Abstract
Influenza imposes a great burden on society, not only in its seasonal appearance that affects both humans and domesticated animals but also through the constant threat of potential pandemics. Migratory birds are considered to be the reservoir hosts for influenza viruses, but other animals must also be considered. The recently identified influenza-like virus genome, from H17N10 in bats, was shown to be markedly different from genomes of other known influenza viruses, as both its surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) do not have canonical functions. However, no studies on other individual proteins from this particular virus have been reported until now. Here, we describe the structure of the N-terminal domain of PA from H17N10 influenza-like virus at 2.7-Å resolution and show that it has a fold similar to those of homologous PA domains present in more familiar influenza A virus strains. Moreover, we demonstrate that it possesses endonuclease activity and that the histidine residue in the active site is essential for this activity. Although this particular influenza virus subtype is probably not infectious for humans (even its virus state has not been confirmed in bats, as only the genome has been sequenced), reassortment of canonical influenza viruses with certain segments from H17N10 cannot be ruled out at this stage. Therefore, further studies are urgently needed for the sake of influenza prevention and control.
INTRODUCTION
Influenza puts a great burden on society through its seasonal occurrence and by keeping the world in a constant state of alertness for the possible occurrence of a pandemic strain (1). Three different lineages of influenza virus, a segmented negative-strand RNA virus within the family Orthomyxoviridae, are known to infect humans; these are designated A, B, and C. The last two are adapted to humans, while only some of all known influenza A viruses are circulating in humans (1). The largest animal reservoir for influenza A virus is formed by wild birds (2), with other animals such as poultry and pigs functioning as reassortment vessels (“mixing vessels”) from which reassorted influenza A virus strains occasionally infect humans, as demonstrated by recent H7N9 cases (3). Such viruses have shown low human-to-human transmission, but the question is not whether but rather when will viruses arise that combine high lethality with human-to-human transmissibility. For two avian H5N1 strains from recent studies, either four or five mutations were shown to be enough to make them transmittable between ferrets, the model for human transmission (4, 5).
The recent discoveries of full-length genomes of influenza-like viruses H17N10 (6) and H18N11 (7) in bats, which are known as reservoirs for zoonotic pathogens (8), demonstrated that these mammals must also be considered with respect to influenza. Crystal structures for the surface glycoproteins hemagglutinin (HA) (7, 9, 10) and neuraminidase (NA) (7, 11, 12) were quickly resolved for these viruses, and, surprisingly, both bat-derived influenza virus proteins were shown to be functionally and structurally distinct from all known HA and NA proteins. Their HA protein did not bind to any sialic acid- or other sugar-containing moiety tested, and their NA did not hydrolyze any substrate containing sialic acids. Therefore, the question was posed whether this influenza-like virus has evolved to bind to other receptors, perhaps specifically adapted to its host (9).
Functional studies of the other proteins (nonglycoproteins) from these bat-derived influenza-like viruses have been limited thus far. It was, however, demonstrated that the ribonucleoprotein complex of H17N10, consisting of PB1, PB2, PA, and NP, is able to transcribe RNA in a minigenome reporter assay and that this activity was abrogated when PB1 was removed from the complex (6). According to previous studies on other influenza virus subtypes, NP binds to viral RNA destined for transcription by PB1 and PB2, which together form the polymerase unit (13). PB1 binds to the C-terminal domain of PA (14, 15), while the N-terminal domain of PA (PAn) can function as an endonuclease to generate small RNA primers that are essential for initiation of the viral gene transcription (16, 17). Recently, it has been shown that both PAn (18) and full-length PA (19) cleave the host-derived RNA in a sequence-specific manner.
Structural and functional studies on any individual subunits in the ribonucleoprotein complex of H17N10 have been lacking so far. Here, we describe the crystal structure of PAn derived from the sequence of the H17N10 and show it to be highly similar to that of PAn from other influenza viruses. Furthermore, we show that H17N10 PAn possesses endonuclease activity, which is dependent on the presence of manganese ions. We also demonstrate that it has activity comparable to that of PAn molecules from avian and human origins. Moreover, the activity of bat-derived influenza-like virus PAn is lost after replacement of the key active residue histidine at position 41 with an alanine. These observations demonstrate the functional competence of H17N10 PAn as a influenza virus endonuclease, indicating the requirement for close monitoring of potential reassorting events involving H17N10 gene segments.
MATERIALS AND METHODS
Multiple alignments and phylogenetic analysis.
Multiple alignments and a neighbor-joining phylogenetic tree were created for the H17N10-derived PAn protein sequence in combination with the protein sequences previously used for crystallization studies of PAn from four different influenza A viruses (16, 17, 20, 21) and from a common influenza B and C virus strain using ClustalW and Espript. The GenBank accession number for A/goose/Guangdong/1/96(H5N1) is YP308666, that for A/Vietnam/1203/2004(H5N1) is ADD97094, that for A/California/04/2009(H1N1) is ACP41104, that for A/Victoria/3/1975(H3N2) is AFM71974, that for B/Memphis/3/1989 is AY582032, and that for C/Ann Arbor/1/1950 is YP089654.
Cloning procedure.
The first 771 nucleotides (nt) of the 5′ part of the PA gene from influenza virus A/little yellow-shouldered bat/Guatemala/060/2010(H17N10) (GenBank accession number CY103891) were synthesized and cloned into pGEX by Generay Biotech Co, Ltd. (Shanghai, China). This construct was used as a template to amplify the first 618 nucleotides of wild-type PAn by PCR with forward primer 5′-GGAATTCCATATGGAAAACTTCGTTCGTACCAACTTCAACCCCATG-3′ and reverse primer 5′-CCGCTCGAGTGAAAATTCCTCCTCCAGTGTTTCTTCTCC-3′ (restriction sites are underlined). Nucleotides C121 and A122 were mutated to G and C, respectively, by overlapping PCR with forward primer 5′-CAAGTTTGCAGCGATATCTACAGCCATGGAGGTATGCT-3′ and reverse primer 5′-AGCATACCTCCATGGCTGTAGATATCGCTGCAAACTTG-3′ (mutated nucleotides are in bold) in combination with the aforementioned primers to create mutant PAn H41A (with a histidine mutation to alanine at amino acid position 41). The first 615 nucleotides of the 5′ part of the PA gene from influenza viruses A/California/4/2009(H1N1) (GenBank accession number FJ966081) and A/great black-headed gull/1/Qinghai/2005(H5N1) (GenBank accession number DQ100553) were both amplified from full-length constructs with forward primer 5′-GGAATTCCATATGGAAGACTTCGTTCGTCAGTGCTTCAACCCGATG-3′ and reverse primer 5′-CCGCTCGAGAAATTTTTCTTCAATTGTCTCTTCGCCTCT-3′ (restriction sites are underlined). Forward primers were generated for optimal codon usage in Escherichia coli using OPTIMIZER at http://genomes.urv.es/OPTIMIZER (22). All genes were cloned into pET21a (Novagen) using the introduced restriction sites NdeI and XhoI and containing a C-terminal 6-His tag for purification purposes.
Protein expression and purification.
Single colonies of Escherichia coli strain BL21(DE3) transformed with the constructs described above were grown overnight at 37°C in LB medium with 100 μg/ml ampicillin. After a 1:100 dilution of the overnight culture into fresh medium, bacteria were grown to an optical density at 600 nm (OD600) of ∼0.6 and expression of PAn proteins was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 to 6 h. During all following steps, materials were kept at 4°C. Cells were harvested by centrifugation for 10 min at 6,000 rpm in a Beckman JA-10 centrifuge. Cell pellets were resuspended in a buffer containing 20 mM Tris-HCl and 50 mM NaCl, pH 8.0. Resuspended cells were sonicated in two rounds of 99 6-s bursts of 400 W with a 12-s interval in a Scientz JY 92-II sonicator using a 1-cm-diameter probe. Inclusion bodies (IBs) were recovered by centrifugation for 20 min at 15,000 rpm in a Beckman JA-25.50 centrifuge. Two detergent washes with a buffer containing 50 mM Tris-HCl, 0.5% Triton X-100, 300 mM NaCl, 10 mM EDTA, and 1 mM dithiothreitol (DTT), pH 8.0, were carried out to remove cell debris and membrane components. Between centrifugation steps of 10 min at 15,000 rpm in a Beckman JA-25.50, the IBs were homogenized in the buffer and sonicated 30 times for 3 s at 200 W with 10-s intervals. Detergent and salt were removed by a similar wash in the same buffer without Triton X-100. Finally, the IBs were solubilized in denaturing buffer (6 M guanidine hydrochloric, 50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10 mM EDTA, 10% [vol/vol] glycerol, and 10 mM DTT) at a final concentration of 30 mg/ml.
Around 90 mg of IBs was renatured by dilution refolding in 500 ml refolding buffer (100 mM Tris-HCl, 2 mM EDTA, 400 mM l-arginine-HCl, 0.5 mM oxidized glutathione, and 5 mM reduced glutathione). The solution containing the refolded proteins was then concentrated using a stirred cell and 10-kDa ultracentrifugal filter devices (both from Millipore) for further purification by fast protein liquid chromatography (FPLC).
Crystallization, data collection, and structure determination.
For crystallization, PAn was purified by size exclusion chromatography, using a calibrated HiLoadTM Superdex 75 16/60 PG column with AKTA FPLC (both from GE Healthcare) and 20 mM Tris (pH 8)–50 mM NaCl buffer. Proteins were concentrated to 5 mg/ml in a buffer of 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 2.5 mM MnCl2. PAn crystals were grown by mixing 1 μl of protein with 1 μl of reservoir solution using the sitting-drop vapor diffusion technique with a reservoir solution (0.1 ml) of 0.1 M sodium chloride, 0.1 M Bis-Tris (pH 6.5), and 1.5 M ammonium sulfate (Hampton Research, Aliso Viejo, CA, USA) at 18°C. The resulting crystals were cryoprotected by soaking in 1 μl well solution mixed with 2 μl 50% polyethylene glycol (PEG) 6000 and then flash-cooled in a cold nitrogen gas stream at 100 K. X-ray diffraction data were collected on Shanghai Synchrotron Radiation Facility (SSRF) beamline 17U, using a MAR 225 charge-coupled device (CCD) detector. The data were processed and scaled using HKL2000 (23). Crystallographic data statistics are shown in Table 1. The structure was solved at 2.7-Å resolution by the molecular replacement method using the structure of PAn from influenza virus A/Victoria/3/1975(H3N2) (17) as the search model (Protein Data Bank [PDB] code 2W69). A starting model was built by PHASER (24). Subsequent refinement was performed using Refmac5 in the CCP4 suite (25) by rigid-body refinement and maximum-likelihood procedures. The structure was then subjected to a series of iterative cycles of manual rebuilding in Coot (26) and refinement with Phenix.refine (27). The PROCHECK (28) program was used to monitor the structural stereochemistry during the course of model building and refinement. All figures were generated by Pymol (http://www.pymol.org/) and Espript (29).
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter | Value for PAn from H17N10a |
|---|---|
| Data collection | |
| Space group | P6122 |
| Cell dimensions | |
| a, b, c (Å) | 74.3, 74.3, 401.159 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50–2.70 (2.80–2.70) |
| Rmerge | 0.112 (0.654) |
| I/σ I | 27.7 (5.5) |
| Completeness (%) | 99.5 (100) |
| Redundancy | 15.6 (16.4) |
| Refinement | |
| Resolution (Å) | 42.8–2.7 |
| No. of reflections | 19,228 |
| Rwork/Rfree | 0.2403/0.2782 |
| No. of atoms | |
| Protein | 4,902 |
| Ligand/ion | 3 |
| Water | 22 |
| B factors | |
| Protein | 62.6 |
| Ligand/ion | 46.9 |
| Water | 60.4 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.551 |
| Ramachandran analysis (%) | |
| Most favored | 86.8 |
| Additional allowed | 8.1 |
| Generally allowed | 5.1 |
| Disallowed | 0 |
Values in parentheses are for the highest-resolution shell.
Endonuclease activity measurements.
For endonuclease assays, PAn was purified using a 5-ml HisTrap HP column (GE Healthcare) with AKTA FPLC by eluting with 50 to 300 mM imidazole in 20 mM Tris (pH 8)–50 mM NaCl buffer. Fractions containing PAn were pooled and dialyzed against 10 mM Tris (pH 8.0)–50 mM NaCl, using 10-kDa filtration devices (Millipore). Collected proteins were concentrated to 5 to 50 mg/ml and subjected to analysis by SDS-PAGE. PAn-specific polypeptides for both the wild-type H17N10 and mutant (H41A) were identified after treatment of the isolated proteins with chymotrypsin and subsequent analysis by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Typically, in a 10-μl reaction volume of reaction buffer consisting of 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 1.5 mM MnCl2, 50 ng/μl of M13mp18 circular single-stranded DNA (ssDNA) (New England BioLabs) was used as a substrate and incubated for 60 min at 37°C in the presence or absence of either the wild-type H17N10, H1N1, H5N1, or mutant H41A PAn molecules (1.25 to 20 μM). The reaction products were loaded on a 0.9% agarose gel and stained with ethidium bromide.
To study RNA degradation, first a DNA template with sequence 5′-taatacgactcactatagggTTGGGGAAGCAGTAATGAGAATGGGAGACCTCCACTCACTCCAAAACAGAAACGGAAAATGGCGAGAAC-3′ with the T7 promoter sequence (lowercase) at the 5′ end was synthesized (the first base of the RNA transcript is in bold). This template was in vitro transcribed with the TranscriptAid T7 high-yield transcription kit (Fermentas number k0441), resulting in the RNA transcript 5′-GGGTTGGGGAAGCAGTAATGAGAATGGGAGACCTCCACTCACTCCAAAACAGAAACGGAAAATGGCGAGAAC-3′. From the 72-nt RNA, 140 ng/ml was incubated with a 12 μM concentration of the different PAn molecules for 15 to 120 min at 37°C in a buffer consisting of 20 mM Tris (pH 8), 50 mM NaCl, 1.5 mM MnCl2, and 10 mM β-mercaptoethanol in a final volume of 10 μl. Reactions were stopped with EDTA (25 mM), and products were subsequently analyzed on a 10% acrylamide–7 M urea gel, followed by silver staining (SilverQuest silver staining kit; Invitrogen catalog no. LC6070).
Protein structure accession number.
The atomic coordinates and structure factors of the H17N10 PAn structure were deposited in the Protein Data Bank (www.pdb.org) under PDB ID code 4NFZ.
RESULTS
PAn from H17N10 is highly similar to PAn from other influenza A viruses.
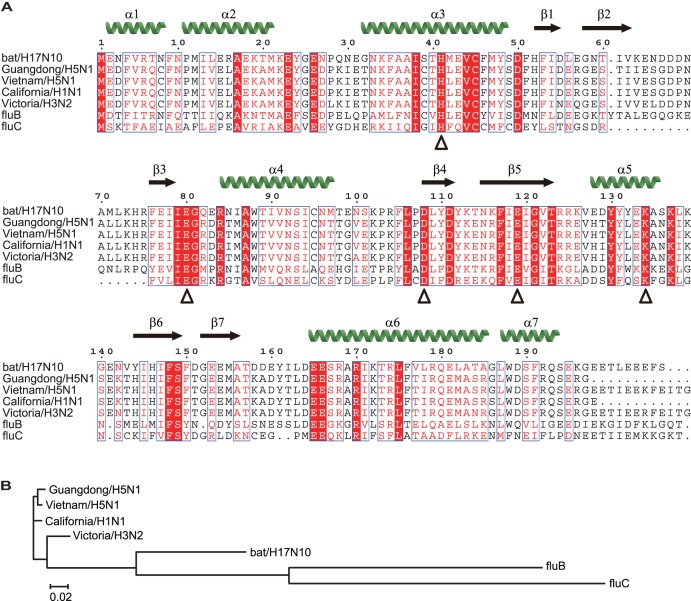
The crystal structures of PAn from A/goose/Guangdong/1/96 (avian H5N1) and A/Victoria/3/1975 (human H3N2) have been described previously (16, 17), as well as those from A/Vietnam/1203/2004 (avian H5N1) and A/California/04/2009 (human pH1N1) in complex with inhibitors (20, 21). Here, we aligned their protein sequence with those of A/yellow-shouldered bat/Guatemala/060/2011 (bat H17N10), B/Memphis/3/1989 (human fluB), and C/Ann Arbor/1/1950 (human fluC) (Fig. 1A). All key active-site residues (H41, E80, D108, E119, and K134) are conserved in PAn from H17N10, suggesting that it may function as an endonuclease like its counterparts from the other influenza viruses. However, it is also interesting to look at the relative distance in the relationship between H17N10 PAn and the other PAn molecules (Fig. 1B). The sequence identity between PAn from the four influenza A viruses is 93 to 98%, while the PAn sequence identity of H17N10 with the four influenza A virus strains is only 71 to 72%. The sequence identity between PAn from influenza B virus and PAn from the four influenza A viruses is much lower (31 to 34%) and is in the same range as the identity with H17N10 PAn (35%). PAn from influenza C virus is at a greater distance, sharing 22 to 28% identity with the PAn from the four influenza A viruses and 21% with H17N10 PAn. Clearly, based on this comparison, H17N10 PAn is evolutionarily distant from PAn of other viruses used in this comparison, justifying experiments to confirm whether it indeed can fold into an active enzyme with endonuclease activity.
FIG 1.
Evolutionary relationship between PAn molecules from several influenza virus strains. (A) Sequence alignment of PAn from influenza virus strains A/yellow-shouldered bat/Guatemala/060/2011 (bat H17N10), A/goose/Guangdong/1/96 (avian H5N1), A/Vietnam/1203/2004 (avian H5N1), A/California/04/2009 (human pH1N1), A/Victoria/3/1975 (human H3N2), B/Memphis/3/1989 (human fluB), and C/Ann Arbor/1/1950 (human fluC). Secondary-structure elements of bat influenza PAn are shown as green α-helices and black β-strands. Residues in a solid red background are identical among the seven sequences. The black triangles indicate key active-site residues. Histidine at position 41 has been mutated to an alanine to create the mutant used in this study. (B) Neighbor-joining phylogenetic tree (without distance corrections) of these strains based on the sequences in panel A, showing their evolutionary relationship with relative distances.
The crystal structure of H17N10 PAn is similar to that of influenza A viruses.
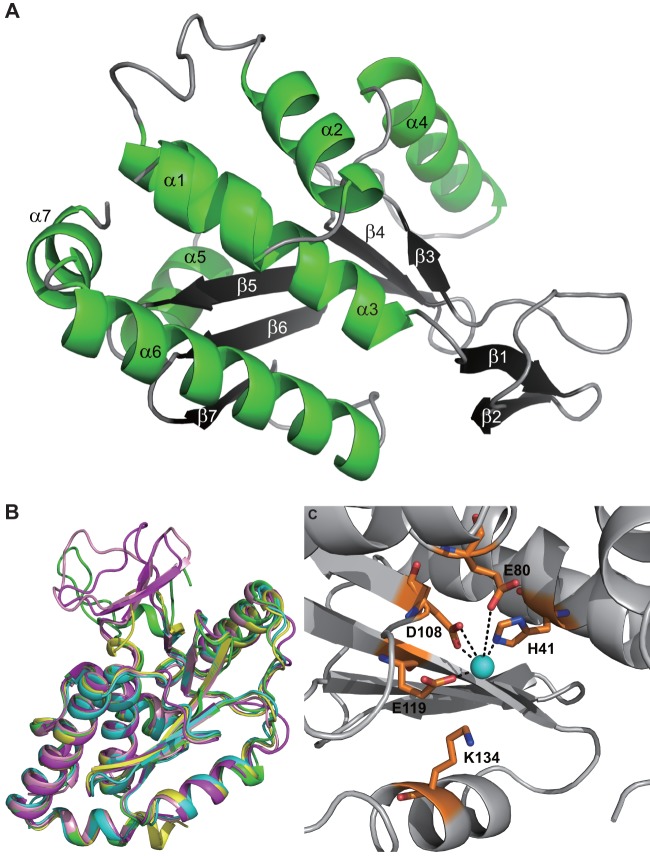
In the solved structure at a resolution of 2.7 Å (detailed statistics are shown in Table 1), three PAn molecules are present in the asymmetric unit. Clear electron densities could be observed for 196 residues (from residue 1 to 196), folding into five α-helices (α1 to α5) and seven β-strands (β1 to β7). Strands β3 to β7 form a center sheet with the helices peripherally aligned around it, together forming an overall globular fold. Two extra strands further decorate the main body structure at the bottom side (Fig. 2A). As expected, this observed fold is highly similar to the structures from the four PAn molecules of influenza A viruses whose structures have been solved and previously published (Fig. 2A). Superimposition of these structures revealed root mean square deviation (RMSD) values ranging from 0.883 to 1.362. Well-aligned structural elements are observed for all the helices and most of the strands and intervening loops, except for the part covering H17N10 PAn residues 49 to 75 (the first two β-strands and the flanking loops), which exhibits a certain conformational variance among different PAn structures (Fig. 2B). The catalytically conserved residues, including H41, E80, D108, E119, and K134, are ordered in close proximity such that they appropriately accommodate a manganese ion in the center (Fig. 2C). These characteristic structural features clearly relate H17N10 PAn to functional endonucleases. Hence, we predicted that H17N10 PAn likely has the capability to cleave RNA and therefore proceeded to examine its functionality.
FIG 2.
Crystal structure of the N-terminal domain of PA from H17N10. (A) Structure of PAn from influenza virus A/yellow-shouldered bat/Guatemala/060/2011 at 2.7 Å with α-helices in green and β-strands in black, labeled as in Fig. 1. (B) Overlay of H17N10 PAn with the four previously described PAn molecules from influenza A virus strains. Coloring is as follows: A/yellow-shouldered bat/Guatemala/060/2011 (bat H17N10) in magenta, A/goose/Guangdong/1/96 (avian H5N1) in cyan. A/Vietnam/1203/2004 (avian H5N1) in yellow, A/California/04/2009 (human pH1N1) in pink, and A/Victoria/3/1975 (human H3N2) in green. (C) The catalytic center of H17N10 PAn, depicting in orange key active-site residues, with N atoms in blue and O atoms in red. Dotted lines indicate interactions of the active-site residues with the manganese ion (cyan sphere).
H17N10 PAn possesses endonuclease activity.
To assess whether the PAn from the H17N10 strain possesses enzyme activity, an endonuclease assay was performed using single-stranded circular DNA as a substrate (17). Incubation with 5 μg of His-trap-purified PAn (20 μM) in the presence of 1.5 mM MnCl2 for 1 h led to almost complete degradation of the substrate, as analyzed on an agarose gel stained with ethidium bromide (Fig. 3A). When PAn and its substrate were incubated in the presence of 20 mM EDTA to chelate divalent cations, the activity was inhibited (Fig. 3). These results indicate that H17N10 PAn depends on the presence of divalent cations such as manganese or magnesium for its activity, as described for PAn from other influenza A viruses (16, 17). We further performed a time course experiment with a 72-nt RNA as the substrate in a similar endonuclease assay. Breakdown of the substrate was nearly complete after 120 min, as analyzed on a 10% acrylamide-urea gel with silver staining (Fig. 3C).
FIG 3.
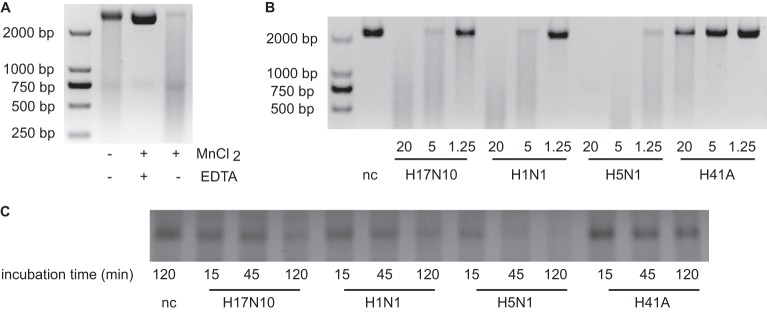
PAn from H17N10 has endonuclease activity. (A) Single-stranded DNA plasmid M13mp18 (50 ng/μl) was incubated with 20 μM wild-type H17N10 PAn for 1 h at 37°C in a buffer consisting of 20 mM Tris (pH 8) and 50 mM NaCl in the absence or presence of 1.5 mM MnCl2 or 20 mM EDTA. Samples were separated by agarose gel electrophoresis, and DNA was subsequently visualized by staining with ethidium bromide. (B) Single-stranded DNA plasmid M13mp18 (50 ng/μl) was incubated for 1 h at 37°C in a buffer with 20 mM Tris (pH 8), 50 mM NaCl, and 1.5 mM MnCl2 with decreasing concentrations (μM) of wild-type H17N10, human H1N1, avian H5N1, and H17N10 mutant H41A PAn molecules. In the negative control (nc), no enzyme was added. (C) A 72-nt RNA (140 ng/μl) was incubated with a 12 μM concentration of the indicated PAn molecules for 15 to 120 min in a buffer consisting of 20 mM Tris (pH 8), 50 mM NaCl, 1.5 mM MnCl2, and 10 mM β-mercaptoethanol. Samples were separated by acrylamide-urea gel electrophoresis, and RNA was subsequently visualized by silver staining. The figures shown are representative of at least two independent experiments.
It has been shown that replacement of the histidine at position 41 by an alanine in PAn from H5N1 leads to inactivation of the enzyme (16). Therefore, this mutation was chosen to create a potential inactive H17N10 PAn. This H41A mutant was expressed and purified by His-trap, as for the wild-type H17N10 PAn. When increasing amounts of the H41A mutant PAn were incubated with the single-stranded DNA, no breakdown of the substrate above background levels was observed, in contrast to the higher breakdown seen with increasing amounts of added wild-type H17N10 PAn (Fig. 3B). In agreement, mutant H41A PAn did not display endonuclease activity on the RNA substrate as seen for wild-type H17N10 PAn (Fig. 3C). These results illustrate that, as with other categorized influenza virus PAn, the histidine located at position 41 in the active site is indispensable for the endonuclease activity of H17N10-derived PAn.
Finally, we compared the activity of H17N10 PAn with those of PAn from the pandemic A/California/4/2009(H1N1) influenza virus and PAn from the avian A/great black-headed gull/1/Qinghai/2005(H5N1) influenza virus in both endonuclease assays with ssDNA (Fig. 3B) or RNA (Fig. 3C) as the substrate. The avian H5N1 PAn appeared to be more active than the H17N10 under the conditions tested, whereas the activity of H1N1 PAn was in the same range as that of H17N10 PAn.
DISCUSSION
The protein sequence of PAn from influenza-like virus H17N10 has a high homology with the that of crystallized PAn molecules from several other influenza A viruses, and the key active residues are all conserved. This suggests that H17N10 PAn is most likely able to generate the short primers needed for the initiation of viral replication via its endonuclease activity. However, for the glycoproteins from H17N10, HA and NA, no canonical function could be demonstrated, despite their structural similarities to previously characterized HA and NA molecules (9–12). Therefore, it was interesting to determine whether PAn from H17N10 would indeed have a function similar to that of categorized influenza virus PAn molecules. In this study, the structure of H17N10 PAn was solved, revealing a protein fold that is highly similar to those described previously, such as those in the PAn structures from viruses A/goose/Guangdong/1/96(H5N1), A/Vietnam/1203/2004(H5N1), A/California/04/2009(H1N1), and A/Victoria/3/1975(H3N2) (16, 17, 20, 21).
The endonuclease assays conducted in this work show that H17N10 PAn possesses this activity. The H41A mutant was inactivated, showing an essential role for the histidine in this enzyme, as described for H5N1 PAn (16, 30). This histidine residue is in vivo very likely to contribute to binding to both the ribose and the nucleoside of the substrate RNA (30). Besides the key active-site residues (Fig. 1A), which are conserved among all PAn molecules from different strains, several other amino acids proposed to be important for interaction with the substrate (30) are conserved as well, namely, Ala17, Ala37, Ile38, Tyr130, and Lys137. Apparently, the amino acid substitutions A20T, L42M, C39S, and R196K in H17N10 PAn are not detrimental for its endonuclease activity, although these residues are conserved among the PAn molecules from the four influenza A viruses that have been characterized thus far and were proposed to be involved in substrate binding (30).
In the work describing the H17N10 influenza virus subtype for the first time, the function of its ribonucleoprotein complex was examined (6). The results from these experiments indicated that human cells are compatible with the functions of the H17N10 ribonucleoprotein complex and that the H17N10 polymerase complex is compatible with human virus NCRs (6). Our work provides both structural and functional evidence for an H17N10 PAn that exhibits endonuclease activities similar to that of PAn molecules derived from human and avian influenza viruses. This indicates that the replication mechanism in the H17N10 subtype is likely similar to that for the more familiar influenza A viruses, with PAn generating primers to initiate the replication by the PB1/PB2 complex, to which PA is bound via its C-terminal domain. Although this work contributes to understanding the life cycle of H17N10, several intriguing questions regarding its infection route still remain. Which receptor is employed by HA for attachment and entry into the bat host cell? Is NA able to degrade the virus-receptor complex upon exit of the virions, similar to the case for the sialic acid-HA complexes in other influenza viruses? Bat-derived cells seem to be a prerequisite to study these phenomena, so the recently described Pteropusalecto kidney cells seem to be an interesting tool in that respect, as they are susceptible to infection by several influenza viruses and reassortment (31). Future studies are needed to fully investigate the compatibility of this and other intriguing influenza-like viruses found in bats with categorized influenza viruses.
ACKNOWLEDGMENTS
B.T. was supported by a Chinese Academy of Sciences Fellowship for Young International Scientists (grant no. 2012Y1SB0010). G.F.G. is a leading principal investigator of the innovative research group of the National Natural Science Foundation of China (NSFC) (grant no. 81321063). Work in G.F.G.'s laboratory is partially supported by the 973 Project of the Ministry of Science and Technology of China (MOST) (grant no. 2011CB504703).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Shutao Sun (MS core facility, IMCAS) for the analysis of the PAn proteins by mass spectrometry. We acknowledge assistance by the staff (especially Jianhua He and Sheng Huang) at the Shanghai Synchrotron Radiation Facility (SSRF beamline BL17U).
B.T. performed the experiments and wrote the manuscript; G.L. and Y.Z. assisted with protein expression, purification, and crystallization experiments and edited the manuscript; J.H. discussed the experiments and edited the manuscript; J.Q. performed the crystal data collection and solved the crystal structure; L.Z. and T.D. facilitated and discussed the RNA degradation experiments; G.F.G. conceived and supervised the research and edited the manuscript.
We declare no competing financial interests.
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Runstadler J, Hill N, Hussein IT, Puryear W, Keogh M. 2013. Connecting the study of wild influenza with the potential for pandemic disease. Infect. Genet. Evol. 17:162–187. 10.1016/j.meegid.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. 10.1016/S0140-6736(13)60938-1 [DOI] [PubMed] [Google Scholar]
- 4.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza a viruses. PLoS Pathog. 9:e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, Mills JN, Timonin ME, Willis CK, Cunningham AA, Fooks AR, Rupprecht CE, Wood JL, Webb CT. 2013. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 280:20122753. 10.1098/rspb.2012.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Shi Y, Lu X, He J, Gao F, Yan J, Qi J, Gao GF. 2013. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep. 3:769–778. 10.1016/j.celrep.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 10.Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. 2013. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. U. S. A. 110:1458–1463. 10.1073/pnas.1218509110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Sun X, Li Z, Liu Y, Vavricka CJ, Qi J, Gao GF. 2012. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 109:18897–18902. 10.1073/pnas.1211037109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, Stevens J, Wilson IA. 2012. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc. Natl. Acad. Sci. U. S. A. 109:18903–18908. 10.1073/pnas.1212579109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JR, Nagata K, Park SY. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803–1811. 10.1038/emboj.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131. 10.1038/nature07225 [DOI] [PubMed] [Google Scholar]
- 15.He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126. 10.1038/nature07120 [DOI] [PubMed] [Google Scholar]
- 16.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913. 10.1038/nature07720 [DOI] [PubMed] [Google Scholar]
- 17.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. 10.1038/nature07745 [DOI] [PubMed] [Google Scholar]
- 18.Datta K, Wolkerstorfer A, Szolar OH, Cusack S, Klumpp K. 2013. Characterization of PA-N terminal domain of influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 41:8289–8299. 10.1093/nar/gkt603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble E, Cox A, Deval J, Kim B. 2012. Endonuclease substrate selectivity characterized with full-length PA of influenza A virus polymerase. Virology 433:27–34. 10.1016/j.virol.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OH, Ruigrok RW, Cusack S. 2012. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8:e1002831. 10.1371/journal.ppat.1002831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ, Webb TR, White SW. 2012. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8:e1002830. 10.1371/journal.ppat.1002830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. 2007. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35:W126–131. 10.1093/nar/gkm219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- 29.Gouet P, Courcelle E, Stuart DI, Metoz F. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308. 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- 30.Zhao C, Lou Z, Guo Y, Ma M, Chen Y, Liang S, Zhang L, Chen S, Li X, Liu Y, Bartlam M, Rao Z. 2009. Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center. J. Virol. 83:9024–9030. 10.1128/JVI.00911-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dlugolenski D, Jones L, Tompkins SM, Crameri G, Wang LF, Tripp RA. 2013. Bat cells from Pteropus alecto are susceptible to influenza A virus infection and reassortment. Influenza Other Respir. Viruses 7:900–903. 10.1111/irv.12128 [DOI] [PMC free article] [PubMed] [Google Scholar]