. In the present study, using the mouse left carotid artery wire injury model, we unexpectedly observed that the expression of IRF8 was greatly enhanced in smooth muscle cells (SMCs) by injury. Compared with the wild-type controls, IRF8 global knockout mice exhibited reduced neointimal lesions and maintained SMC marker gene expression. We further generated SMC-specific IRF8 transgenic mice using an SM22α-driven IRF8 plasmid construct. In contrast to the knockout mice, mice with SMC-overexpressing IRF8 exhibited a synthetic phenotype and enhanced neointima formation. Mechanistically, IRF8 inhibited SMC marker gene expression through regulating serum response factor (SRF) transactivation in a myocardin-dependent manner. Furthermore, a coimmunoprecipitation assay indicated a direct interaction of IRF8 with myocardin, in which a specific region of myocardin was essential for recruiting acetyltransferase p300. Altogether, IRF8 is crucial in modulating SMC phenotype switching and neointima formation in response to vascular injury via direct interaction with the SRF/myocardin complex.
INTRODUCTION
Phenotypic switching of vascular smooth muscle cells (VSMCs) and subsequent proliferation contribute to various vascular diseases, including atherosclerosis, in-stent restenosis, transplant vasculopathy, and vein bypass graft failure (1). In response to vascular injury, quiescent smooth muscle cells (SMCs) lose contractile characteristics and gain a proliferative synthetic phenotype to produce an extracellular matrix to form neointimal lesions (2, 3). The phenotypic switching of SMCs is intended to repair the injury of the vessel or to stabilize atheromatous plaque. However, this response leads to the narrowing or even occlusion of blood vessels. Ways to effectively inhibit SMC phenotypic switching and prevent the neointima formation remain to be determined. The current mechanistic understanding of SMC phenotypic switching largely focuses on the regulation of SMC-specific genes, including smooth muscle α-actin (α-SMA), SM22α, smoothelin, and desmin genes (4). In recent years, several regulatory mechanisms have been elucidated to regulate the expression of these genes, among which those dependent on the conserved G/C-repressive elements (5) and CArG [CC(A/T)6GG] elements (6) present in the promoters of SMC-specific genes are the most prominent. Hence, the regulation of serum response factor (SRF), which binds to CArG elements, is of particular interest (7, 8). Platelet-derived growth factor BB (PDGF-BB) induces Krüppel-like zinc finger factor type 4 (KLF4) expression (4) and Elk-1 phosphorylation (9). KLF4 was reported to reduce SRF binding through inhibiting the expression of the coactivator myocardin and recruiting histone deacetylases (HDACs) (10, 11). There is also evidence that phosphorylated Elk-1 (9) and HERP1 (12) inhibit SRF transactivation through interrupting the interaction of SRF with myocardin. Because the phenotypic switching of SMCs is induced by numerous environmental cues and is actively regulated, the factors noted above are by no means the only regulators. To further elucidate the mechanism of SMC phenotypic switching, more detailed molecular mechanisms on the transcription factors involved could be needed.
Interferon regulatory factors (IRFs) are a family of transcription factors that play central roles in innate immunity and immune cell differentiation (13–15). Our previous work has demonstrated that IRFs are also involved in the pathophysiological processes of heart diseases (16, 17) and metabolic disorders (18). Interferon regulatory factor 8 (IRF8; also known as interferon consensus sequence binding protein [ICSBP]) is one of the nine members of this family in mammals (14). IRF8 is primarily expressed in hematopoietic cells (19) and is critical for the development of some lineages (13). Recent studies have shed new light on the involvement of IRF8 in cardiovascular diseases. Hematopoietic IRF8 deficiency was reported to accelerate atherosclerosis (20). IRF8 was also reported to regulate the expression of macrophage arginase 1, which inversely correlates with atherosclerotic progression (21). Therefore, we hypothesize that IRF8 may also play a role in injury-induced intima growth. To determine the role of IRF8 in the regulation of SMC phenotypic switching and neointima formation, we performed carotid artery wire injury on IRF8−/−, IRF8 SMC-specific transgenic (TG), and wild-type (WT) mice. Surprisingly, we found that IRF8 expression was significantly increased in SMCs after vascular injury. IRF8 promoted the phenotypic switching of SMCs and exacerbated the formation of neointima. In addition, we found that the effect of IRF8 on neointima formation is mediated by its interaction with myocardin.
MATERIALS AND METHODS
Mice.
All of the animal protocols were approved by the Animal Care and Use Committee of Renmin Hospital of Wuhan University. IRF8-deficient mice (B6.129P2-Irf8tm1Hor/Kctt) were purchased from the European Mouse Mutant Archive (EMMA; EM 02414). IRF8−/− mice were confirmed using PCR analysis with the primers 5′-CATGGCACTGGTCCAGATGTCTTCC-3′, 5′-CTTCCAGGGGATACGGAACATGGTC-3′, and 5′-CGAAGGAGCAAAGCTGCTGCTATTGGCC-3′. The IRF8−/− mice with a B6.129 background were backcrossed to a C57BL/6 mouse for at least six generations to yield IRF8−/− mice. SM22-Cre [B6.Cg-Tg(Tagln-cre)1Her/J; stock number 017491] mice and IRF8flox/flox [B6(Cg)-Irf8tm1.1Hm/J; stock number 014175] mice were purchased from The Jackson Laboratory. SM22-Cre mice and IRF8flox/flox mice were crossed to generate IRF8flox/+-SM22-Cre mice, and then IRF8flox/+-SM22-Cre male and female mice were crossed to each other to produce IRF8flox/flox-SM22-Cre (SMC IRF8-knockout [SMC-IRF8-KO]) mice. To obtain SMC-specific transgenic mice, the SMC-specific transgenic vector was first constructed with a mouse SM22a promoter, a 5′ hemagglutinin (HA) tag, and a simian virus 40 (SV40) poly(A) signal. The mouse full-length IRF8 cDNA (Openbiosystem) was cloned into the SMC-specific transgenic vector between the EcoRI sites by a ligation-independent cloning (LIC) method, as previously reported (22). Vascular SMC-specific IRF8 transgenic mice were generated in a C57BL/6 background through the microinjection of mouse IRF8 cDNA under the control of a minimal SM22α promoter (a region of the murine SM22a promoter that contains 1,275 bp). IRF8 cDNA was obtained and amplified by PCR using the primers 5′-GCCACCATGGAATTCTGTGACCGGAACGGCGGG-3′ and 5′-GTATGGGTAGAATTCGACGGTGATCTGTTGATTTTC-3′. IRF8 overexpression was assessed by Western blotting analyses.
Carotid artery wire injury mode.
Ten- to 12-week-old male mice underwent carotid artery injury surgery. The mouse model of carotid artery wire injury has been described previously (23). Briefly, the mice were anesthetized by intraperitoneal injection of sodium pentobarbital (80 mg/kg of body weight), and 1% lidocaine hydrochloride was injected subcutaneously for local analgesia. The left carotid artery was exposed by a blunt-end dissection. The external carotid artery was ligated with an 8-0 suture immediately proximal from the point of bifurcation. Internal and common carotid arteries were clamped to intercept blood flow, and then a transverse incision was made immediately proximal from the suture around the external carotid artery. A guide wire (diameter, 0.38 mm; no. C-SF-15-15; Cook, Bloomington, IN) was then introduced into the arterial lumen toward the aortic arch and then withdrawn 5 times in a rotating motion. After removal of the guide wire, the vascular clamps were removed and blood flow was restored. The skin incision was then closed. Sham controls underwent the same procedures, except for the dissection and injury. The animals were then processed for morphological and biochemical studies at specific time points after surgery.
Histological and morphometric analysis.
At 0, 7, 14, or 28 days postinjury, mice were killed by an overdose of sodium pentobarbital (150 mg/kg) by intraperitoneal injection. Their carotid arteries were harvested after circulating perfusion and fixed with 4% paraformaldehyde dissolved in phosphate-buffered saline (PBS). The arteries were further fixed for 16 h and then paraffin embedded. Serial 3-μm sections were produced from the entire approximately 300-μm region at the bifurcation of the left carotid artery. For morphometric purposes, we stained sections with hematoxylin-eosin (H&E) after deparaffinization and rehydration. To determine the extent of neointima formation, intima areas and the intima-to-media (I/M) ratios were determined, using Image Pro Plus software (version 6.0; Media Cybernetics), by a single observer blinded to the treatment protocols. A mean value was generated from five sections of each artery sample.
Immunofluorescence staining.
After a 5-min high-pressure antigen retrieval process (100× sodium citrate buffer, pH 6.0), arterial sections were blocked in PBS containing 10% goat serum for 60 min and incubated overnight with primary antibodies at 4°C. Rat VSMCs were seeded on coverslips placed in 6-well plates. Cells were fixed with 4% fresh paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After washing with PBS containing 10% goat serum for 60 min, the slides were incubated overnight with primary antibodies at 4°C. The antibodies used were directed against IRF8 (1:100; rabbit; catalog no. sc-13043; Santa Cruz), α-SMA (1:100; mouse; catalog no. ab7817; Abcam), proliferating cell nuclear antigen (PCNA; 1:100; mouse; catalog no. 2586; Cell Signaling Technology), cyclin D1 (1:25; rabbit; catalog no. 2978; Cell Signaling Technology), α-SMA (1:100; rabbit; catalog no. ab5694; Abcam), SM22α (1:100; rabbit; catalog no. ab14106; Abcam), smoothelin (1:100; rabbit; catalog no. sc-28562; Santa Cruz), and osteopontin (OPN; 1:100; rabbit; catalog no. BS1264; Bioworld). The sections were washed with PBS and incubated with the appropriate secondary antibodies for 1 h at 37°C. The secondary antibodies used were Alexa Fluor 568-conjugated goat anti-rabbit IgG (catalog no. A11011; Invitrogen, Carlsbad, CA), Alexa Fluor 488-conjugated goat anti-mouse IgG (catalog no. A11001; Invitrogen, Carlsbad, CA), Alexa Fluor 568-conjugated goat anti-mouse IgG (catalog no. A11004; Invitrogen, Carlsbad, CA), and Alexa Fluor 488-conjugated goat anti-rabbit IgG (catalog no. A11008; Invitrogen, Carlsbad, CA). The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Image acquisition was performed using a fluorescence microscope (DX51; Olympus Japan) and DP2-BSW software (version 2.2). The integrated optical density (IOD) values were measured using Image Pro Plus software (version 6.0; Media Cybernetics).
Cell culture and adenovirus infection.
Human aortic smooth muscle cells (HASMCs), the A7r5 rat embryonic aortic vascular smooth muscle cell line, and mouse VSMC line MOVAS were obtained from the American Type Culture Collection (ATCC). Primary vascular smooth muscle cells were isolated from the thoracic aortas of male C57BL/6 mice and Sprague-Dawley rats by enzymatic digestion. All of the cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium containing 10% fetal bovine serum (FBS; SV30087.02; HyClone) and 1% penicillin-streptomycin and maintained at 37°C in a humidified atmosphere containing 5% CO2. The cells used in the experiments were from passages 3 to 5. To overexpress IRF8, the entire coding region of the rat IRF8 gene under the control of the cytomegalovirus promoter was encompassed by replication-defective adenoviral vectors. A similar adenoviral vector encoding the green fluorescent protein (AdGFP) gene was used as a control. Three rat IRF8 short hairpin RNA (shIRF8) constructs were obtained from SABiosciences (KR59496G) to knock down IRF8 expression. The construct decreasing IRF8 levels to the greatest extent was used for all further experiments. Adenovirus containing short hairpin RNA (AdshRNA) was the nontargeting control. Rat VSMCs and A7r5 cells were infected with the adenoviruses described above in diluted medium at a multiplicity of infection of 100 for 24 h.
BrdU assessment of cell proliferation.
DNA synthesis in the SMCs was assessed by measuring the incorporation of 5′-bromo-2′-deoxyuridine (BrdU). Mouse SMCs (5 × 103/well) were grown to 60% confluence and growth arrested in a 96-well microplate. After the cells were serum starved for 24 h, quiescent cells were treated with 20 ng/ml of PDGF-BB (purchased from ProSpec [Rehovot, Israel]) for 48 h. BrdU was added for the last 2 h of treatment. BrdU incorporation was determined using a cell proliferation enzyme-linked immunosorbent assay kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocol.
Migration assay.
An SMC migration assay was performed using the modified Boyden chamber method as previously described (24). Briefly, mouse SMCs were grown to 70% to 80% confluence and then maintained in a quiescent state in serum-free medium for 24 h. Cells were trypsinized, washed with PBS, and resuspended in DMEM containing 20% FBS. Approximately 5 × 104 cells/well were added to the upper chamber of a 24-well transwell dish (with a 6.5-mm polycarbonate membrane with 8-mm pores; Corning, Corning, NY) and allowed to attach for 30 min. SMCs were exposed to the medium containing or not containing PDGF-BB (20 ng/ml), which was added to the lower chamber as the chemoattractant. After 6 h of incubation, cells remaining on the upper membrane were removed using a cotton bud, while those that had migrated to the lower side of the surface were fixed, stained with 0.1% crystal violet–20% methanol, and counted. The average number of cells that had migrated was calculated from five randomly chosen high-power fields (×200) in three independent experiments. Images were quantified using Image Pro Plus software.
Western blotting.
Western blotting procedures were as described previously (25). In brief, cellular and mouse tissue proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). After a complete homogenization on ice, the samples were centrifuged and the supernatants obtained were fractionated by 10% SDS-PAGE and transferred to an Immobilon-FL membrane (Millipore). After blocking with Tris-buffered saline (TBS) containing 5% nonfat milk, the membranes were probed with primary antibodies directed against IRF8 (1:200; catalog no. sc-13043; Santa Cruz), PCNA (1:1,000; mouse; catalog no. 2586; Cell Signaling Technology), cyclin D1 (1:1,000; rabbit; catalog no. 2978; Cell Signaling Technology), matrix metalloproteinase 9 (MMP9; 1:200; goat; catalog no. sc-6841; Santa Cruz), α-SMA (1:1,000; mouse; catalog no. ab7817; Abcam), SM22α (1:300; rabbit; catalog no. ab14106; Abcam), smoothelin (1:200; rabbit; catalog no. sc-28562; Santa Cruz), desmin (1:1,000; rabbit; catalog no. 4024; Cell Signaling Technology), OPN (1:500; rabbit; catalog no. BS1264; Bioworld), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 1:10,000; mouse; catalog no. MB001; Bioworld).
Plasmid constructs.
Enhanced green fluorescent protein (EGFP)-myc-IRF8 and glutathione S-transferase (GST)–IRF8 constructs were generated by amplifying the coding region of the IRF8 gene using primers IRF8-5′ and IRF8-3′ from HA-IRF8 and subcloning them into pEGFP-myc-C1 and pGEX-4T-1, respectively. To obtain the IRF8 fragments consisting of residues 1 to 125, 110 to 200, and 200 to 424, HA-IRF8 was PCR amplified using primers IRF8-5′ and IRF8-N-3′, IRF8-Pro-5′ and IRF8-Pro-3′, and IRF8-C-5′ and IRF8-3′, respectively. The products were digested with EcoRI and XhoI and ligated into pEGFP-myc-C1 to create an in-frame fusion with EGFP-myc. The Flag-myocardin and mCherry-myocardin constructs were generated by amplifying the coding region of the myocardin gene using primers MyoD-5′ and MyoD-3′ from human cDNAs and cloning them into psi-Flag-C1 and psi-mCherry-C1, respectively. To obtain the myocardin fragments consisting of residues 1 to 738 and 738 to 938, Flag-myocardin was PCR amplified using primers MyoD-5′ and MyoD-N-3′ and primers MyoD-C-5′ and MyoD-3′, respectively. The products were digested with XhoI and ligated into psi-Flag to create an in-frame fusion with a Flag tag. The primers for producing these constructs are shown in Table S1 in the supplemental material. All of the plasmids were verified by sequencing.
Immunoprecipitation.
For immunoprecipitation, cultured 293T cells were cotransfected with Flag-myocardin and EGFP-myc-IRF8 for 48 h and lysed in immunoprecipitation (IP) buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl,1 mM EDTA, 0.5% NP-40) supplemented with a protease inhibitor cocktail (Roche). The cell homogenates were incubated for 20 min at 4°C with constant agitation and then pelleted by centrifugation (13,000 × g for 15 min at 4°C). For each IP, 500 μl of the sample was incubated with 10 μl of protein A/G-agarose beads (catalog no. 11719394001 and 11719386001; Roche) and 1 μg of antibody on a rocking platform (overnight at 4°C) according to the manufacturer's recommendations. Finally, the immunoprecipitates were washed five to six times with cold IP buffer before adding 1× PAGE loading buffer. Cell lysates and immunoprecipitates were immunoblotted using the indicated primary antibodies, the corresponding secondary antibodies, and a SuperSignal chemiluminescence kit (Millipore).
GST pulldown assay.
The GST-IRF8 encoded by the pGEX-4T-1 construct was expressed in Escherichia coli Rosetta(DE3) cells. For the pulldown assay, 10 ml of E. coli (after IPTG [isopropyl-β-d-thiogalactopyranoside] induction) was harvested, and the purified GST fusion protein was immobilized on glutathione-Sepharose 4B beads (GE Healthcare Bio-Sciences AB). The GST-IRF8 beads were incubated with lysates of Flag-myocardin-transfected 293T cells in IP buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40 supplemented with a protease inhibitor cocktail) for 4 h at 4°C. The GST tag was used as the negative control under the same conditions. The samples were analyzed by Western blotting using anti-Flag antibodies, after being washed four times with IP lysis buffer (no cocktail).
Confocal microscopy.
293T cells were seeded in a 24-well plate containing coverslips. After pmCherry-myocardin and EGFP-myc-IRF8 cotransfection for 48 h, the cells were fixed in 4% fresh paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and then incubated in Image-IT FX signal enhancer (catalog no. I36933; Invitrogen) for 30 min. The cells were washed with TBS-Tween three times and stained with DAPI (1 g/ml, 15 min). Finally, the slides were mounted with mounting solution (catalog no. D2522; Sigma). Images were obtained using a confocal laser scanning microscope (Fluoview 1000; Olympus).
Luciferase reporter assays.
The SM22α-luciferase (SM22α-luc) plasmid was obtained by subcloning the SM22α promoter into the pGL3-basic vector (Promega). The CArG-luciferase (CArG-luc) plasmid was obtained by inserting three CArG consensus sequences into the pGL3-promoter vector (Promega). The SRF-luc plasmid was obtained by cloning the SRF promoter into the pGL3-basic vector (Promega). Rat aortic smooth muscle cells (RASMCs) and A7r5 cells were cultured in a 24-well plate. The A7r5 cells were infected with the indicated recombinant adenoviruses. Twenty-four hours later, the cells were harvested and lysed with 100 μl of passive lysis buffer (PLB; Promega). After the cell debris was removed by centrifugation, the supernatant was used for luciferase assays, which were performed using a single-mode SpectraMax microplate reader according to the manufacturer's instructions. The luciferase activities were normalized according to the protein content of the samples.
ChIP assay.
For the chromatin immunoprecipitation (ChIP) assay, MOVAS cells were transfected with HA-tagged IRF8 plasmid or control vector pcDNA3.1. After 48 h, the cells were fixed with 1% formaldehyde, washed with ice-cold PBS, and harvested. Then, the sonicated DNA fragments was subjected to immunoprecipitation using anti-SRF(H300)X (catalog no. SC-13029X; Santa Cruz) and normal rabbit IgG (catalog no. 2729S; Cell Signaling Technology), followed by using protein G-magnetic beads (catalog no. 10004D; Invitrogen). Precipitated DNA was recovered via phenol-chloroform extraction and amplified by reverse transcription-PCR for 40 cycles using primer sets specific for the indicated specific promoter regions of the α-SMA, SM22α, and smoothelin genes. The PCR primers for ChIP assays are provided in Table S2 in the supplemental material.
Statistical analyses.
Data analysis was performed using SPSS (version 17.0) software (SPSS, Inc., Chicago, IL). The data are presented as the mean values ± standard deviations (SDs). The differences among three or more groups were determined using a one-way or two-way analysis of variance (ANOVA) with repeated measures, followed by Bonferroni's post hoc test. A probability value of less than 0.05 was considered statistically significant.
RESULTS
Vascular injury induces IRF8 expression in smooth muscle cells.
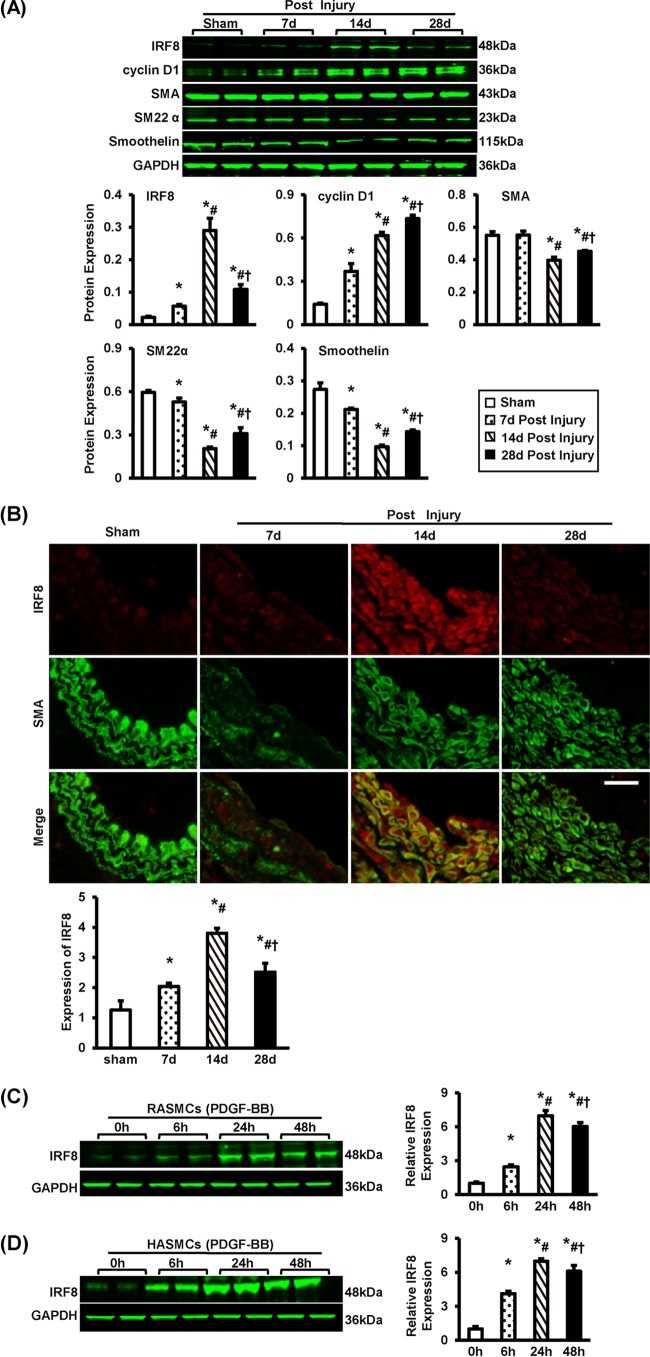
To investigate the role of IRF8 in the pathophysiology of intimal hyperplasia, we performed left carotid artery wire injury on mice and then examined the expression of IRF8. Four weeks after the surgery, the intimal area was greatly increased (see Fig. S1A in the supplemental material). Most of the cells in the neointima were SMCs, as shown by the staining of α-smooth muscle actin (α-SMA) (see Fig. S1B in the supplemental material). The Western blot analyses showed that IRF8 was expressed at a low level in the uninjured carotid arteries. However, the expression of IRF8 was mildly increased at 7 days after injury, was markedly elevated on day 14 postinjury, and returned to a lower level at day 28 (Fig. 1A). Interestingly, we observed that IRF8 was highly expressed in the cells labeled with the α-SMA antibody, particularly at 14 days after injury (Fig. 1B), indicating that IRF8 expression can be greatly induced in SMCs after injury.
FIG 1.
IRF8 expression in smooth muscle cells is induced by vascular injury. (A) The levels of IRF8, cyclin D1, α-SMA, SM22α, and smoothelin proteins in left carotid arteries were determined by Western blotting. The arteries were harvested from uninjured mice (that underwent a sham operation) and injured mice at 7, 14, and 28 days after surgery. The protein levels were quantified. Blots are representative of three independent experiments. (B) Immunofluorescent staining showing the expression and localization of IRF8 (red) in arteries at different time points postinjury. The arterial smooth muscle cells are indicated by α-SMA staining (green). The relative optical density (OD) values of IRF8 fluorescence are also provided. (n = 3 to 5 per group at each time point). Bar, 20 μm. (C and D) IRF8 protein levels in RASMCs (C) and HASMCs (D) were detected by Western blotting before and at 6, 24, and 48 h after PDGF-BB administration. Protein levels were also quantified. Blots are representative of three independent experiments. In panels A, C, and D, the protein levels were normalized to the level of GAPDH. In panels A to D, all values are presented as means ± SDs, and the statistical significance is indicated. (A and B) *, P < 0.05 compared with the sham operation group; #, P < 0.05 compared with the group at 7 days postinjury; †, P < 0.05 compared with the group at 14 days postinjury. (C and D) *, P < 0.05 compared with the group not treated with PDGF-BB; #, P < 0.05 compared with the group at 6 h poststimulation; †, P < 0.05 compared with the group at 24 h poststimulation.
Because PDGF-BB is a mitogen and chemoattractant for SMCs, it is widely used to mimic the process of intima hyperplasia in vitro (26, 27). To confirm IRF8 expression as a response to injury and IRF8 localization, we stimulated two types of SMCs, primary rat aortic smooth muscle cells (RASMCs) and human aortic smooth muscle cells (HASMCs), with PDGF-BB. The Western blotting results showed that, before PDGF-BB administration, IRF8 was scarcely expressed in either cell type. After PDGF-BB (20 ng/ml) was added to the culture medium, IRF8 expression was greatly induced and peaked approximately 24 h after PDGF-BB administration (Fig. 1C and D). Taken together, our results demonstrate that IRF8 can be induced in response to vascular injury in SMCs.
IRF8 deficiency represses neointima formation and SMC phenotypic switching in response to arterial injury.
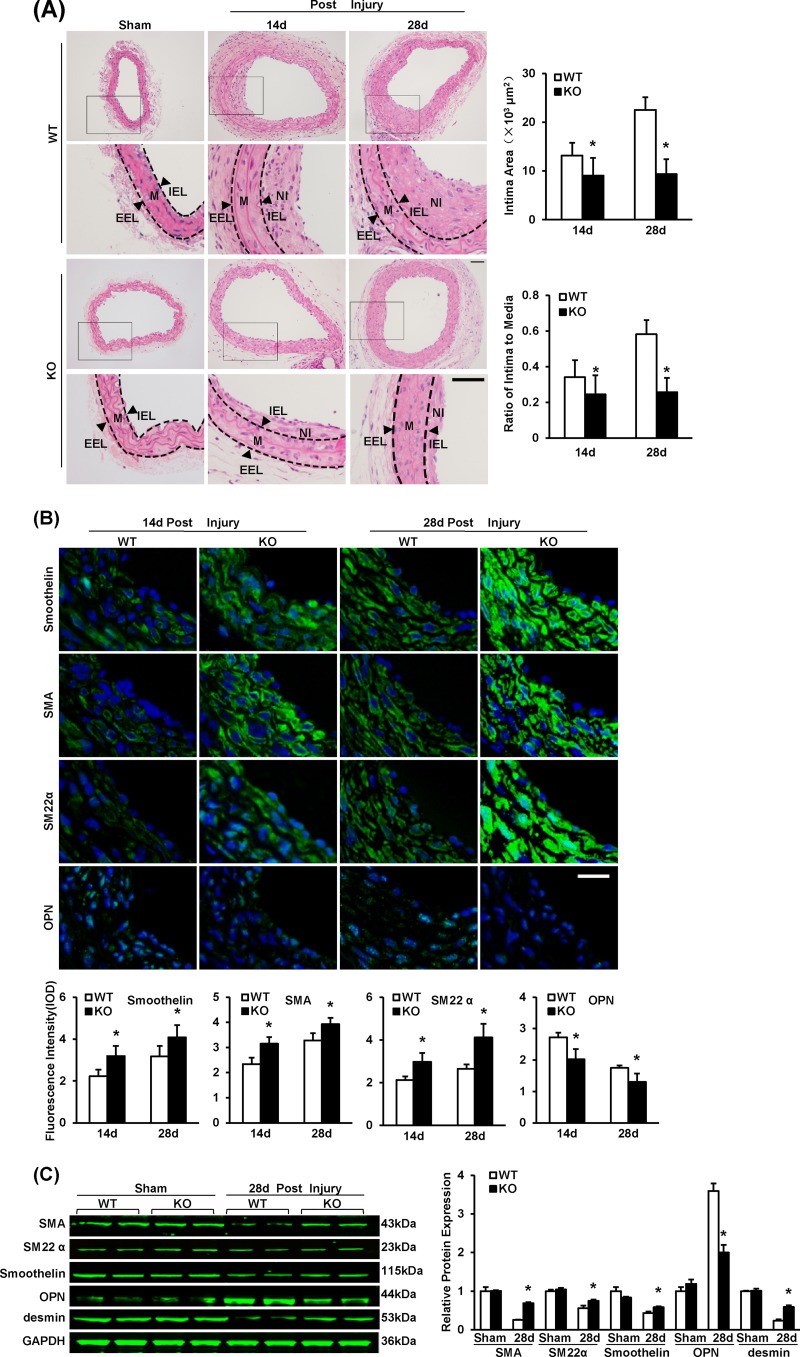
To investigate whether IRF8 modulates neointima formation, we performed carotid wire injury on IRF8−/− (KO) mice and IRF8+/+ (WT) mice. At 14 days after injury, a tendency toward a lower intima/media ratio was observed in the IRF8−/− mice. At 28 days after injury, the intima/media ratio was significantly lower in the IRF8−/− mice than the IRF8+/+ mice (Fig. 2A). To directly exclude the effects of the loss of function of IRF8 in immune cells, carotid wire injury was carried out on SMC-IRF8-KO mice and SM22-Cre control mice, and the phenotype were examined on day 14 postinjury. The SMC-IRF8-KO mice showed less of an increase in the neointima size and the intima/media ratio than the SM22-Cre control mice (see Fig. S2A in the supplemental material), while no significant difference in intimal hyperplasia in the sham-operated arteries was observed between the two genotypes (see Fig. S2A in the supplemental material). These results are consistent with data from IRF8 global knockout mice (Fig. 2A). This suggests that IRF8 deficiency attenuates intimal hyperplasia.
FIG 2.
IRF8 deficiency represses neointima formation and SMC phenotypic switching in response to artery injury. (A) HE-stained sections showing the structures of WT and IRF8−/− (KO) carotid arteries from mice that underwent a sham operation or wire injury surgery (at 14 and 28 days after surgery). Black-framed areas in the upper panels are magnified and shown in the lower panels. Arrowheads, internal elastic lamina (IEL) and external elastic lamina (EEL), highlighted in dotted lines. The internal elastic lamina indicates the intimal-medial boundary, while the external elastic lamina represents the medial-adventitial boundary. M, media; NI, neointima. Intimal areas and intima/media ratios were quantified (n = 9 to 12 per group at each time point). Bars, 50 μm. (B) Immunofluorescent staining of smoothelin, α-SMA, SM22α, and OPN (green in different rows) in WT and IRF8−/− arteries at 14 days and 28 days after injury. DAPI (blue) staining indicates nuclei. The optical density values of smoothelin, α-SMA, SM22α, and OPN fluorescence are also provided (n = 3 to 6 per group at each time point). Bar, 20 μm. (C) The levels of α-SMA, SM22α, smoothelin, OPN, and desmin protein in WT and IRF8−/− arteries were determined by Western blotting. Arteries harvested from the sham operation group and the groups at 28 days postinjury were used. The expression levels were normalized to the expression level of GAPDH and quantified. Blots are representative of three independent experiments. In panels A to C, all values are presented as means ± SDs, and statistical significance is indicated. *, P < 0.05 compared with the WT group.
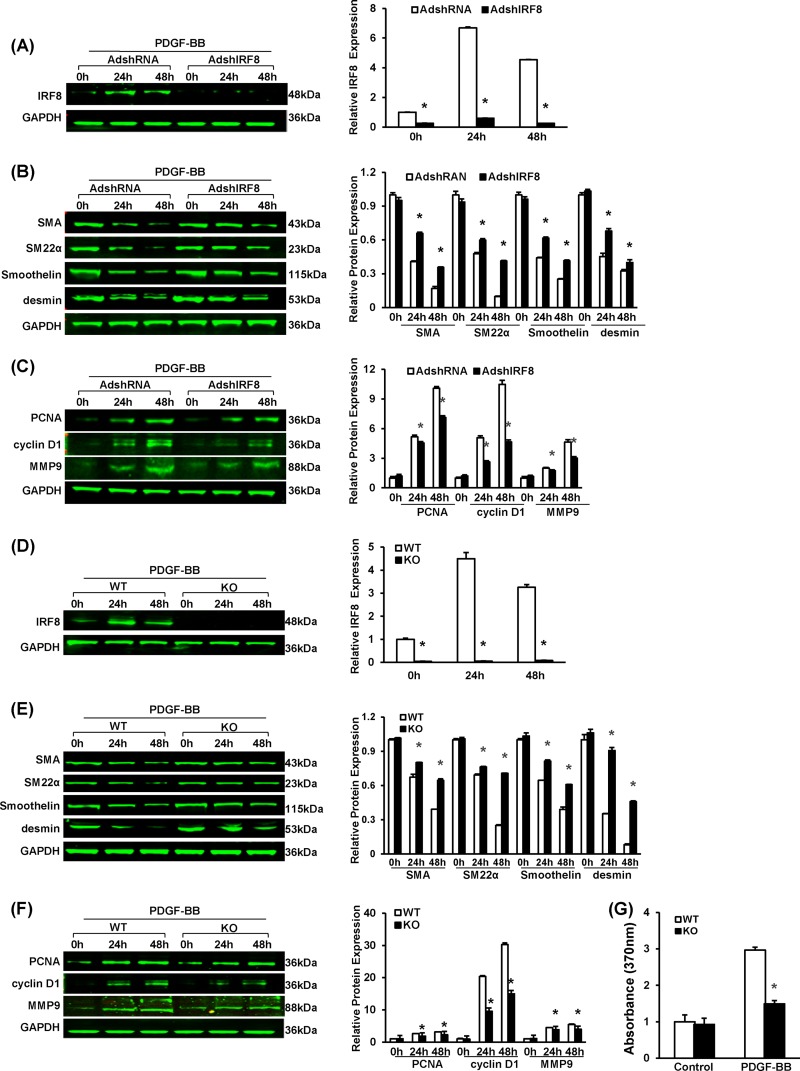
To investigate the effect of IRF8 on the phenotypic switching of SMCs, we analyzed the expression of SMC-specific genes, including those for α-SMA, SM22α, smoothelin, and desmin (28), by immunofluorescent staining and Western blotting. In SMCs of the mice, the expression of these genes was greatly downregulated in response to arterial injury, indicating a switch from a quiescent contractile phenotype to an active synthetic phenotype. However, in the IRF8-deficient SMCs, the increase in the expression of these genes was less prominent on day 14 after injury and the SMC-specific gene expression was significantly higher than that in wild-type cells at day 28 (Fig. 2B and C). Osteopontin, which is expressed in synthetic SMCs rather than in contractile SMCs, is another marker for SMC phenotypic switching (28) and was highly expressed in wild-type SMCs but not IRF8−/− cells (Fig. 2B and C). Previous studies have demonstrated that except for SMC-specific contractile protein reduction, the hallmarks of SMC phenotypic switching also include changes of morphological characteristics (29). Immunofluorescence staining indicated that, in the absence of PDGF-BB stimulation, rat VSMCs kept a spindle-like shape, indicating a differentiated state. However, PDGF-BB elicited a quickly flattened morphology that suggested a dedifferentiated state (see Fig. S2B in the supplemental material). PDGF-BB suppresses the expression of smooth muscle genes in RASMCs, whereas this effect was inhibited by knocking down IRF8 expression using short hairpin RNA (shRNA) (Fig. 3A and B). The preservation of SMC-specific gene expression was also confirmed in the IRF8-deficient primary mouse SMCs stimulated with PDGF-BB in vitro (Fig. 3D and E).
FIG 3.
IRF8 deficiency represses SMC proliferation, migration, and phenotypic switching upon PDGF-BB stimulation. (A) Western blotting validated the efficacy of adenovirus-mediated shRNA interference in lowering IRF8 expression in RASMCs upon PDGF-BB (20 ng/ml) stimulation. Cells were harvested before and at 24 and 48 h after PDGF-BB administration. (B) α-SMA, SM22α, smoothelin, and desmin protein levels in RASMCs infected with AdshRNA and adenovirus containing a short hairpin sequence targeting IRF8 (AdshIRF8) were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (C) PCNA, cyclin D1, and MMP9 protein levels in RASMCs infected with AdshRNA and adenovirus containing a short hairpin sequence targeting IRF8 were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (D) Western blotting to determine the IRF8 levels in WT and IRF8−/− (KO) mouse arterial smooth muscle cells in response to PDGF-BB (20 ng/ml) stimulation. Cells were harvested before and at 24 and 48 h after PDGF-BB administration. (E) α-SMA, SM22α, smoothelin, and desmin protein levels in mouse SMCs were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (F) PCNA, cyclin D1, and MMP9 protein levels in mouse arteries were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. In panels A to F, the corresponding protein levels were normalized to the level of GAPDH and quantified. These blots are representative of three blots obtained from three independent experiments. All values are presented as the means ± SDs, and statistical significance is indicated. *, P < 0.05 compared with the control group. (G) BrdU incorporation was measured by determination of the absorption at 370 nm to assess SMC proliferation upon PDGF-BB (20 ng/ml) stimulation. Values are presented as the means ± SDs, and statistical significance is indicated, *, P < 0.05 compared with the PDGF-BB-treated WT group (n = 4).
In contrast to contractile SMCs, synthetic cells generally have prominent proliferative and migratory properties. Therefore, we examined the expression of some key markers of proliferation (proliferating cell nuclear antigen [PCNA] and cyclin D1) and migration (matrix metalloproteinase 9 [MMP9]). As expected, the IRF8−/− SMCs expressed these genes more slowly in response to injury (see Fig. S2C to E in the supplemental material). The IRF8-knockdown RASMCs and the IRF8−/− SMCs also exhibited lower levels of PCNA, cyclin D1, and MMP9 expression after PDGF-BB administration (Fig. 3C and F). We also examined the SMC proliferative response ex vivo upon PDGF-BB stimulation by measuring the incorporation of 5′-bromo-2′-deoxyuridine (BrdU). The results showed that IRF8-deficient SMCs incorporated less BrdU than WT SMCs, which indicates a lower rate of proliferation (Fig. 3G). The VSMC migration assay indicated that treatment with PDGF-BB (20 ng/ml) caused an almost 6-fold increase in the migration of WT VSMCs; however, IRF8 deficiency significantly reduced PDGF-BB-induced migration (see Fig. S2E in the supplemental material).
IRF8 overexpression promotes neointima formation and SMC phenotypic switching in response to arterial injury.
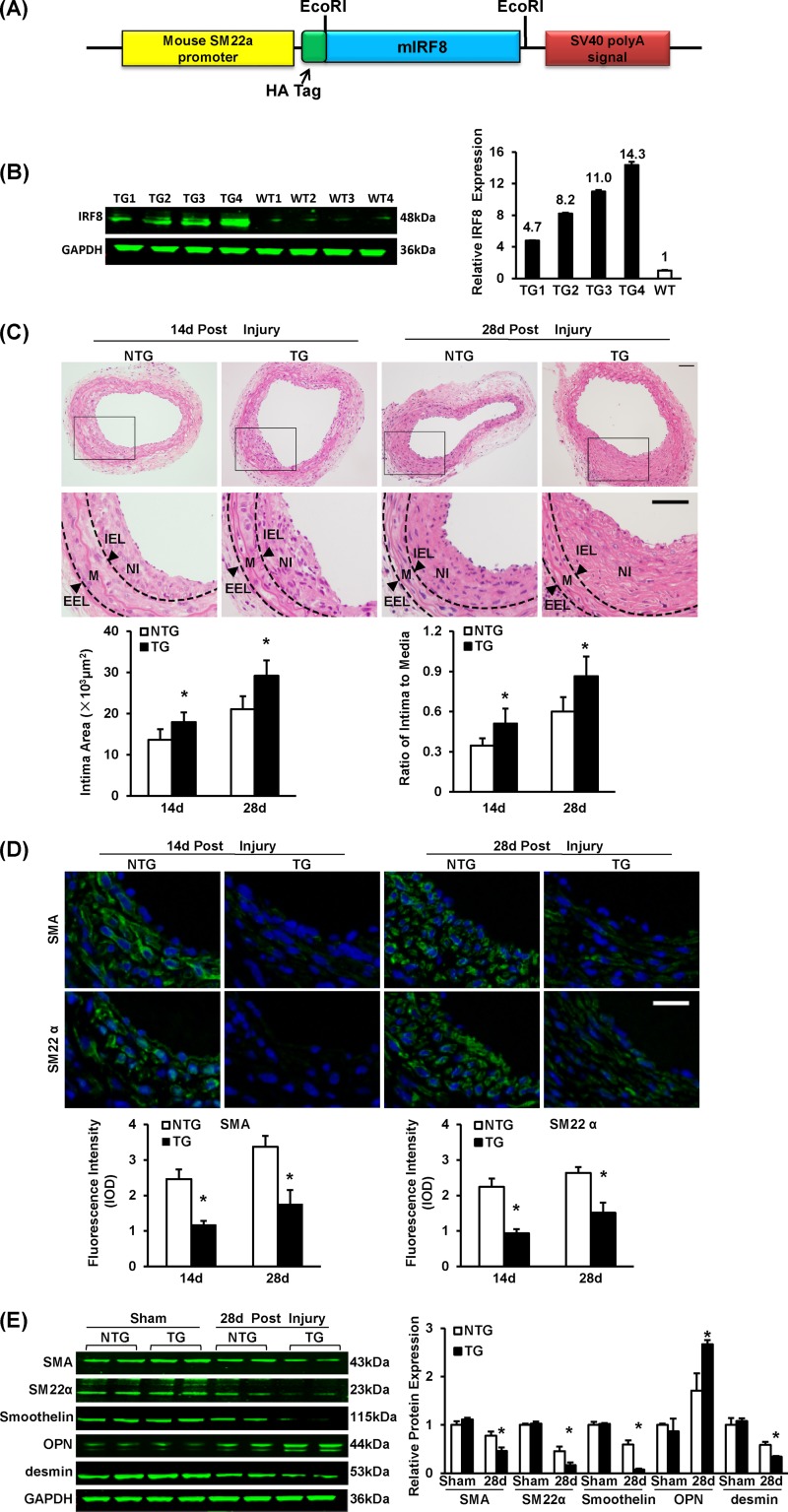
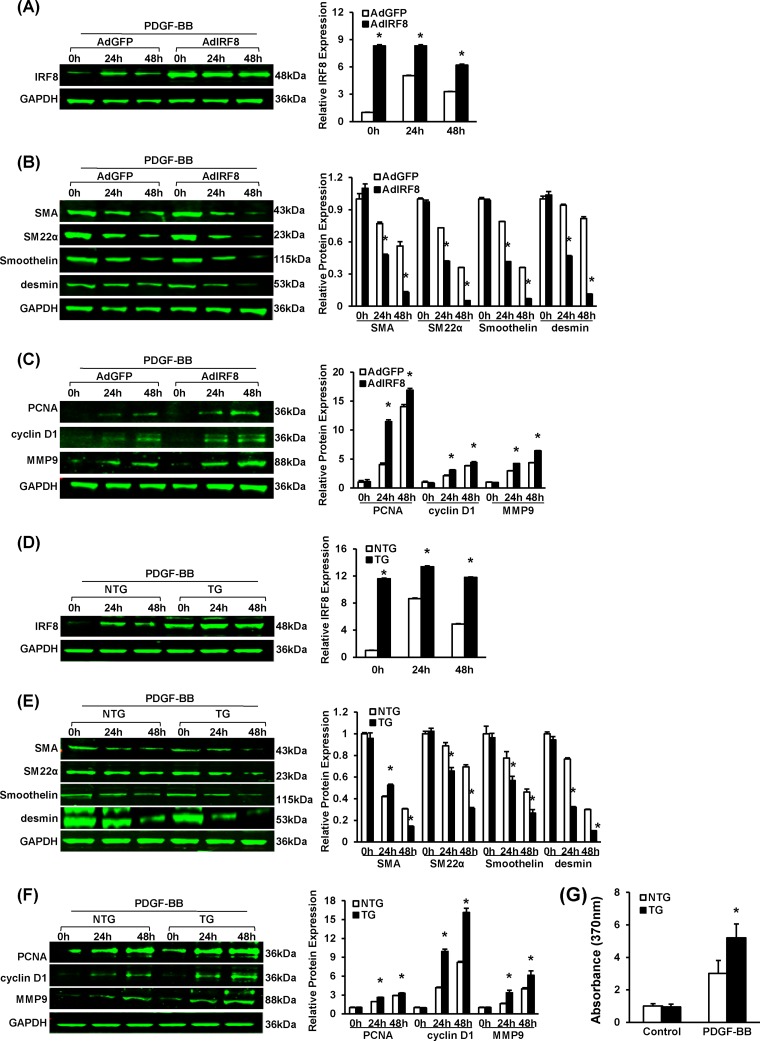
Since IRF8 ablation showed a repressive effect on neointima formation, we next sought to investigate whether IRF8 overexpression promotes neointima growth. To focus on SMCs, we generated SMC-specific IRF8 transgenic (TG) mice in a C57BL/6 background. The TG construct contained mouse IRF8 cDNA under the control of an SMC-specific mouse minimal SM22 promoter (Fig. 4A). Four lines of IRF8 TG mice were successfully generated. Mice from line 4 exhibited a 14.3-fold increase in the IRF8 expression level (Fig. 4B), which was comparable to the level of IRF8 induced in normal SMCs at 14 days after vascular injury, and were therefore used in the subsequent phenotypic evaluations. In contrast to the results obtained using IRF8−/− mice, the intima/media ratio was higher in IRF8 TG mice than in nontransgenic controls (Fig. 4C). By immunofluorescent staining and Western blotting, we observed that the decrease of SMC-specific gene (α-SMA, SM22α, smoothelin, and desmin gene) expression and the increase of osteopontin after injury were more significant in the SMCs derived from the IRF8 TG mice (Fig. 4D and E). These results indicate that IRF8 promotes SMC phenotypic switching. The expression levels of PCNA, cyclin D1, and MMP9 were slightly higher in the SMCs of the transgenic mice than in those of the controls (see Fig. S3A and B in the supplemental material).
FIG 4.
SMC-specific IRF8 overexpression promotes neointima formation and SMC phenotypic switching in response to wire injury. (A) Schematic of the SMC-specific TG vector structure. The vector was first constructed with a mouse SM22α promoter, a 5′ HA tag, and an SV40 poly(A) signal. The mouse full-length IRF8 cDNA (mIRF8) was cloned into the vector between the EcoRI sites. (B) Arterial IRF8 protein levels in TG and WT mice determined by Western blotting. (C) HE-stained sections show the structures of nontransgenic (NTG) and SMC-specific IRF8 TG mouse carotid arteries at 14 days and 28 days after wire injury surgery. Black-framed areas in the upper panels are magnified and shown in the lower panels. Arrowheads, internal elastic lamina (IEL) and external elastic lamina (EEL), highlighted in dotted lines. The internal elastic lamina indicates the intimal-medial boundary, while the external elastic lamina represents the medial-adventitial boundary; M, media; NI, neointima. Intimal areas and intima/media ratios were quantified (n = 6 to 12 per group at each time point). Bars, 50 μm. (D) Immunofluorescent staining of α-SMA and SM22α (green in different rows) in nontransgenic and TG mouse arteries at 14 and 28 days after injury. DAPI (blue) staining marks the nuclei. The optical density values of α-SMA and SM22α fluorescence are also provided (n = 3 or 4 per group at each time point). White bar, 20 μm. (E) Levels of α-SMA, SM22α, smoothelin, OPN, and desmin protein in nontransgenic and TG mouse arteries were determined by Western blotting. Arteries harvested from the sham operation group and the injured group at 28 days postinjury were used. The expression levels were normalized to the expression level of GAPDH and quantified. Blots are representative of three independent experiments. In panels B to E, all of the values are presented as means ± SDs, and statistical significance is indicated. *, P < 0.05 compared with the nontransgenic group.
In agreement with the in vivo results, studies of RASMCs and mouse SMCs treated with PDGF-BB confirmed that IRF8 promotes SMC phenotypic switching and neointima growth. Although IRF8 expression can be induced by PDGF-BB in normal SMCs, at all time points after stimulation, RASMCs infected with adenovirus containing IRF8 and IRF8-overexpressing SMCs exhibited higher IRF8 levels than control cells (Fig. 5A and D). Compared with the level of expression by normal SMCs, the level of expression of SMC-specific genes (α-SMA, smoothelin, SM22α, and desmin genes) was lower, whereas the level of expression of proliferation- and migration-related genes (PCNA, cyclin D1, and MMP9 genes) was higher in the IRF8-overexpressing RASMCs and IRF8 TG SMCs (Fig. 5B, C, E, and F). Upon PDGF-BB stimulation, the IRF8-overexpressing SMCs consistently exhibited a higher proliferation level than the control cells, as shown by more BrdU labeling (Fig. 5G).
FIG 5.
SMC-specific IRF8 overexpression promotes SMC phenotypic switching upon PDGF-BB stimulation. (A) Western blotting validated the efficacy of adenovirus-mediated IRF8 overexpression in RASMCs upon PDGF-BB (20 ng/ml) stimulation. Cells were harvested before and at 24 and 48 h after PDGF-BB administration. (B) α-SMA, SM22α, smoothelin, and desmin protein levels in RASMCs infected with AdGFP and adenovirus expressing IRF8 (AdIRF8) were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (C) PCNA, cyclin D1, and MMP9 protein levels in RASMCs infected with AdGFP and adenovirus expressing IRF8 were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (D) Western blotting validated the IRF8 levels in nontransgenic and IRF8 TG mouse arterial smooth muscle cells in response to PDGF-BB (20 ng/ml) stimulation. Cells were harvested before and at 24 and 48 h after PDGF-BB administration. (E) α-SMA, SM22α, smoothelin, and desmin protein levels in nontransgenic and TG mouse SMCs were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. (F) PCNA, cyclin D1, and MMP9 protein levels in nontransgenic and TG mouse arteries were determined by Western blotting at different time points after PDGF-BB (20 ng/ml) administration. In panels A to F, the corresponding protein levels were normalized to the GAPDH level and quantified. These blots are representative of three blots obtained from three independent experiments. All of the values are presented as the means ± SDs, and the statistical significance is indicated. *, P < 0.05 compared with the AdGFP-treated or nontransgenic group. (G) BrdU incorporation was measured by determination of the absorption at 370 nm to assess nontransgenic and TG mouse VSMC proliferation upon PDGF-BB (20 ng/ml) stimulation. The values presented are the means ± SDs, and the statistical significance is indicated. *, P < 0.05 compared with the PDGF-BB-treated nontransgenic group (n = 4).
IRF8 inhibits smooth muscle gene expression through the SRF/CArG axis.
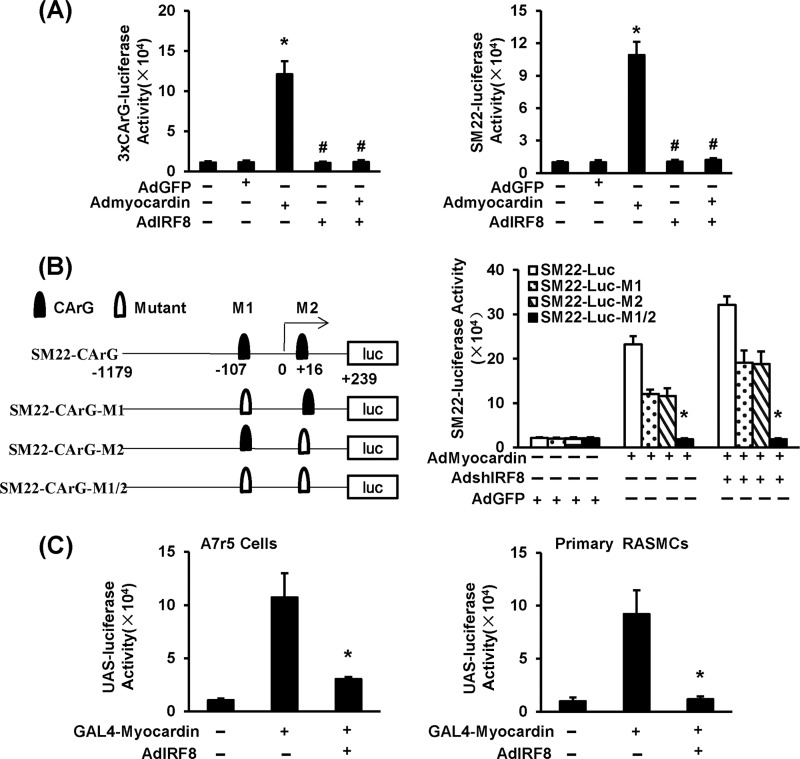
Because both gain- and loss-of-function studies confirmed an inverse relationship between IRF8 expression and SMC-specific gene expression, we were interested in elucidating the mechanisms for this phenomenon. Evidence has firmly established that the CArG box, which is recognized by SRF, present within the promoters of SMC genes, plays a pivotal role in controlling their transcription (30, 31). Thus, we reasoned that IRF8 might inhibit SMC gene transcription through interfering with the SRF/CArG axis. Therefore, we examined the transactivation of SRF using a luciferase reporter driven by three tandem repeats of a consensus SRF response element (3×CArG-luc) in RASMCs. We also cloned and inserted the mouse SM22α promoter sequence in front of the luciferase reporter gene to determine the transcriptional inhibition of SM22α by IRF8. As expected, in both systems, luciferase activity was greatly induced after infection with a myocardin (coactivator of SRF)-containing adenovirus (Fig. 6A). When we coinfected cells with the myocardin-containing virus and the IRF8-containing virus, the activity of 3×CArG-luc could not be induced (Fig. 6A). These results indicate that IRF8 inhibits SRF/CArG-mediated transcription.
FIG 6.
IRF8 inhibits smooth muscle gene expression through the SRF/CArG axis. (A) Adenovirus constructs containing 3×CArG-luc (left) and SM22α-luc (right) were used to infect RASMCs to investigate SRF/CArG-dependent gene expression. After AdGFP, adenovirus containing myocardin (Admyocardin), and adenovirus expressing IRF8 AdIRF8) were introduced into RASMCs separately or jointly, luciferase activity was measured. The data represent mean values ± SDs. *, P < 0.05 compared with the AdGFP-treated control; #, P < 0.05 compared with the group positive for adenovirus containing myocardin and negative for adenovirus expressing IRF8. (B) (Left) Schematic showing reporter constructs produced by mutating different CArG elements identified on the SM22α promoter. CArG, original CArG elements; Mutant, a mutated CArG element. (Right) Different reporter constructs were introduced into RASMCs for luciferase activity analysis. Adenovirus containing a short hairpin sequence targeting IRF8 was used to knock down endogenous IRF8 expression. The data represent mean values ± SDs. *, P < 0.05 compared with the WT SM22-luc group. (C) A construct encoding Gal4-myocardin fusion protein was generated. A7r5 cells and RASMCs were infected with an adenovirus containing UAS-luc and then coinfected with Gal4-myocardin- or IRF8-containing adenoviruses. The corresponding luciferase activities were analyzed. The data represent mean values ± SDs. *, P < 0.05 compared with GAL4-myocardin-positive, IRF8-negative group.
To further confirm this conclusion, we mutated the CArG elements that were identified at approximately residues −107 and +16 of the SM22α transcription start site and constructed the corresponding mutant SM22α-luc plasmids. In myocardin-containing adenovirus-infected cells, the mutant SM22α-luc activity was lower than that of wild-type SM22α-luc (Fig. 6B). Knockdown of IRF8 using shRNA increased the activities of wild-type SM22α-luc and SM22α-luc with a single mutation but did not affect the activity of SM22α-luc with a double mutation (Fig. 6B). Chromatin immunoprecipitation was applied to further evaluate the effect of IRF8 on SRF transactivation, with the results indicating that significantly less SRF was bound to CArG elements in HA-IRF8 MOVAS cells than in pcDNA3.1 control cells, which directly indicated that IRF8 impairs SRF binding to CArG elements (see Fig. S4A in the supplemental material). This finding suggests that the inhibition of SM22α transcription by IRF8 is dependent on the CArG element.
These results prompted us to investigate whether IRF8, as a transcription factor, inhibits SRF transcription. Using SRF promoter-luciferase assays, we did not observe any significant change in SRF-luciferase activity upon interfering with the IRF8 level in A7r5 cells and RASMCs (data not shown). Thus, we speculated that IRF8 might directly inhibit SRF activity through protein-protein interaction. Unfortunately, no explicit interaction was observed in immunoprecipitation (IP) or GST pulldown experiments (data not shown). The results presented above indicate that IRF8 most likely blunts SRF/CArG in an indirect manner. Because myocardin, a master coactivator of SRF, is essential for SMC development and phenotypic maintenance (31–33), we speculated that IRF8 might regulate SRF/CArG activity through myocardin. To examine the function of myocardin in an SRF/CArG-independent manner, we employed the Gal4-upstream activating sequence (UAS) system and constructed a chimeric transcription factor with the Gal4 DNA-binding domain (DBD) fused to myocardin. UAS-luciferase (UAS-luc) activity was significantly induced by Gal4-myocardin-containing adenovirus infection in A7r5 cells and RASMCs (Fig. 6C). When we coinfected with the IRF8-containing adenovirus, however, the induction of UAS-luc activity by Gal4-myocardin was largely inhibited (Fig. 6C). Hence, we demonstrated that IRF8 inhibits SMC gene transcription in a myocardin-dependent way.
IRF8 inhibits smooth muscle gene expression through the interaction with myocardin.
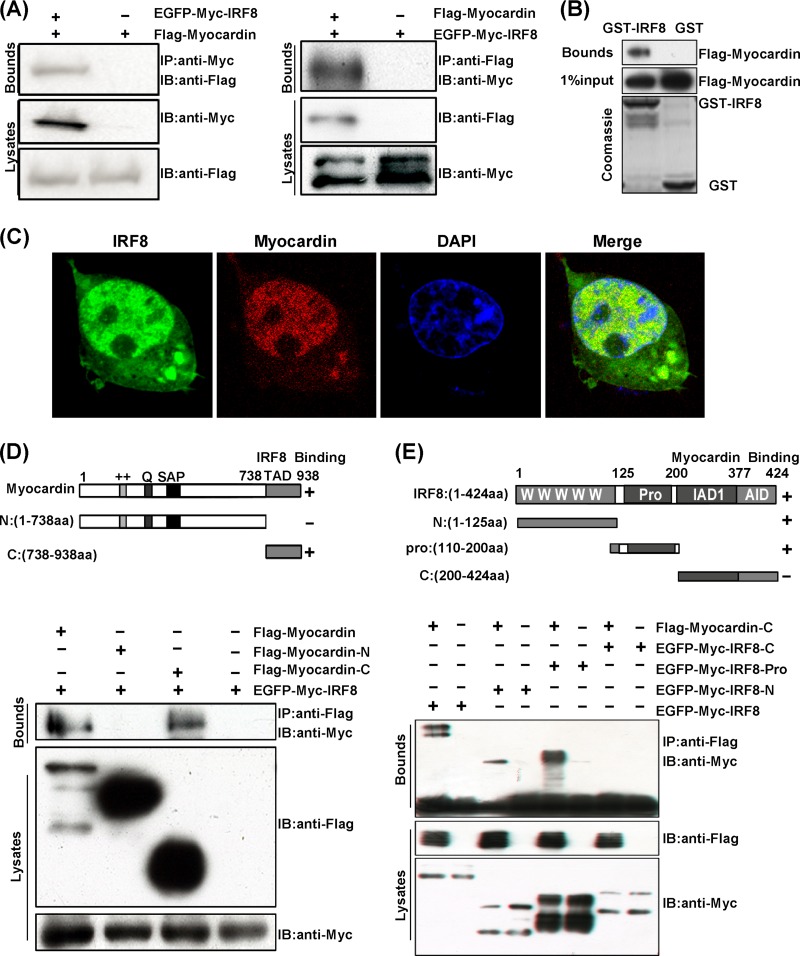
The fact that the induction of UAS-luc activity by the Gal4-myocardin fusion protein was blocked by IRF8 raises the question whether IRF8 directly affects the structure or activity of myocardin. Therefore, we sought to investigate the possible protein-protein interaction between myocardin and IRF8. We transfected 293T cells with plasmids expressing EGFP-myc-IRF8 and Flag-myocardin and then performed IP experiments. We observed that EGFP-myc-IRF8 coimmunoprecipitated with Flag-myocardin, and vice versa (Fig. 7A). This interaction was confirmed by a GST pulldown experiment, which showed that Flag-myocardin was pulled down with GST-tagged IRF8 but not GST per se (Fig. 7B). Moreover, we transfected 293T cells with pEGFP-IRF8 and pmCherry-myocardin and then examined the localization of each protein using fluorescent confocal microscopy. We observed that both IRF8 and myocardin predominantly localized in the nucleus, and they merged quite well (Fig. 7C). To identify the specific regions of IRF8 and myocardin that interacted, we generated various truncations of IRF8 and myocardin. The IP results showed that only the C-terminal transcriptional activation domain (TAD; amino acids [aa] 738 to 938) of myocardin interacted with IRF8 (Fig. 7D). Mapping the IRF8 regions showed that the N-terminal DBD (aa 1 to 125) and the intermediate region (aa 125 to 200) of IRF8 interacted with myocardin, but the C-terminal IRF association domain (IAD; aa 200 to 377) and autoinhibitory domain (AID; aa 377 to 424) did not (Fig. 7E).
FIG 7.
IRF8 inhibits smooth muscle gene expression through its interaction with myocardin. (A) Coimmunoprecipitation of IRF8 and myocardin. Cell lysates from 293T cells transfected with EGFP-Myc-tagged IRF8 and Flag-tagged myocardin were prepared. These lysates were subjected to IP using a Myc antibody and analyzed by immunoblotting (IB) using Myc or Flag antibodies (left). In addition, these lysates were subjected to IP with Flag antibody and then immunoblotted using Myc or Flag antibodies (right). (B) GST pulldown assays of Flag-tagged myocardin with GST or GST-IRF8. (C) Colocalization of IRF8 and myocardin in the nucleus is shown. pmCherry-myocardin (red) and pEGFP-IRF8 (green) were transfected into 293T cells, and fluorescence analysis was performed. The cells were visualized by confocal microscopy. The nuclei were stained with DAPI (blue). (D) (Top) Schematic representation of two types of myocardin-deletion mutants. Myocardin-N contains the N-terminal 738 aa, while myocardin-C contains the C-terminal transcription activation domain (TAD; aa 738 to 938). (Bottom) Mapping the IRF8-binding region of myocardin. Cell lysates from 293T cells transfected with EGFP-Myc-tagged IRF8 and Flag-tagged myocardin-deletion mutants were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-Flag and anti-Myc antibodies. (E) (Top) Schematic representation of three types of IRF8-deletion mutants. IRF8-N contains the N-terminal 125 aa of IRF8 (the “W” repeat represents the conserved DNA binding domain [DBD]), IRF8-pro contains the intermediate aa 110 to 200 of IRF8, and IRF8-C contains the C-terminal IRF association domain (IAD; aa 200 to 377) and autoinhibitory domain (AID; aa 377 to 424). (Bottom) Map of the myocardin-binding region of IRF8. 293T cells were transfected with Flag-tagged myocardin (C-terminal) and EGFP-Myc-tagged IRF8-deletion mutants.
To determine whether IRF8 inhibits smooth muscle gene expression through its interaction with myocardin, we investigated the effect of an IRF8-deletion mutant which lacks the N-terminal and intermediate regions required for the interaction with myocardin on 3×CArG-luc and SM22α-luc activity. As described above, wild-type IRF8 inhibited the induction of 3×CArG-luc and SM22α-luc activities by myocardin (Fig. 6A; see Fig. S4B in the supplemental material). However, mutant IRF8 failed to repress the luciferase activity induced by myocardin (see Fig. S4B in the supplemental material). This result indicates that the interaction between IRF8 and myocardin is critical for the inhibition of SMC gene expression. It is noteworthy that the C-terminal transcriptional activation domain of myocardin, which interacts with IRF8, functions to recruit acetyltransferase p300 to activate transcription (34). Hence, it is possible that IRF8 competitively inhibits the recruitment of p300. Using luciferase reporter assays, we observed that p300, in collaboration with myocardin, induced 3×CArG-luc and SM22α-luc activity in a dose-dependent way (see Fig. S4C and D in the supplemental material). Similarly, when the amount of transfected p300 remained constant and a higher dose of IRF8 was added, the decrease of luciferase activity was more significant (see Fig. S4C and D in the supplemental material). In summary, IRF8 inhibits smooth muscle gene transcription through its interaction with myocardin.
DISCUSSION
In the present study, we demonstrated for the first time that IRF8 mediates the phenotypic switching of SMCs in response to arterial wire injury. Through in vivo and in vitro studies, we found that the expression of IRF8 was upregulated in SMCs after injury or PDGF-BB stimulation. A lack of IRF8 in mice retarded SMC phenotypic switching and intimal hyperplasia following injury, whereas SMC-specific overexpression aggravated these responses. We further identified IRF8 to be a potential antagonist competing with acetyltransferase p300 to bind to the muscle-specific coactivator myocardin. In this manner, IRF8 downregulates SRF transactivation, reduces the expression of smooth muscle-specific genes, and facilitates the switch of SMCs from a contractile to a synthetic phenotype. These findings provide the basic information needed to understand the molecular mechanisms of SMC growth and neointimal hyperplasia.
IRF8, also known as interferon consensus sequence binding protein (ICSBP), was first cloned and identified as a protein that binds to interferon-stimulated response element (ISRE) motif in the promoter region of major histocompatibility complex class I H-2LD gene (19). The expression of IRF8 was thought to be restricted to lymphocyte and monocyte/macrophage lineages (19, 35). Evidence that has accumulated throughout the past 2 decades has demonstrated that IRF8 plays critical roles in the differentiation and development of dendritic cells, macrophages, and some lineages of lymphocytes (13). IRF8-deficient mice were reported to present with a chronic myeloid leukemia (CML)-like syndrome and were susceptible to viral and parasitic infections (36–38). IRF8 mutations were also identified to cause human dendritic cell immunodeficiency (39). In the present study, we discovered a role of IRF8 in vascular physiology and the SMC phenotype not reported previously. IRF8 expression in SMCs can be induced by injury in vivo or by PDGF-BB stimulation in vitro. Using global knockout, SMC-specific knockout, and SMC-specific transgenic mice, we demonstrated that IRF8 inhibits the expression of SMC-specific genes and promotes the phenotypic switching of SMCs, thereby facilitating intimal hyperplasia. Probably due to the extremely low level of IRF8 in normally contractile SMCs, little attention has previously been paid to the potential roles of IRF8 in SMCs. Therefore, the findings in the present work broaden our knowledge of the tissue distribution of IRF8 as well as its versatility.
In recent years, the roles of IRF8 in cardiovascular diseases have been revealed. By comparing allele frequencies between systemic lupus erythematosus patients with and without coronary heart disease, single nucleotide polymorphisms located in the IRF8 gene were identified to be associated with the presence of carotid plaques and increased intima-media thickness (40). Thus, these findings suggest a potential involvement of IRF8 in neointima formation and the development of vascular occlusive diseases. Döring et al. reported that the CML phenotype caused by hematopoietic IRF8 deficiency contributed to accelerated atherosclerosis (20). The expression of arginase 1 (Arg1), a marker of anti-inflammatory M2-type macrophages, is inversely correlated with atherosclerosis progression (41). Pourcet et al. found that Arg1 expression could be induced by IRF8 in synergy with PU.1, which indicates an antiatherosclerotic feature of IRF8 (21). Unlike the roles of IRF8 in hematopoietic cells, we found that, in SMCs, IRF8 modulated the cell physiology and phenotype to promote neointima formation. Hence, targeting IRF8 in SMCs holds promise as a therapeutic strategy to treat vasculoproliferative diseases.
Neointima formation or vascular remodeling results from the interplay between vascular cells and environmental stimuli (1). During this process, SMCs undergo a phenotypic switching; i.e., they partially lose the expression of normal SMC marker genes and gain a less differentiated state of proliferation, migration, and secretion (42). The mechanisms of SMC-specific gene inhibition are diverse, but many of them converge to the regulation of SRF, a master regulator of smooth muscle genes (4). Upon phosphorylation, the Ets domain protein Elk1 prevents the interaction of SRF with its coactivator myocardin through binding to SRF (9). KLF4 promotes SMC phenotypic switching by recruiting HDACs to SRF to downregulate SRF transactivation (10). FoxO4 represses SMC differentiation by interacting with myocardin (43) to indirectly regulate SRF activity. In the present study, we demonstrated that IRF8 represses the expression of SMC differentiation genes and promotes the phenotypic switching of SMCs in response to injury. Considering that SRF and its coactivator, myocardin, play essential roles in muscle cell differentiation and SMC phenotypic modulation (32), we further examined the effect of IRF8 on SRF expression. Through SRF-luciferase reporter assays, we excluded the possibility that IRF8 transcriptionally inhibits SRF expression. Moreover, we failed to detect an explicit interaction between IRF8 and SRF. In investigating whether IRF8 affected the function of myocardin, we successfully coimmunoprecipitated IRF8 and myocardin. By mapping the interaction using different truncation mutants of myocardin, we identified that the site which is indispensable for recruiting acetyltransferase p300 is within the region occupied by IRF8. Therefore, IRF8 downregulates SRF/myocardin transactivation at least in part by competitively inhibiting the recruitment of p300. The elucidation of IRF8's involvement adds a new layer in understanding how the SRF/myocardin-dependent genes are tightly regulated.
In summary, we identified that IRF8 is a novel mediator in response to arterial injury in SMCs. IRF8 facilitated SMC phenotypic switching and neointima formation. We further demonstrated that the mechanism by which IRF8 downregulates the expression of SMC-specific genes involves its interaction with myocardin. Our findings broaden the knowledge of the tissue distribution of IRF8. More importantly, the emerging immunity-independent functions of IRF8 will undoubtedly shed new light on the treatment of cardiovascular diseases.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate Xiang-jie Kong for his technical assistance with performing carotid arterial injuries.
This study was supported by the National Natural Science Foundation of China (grant numbers 81170086, 81100230, 81070089, and 81370365), the National Science and Technology Support Project (grant numbers 2011BAI15B02, 2012BAI39B05, and 2013YQ030923-05), the National Basic Research Program of China (grant number 2011CB503902), the Ministry of Education New Century Outstanding Talents Support Program (grant number NCET-10-0641), the Independent Scientific Research Project of Wuhan University (grant number 2012302020215), and the key project of the National Natural Science Foundation (grant number 81330005).
Footnotes
Published ahead of print 18 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01070-13.
REFERENCES
- 1.Dzau VJ, Braun-Dullaeus RC, Sedding DG. 2002. Vascular proliferation and atherosclerosis: new perspectives and therapeutic strategies. Nat. Med. 8:1249–1256. 10.1038/nm1102-1249 [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84:767–801. 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- 3.Gibbons GH, Dzau VJ. 1994. The emerging concept of vascular remodeling. N. Engl. J. Med. 330:1431–1438. 10.1056/NEJM199405193302008 [DOI] [PubMed] [Google Scholar]
- 4.Kawai-Kowase K, Owens GK. 2007. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 292:C59–C69. 10.1152/ajpcell.00394.2006 [DOI] [PubMed] [Google Scholar]
- 5.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. 2004. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ. Res. 95:981–988. 10.1161/01.RES.0000147961.09840.fb [DOI] [PubMed] [Google Scholar]
- 6.Regan CP, Adam PJ, Madsen CS, Owens GK. 2000. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J. Clin. Invest. 106:1139–1147. 10.1172/JCI10522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miano JM. 2003. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35:577–593. 10.1016/S0022-2828(03)00110-X [DOI] [PubMed] [Google Scholar]
- 8.Camoretti-Mercado B, Dulin NO, Solway J. 2003. Serum response factor function and dysfunction in smooth muscle. Respir. Physiol. Neurobiol. 137:223–235. 10.1016/S1569-9048(03)00149-6 [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428:185–189. 10.1038/nature02382 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. 2005. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 280:9719–9727. 10.1074/jbc.M412862200 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T, Gan Q, Owens GK. 2008. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am. J. Physiol. Cell Physiol. 295:C1175–C1182. 10.1152/ajpcell.00288.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. 2005. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler. Thromb. Vasc. Biol. 25:2328–2334. 10.1161/01.ATV.0000185829.47163.32 [DOI] [PubMed] [Google Scholar]
- 13.Tamura T, Yanai H, Savitsky D, Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26:535–584. 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- 14.Savitsky D, Tamura T, Yanai H, Taniguchi T. 2010. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol. Immunother. 59:489–510. 10.1007/s00262-009-0804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658. 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Bian ZY, Zhang R, Zhang Y, Liu C, Yan L, Zhang SM, Jiang DS, Wei X, Zhu XH, Chen M, Wang AB, Chen Y, Yang Q, Liu PP, Li H. 2013. Interferon regulatory factor 3 is a negative regulator of pathological cardiac hypertrophy. Basic Res. Cardiol. 108:326. 10.1007/s00395-012-0326-9 [DOI] [PubMed] [Google Scholar]
- 17.Jiang DS, Bian ZY, Zhang Y, Zhang SM, Liu Y, Zhang R, Chen Y, Yang Q, Zhang XD, Fan GC, Li H. 2013. Role of interferon regulatory factor 4 in the regulation of pathological cardiac hypertrophy. Hypertension 61:1193–1202. 10.1161/HYPERTENSIONAHA.111.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XA, Zhang R, Jiang D, Deng W, Zhang S, Deng S, Zhong J, Wang T, Zhu LH, Yang L, Hong S, Guo S, Chen K, Zhang XF, She Z, Chen Y, Yang Q, Zhang XD, Li H. 2013. Interferon regulatory factor 9 protects against hepatic insulin resistance and steatosis in male mice. Hepatology 58:603–616. 10.1002/hep.26368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Driggers PH, Ennist DL, Gleason SL, Mak WH, Marks MS, Levi BZ, Flanagan JR, Appella E, Ozato K. 1990. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. U. S. A. 87:3743–3747. 10.1073/pnas.87.10.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Döring Y, Soehnlein O, Drechsler M, Shagdarsuren E, Chaudhari SM, Meiler S, Hartwig H, Hristov M, Koenen RR, Hieronymus T, Zenke M, Weber C, Zernecke A. 2012. Hematopoietic interferon regulatory factor 8-deficiency accelerates atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 32:1613–1623. 10.1161/ATVBAHA.111.236539 [DOI] [PubMed] [Google Scholar]
- 21.Pourcet B, Feig JE, Vengrenyuk Y, Hobbs AJ, Kepka-Lenhart D, Garabedian MJ, Morris SM, Jr, Fisher EA, Pineda-Torra I. 2011. LXRalpha regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ. Res. 109:492–501. 10.1161/CIRCRESAHA.111.241810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Li S, Zhang H, Wan Z, Zhang X, Du R. 2012. A one-step cloning method for the construction of somatic cell gene targeting vectors: application to production of human knockout cell lines. BMC Biotechnol. 12:71. 10.1186/1472-6750-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhel DG, Zhu B, Witte DP, Hui DY. 2002. Distinction in genetic determinants for injury-induced neointimal hyperplasia and diet-induced atherosclerosis in inbred mice. Arterioscler. Thromb. Vasc. Biol. 22:955–960. 10.1161/01.ATV.0000017994.77066.75 [DOI] [PubMed] [Google Scholar]
- 24.Cho A, Reidy MA. 2002. Matrix metalloproteinase-9 is necessary for the regulation of smooth muscle cell replication and migration after arterial injury. Circ. Res. 91:845–851. 10.1161/01.RES.0000040420.17366.2E [DOI] [PubMed] [Google Scholar]
- 25.Li H, He C, Feng J, Zhang Y, Tang Q, Bian Z, Bai X, Zhou H, Jiang H, Heximer SP, Qin M, Huang H, Liu PP, Huang C. 2010. Regulator of G protein signaling 5 protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Proc. Natl. Acad. Sci. U. S. A. 107:13818–13823. 10.1073/pnas.1008397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blank RS, Owens GK. 1990. Platelet-derived growth factor regulates actin isoform expression and growth state in cultured rat aortic smooth muscle cells. J. Cell. Physiol. 142:635–642. 10.1002/jcp.1041420325 [DOI] [PubMed] [Google Scholar]
- 27.Jawien A, Bowen-Pope DF, Lindner V, Schwartz SM, Clowes AW. 1992. Platelet-derived growth factor promotes smooth muscle migration and intimal thickening in a rat model of balloon angioplasty. J. Clin. Invest. 89:507–511. 10.1172/JCI115613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rensen SS, Doevendans PA, van Eys GJ. 2007. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 15:100–108. 10.1007/BF03085963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rzucidlo EM, Martin KA, Powell RJ. 2007. Regulation of vascular smooth muscle cell differentiation. J. Vasc. Surg. 45(Suppl A):A25–A32. 10.1016/j.jvs.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 30.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. 2006. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J. Clin. Invest. 116:36–48. 10.1172/JCI26505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. 2003. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. U. S. A. 100:9366–9370. 10.1073/pnas.1233635100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida T. 2003. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92:856–864. 10.1161/01.RES.0000068405.49081.09 [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Kitchen CM, Streb JW, Miano JM. 2002. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 34:1345–1356. 10.1006/jmcc.2002.2086 [DOI] [PubMed] [Google Scholar]
- 34.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. 2005. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol. Cell. Biol. 25:364–376. 10.1128/MCB.25.1.364-376.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takaoka A, Tamura T, Taniguchi T. 2008. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci. 99:467–478. 10.1111/j.1349-7006.2007.00720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC, III, Ozato K, Horak I. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87:307–317. 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- 37.Scharton-Kersten T, Contursi C, Masumi A, Sher A, Ozato K. 1997. Interferon consensus sequence binding protein-deficient mice display impaired resistance to intracellular infection due to a primary defect in interleukin 12 p40 induction. J. Exp. Med. 186:1523–1534. 10.1084/jem.186.9.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giese NA, Gabriele L, Doherty TM, Klinman DM, Tadesse-Heath L, Contursi C, Epstein SL, Morse HC., III 1997. Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J. Exp. Med. 186:1535–1546. 10.1084/jem.186.9.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, Menon G, Trouillet C, McDonald D, Carey P, Ginhoux F, Alsina L, Zumwalt TJ, Kong XF, Kumararatne D, Butler K, Hubeau M, Feinberg J, Al-Muhsen S, Cant A, Abel L, Chaussabel D, Doffinger R, Talesnik E, Grumach A, Duarte A, Abarca K, Moraes-Vasconcelos D, Burk D, Berghuis A, Geissmann F, Collin M, Casanova JL, Gros P. 2011. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 365:127–138. 10.1056/NEJMoa1100066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard D, Svenungsson E, Sandling JK, Berggren O, Jonsen A, Bengtsson C, Wang C, Jensen-Urstad K, Granstam SO, Bengtsson AA, Gustafsson JT, Gunnarsson I, Rantapaa-Dahlqvist S, Nordmark G, Eloranta ML, Syvanen AC, Ronnblom L. 2013. Coronary heart disease in systemic lupus erythematosus is associated with interferon regulatory factor 8 gene variants. Circ. Cardiovasc. Genet. 6:255–263. 10.1161/CIRCGENETICS.113.000044 [DOI] [PubMed] [Google Scholar]
- 41.Teupser D, Burkhardt R, Wilfert W, Haffner I, Nebendahl K, Thiery J. 2006. Identification of macrophage arginase I as a new candidate gene of atherosclerosis resistance. Arterioscler. Thromb. Vasc. Biol. 26:365–371. 10.1161/01.ATV.0000195791.83380.4c [DOI] [PubMed] [Google Scholar]
- 42.Nguyen AT, Gomez D, Bell RD, Campbell JH, Clowes AW, Gabbiani G, Giachelli CM, Parmacek MS, Raines EW, Rusch NJ, Speer MY, Sturek M, Thyberg J, Towler DA, Weiser-Evans MC, Yan C, Miano JM, Owens GK. 2013. Smooth muscle cell plasticity: fact or fiction? Circ. Res. 112:17–22. 10.1161/CIRCRESAHA.112.281048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu ZP, Wang Z, Yanagisawa H, Olson EN. 2005. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 9:261–270. 10.1016/j.devcel.2005.05.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.