Abstract
Simian hemorrhagic fever virus (SHFV) causes a fatal hemorrhagic fever in macaques but an asymptomatic, persistent infection in baboons. To investigate factors contributing to this differential infection outcome, the targets of SHFV infection, macrophages (MΦs) and myeloid dendritic cells (mDCs), were differentiated from macaque and baboon peripheral blood monocytes and used to compare viral replication and cell responses. SHFV replicated in >90% of macaque MΦs but in only ∼10% of baboon MΦs. Although SHFV infected ∼50% of macaque and baboon mDCs, virus replication was efficient in macaque but not in baboon mDCs. Both types of macaque cultures produced higher virus yields than baboon cultures. A more efficient type I interferon response and the production of proinflammatory cytokines, including interleukin-1β (IL-1β), IL-6, IL-12/23(p40), tumor necrosis factor alpha (TNF-α), and macrophage inflammatory protein 1α (MIP-1α), in response to SHFV infection were observed in macaque but not baboon cultures, suggesting less efficient counteraction of these responses by viral proteins in macaque cells. Baboon cultures produced higher levels of IL-10 than macaque cultures both prior to and after SHFV infection. In baboon but not macaque cell cultures, SHFV infection upregulated IL-10R1, a subunit of the IL-10 receptor (IL-10R), and also SOCS3, a negative regulator of proinflammatory cytokine production. Incubation of macaque cultures with human IL-10 before and/or after SHFV infection decreased production of IL-6, IL-1β, and MIP-1α but not TNF-α, suggesting a role for IL-10 in suppressing SHFV-induced proinflammatory cytokine production in macaques.
INTRODUCTION
Simian hemorrhagic fever virus (SHFV) was isolated in 1964 as the causative agent of outbreaks of a fatal hemorrhagic fever disease in macaque colonies in the United States, Russia, and Europe (1, 2). These SHFV outbreaks are thought to have been initiated by accidental transmission of SHFV present in the blood of a persistently infected, asymptomatic African nonhuman primate (NHP) to a disease-susceptible macaque (3). It was subsequently estimated that 1 to 10% of wild-caught African NHPs, such as patas (Erythrocebus patas), baboons (Papio papio), and African greens (Cercopithecus aethiops), are persistently infected with SHFV (3).
SHFV infections in macaques are characterized by mortality approaching 100% by 3 weeks. Initial symptoms include fever and facial edema followed by depression, anorexia, and dehydration and, finally, signs of coagulation defects, such as skin petechiae, epitaxis, melena, retrobulbar hemorrhages, and subcutaneous hematomas, appear (4, 5). Necropsy reveals hemorrhage in the spleen and necrosis of the liver, adrenal glands, and lymphatic tissue (4). The symptoms of an SHFV infection closely resemble those induced in macaques by infection with other hemorrhagic fever viruses, such as Ebola, Marburg, and Lassa (6, 7). In response to infection with these viruses, macrophages (MΦs) and dendritic cells (DCs) release proinflammatory cytokines that induce tissue factor and subsequent disseminated intravascular coagulopathy (7–9). Marked impairment of the adaptive immune response due to impaired DC function and lymphocyte apoptosis occurs as a consequence of Ebola virus infection (7).
SHFV is a member of the family Arteriviridae that also includes equine arteritis virus (EAV), porcine reproduction and respiratory syndrome virus (PRRSV), and lactate dehydrogenase-elevating virus (LDV). Arterivirus genomes are polycistronic, single-stranded, positive-sense RNAs with a 5′ type I cap and a 3′ poly(A) tail (10). The SHFV genome is 15.7 kb in length. Arteriviruses have highly restricted host ranges and cell tropisms. Only MΦs and DCs are infected in horses and donkeys by EAV, in pigs by PRRSV, in mice by LDV, or in NHPs by SHFV (11). Both EAV and PRRSV infections can cause disease symptoms, including fever, anorexia, tissue necrosis, inflammation of the respiratory tract, spontaneous abortions, or delivery of weak offspring (10). LDV typically causes asymptomatic, lifelong, persistent infections (10). Due to the significant agricultural impact of the diseases caused by EAV and PRRSV, the majority of the research done on arterivirus infections has been focused on these two viruses.
SHFV replication and virus-induced cytokine production in primary MΦs and mDCs from disease-resistant baboons and disease-susceptible macaques were compared. Although viral replication was efficient in both macaque and baboon MΦs, a majority of macaque MΦs but only ∼10% of baboon MΦs were infected. In contrast, similar numbers of macaque and baboon myeloid DCs (mDCs) were infected by SHFV, but virus replication was less efficient in the baboon cells. Both types of macaque cultures produced higher virus yields than the corresponding baboon cultures. Proinflammatory cytokines were produced in response to SHFV infection by both types of macaque cells but not by baboon cells. Interleukin-10 (IL-10) was detected in the culture fluids of both uninfected and infected baboon cells. SHFV infection of baboon but not macaque cells resulted in the upregulation of IL-10R1, a subunit of the IL-10 receptor (IL-10R) and SOCS3 (suppressor of cytokine signaling 3), a negative regulator of cytokine production. Incubation of infected macaque mDCs or MΦs with recombinant human IL-10 (rhIL-10) resulted in decreased production of IL-6, IL-1β, and macrophage inflammatory protein 1α (MIP-1α) but not tumor necrosis factor alpha (TNF-α). These data suggest that IL-10 may contribute to suppressing proinflammatory cytokine production in response to SHFV infection.
MATERIALS AND METHODS
Cells.
Blood was obtained from baboons (Southwest National Primate Research Center, San Antonio, TX) or rhesus macaques (Yerkes Regional Primate Research Center, Atlanta, GA) under approved IACUC protocols that covered tissue sharing by each institution. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Ficoll 400 (Mediatech, Inc., Manassas VA) density gradient centrifugation according to standard protocols. Monocytes were seeded at 106 cells/well in a 24-well plate or at 106 cells/well on an eight-chamber slide and allowed to adhere for 2 h before a gentle washing with Hanks buffered saline solution (HBSS; Gibco). Immature mDCs were cultured from adherent cells by incubation with RPMI 1640 culture medium (Gibco) supplemented with 10% autologous serum or 10% fetal bovine serum (FBS), 50 U/ml of penicillin, 50 μg/ml of streptomycin, human recombinant granulocyte-macrophage colony-stimulating factor (1,000 U/ml; R&D Systems), and recombinant human interleukin-4 (500 U/ml) for 11 days at 37°C in a 5% CO2 atmosphere. MΦs were cultured from adherent cells by incubation with RPMI 1640 culture medium supplemented with 10% autologous serum or 10% FBS, 50 U/ml of penicillin, 50 μg/ml of streptomycin, and human recombinant macrophage colony-stimulating factor (5,000 U/ml; R&D Systems) for 11 days at 37°C in a 5% CO2 atmosphere. Two-thirds of the culture medium was replaced with fresh growth medium every 3 days to replenish growth factors. Cells were stained with fluorescently labeled antibodies directed against the surface markers HLA-DR, DC-SIGN, CD11c, and CD83 for mDCs and CD91 and CD163 for MΦs (BD Bioscience). Cell identity and numbers were analyzed using a FACSCanto flow cytometer and FACSDiva software (BD Bioscience). Ninety percent or more of cells in the mDC cultures were HLA-DR-positive (HLA-DR+) DC-SIGN+ CD11c+ CD83−, and >95% of cells in the MΦs cultures were CD91+ CD163+. All baboon blood samples were assayed using an SHFV-specific reverse transcription-PCR (RT-PCR) assay to detect samples from persistently infected animals. Cells from virus-positive baboons were not used for experiments.
MA104 cells were obtained from O. Nainin, Centers for Disease Control and Prevention, and grown in minimum essential medium (MEM; Gibco) supplemented with 10% FBS, 1% l-glutamine, and 1% gentamicin at 37°C in a 5% CO2 atmosphere.
Virus.
SHFV, strain LVR 42−0/M6941 (SHFV-LVR), obtained from the American Type Culture Collection, was sequentially plaque purified three times and then amplified once on MA104 cell monolayers. Pools of SHFV-LVR were prepared by infecting confluent MA104 monolayers at a multiplicity of infection (MOI) of 0.2, harvesting culture fluid at 32 h after infection, clarifying the culture fluid by centrifugation, and freezing aliquots at −80°C. Virus pools contained ∼107 PFU/ml. Primary cell cultures were infected at an MOI of 1 or 10. After virus adsorption for 1 h at 37°C in a 5% CO2 atmosphere, the inoculum was removed, and cells were washed three times with Hank's balanced salt solution (HBSS) and then incubated with cell-specific medium at 37°C in a 5% CO2 atmosphere.
Plaque assays were performed on confluent monolayers of MA104 cells in six-well plates. After adsorption for 1 h at room temperature, the virus inoculum was removed, and the wells were overlaid with 1% SeaKem ME agarose (Bio-Whittaker Molecular Applications) mixed 1:1 with 2× MEM containing 5% fetal calf serum (FCS) and incubated at 37°C for 72 h. After removal of the agarose, the cells were stained with 0.05% crystal violet in 10% ethanol. Each virus dilution was assayed in duplicate.
Preparation of UV-inactivated virus.
An aliquot of SHFV was inactivated by exposure to UV light (4.75 J/cm2) for 10 min at a distance of 1 cm. The complete loss of detectable infectivity after UV exposure was confirmed by plaque assay.
Quantification of secreted cytokines.
Culture fluid was collected from mock-infected and SHFV-infected cultures at various times after infection. Proinflammatory cytokine levels were quantified using a Luminex microsphere-based enzyme-linked immunosorbent assay (ELISA) as previously described (12). Briefly, anti-human cytokine polyclonal antibodies were conjugated to polystyrene xMAP microspheres using an xMAP antibody coupling kit (Luminex). A total of 500 coupled microspheres per cytokine were incubated at 4°C for 16 h with 25 μl of culture fluid or phosphate-buffered saline (PBS), pH 7.4 (Sigma), containing 1% bovine serum albumin (BSA) and a nonhuman primate (NHP) cytokine standard (alpha interferon [IFN-α] from Mabtech, IL-10 from BioLegend, and the remaining cytokines from Millipore) diluted to concentrations ranging from 400 pg/ml to 0.128 pg/ml. The microspheres were then washed three times in PBS–1% BSA, pH 7.4, and incubated with a biotinylated anti-human antibody for each cytokine (Table 1) at 25°C for 1 h. Microspheres were washed three times in PBS–1% BSA, pH 7.4, incubated with 5 μg/ml streptavidin–R-phycoerythrin (Sigma) at 25°C for 30 min, washed three times in PBS–1% BSA, pH 7.4, resuspended in Luminex xMAP systems fluid, and analyzed on a Luminex 100 analyzer (Qiagen). At least 100 events were measured for each cytokine. Alternatively, proinflammatory cytokines in culture fluids were screened using a 14-plex nonhuman primate cytokine Milliplex panel (Millipore) according to the manufacturer's protocol. Although absolute cytokine levels varied between animals, a similar effect of SHFV infection was observed in all cultures of a particular type from one of the animal species tested. The data obtained from cultures prepared from different animals of the same species were averaged and are shown as the fold change compared to data from time-matched control cultures.
TABLE 1.
Antibody clones used in the Luminex ELISAs
| Cytokine | Capture antibody | Biotinylated detection antibody |
|---|---|---|
| TNF-α | MAb1a | MAb11a |
| IFNα | MT1-3-5b | MT2/4/6b |
| IL-1β | JK1B-1a | JK1B-2a |
| IL-6 | MQ2-13A5a | MQ2-39C3a |
| IL-10 | JES3-9D7a | JES3-12G8a |
| IL-12/23(p40) | MT86/221b | MT618b |
| MIP-1α | AF-270-NAc | BAF270c |
| MIP-1β | AF-271-NAc | BAF271c |
| RANTES | AF-278-NAc | BAF-278c |
Antibody purchased from BioLegend, San Diego, CA.
Antibody purchased from Mabtech, Inc., Mariemont, OH.
Antibody purchased from R&D Systems, Inc., Minneapolis, MN.
qRT-PCR.
Total cellular RNA from SHFV-infected or mock-infected primary cells was isolated using an RNeasy Minikit (Qiagen) according to the manufacturer's protocol. Specific primer mixes and TaqMan minor groove binder (MGB) probes were used to detect rhesus macaque IFN-β mRNA (IFN-β Rh03648734_s1) and rhesus macaque IL-10 mRNA (IL-10 Rh02621709_m1) (Applied Biosystems). Quantitative real-time RT-PCR (qRT-PCR) was performed for each target gene mRNA and for the endogenous control (eukaryotic 18S rRNA VIC/TAMRA dye-labeled) (Applied Biosystems) in a single-plex format with 100 ng of cellular RNA and a TaqMan one-step RT-PCR Master Mix reagent kit (Applied Biosystems). The cycling parameters were 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and then 60°C for 1 min on an Applied Biosystems 7500 sequence detection system. Multiple independent experiments were performed, and each sample was assayed in triplicate. The triplicate threshold cycle (CT) values were analyzed with Microsoft Excel using the comparative CT (ΔΔCT) method of the SDS Applied Biosystems software, which also applied statistical analysis to the data (TINV test in Microsoft Excel). The values were normalized to those for 18S rRNA in the same sample and are presented as the relative fold change compared to the mock-infected 24-h calibrator sample value in relative quantification units (RQUs). Error bars represent the standard error of the mean (13) and indicate the calculated minimum (RQmin) and maximum (RQmax) of the mRNA expression levels based on an RQmin/max of the 95% confidence level.
SHFV genomic RNA in infected cell lysates was quantified by real-time RT-PCR. A primer-probe set targeting the nsp9 (helicase) region of the SHFV genome was designed from the sequence of strain LVR (GenBank accession number AF180391.1). The primer and probe sequences used were as follows: forward primer, 5′-CGTACACCCGCCGTCTCT-3′; TaqMan probe, 5′-6FAM-TTGACGTTCTCACAAAGG-MGBNFQ-3′ (where 6FAM is 6-carboxyfluorescein and MGBNFQ is minor groove binder nonfluorescent quencher); and reverse primer, 5′-CGGCAAGTGGCATCCAA-3′. The reaction mixture contained 200 ng of cellular RNA, the primer pair (1 μM), and the probe (0.2 μM) in a total volume of 20 μl. Intracellular genomic SHFV RNA was quantified using a standard curve generated with serial dilutions of a known concentration of in vitro synthesized SHFV RNA. The in vitro synthesized SHFV RNA was transcribed with an SP6 mMessage mMachine kit (Ambion) from an SHFV nsp9 cDNA template; the DNA template was digested with Turbo DNase at 37°C for 5 min, and the product RNA was purified with lithium chloride, precipitated with ethanol, washed with 70% ethanol, resuspended in RNase-free water, and quantified by UV spectrophotometry.
Confocal microscopy.
Cultures of MΦs or mDCs grown in eight-chamber slides were mock infected or infected with SHFV-LVR at an MOI of 10. At various times after infection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 5 min, permeabilized with 0.01% Triton-X in PBS for 5 min, and blocked in 5% horse serum overnight at 4°C. Cells were incubated with murine anti-double-stranded RNA (dsRNA) antibody, washed with PBS, and then incubated with Alexa Fluor 594-conjugated rabbit anti-mouse IgG (Invitrogen) and Hoechst 33342 nuclear stain (0.5 μg/ml; Invitrogen). Cells were washed with PBS before a coverslip was mounted using ProLong Gold antifade reagent (Invitrogen). Cells were visualized with a 40× oil immersion objection on a LSM700 laser scanning confocal microscope (Zeiss), and images were analyzed with Zeiss LSM software, version 4.2.
Western blotting.
Cell lysates were collected in radioimmunoprecipitation assay (RIPA) buffer (1×PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing Halt protease inhibitor cocktail (Thermo Scientific). Following separation by SDS-PAGE, cell proteins were electrophoretically transferred to a nitrocellulose membrane. Membranes were blocked with phosphate buffered saline containing either 5% bovine serum albumin or 5% nonfat dry milk and 0.1% Tween 20 before incubation with either anti-SOCS3 (L210) antibody (Cell Signaling Technology) or anti-IL-10R1 antibody (clone F39; Biolegend) in the presence of blocking buffer. Actin was used as a loading control and was detected with anti-actin antibody (C-11) (Santa Cruz Biotechnology). Blots were washed, incubated with anti-sheep or anti-mouse horseradish peroxidase-conjugated antibody (Santa Cruz), washed again, and processed for chemiluminescence using a SuperSignal West Pico detection kit (Pierce Scientific) according to the manufacturer's protocol.
RESULTS
Analysis of viral replication kinetics in primary NHP cell cultures.
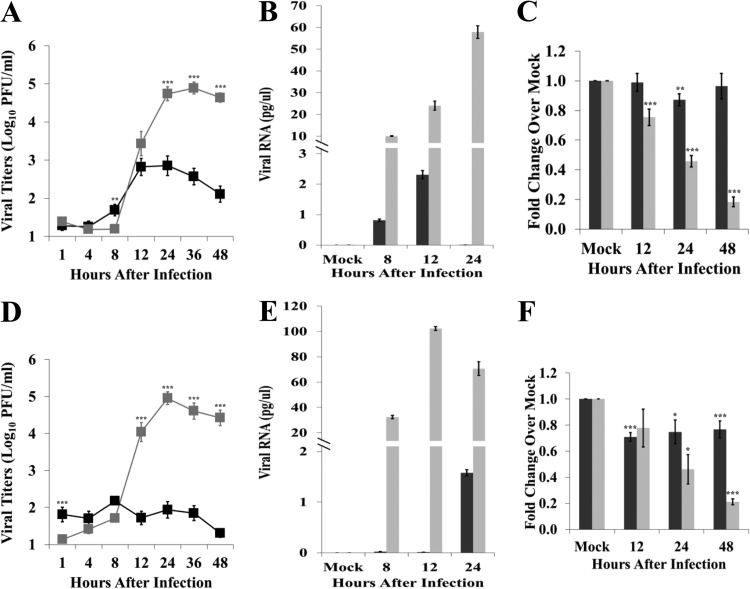
Although it was previously reported that peak SHFV yields produced by primary cultures of MΦs from disease-susceptible macaques were 10-fold higher than those from disease-resistant patas (14), the numbers of infected MΦs in these two types of cultures were not determined, and viral replication in mDCs was not analyzed. MΦs and mDCs were separately cultured from PBMCs isolated from baboon and rhesus macaque whole-blood samples as described in Materials and Methods. Comparable numbers of cells were generated in the cultures prepared from the two types of NHPs (data not shown). The cells were infected with SHFV at an MOI of 1. Viral titers in culture fluids were determined by plaque assay on MA104 cells. Macaque MΦ cultures produced a peak titer of 8 × 104 PFU/ml by 24 h after infection while baboon MΦ cultures produced a peak titer of 7 × 102 PFU/ml by 12 h after infection (Fig. 1A). The levels of intracellular viral RNA were analyzed by qRT-PCR. Intracellular viral RNA levels were higher in macaque MΦs than in baboon MΦs at all times analyzed (Fig. 1B). Macaque mDCs produced a peak titer of 9 × 104 PFU/ml by 24 h after infection while baboon mDCs produced a peak titer of 2 × 102 PFU/ml by 8 h after infection (Fig. 1D). Intracellular viral RNA levels were higher in macaque mDCs than in baboon mDCs at all times analyzed (Fig. 1E). The detection of an increase in intracellular SHFV plus-strand RNA at 24 h confirmed that a low level of virus replication was occurring in the baboon DCs. An MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay was performed to assess the effect of SHFV infection on cell viability. Macaque MΦ viability decreased by 25% at 12 h and continued to decrease with time after infection while baboon MΦ viability decreased by 13% at 24 h and did not decrease further by 48 h (Fig. 1C). Macaque mDCs also showed a progressive decline in viability with time after infection. Baboon mDC viability decreased by ∼25% at 12 h but did not decrease further through 48 h (Fig. 1F). These results indicate that SHFV replicates more efficiently in both types of macaque cells and significantly decreases macaque cell viability.
FIG 1.
Kinetics of SHFV replication in primary cells isolated from disease-resistant and disease-susceptible primates. MΦs (A, B, and C) and mDCs (D, E, and F) isolated from baboon (black) or macaque (gray) PBMCs were mock infected or infected with SHFV-LVR at an MOI of 1. (A and D) Culture fluid was collected at the indicated times after infection, and viral yields were quantified by plaque assay on MA104 cells. Each data point is the average of duplicate titrations done on samples collected from cells isolated from 14 baboons or 28 macaques. (B and E) Total cellular RNA was isolated from cell lysates prepared at the indicated times after infection, and the amount of viral genome RNA was quantified by qRT-PCR using a helicase region primer and probes as described in Materials and Methods. Each sample was assayed in triplicate. Values were first normalized to the level of the 18S rRNA in the same sample, and then the amount of viral RNA was determined by comparison to a standard curve generated with a known amount of in vitro synthesized viral RNA. Error bars represent the standard error of mean (n = 6). (C and F) Cell viability was determined by MTT assay. Values shown are averages from six independent experiments. Each sample was assayed in triplicate. Error bars indicate the standard error of mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
Analysis of the number of productively infected cells.
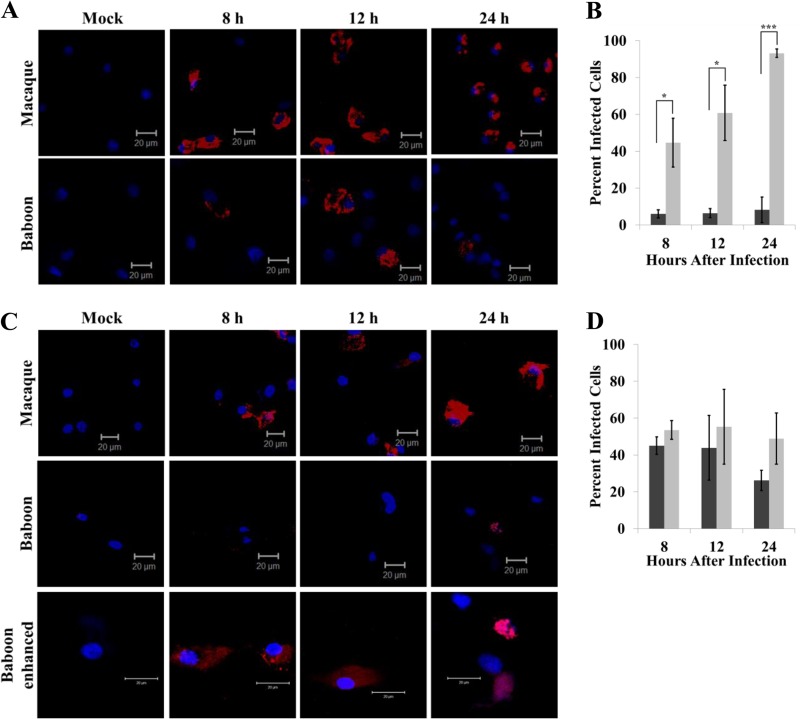
The differential viral production and cell viability observed between the macaque and baboon cells could be due to differences in virus replication efficiencies and/or to differences in the numbers of cells infected. Cell cultures were infected with SHFV at an MOI of 10. The number of infected cells and the relative intensity of intracellular viral dsRNA in the infected cells were assessed by immunofluorescence. A higher MOI was used for these experiments to ensure that a maximum number of infected cells was obtained and to enhance the detection of the weaker dsRNA signal in the infected baboon mDCs. Perinuclear dsRNA staining was observed in the majority of macaque MΦ, the number of infected cells was observed to increase with time after infection, and cell rounding indicative of cytopathology was detected by 24 h (Fig. 2A and B). In contrast, dsRNA was detected in only ∼5 to 10% of the baboon MΦs, but the dsRNA signal was as strong as in the infected macaque MΦs. The data suggest that SHFV is able to infect and replicate efficiently in only a subset of the baboon MΦs while the majority of macaque MΦs are permissive to SHFV infection. Similar percentages of infected baboon and macaque mDCs were detected at 8 and 12 h after infection (Fig. 2D). However, the intensity of the dsRNA staining was much higher in macaque mDCs than in baboon mDCs (Fig. 2C). The data indicate that although similar numbers of baboon and macaque mDCs were susceptible to infection, virus replication was less efficient in baboon mDCs than in macaque mDCs. Interestingly, the number of baboon DCs with detectable dsRNA staining decreased by 24 h even though no reduction in cell number was observed at this time (data not shown), suggesting the possibility that virus replication decreases with time after infection in these cells.
FIG 2.
Number of primary NHP cells infected by SHFV. MΦs (A) and mDCs (C) cultured from baboons or macaques were mock infected or infected with SHFV at an MOI of 10. Cells were fixed at the indicated times after infection, permeabilized, stained with anti-dsRNA antibody, and visualized by laser scanning confocal microscopy. Images shown are representative of three independent experiments. The same microscope settings were used to obtain all of the images compared. The percentages of MΦs (B) and mDCs (D) containing viral dsRNA were determined. The total number of cells and the number of infected cells were counted in each of four fields for each sample. Black bars, baboon; gray bars, macaque. The values are averages obtained from cells isolated from three macaques and three baboons. Error bars indicate the standard error of mean. *, P ≤ 0.05; ***, P ≤ 0.005. Both the intensity of the signal and the image size were increased for the last row of images.
Analysis of type I IFN induction in SHFV-infected NHP cell cultures.
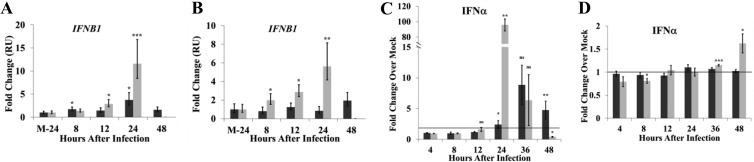
Several PRRSV proteins are able to suppress type I interferon (IFN) production and signaling (15–17). To analyze the kinetics of the type I IFN response to SHFV by disease-resistant and disease-susceptible cell cultures, primary MΦ and mDC cultures were mock infected or infected with SHFV at an MOI of 1, and intracellular IFN-β mRNA levels were quantified by qRT-PCR at different times after infection. IFN-β mRNA levels in macaque MΦs increased 3-fold by 12 h and 11.6-fold by 24 h after infection (Fig. 3A). In baboon MΦs, IFN-β mRNA levels increased 1.7-fold by 8 h and 3.7-fold by 24 h after infection (Fig. 3A). IFN-β mRNA levels in SHFV-infected macaque mDCs progressively increased between 8 and 24 h after infection while in baboon mDCs the levels did not increase significantly (Fig. 3B).
FIG 3.
Induction of type I interferon in SHFV-infected primary NHP cell cultures. Cultured MΦs and mDCs were mock infected or infected with SHFV at an MOI of 1. Total cell RNA was isolated from MΦs (A) or mDCs (B) and used to assay IFN-β mRNA levels by real-time qRT-PCR. Each sample was assayed in triplicate. Values were normalized to the level of 18S rRNA in the same sample. The values represent fold change over the amount of IFN-β mRNA in 24-h mock-infected (M-24) samples expressed as relative quantification units (RUs) and are the average of seven independent experiments. The error bars represent the calculated standard error of mean and are based on an RQmin/max of the 95% confidence level. Culture fluids from MΦ (C) or mDC (D) cultures were analyzed for IFN-α protein by a multiplexed ELISA. The values are average fold change in IFN-α levels in SHFV-infected cell culture fluids compared to those in an autologous, time-matched, mock-infected culture fluid. Values shown are averages from cultures prepared from four baboons (black bars) and four macaques (gray bars). Error bars represent the standard error of mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ns, not significant.
IFN-α protein in culture fluids was quantified by ELISA. IFN-α levels in macaque MΦ culture fluids increased 96-fold compared to those from time-matched, autologous mock-infected cultures at 24 h and then decreased (Fig. 3C). In baboon MΦ culture fluids, IFN-α levels were lower, and the peak level was not observed until 36 h (Fig. 3C). IFN-α levels increased in macaque mDC culture fluids at 36 and 48 h after infection, but no increase in IFN-α levels was observed in baboon mDC culture fluids at any time analyzed (Fig. 3D). These data indicate that SHFV infection efficiently upregulated IFN-β gene expression and extracellular IFN-α protein in both macaque mDCs and MΦs while infected baboon MΦs showed a lower type I IFN response, and baboon mDCs did not produce a detectable response.
Analysis of proinflammatory cytokine production by SHFV-infected primary cell cultures.
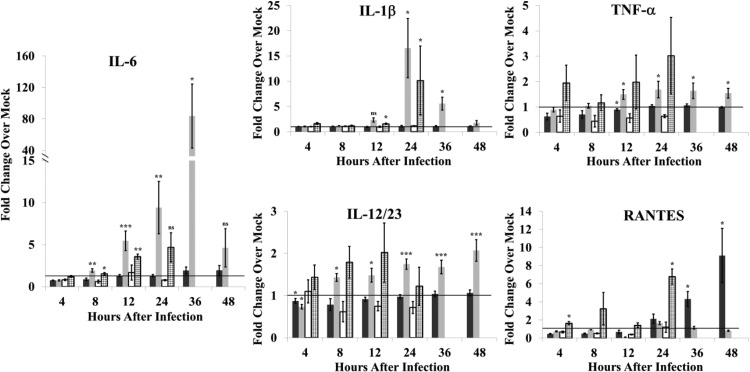
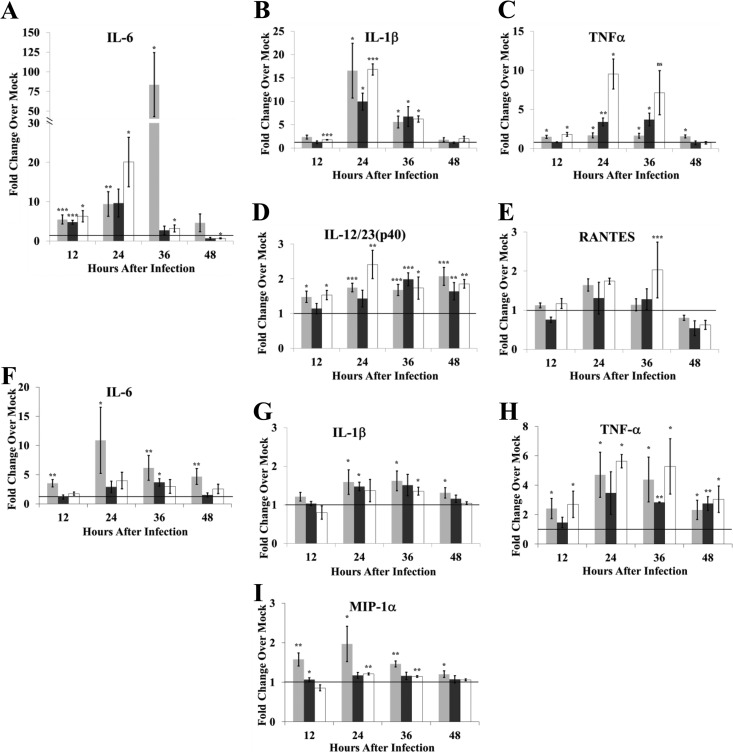
Previous studies showed that infection of primates with either Ebola virus or Lassa virus induced the production of several proinflammatory cytokines, including TNF-α, IL-6, IL-8, IL-1β, MIP-1α, and MIP-1β (18, 19). Upregulation of proinflammatory cytokines by other arteriviruses has also been reported. The pathogenicity of virulent strains of PRRSV was reported to correlate with inflammatory cytokine production (20). EAV infection of equine MΦs induced upregulation of the mRNAs of a similar set of cytokines (21). Although PRRSV infection did not induce proinflammatory cytokine gene expression in porcine mDCs and MΦs, expression of individual PRRSV proteins indicated that some viral proteins suppressed NF-κB activation (22) or TNF-α promoter expression (23, 24) while others activated endogenous NF-κB and the production of proinflammatory cytokines (25–27). Induction of inflammatory cytokine production by SHFV infection in cells from disease-resistant and disease-susceptible NHPs was analyzed. Baboon and macaque MΦ and DC cultures were mock infected or infected with SHFV (MOI of 1), and cytokines in culture fluids were quantified by ELISA. Because the absolute amount of a cytokine varied in cultures from different animals, data were collected from a large number of replicate cultures, and the values shown are average fold change compared to autologous controls. No significant change in the levels of IFN-γ, IL-2, IL-5, IL-8, IL-13, monocyte chemotactic protein 1 (MCP-1), MIP-1α, or MIP-1β was observed in culture fluids from SHFV-infected macaque or baboon MΦ cultures (data not shown). However, the levels of IL-1β, IL-6, and IL-12/23(p40) increased significantly in the culture fluids from SHFV-infected macaque MΦs (Fig. 4). TNF-α levels also increased significantly in SHFV-infected macaque MΦ culture fluids, but this increase was less than 2-fold. In contrast, the levels of IL-1β, TNF-α, and IL-12/23(p40) in baboon MΦ culture fluids did not increase by 48 h after infection, and only a minimal increase (<2-fold) in IL-6 at 36 and 48 h after infection was observed. Infected baboon MΦ cultures produced increased levels of RANTES at 36 and 48 h while macaque MΦ cultures did not. To determine whether viral replication was required to induce proinflammatory cytokine production in MΦs, cytokine levels in culture fluid were quantified after incubation of cells with UV-inactivated virus. No increase in cytokine levels was detected in culture fluids from baboon MΦ cultures incubated with UV-inactivated virus. However, a 3.7-fold increase at 12 h and a 4.7-fold increase at 24 h in IL-6 levels as well as a 10.6-fold increase in IL-1β and a 6.8-fold increase in RANTES levels at 24 h were detected in culture fluids from macaque MΦs incubated with UV-inactivated virus. These data indicate that signaling leading to proinflammatory cytokine production can be triggered by UV-inactivated virus in macaque MΦs but that virus replication is required to amplify and sustain cytokine production.
FIG 4.
Proinflammatory cytokine production in response to SHFV infection by MΦs isolated from disease-resistant and disease-susceptible NHPs. Fluids from MΦ cultures that were mock infected, infected with SHFV (MOI of 1.0), or incubated with UV-inactivated SHFV virus were analyzed for proinflammatory cytokines by multiplexed ELISA. Values shown represent average fold change in cytokine level in an SHFV-infected culture fluid compared to that in an autologous time-matched mock-infected culture fluid. The data were averaged from 20 baboon cultures infected with SHFV (black bars), 21 macaque cultures infected with SHFV (gray bars), 3 baboon cultures incubated with UV-inactivated SHFV (white bars), or 3 macaque cultures incubated with UV-inactivated SHFV (stippled bars). Error bars represent the standard error of mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ns, not significant.
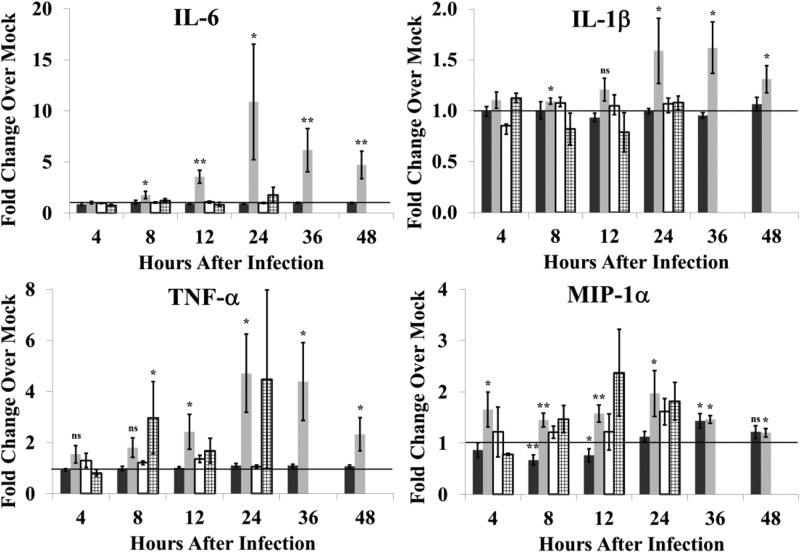
The production of proinflammatory cytokines in response to SHFV infection by baboon and macaque mDCs was also analyzed. For both macaque and baboon cultures, no significant changes in the levels of IFN-γ, IL-2, IL-5, IL-8, IL-12/23(p40), IL-13, MCP-1, MIP-1β, or RANTES were observed at any time after SHFV infection compared to levels from time-matched, autologous, mock-infected culture fluids (data not shown). SHFV infection induced IL-6, IL-β, MIP-1α, and TNF-α production by macaque mDCs but induced only a 1.5-fold increase in MIP-1α production at 36 h in baboon mDCs (Fig. 5). Incubation of either macaque or baboon mDC cultures with UV-inactivated virus did not increase the levels of IL-6, MIP-1α, and IL-1β in culture fluids. However, a 4.5-fold increase in the level of TNF-α at 24 h and a 2.5-fold increase in MIP-1α levels at 12 h were detected in fluids from macaque mDC cultures after incubation with UV-inactivated virus. These data indicate that viral replication is required for production of IL-6 and IL-1β but not TNF-α and MIP-1α by macaque mDCs.
FIG 5.
Proinflammatory cytokine production in response to SHFV infection by mDCs isolated from disease-resistant and disease-susceptible NHPs. Fluids from mDC cultures that were mock infected, infected with SHFV (MOI of 1.0), or incubated with UV-inactivated SHFV virus were analyzed for proinflammatory cytokines by multiplexed ELISA. Values shown represent average fold change in cytokine level in an SHFV-infected culture fluid compared to that in an autologous time-matched mock-infected culture fluid. The data were averaged from 25 baboon cultures infected with SHFV (black bars), 22 macaque cultures infected with SHFV (gray bars), 4 baboon cultures incubated with UV-inactivated SHFV (white bars), or 4 macaque cultures incubated with UV-inactivated SHFV (stippled bars). Error bars represent the standard error of mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ns, not significant.
Analysis of IL-10 signaling in SHFV-infected primary cell cultures.
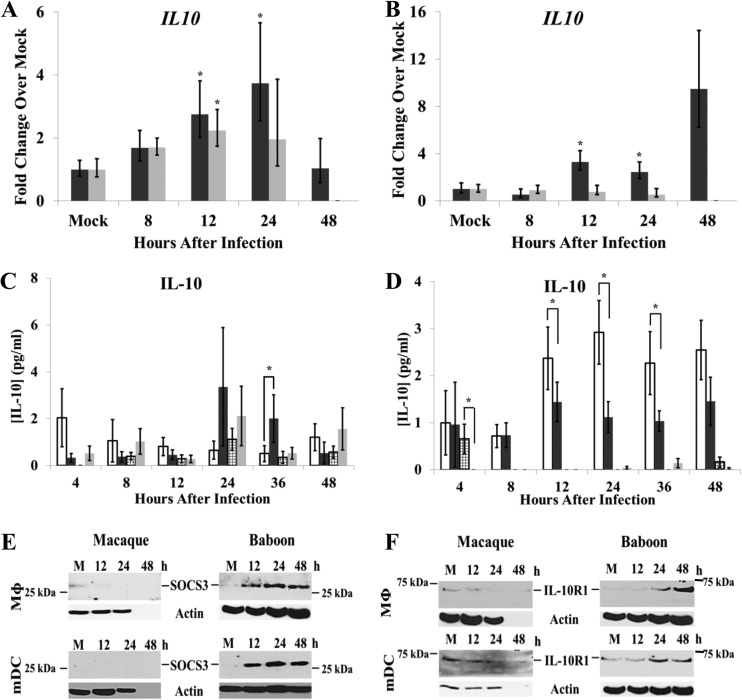
EAV infection of equine MΦs was previously reported to induce the expression of proinflammatory cytokine genes, but PRRSV infection of porcine mDCs and MΦs did not (28, 29). However, PRRSV infection induced upregulation of the regulatory cytokine interleukin-10 (IL-10) (15, 30, 31). To determine whether IL-10 plays a role in modulating the proinflammatory cytokine response to SHFV infection in baboon cells but not in macaque cells, the upregulation of IL-10 mRNA by SHFV infection was examined by qRT-PCR. IL-10 mRNA levels increased with time after infection in both SHFV-infected baboon and macaque MΦs. Peak levels (2.2-fold increase) were detected in macaque MΦs at 12 h while in baboon MΦs peak levels (3.7-fold increase) were observed at 24 h after infection (Fig. 6A). In baboon mDCs, IL-10 mRNA expression increased by 3-fold at 12 h and by 9.5-fold at 48 h after infection while no increase in IL-10 mRNA expression was observed in infected macaque mDCs (Fig. 6B).
FIG 6.
IL-10 production in primary NHP cells. Total cell RNA was isolated from cultured MΦs (A) and mDCs (B) at different times after mock infection or infection with SHFV (MOI of 1.0), and IL-10 mRNA levels were analyzed by real-time qRT-PCR. Each sample was assayed in triplicate. Each value was normalized to the level of 18S rRNA in the same sample. The data shown are averages of values from cell cultures prepared from seven baboons (black bars) and six macaques (gray bars). IL-10 protein levels in culture fluids collected from MΦs (C) and mDCs (D) were quantified by ELISA. Values shown are averages of data obtained from cultures from 10 baboons (white bars, mock infection; black bars, infected) and 8 macaques (stippled bars, mock infection; gray bars, infected). Error bars represent the standard error of mean. *, P ≤ 0.05. SOCS3 (E) and IL-10R1 (F) protein levels were detected by Western blotting in mock- and SHFV-infected (MOI of 1) MΦ and mDC lysates. Actin was used as a loading control. The data shown are representative of data from three independent experiments.
IL-10 protein production has been reported to be posttranscriptionally regulated (32–35). To determine whether the increase in IL-10 mRNA expression observed in response to SHFV infection resulted in increased levels of secreted IL-10 protein, IL-10 protein levels in culture fluids were quantified by ELISA. IL-10 protein levels ranged from <0.2 to 1.1 pg/ml at different times after mock infection in macaque MΦ culture fluids, with the highest level observed at 24 h (Fig. 6C). Culture fluids from SHFV-infected macaque MΦ cultures contained IL-10 protein levels ranging between <0.2 and 2.1 pg/ml, with the peak level at 24 h after infection. The results indicate that the IL-10 levels were similar in culture fluids from mock- and SHFV-infected macaque MΦs. IL-10 protein levels in culture fluids from mock-infected baboon MΦs ranged from 0.5 to 2.1 pg/ml, with the peak level observed at 4 h, and ranged from 0.3 to 3.4 pg/ml, with the peak level observed at 24 h after infection, in SHFV-infected baboon MΦ culture fluids. However, the level of IL-10 was significantly increased in infected baboon MΦ culture fluids only at 36 h. These results indicate that although an SHFV infection can upregulate the expression of IL-10 mRNA in both macaque and baboon MΦs, secreted IL-10 protein levels increased only in culture fluids from infected baboon MΦs.
Very low IL-10 protein levels (>0.2 pg/ml) were observed at 4 h in culture fluids from mock-infected macaque mDCs, and the levels were even lower in fluids from infected macaque mDCs at all times after infection (Fig. 6D). In contrast, in mock-infected baboon mDC culture fluids, IL-10 protein levels ranged from 1.0 to 2.9 pg/ml, with the peak level at 24 h; in SHFV-infected baboon mDC culture fluids, IL-10 levels ranged between 0.7 and 1.5 pg/ml, with the highest levels observed at 12 and 48 h after infection. The levels of IL-10 protein in mock-infected baboon mDC culture fluids were significantly higher than those in SHFV-infected baboon mDC culture fluids between 12 and 36 h after infection. These results suggest that although SHFV infection increased IL-10 mRNA expression in baboon mDCs, IL-10 protein levels in the culture fluids of these cells decreased compared to those in mock-infected culture fluids. The levels of IL-10 protein detected in the baboon MΦs and mDC culture fluids were higher than those previously reported in immature cultured human mDCs (36) on a per-cell basis; unstimulated human mDC cultures produced 20 to 50 pg/ml per 106 cells. The NHP cultures used in our study contained only 104 cells due to the smaller samples of blood that can be obtained from the NHPs.
The high levels of IL-10 observed in mock-infected baboon mDC culture fluids suggested that these cultures were producing IL-10 prior to infection. To more accurately assess the basal levels of IL-10 produced by the various types of cultures, IL-10 levels were measured in culture fluids prior to infection. As described in Materials and Methods, both MΦs and mDCs were cultured with two-thirds of the medium being replaced every 3 days with fresh culture medium, and 11-day culture fluids were collected and assayed by ELISA. Macaque MΦ culture fluids contained IL-10 levels of 0.5 pg/ml ± 0.2 pg/ml (n = 6) while baboon MΦ culture fluids contained 1.7 pg/ml ± 0.2 pg/ml (n = 6). Macaque mDC culture fluids contained <0.2 pg/ml (n = 8) of IL-10 while baboon mDC culture fluids contained 5.4 pg/ml ± 3 pg/ml (n = 9) of IL-10.
IL-10 is a regulatory cytokine that can suppress proinflammatory cytokine production (37). Extracellular IL-10 binds to a heterodimeric cell surface receptor composed of the IL-10R1 and IL-10R2 subunits. IL-10R2 is constitutively expressed and is present at high levels on the surfaces of monocytic cells while IL-10R1 is expressed at low levels (<100 units/cell surface) until its expression is upregulated by various stimuli (37). IL-10 binding to its receptor signals the induction of suppressor of cytokine signaling 3 (SOCS3) expression, and SOCS3 suppresses cytokine production. To determine whether the high basal levels of IL-10 produced by baboon mDC and MΦ cultures were sufficient to induce SOCS3 expression, cell lysates from mock- or SHFV-infected (MOI of 1) macaque and baboon cell cultures were collected at various times after infection and analyzed by Western blotting. Even though the culture fluids from mock-infected baboon MΦs and mDCs contained IL-10, SOCS3 was not detected in the lysates of these cells (Fig. 6E). However, SOCS3 levels increased with time after SHFV infection in both baboon MΦs and mDCs. In contrast, a low level of SOCS3 was observed in mock-infected macaque MΦ lysates, but no SOCS3 was detected in either mock-infected macaque mDC lysates or in any of the infected macaque cell lysates (Fig. 6E). To determine whether the lack of SOCS3 expression in mock-infected baboon cells correlated with low IL-10R1 expression and reduced IL-10 signaling, IL-10 levels in mock- and SHFV-infected cell lysates were analyzed by Western blotting using anti-IL-10R1 antibody. IL-10R1 was not detected in mock-infected baboon MΦs but was present at a low level in mock-infected baboon mDCs. The levels of IL-10R1 increased with time after infection in both types of baboon cells (Fig. 6F). In contrast, the levels of IL-10R1 were low in mock-infected macaque MΦs and mDCs and decreased to undetectable levels with time after infection (Fig. 6F). The data indicate that in baboon but not macaque cells, SHFV infection induces the upregulation of IL-10R1 expression. Increased IL-10R1 levels would be expected to enhance IL-10 signaling through its receptor, leading to sustained upregulation of SOCS3 expression. However, SOCS3 levels were increased in baboon cells by 12 after infection, a time when IL-10R1 levels were still low, suggesting that SHFV infection may upregulate SOCS3 through a mechanism that is independent of IL-10 signaling. In contrast, in macaque cells, SHFV infection downregulated IL-10R1 expression and did not upregulate SOCS3 expression.
Analysis of modulation of host responses to SHFV infection in disease-susceptible cells by exogenous IL-10.
Based on the finding that disease-resistant baboon mDCs produce higher levels of IL-10 both before and after SHFV infection than disease-susceptible macaque mDCs and produce significantly smaller amounts of proinflammatory cytokines in response to infection, we hypothesized that IL-10 plays a role in suppressing SHFV-induced proinflammatory cytokine production in baboon cultures. The observation that detectable levels of IL-10R1 were expressed by both mock-infected macaque MΦs and mDCs suggested that addition of exogenous IL-10 to these cultures might suppress proinflammatory cytokine production in response to SHFV infection. Macaque MΦ and mDC cultures were treated with 5 pg/ml of rhIL-10 for 47 h starting immediately after the 1-h virus adsorption period or for 1 h prior to viral adsorption (MOI of 1) and then for 47 h after virus adsorption. Proinflammatory cytokines in culture fluids from rhIL-10-treated and untreated mock- and SHFV-infected cultures were quantified by ELISA. Both protocols of rhIL-10 treatment significantly reduced the levels of IL-6 produced by macaque MΦs at 36 and 48 h after SHFV infection (Fig. 7A). However, the levels of IL-1β, TNF-α, IL-12/23(p40), and RANTES were not altered (Fig. 7B to E). Treatment of infected macaque mDCs with rhIL-10 decreased IL-6 (Fig. 7F) and MIP-1α levels (Fig. 7I) but not IL-1β or TNF-α levels (Fig. 7G and H) in culture fluids. Treatment of macaque MΦ or mDC cultures with rhIL-10 (5, 10, or 20 pg/ml) did not alter viral yields (assessed by plaque assay) or cell viability (analyzed by MTT assay) (data not shown). The data indicate that IL-10 treatment was able to suppress SHFV-induced production of some proinflammatory cytokines, such as IL-6, but not that of others, such as TNF-α, in macaque MΦ and mDC cultures.
FIG 7.
Effects of IL-10 treatment on SHFV-induced proinflammatory cytokine production. (A to E) Macaque MΦ cultures were left untreated (gray bars), treated with rhIL-10 after infection (black bars), or treated with rhIL-10 for 1 h before infection and after infection (white bars). (F to I) Macaque mDC cultures were left untreated (gray bars), treated with rhIL-10 after infection (black bars), or treated with rhIL-10 for 1 h before infection and after infection (white bars). Proinflammatory cytokines present in culture fluids collected at the indicated times after infection were assayed by multiplexed ELISA. The values represent fold change in cytokine levels in SHFV-infected culture fluids compared to the levels in autologous time- and treatment-matched mock-infected culture fluid. The values are averages of data from six macaque cultures. Error bars represent the standard error of mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ns, not significant.
To determine whether inhibition of IL-10 signaling in baboon cultures would increase SHFV-induced proinflammatory cytokine expression, baboon MΦ and mDC cultures were treated with 5, 10, or 20 pg/ml of anti-IL-10 antibody or 10 pg/ml of a control isotype IgA for 47 h starting after the 1-h virus adsorption period or for 1 h prior to virus adsorption and then for 47 h afterward. No significant increase in the levels of any of the proinflammatory cytokines was observed in culture fluids from infected baboon MΦs or mDCs (data not shown). Neither viral replication nor cell viability was altered by anti-IL-10 antibody treatment of infected baboon cells (data not shown). These data suggest that effective counteraction/suppression of an SHFV-induced inflammatory response in baboon cells does not require IL-10 signaling.
DISCUSSION
The arteriviruses PRRSV and LDV were previously reported to productively infect only 5 to 10% of the cells in pig or mouse MΦ cultures, respectively (38–40). Similarly, SHFV was found to infect ∼5 to 10% of baboon MΦs. Based on the intensity of the dsRNA staining, SHFV replication was efficient in this subset of baboon MΦs. In contrast, in MΦ cultures prepared from macaques which are not natural hosts for SHFV, ∼90% of the cells became infected, efficiently replicated viral RNA, and showed evidence of cytopathology. The higher virus yields produced by macaque MΦ cultures were likely due to the higher number of cells infected in these cultures. The larger number of susceptible macaque MΦs was not due to a higher number of cells expressing the arterivirus coreceptor CD163 since this was one of the cell surface molecules analyzed by flow cytometry, and >90% of both the baboon and macaque MΦs expressed this surface marker (41, 42). The expression of the other known arterivirus coreceptor, sialoadhesin (43), on baboon and macaque cells could not be compared because an antibody that cross-reacts with NHP sialoadhesin is not currently available. In contrast to what was observed with MΦs, SHFV infected about 50% of the cells in both baboon and macaque mDC cultures. However, SHFV replication was less efficient in baboon mDCs. How virus replication is restricted in baboon mDCs is not currently known.
Several African NHP species, including baboons, patas, and African greens, were previously reported to be natural hosts of SHFV (1, 3). SHFV causes a persistent, asymptomatic infection in these animals. Analysis of persistent viremia in baboons indicated that virus titers are very low, namely, 10 to 100 PFU per ml of serum (H. A. Vatter and M. A. Brinton, unpublished data). In contrast, SHFV-infected, asymptomatic colobus monkeys appear to have a high-titer viremia (44, 45). SHFV isolates from colobus show extensive sequence variation from the patas isolate used in the current study (SHFV strain LVR) and also from new SHFV isolates from persistently infected baboons that are similar to LVR (H. A. Vatter, E. F. Donaldson, and M. A. Brinton, unpublished data). However, neither the replication efficiency of the colobus SHFV isolates in MΦs and mDCs from other NHP species nor the ability of these isolates to cause hemorrhagic disease in macaques has yet been analyzed.
Macaques were previously reported to produce proinflammatory cytokines in response to infection with hemorrhagic fever viruses that resulted in bystander T cell apoptosis as well as increased tissue factor expression leading to increased vascular permeability and coagulation defects (7, 9, 46, 47). The similar clinical manifestations observed in SHFV-infected macaques suggested that proinflammatory cytokines were produced (3, 5, 14, 48, 49). Data from the present study show that SHFV infection induces the production of proinflammatory cytokines, including IL-6 and TNF-α, in macaque but not baboon MΦs and mDCs. Previous studies of the effects of individually expressed PRRSV proteins showed that some suppress/counteract while others activate cell innate immune responses (16). Appropriate regulation of cell innate responses by SHFV proteins is expected to occur in cells from natural African NHP hosts, resulting in survival of the host, while inadequate/inappropriate regulation appears to occur in cells from a nonnatural host such as a macaque. Although the type I IFN response of SHFV-infected macaque cells was more efficient than that of infected baboon cells, it was not sufficient to effectively suppress virus replication. Viral hemorrhagic fever/bacterial septic shock can result when a pathogen evokes an inflammatory response and overcomes the host's innate barriers. Typically, this results in massive systemic release of inflammatory mediators that can result in the death of the host (7).
The levels of the immunoregulatory cytokine IL-10 were found to differ in baboon and macaque MΦ and mDC culture fluids. Baboon culture fluids contained higher levels of IL-10 both prior to and after SHFV infection than macaque cell culture fluids. It was previously reported that porcine mDCs produced IL-10 but not proinflammatory cytokines in response to infection with PRRSV (30, 31, 50) while virulent strains of EAV induced proinflammatory cytokines but not IL-10 in equine MΦs (21, 28). These data led to the suggestion that IL-10 suppressed proinflammatory cytokine production in PRRSV-infected mDCs. Data from other virus models support this suggestion. Reduced proinflammatory cytokine and chemokine production as well as reduced inflammatory cell infiltration into the central nervous system (CNS) was observed after IL-10 expression from a recombinant neurotropic coronavirus (51). Suppression of proinflammatory cytokine production and decreased liver pathology were observed when IL-10 knockout mice were infected with murine cytomegalovirus, but there was no effect on virus replication (52).
Basal IL-10 production in human cells was shown to be due to a constitutive level of IL-10 mRNA expression. In unstimulated cells, IL-10 protein production is suppressed by destabilizing motifs in the 3′ untranslated region of the IL-10 mRNA that lead to rapid mRNA degradation, and miR-106a has also been reported to suppress IL-10 mRNA translation (32–35). Although the expression of IL-10 mRNA was upregulated in both macaque and baboon cells by SHFV infection, IL-10 protein was detected in culture fluids only from baboon cells, indicating that posttranscriptional suppression of IL-10 mRNA is more effective in macaque cells. However, the observation of lower IL-10 protein levels in SHFV-infected than in mock-infected baboon mDCs suggested that SHFV infection may increase IL-10 mRNA degradation/silencing in baboon cells.
Several previous studies showed that incubation of human or murine MΦ and DC cultures with IL-10 suppressed the production of proinflammatory cytokines, including IL-6, IL-1β, and TNF-α, in response to a viral infection or a sustained inflammatory response (37). Although incubation of macaque cells with rhIL-10 resulted in decreased IL-6 and MIP-1α (mDCs only) production in response to SHFV infection, no significant decrease in TNF-α, IL-1β, or IL-12/23(p40) production was observed. The use of human instead of macaque IL-10 might be the cause of the more limited suppression observed. Unfortunately, macaque IL-10 is currently not commercially available.
SOCS3 suppresses proinflammatory cytokine production. SOCS3 expression is upregulated by IL-10 signaling through its receptor composed of IL-10R1 and IL-10R2 heterodimers. Despite the high basal levels of IL-10 in baboon MΦ and mDC culture fluids, SOCS3 expression was observed only after SHFV infection. The high levels of SOCS3 expression in both types of baboon cells at 12 h after infection, a time when IL-10R1 levels were still low, and the inability of anti-IL-10 antibody to upregulate proinflammatory cytokine production in SHFV-infected baboon cells suggest that SHFV infection may be able to upregulate SOCS3 expression in an IL-10-independent manner. PRRSV proteins have been reported to regulate innate and inflammatory responses in infected cells (16). The functions of individual SHFV proteins have not yet been tested. The data obtained in the current study suggest that differential effects of SHFV proteins in target cells from a natural host such as a baboon and ones from a nonnatural host such as a macaque may determine the outcome of the infection.
ACKNOWLEDGMENTS
This work was supported by Public Health Service research grant AI073824 to M.A.B from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. H.A.V. was supported by a Molecular Basis of Disease Fellowship from Georgia State University. This project used biological materials from the Southwest National Primate Research Center (P51 OD011133) and also from the Yerkes National Primate Research Center Comparative AIDS Core (P51OD011132).
We thank Han Di for reading the manuscript and Mausumi Basu for assistance with preparation of the figures.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Gravell M, London WT, Rodriguez M, Palmer AE, Hamilton RS. 1980. Simian haemorrhagic fever (SHF): new virus isolate from a chronically infected patas monkey. J. Gen. Virol. 51:99–106. 10.1099/0022-1317-51-1-99 [DOI] [PubMed] [Google Scholar]
- 2.Tauraso NM, Shelokov A, Palmer AE, Allen AM. 1968. Simian hemorrhagic fever. 3. Isolation and characterization of a viral agent. Am. J. Trop. Med. Hyg. 17:422–431 [PubMed] [Google Scholar]
- 3.London WT. 1977. Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature 268:344–345. 10.1038/268344a0 [DOI] [PubMed] [Google Scholar]
- 4.Allen AM, Palmer AE, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. II. Studies in pathology. Am. J. Trop. Med. Hyg. 17:413–421 [DOI] [PubMed] [Google Scholar]
- 5.Palmer AE, Allen AM, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am. J. Trop. Med. Hyg. 17:404–412 [PubMed] [Google Scholar]
- 6.Mahanty S, Bray M. 2004. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 4:487–498. 10.1016/S1473-3099(04)01103-X [DOI] [PubMed] [Google Scholar]
- 7.Bray M. 2005. Pathogenesis of viral hemorrhagic fever. Curr. Opin. Immunol. 17:399–403. 10.1016/j.coi.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 8.Levi M, van der Poll T, ten Cate H. 2006. Tissue factor in infection and severe inflammation. Semin. Thromb. Hemost. 32:33–39. 10.1055/s-2006-933338 [DOI] [PubMed] [Google Scholar]
- 9.Geisbert TW, Hensley LE, Jahrling PB, Larsen T, Geisbert JB, Paragas J, Young HA, Fredeking TM, Rote WE, Vlasuk GP. 2003. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet 362:1953–1958. 10.1016/S0140-6736(03)15012-X [DOI] [PubMed] [Google Scholar]
- 10.Snijder EJ, Kikkert M. 2013. Arteriviruses, p 859–879 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 11.Snijder EJ, Meulenberg JJM. 1998. The molecular biology of arterivirus. J. Gen. Virol. 79:961–979 [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson KL, Villinger F, Miranda ME, Ksiazek TG, Peters CJ, Rollin PE. 2001. Multiplex analysis of cytokines in the blood of cynomolgus macaques naturally infected with Ebola virus (Reston serotype). J. Med. Virol. 65:561–566. 10.1002/jmv.2073 [DOI] [PubMed] [Google Scholar]
- 13.Denner J, Eschricht M, Lauck M, Semaan M, Schlaermann P, Ryu H, Akyuz L. 2013. Modulation of cytokine release and gene expression by the immunosuppressive domain of gp41 of HIV-1. PLoS One 8:e55199. 10.1371/journal.pone.0055199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravell M, London WT, Leon ME, Palmer AE, Hamilton RS. 1986. Differences among isolates of simian hemorrhagic fever (SHF) virus. Proc. Soc. Exp. Biol. Med. 181:112–119. 10.3181/00379727-181-42231 [DOI] [PubMed] [Google Scholar]
- 15.Song S, Bi J, Wang D, Fang L, Zhang L, Li F, Chen H, Xiao S. 2013. Porcine reproductive and respiratory syndrome virus infection activates IL-10 production through NF-κB and p38 MAPK pathways in porcine alveolar macrophages. Dev. Comp. Immunol. 39:265–272. 10.1016/j.dci.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 16.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 154:61–76. 10.1016/j.virusres.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, Nelson EA, Fang Y. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97. 10.1016/j.virol.2009.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensley LE, Young HA, Jahrling PB, Geisbert TW. 2002. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol. Lett. 80:169–179. 10.1016/S0165-2478(01)00327-3 [DOI] [PubMed] [Google Scholar]
- 19.Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797–2801 [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Liu J, Bai J, Wang X, Li Y, Jiang P. 2013. Comparative expression of Toll-like receptors and inflammatory cytokines in pigs infected with different virulent porcine reproductive and respiratory syndrome virus isolates. Virol. J. 10:135. 10.1186/1743-422X-10-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore BD, Balasuriya UB, Watson JL, Bosio CM, MacKay RJ, MacLachlan NJ. 2003. Virulent and avirulent strains of equine arteritis virus induce different quantities of TNF-alpha and other proinflammatory cytokines in alveolar and blood-derived equine macrophages. Virology 314:662–670. 10.1016/S0042-6822(03)00506-3 [DOI] [PubMed] [Google Scholar]
- 22.Song C, Krell P, Yoo D. 2010. Nonstructural protein 1alpha subunit-based inhibition of NF-kappaB activation and suppression of interferon-beta production by porcine reproductive and respiratory syndrome virus. Virology 407:268–280. 10.1016/j.virol.2010.08.025 [DOI] [PubMed] [Google Scholar]
- 23.Subramaniam S, Kwon B, Beura LK, Kuszynski CA, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus non-structural protein 1 suppresses tumor necrosis factor-alpha promoter activation by inhibiting NF-κB and Sp1. Virology 406:270–279. 10.1016/j.virol.2010.07.016 [DOI] [PubMed] [Google Scholar]
- 24.Subramaniam S, Beura LK, Kwon B, Pattnaik AK, Osorio FA. 2012. Amino acid residues in the non-structural protein 1 of porcine reproductive and respiratory syndrome virus involved in down-regulation of TNF-alpha expression in vitro and attenuation in vivo. Virology 432:241–249. 10.1016/j.virol.2012.05.014 [DOI] [PubMed] [Google Scholar]
- 25.Fang Y, Fang L, Wang Y, Lei Y, Luo R, Wang D, Chen H, Xiao S. 2012. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-κB activation. Virol. J. 9:83. 10.1186/1743-422X-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo R, Fang L, Jin H, Jiang Y, Wang D, Chen H, Xiao S. 2011. Antiviral activity of type I and type III interferons against porcine reproductive and respiratory syndrome virus (PRRSV). Antiviral Res. 91:99–101. 10.1016/j.antiviral.2011.04.017 [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, Hou Q, Zhong Z, Li X, Chen H, Li W, Wen J, Wang L, Liu W, Zhong F. 2013. Porcine reproductive and respiratory syndrome virus activates inflammasomes of porcine alveolar macrophages via its small envelope protein E. Virology 442:156–162. 10.1016/j.virol.2013.04.007 [DOI] [PubMed] [Google Scholar]
- 28.Balasuriya UBR, MacLachlan NJ. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107–129. 10.1016/j.vetimm.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 29.Chang HC, Peng YT, Chang HL, Chaung HC, Chung WB. 2008. Phenotypic and functional modulation of bone marrow-derived dendritic cells by porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 129:281–293. 10.1016/j.vetmic.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 30.Suradhat S, Thanawongnuwech R. 2003. Upregulation of interleukin-10 gene expression in the leukocytes of pigs infected with porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 84:2755–2760. 10.1099/vir.0.19230-0 [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Fuertes L, Campos E, Domenech N, Ezquerra A, Castro JM, Dominguez J, Alonso F. 2000. Porcine reproductive and respiratory syndrome (PRRS) virus down-modulates TNF-alpha production in infected macrophages. Virus Res. 69:41–46. 10.1016/S0168-1702(00)00172-6 [DOI] [PubMed] [Google Scholar]
- 32.Powell MJ, Thompson SA, Tone Y, Waldmann H, Tone M. 2000. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J. Immunol. 165:292–296 [DOI] [PubMed] [Google Scholar]
- 33.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. 2000. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 165:286–291 [DOI] [PubMed] [Google Scholar]
- 34.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. 2008. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J. Interferon Cytokine Res. 28:679–691. 10.1089/jir.2008.0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. 2009. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc. Natl. Acad. Sci. U. S. A. 106:5761–5766. 10.1073/pnas.0808743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corinti S, Albanesi C, la Sala A, Pastore S, Girolomoni G. 2001. Regulatory activity of autocrine IL-10 on dendritic cell functions. J. Immunol. 166:4312–4318 [DOI] [PubMed] [Google Scholar]
- 37.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683–765. 10.1146/annurev.immunol.19.1.683 [DOI] [PubMed] [Google Scholar]
- 38.Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 142:2483–2497. 10.1007/s007050050256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalchyk K, Plagemann PG. 1985. Cell surface receptors for lactate dehydrogenase-elevating virus on subpopulation of macrophages. Virus Res. 2:211–229. 10.1016/0168-1702(85)90010-3 [DOI] [PubMed] [Google Scholar]
- 40.Ritzi DM, Holth M, Smith MS, Swart WJ, Cafruny WA, Plagemann GW, Stueckemann JA. 1982. Replication of lactate dehydrogenase-elevating virus in macrophages. 1. Evidence for cytocidal replication. J. Gen. Virol. 59:245–262. 10.1099/0022-1317-59-2-245 [DOI] [PubMed] [Google Scholar]
- 41.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 81:7371–7379. 10.1128/JVI.00513-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch SK, Calvert JG. 2010. A brief review of CD163 and its role in PRRSV infection. Virus Res. 154:98–103. 10.1016/j.virusres.2010.07.018 [DOI] [PubMed] [Google Scholar]
- 43.Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. 2008. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2943–2953. 10.1099/vir.0.2008/005009-0 [DOI] [PubMed] [Google Scholar]
- 44.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6:e19056. 10.1371/journal.pone.0019056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J. Virol. 87:688–691. 10.1128/JVI.02433-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruf W. 2004. Emerging roles of tissue factor in viral hemorrhagic fever. Trends Immunol. 25:461–464. 10.1016/j.it.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 47.Bray M, Geisbert TW. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell Biol. 37:1560–1566. 10.1016/j.biocel.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 48.Abildgaard C, Harrison J, Espana C, Spangler W, Gribble D. 1975. Simian hemorrhagic fever: studies of coagulation and pathology. Am. J. Trop. Med. Hyg. 24:537–544 [DOI] [PubMed] [Google Scholar]
- 49.Johnson RF, Dodd LE, Yellayi S, Gu W, Cann JA, Jett C, Bernbaum JG, Ragland DR, St Claire M, Byrum R, Paragas J, Blaney JE, Jahrling PB. 2011. Simian hemorrhagic fever virus infection of rhesus macaques as a model of viral hemorrhagic fever: clinical characterization and risk factors for severe disease. Virology 421:129–140. 10.1016/j.virol.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz I, Darwich L, Pappaterra G, Pujols J, Mateu E. 2005. Immune responses of pigs after experimental infection with a European strain of Porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 86:1943–1951. 10.1099/vir.0.80959-0 [DOI] [PubMed] [Google Scholar]
- 51.Trandem K, Jin Q, Weiss KA, James BR, Zhao J, Perlman S. 2011. Virally expressed interleukin-10 ameliorates acute encephalomyelitis and chronic demyelination in coronavirus-infected mice. J. Virol. 85:6822–6831. 10.1128/JVI.00510-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang-Feldman YJ, Lochhead GR, Lochhead SR, Yu C, Pomeroy C. 2011. Interleukin-10 repletion suppresses proinflammatory cytokines and decreases liver pathology without altering viral replication in murine cytomegalovirus (MCMV)-infected IL-10 knockout mice. Inflamm. Res. 60:233–243. 10.1007/s00011-010-0259-4 [DOI] [PMC free article] [PubMed] [Google Scholar]