ABSTRACT
Genotype II.3 (GII.3) noroviruses are a major cause of sporadic gastroenteritis, particularly in children. The greater incidence of GII.3 noroviruses in the pediatric population compared to the adult demographic suggests development of herd immunity to this genotype, possibly as a consequence of limited evolution of immune epitopes. This study aimed to identify and characterize immune epitopes on the GII.3 capsid protein and to determine the level of immune cross-reactivity within the genotype. A panel of seven GII.3 virus-like particles (VLPs), representing norovirus strains isolated during 1975 to 2008, was tested by enzyme-linked immunosorbent assay (ELISA) for reactivity with human sera and a rabbit anti-GII.3 strain-specific polyclonal serum generated against the 2008 GII.3 VLP. Immunoprecipitation of protease-digested GII.3 VLPs and sequencing of bound peptides via mass spectrometry were used to locate epitopes on the capsid. Two epitopes were investigated further using Mimotopes technology. Serum binding studies demonstrated complete intragenotype GII.3 cross-reactivity using both human and rabbit serum. Six immunoreactive regions containing epitopes were located on the GII.3 capsid protein, two within each capsid domain. Epitopes in the S and P1 domains were highly conserved within GII.3 noroviruses. P2 domain epitopes were variable and contained evolutionarily important residues and histo-blood group antigen (HBGA) binding residues. In conclusion, anti-GII.3 antibody-binding epitopes are highly cross-reactive and mostly conserved within GII.3 strains. This may account for the limited GII.3 prevalence in adults and suggests that a GII.3 strain may be a valuable inclusion in a multivalent pediatric targeted VLP vaccine. Exploration of norovirus immune epitopes is vital for effective vaccine design.
IMPORTANCE
INTRODUCTION
Norovirus is the most common cause of gastroenteritis (1), resulting in >90% of viral gastroenteritis cases and 50% of all gastroenteritis outbreaks worldwide (2). Annually in the United States, norovirus is estimated to cause approximately 21 million cases of gastroenteritis (1), and it is the most common cause of gastroenteritis-related emergency department visits (2). In developing countries, norovirus is estimated to cause 1 million hospitalizations and 200,000 deaths in children less than 5 years of age annually (2).
Human noroviruses belong to the family Caliciviridae, genus Norovirus, and contain a positive-sense, single-stranded RNA genome (3, 4). The genome is approximately 7.5 to 7.7 kb in length and consists of three open reading frames (ORF) (5). ORF 1 encodes nonstructural proteins, including the RNA-dependent RNA polymerase (RdRp); ORF 2 encodes viral protein 1 (VP1), the major capsid protein; and ORF 3 encodes VP2, a minor structural protein (6, 7). Based on sequence analysis of regions within the VP1 or RdRp gene, noroviruses are classified into five genogroups, with a sixth group tentatively proposed (8). Genogroup I, II, and IV (GI, GII, and GIV) noroviruses are known to infect humans, and GII noroviruses are predominant. Human noroviruses can be further classified into at least 35 different genotypes (9). Genotype II.4 (GII.4) is the most common genotype circulating worldwide, with at least eight variants having given rise to large global epidemics every 2 to 6 years since 1995 (9). Genotype II.3 (GII.3) is a common cause of sporadic pediatric infections (10–13), and 70% of children show evidence of GII.3 infection by 2 years of age (14). In sporadic infections in infants and young children, GII.3 strains often predominate, and they were particularly prevalent throughout the late 1970s, late 1980s (10), and early 1990s (15, 16) and in the early and late 2000s (13, 17–20).
The major capsid protein, VP1, is characterized by three structural domains, the shell (S) domain and two protruding (P) domains, P1 and P2 (21). The P domain is the most exposed region of the capsid protein and is likely to contain determinants for antigenicity and host cell attachment (21–23). The human histo-blood group antigens (HBGA) act as norovirus host cell attachment factors, and the HBGA binding interface of the norovirus P domain has been elucidated through crystallography and mutagenesis studies (24–26).
Mapping of antibody-binding epitopes on norovirus capsids has proven challenging, as the use of classical antibody escape mapping approaches has been prevented by the absence of a human norovirus cell culture system. However, several studies have mapped antibody-binding epitopes on GI and GII norovirus capsids using techniques such as bioinformatic prediction, screening of monoclonal antibody (MAb) binding to expressed capsid proteins or short peptides, phage display, targeted mutagenesis, and crystallography (22, 23, 27–35). The location and sequence conservation of previously mapped epitopes are highly variable. Unexpectedly, many epitopes have been mapped to highly conserved regions of the S domain and occluded regions of the P1 domain (31, 35, 36), indicating a high level of conformational flexibility in the capsid protein. As expected, many blockade epitopes (epitopes targeted by antibodies that “block” viral binding to HBGAs) have been mapped to the P2 domain (34, 37). Several recent mutagenesis epitope mapping studies have defined specific blockade epitopes on the GII.4 capsid P2 domain (epitopes A [294, 296 to 298, 368, and 372], D [393 to 395], and E [407, 412, and 413]). These GII.4 epitopes have been shown to change over time, suggesting that evolution of GII.4 viruses is driven by herd immunity (34, 37). These and other studies suggest that some MAbs bind epitopes with broad reactivity across genotypes, while other MAbs are strain specific (37, 38).
Locating and characterizing norovirus antibody-binding epitopes is essential to successful norovirus vaccine design. Norovirus virus-like particles (VLPs) are currently being examined in trials as vaccine candidates (39, 40). VLPs are nonreplicating empty particles, morphologically and antigenically identical to native virus, and the nature of their production allows a high level of control over capsid sequence design (41–43). With the aim of eliciting cross-protective immunity, the inclusion of specific epitope sequences that are conserved within and/or between genotypes or genogroups is essential in vaccine design. It is important to locate evolving epitopes so that inclusion of particular residues may be considered in vaccine development.
The high incidence of norovirus GII.3 strains in the pediatric population compared to the adult demographic may be due to the development of herd immunity and limited evolution of immunological epitopes (10). Antibodies produced in response to GII.3 infection have been shown to cross-react with other genotypes; however, there is limited information regarding the level of GII.3 intragenotype cross-reactivity and the location and sequence variability of GII.3 epitopes (31, 44, 45). We hypothesized that the GII.3 capsid contains relatively conserved antibody-binding epitopes and that antibodies produced in response to a GII.3 infection would react with other strains of this genotype. This study aimed to identify and characterize immunological epitopes on the GII.3 capsid protein and to determine the level of immune cross-reactivity within the genotype.
MATERIALS AND METHODS
VLP production.
Seven virus-like particles (VLPs) representing norovirus GII.3 strains isolated from stool samples collected between 1975 and 2008 in Victoria, Australia, and Washington, DC, were produced to create a time-ordered panel of GII.3 VLPs. GII.3 VLPs were expressed using the baculovirus expression system as previously described by Boon and colleagues (10). Briefly, the VP1 genes of norovirus strains DC2005 (GenBank accession number HM072045.1 CHDC2005/1975/US), DC32 (HM072046.1; CHDC32/1976/US), DC4031 (HM072044.1; CHDC4031/1988/US), DC5261 (HM072041.1; CHDC5261/1990/US), AU01 (KC464324; 01–13/477/VP1/2001/AU), AU07 (KC464327; 84/46/VP1/2007/AU), and AU08 (KC464328; 693/425/VP1/2008/AU) were amplified and cloned into the pENTR Gateway entry vector (Invitrogen, Life Technologies, CA) for recombination into baculovirus DNA using the BaculoDirect C-term expression kit (Invitrogen). The recombinant baculovirus strain was transfected into Sf9 cells (Invitrogen) and passaged in suspension culture to express VLPs. Expressed VLPs were purified from culture medium using sucrose cushion and cesium chloride gradient centrifugation and were dialyzed against 1× phosphate-buffered saline (PBS). The expression of VP1 and the presence of assembled VLPs were confirmed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electron microscopy. The VLP concentration was determined using the Pierce 660-nm protein quantification kit (Thermo Scientific, IL); concentrations ranged from 308 μg ml−1 to 1.43 mg ml−1.
VLPs representing genotypes GII.5 (rWR; GenBank accession number AF414423.1; NLV/White River/290/1994/US) and GII.6 (rFV; GenBank accession number AF414407.1; NLV/Florida/269/1993/US) were donated by G. Belliot and J. Noel, CDC, Atlanta, GA.
Stool sample collection and use in this study were considered exempt from the relevant institutional ethics review board approval (National Institutes of Health Institutional Review Board [46] and Royal Children's Hospital Human Research Ethics Committee [47]).
Production of rabbit anti-rAU08 norovirus GII.3 polyclonal serum.
Norovirus GII.3 VLPs representing strain AU08 (rAU08) were used to immunize rabbits that were prescreened for existing norovirus antibodies. Rabbits were immunized subcutaneously with three doses of 200 μg of VLP in Freund's adjuvant at 4-week intervals. The VLPs were emulsified in Freund's complete adjuvant for administration of the first dose and Freund's incomplete adjuvant for administration of the second and third doses. Serum was tested for reactivity against homologous norovirus VLPs following the second and third doses, using a VLP IgG enzyme-linked immunosorbent assay (ELISA). Polyclonal antiserum production was conducted at the Walter and Elisa Hall Research Institute animal facility, Bundoora, Victoria, Australia, as a commercial agreement.
Human serum samples.
Serum samples were collected from three pediatric patients following norovirus gastroenteritis, between February 1978 and July 1980 (C. D. Kirkwood, unpublished data). Norovirus infection was confirmed by electron microscopy of matched stool samples obtained at the time of illness. Serum samples were collected 6 months after illness, and the serum samples analyzed in this study were selected on the basis that they reacted strongly with GII.3 VLP (Kirkwood, unpublished). Serum samples were collected during September 1978 (patient 1), February 1978 (patient 2), and July 1980 (patient 3). Serum sample collection and analyses were approved by the Royal Children's Hospital Human Research Ethics Committee.
Norovirus VLP ELISA.
The immunoreactivity of the time-ordered panel of norovirus GII.3 VLPs and two non-GII.3 VLPs to anti-rAU08-specific polyclonal serum and human serum was examined using ELISA. With the exception of the primary and secondary antibodies, the ELISA protocol was the same for both human and rabbit sera. Briefly, wells of a MaxiSorp 96-well plate (Thermo Scientific) were coated with 150 ng of norovirus VLPs diluted in 60 mM sodium carbonate-bicarbonate buffer, pH 9.6. The plates were incubated for 18 h at 4°C. Wells were washed six times with PBS–0.05% Tween 20 (PBS-T) using the Thermo Scientific Well Wash 4 Mk2 plate washer before blocking with 5% casein–PBS-T for 1 h at 37°C. The primary antibody was either diluted 1:1,000 for human serum samples or serially diluted 3-fold, from 1:27,000 to 1:2,187,000, for the rabbit anti-rAU08 polyclonal serum. Following blocking, plates were washed as before and primary antibody diluted in 1% casein–PBS-T was added and incubated for 3 h at 37°C. Horseradish peroxidase (HRP)-conjugated secondary antibodies (goat anti-rabbit IgG-HRP [1:3,000 dilution] and rabbit anti-human IgG-HRP [1:500]; Dako, Hovedstaden, Denmark) diluted in 1% casein–PBS-T were applied to wells and incubated at 37°C for 1 h. Wells were washed and antibody-binding complexes were detected with the addition of peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB) solution (100 μg ml−1 in 100 mM sodium acetate, 1 mM citric acid, 0.003% [vol/vol] H2O2, and 1% [vol/vol] dimethyl sulfoxide [DMSO]). The colorimetric reaction was developed for 10 min and stopped using 2 M H2SO4. Plates were read at 450 nm on a Titertek Multiscan MCC/340 MKII microtiter plate reader (Labsystems, Uusimaa, Finland). All ELISA data are representative of at least two independent experiments. Rabbit preimmune serum or serum from a norovirus-negative healthy patient was used as a negative control.
For the human sera IgG ELISA, positive-control serum was collected from a patient with norovirus gastroenteritis, as determined by electron microscopy of a matched stool sample. The serum was previously determined to be highly reactive with GII.3 VLPs (Kirkwood, unpublished). A 3-fold serial dilution of the positive-control human serum (seven dilutions starting from 1:166.67) was used to prepare a standard curve in each experiment. An arbitrary concentration of 1,000,000 units of IgG was assigned to neat positive-control serum. Standard curves were constructed in Prism 6 (GraphPad Software, CA), using semilog nonlinear regression, weighted by 1/Y2. Theoretical “bound” IgG concentrations were interpolated from the standard curve based on absorbance values.
Protease selection.
The theoretical cleavage profile of the norovirus capsid protein was performed using the MS-digest application on the online server Protein Prospector (http://prospector.ucsf.edu; The University of California).
The protease Glu-C predicted the highest sequence coverage of peptides within the detectable range, between 800 Da and 4,000 Da.
Protease digestion.
Protease digestion of rAU08 VLPs was performed using Glu-C endoproteinase (Pierce, Thermo Scientific), by following the manufacturer's instructions. Briefly, VLP protein (15 μg or 45 μg) was reduced in 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate [(NH4)HCO3], pH 8, for 1 h at 60°C. The reduced protein was incubated in 20 mM iodoacetamide at room temperature for 30 min, protected from light, before the alkylation reaction was quenched in 10 mM DTT. Glu-C was added to a final concentration of 0.05 μg μl−1, and the reaction mixture was incubated overnight at 37°C. The digest was stored at 4°C or used immediately for immunoprecipitation.
Immunoprecipitation of bound peptides.
Immunoprecipitation of digested rAU08 VLP was carried out using DYNAL Dynabeads, protein G (Invitrogen), according to the manufacturer's instructions. Briefly, 5 to 10 μg of anti-rAU08 rabbit polyclonal serum was immobilized onto Dynabeads, prior to incubation with 15 to 20 μg of digested VLP at room temperature and pH 7.4 for 90 min. The bead/antibody/peptide complex was washed three times with 1× PBS before the complex was dissociated in 50 mM glycine, pH 2.8, during a 2-min rotating incubation at room temperature. The beads were removed and the eluent was collected and stored at 4°C prior to mass spectrometry (MS) analyses. Identical experiments using preimmune rabbit polyclonal serum were conducted in parallel as controls, and peptides detected by preimmune serum were considered nonspecific.
Sequencing of bound VLP peptides using MS.
Electrospray ionization-mass spectrometry (ESI-MS) was performed using a micrOTOF-Q-MS instrument (Bruker-Daltonics, Bremen, Germany) linked to a high-performance liquid chromatography (HPLC) instrument (Ultimate 3000, Thermo Fisher Scientific). VLP peptides were injected onto a Dionex Acclaim Pepmap100 nanotrap trapping column (Thermo Scientific) with 2% acetonitrile–0.1% formic acid (aqueous) as A-buffer. Following a 6-min isocratic wash with A-buffer, the sample was eluted and separated on a resolving column (Dionex Acclaim Pepmap RSLC; Thermo Scientific) using a gradient of 98% acetonitrile–0.1% formic acid (aqueous) as B-Buffer over 70 min at a flow rate of 300 nl min−1. The eluent from the column was directly electrosprayed into the mass spectrometer. Mass data were continuously acquired and for each MS spectrum, MS/MS spectra were recorded for the three most intense peaks. The data were annotated and deconvoluted using DataAnalysis software (Bruker-Daltonics). To identify the detected peptides, mass data were aligned to the capsid protein sequence using BioTools (Bruker-Daltonics) together with manual inspection and peak assignment.
Alignment of capsid protein sequences.
Seventy-four GII.3 VP1 gene sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) from strains collected between 1975 and 2010 were translated and aligned in order to determine the extent of GII.3 epitope conservation and to define a consensus sequence for each GII.3 lineage (47). A representative VP1 gene sequence of each GII genotype and five GII.4 variants, as defined by the Norovirus genotyping tool (48), were obtained from GenBank, translated, and aligned with the GII.3 lineage consensus sequences. Alignments were constructed using ClustalW as part of the Molecular Evolutionary Genetics Analysis (MEGA) program, version 5.1 (49).
Mimotope peptides.
A custom peptide library of 14 overlapping peptides (MP1 to MP14), 10 amino acids (aa) in length, with a 2-amino-acid offset was designed to further minimize potential GII.3 capsid immunoreactive regions detected by MS analysis (Mimotopes Pty Ltd., Melbourne, VIC, Australia) (Table 1). Mimotope peptides were biotin linked at the N terminus with a four-residue linker sequence (SGSG), and an amide group was incorporated at the C terminus. Peptides were prepared to a final stock concentration of 1.4 μmol ml−1 in 50% acetonitrile. Due to a highly negative overall charge, peptides MP7 and MP8 were dissolved in 50% acetonitrile and 28 mM (NH4)HCO3 solution and peptide MP6 was dissolved in 40% acetonitrile and 285 mM (NH4)HCO3 solution. Stock peptide solutions were aliquoted and stored at −30°C.
TABLE 1.
Minimization of P2 domain immunoreactive regions using rabbit anti-rAU08 polyclonal serum and Mimotope-supplied peptides
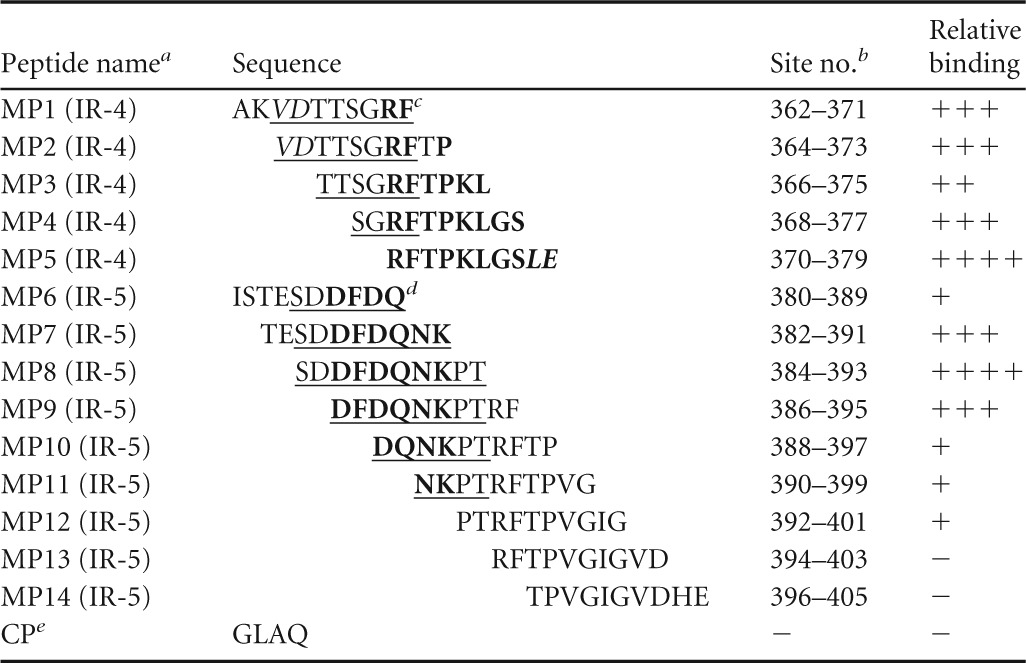
Name and number designated to each Mimotope peptide (MP). The P2 domain immunoreactive region for which the Mimotope was designed is indicated in parentheses, where “(IR-4)” indicates aa 362 to 379 and “(IR-5)” indicates aa 380 to 405.
Site no. refers to amino acid residue number of the capsid protein according to GII.3 strain AU08 numbering.
For immunoreactive region IR-4 (aa 362 to 379), bold residues represent residues that are required to form the high-affinity epitope, based on ELISA results. Underlined residues represent those that likely form a second epitope with reduced binding affinity, based on ELISA results and homology modeling. Structurally internal residues that are likely important for epitope structural formation are italicized.
For immunoreactive region IR-5 (aa 380 to 405), residues forming the minimal binding region of the epitope are underlined and the residues that are essential to binding are in bold.
CP is the negative-control peptide (CP) supplied by Mimotopes.
Mimotope ELISA.
The IgG reactivity of rabbit anti-rAU08 polyclonal serum with the Mimotope library was analyzed by ELISA according to the supplied protocol (Mimotopes). Briefly, preblocked, streptavidin-coated plates (Pierce, Thermo Scientific) were washed four times with PBS–0.1% Tween 20 (PBS-T) using the Thermo Scientific Well Wash 4 Mk2 plate washer. Unless otherwise specified, all subsequent washing steps were identical and all incubations were at room temperature. Wells were saturated with 10 pmol of Mimotope peptides diluted in PBS-T and incubated for 1 h. After washing, rabbit serum (anti-rAU08 polyclonal antiserum or rabbit preimmune serum) diluted at 1:2,000 and 1:8,000 in PBS-T was applied, and plates were incubated for 1 h. Plates were washed, and bound IgG was detected with HRP-conjugated goat anti-rabbit IgG (Abcam, Cambridge, United Kingdom), diluted 1:120,000 in PBS-T–1% sheep serum–1% casein. Plates were incubated with agitation for 1 h and washed four times with PBS-T, followed by two additional washes with PBS only. The presence of bound conjugate was detected by addition of TMB solution for 10 min. The reaction was stopped with 2 M H2SO4, and plates were read at 450 nm on a Titertek Multiscan MCC/340 MKII microtiter plate reader (Labsystems).
Homology modeling and mapping of immunoreactive regions on capsid dimer.
The tertiary structure of the capsid protein of strain AU08 was modeled based on solved homologous capsid structures using the I-TASSER online server (50). A dimer of duplicate capsid monomers was formed based on the Norwalk virus capsid structure (PDB identifier [ID]: 1IHM), using PyMol, version 1.3 (51). Homology models were viewed and immunoreactive regions highlighted using PyMol, version 1.3.
Statistics.
The overall effects of different VLPs or peptides on serum binding capacity were measured using statistical comparison tests available in Prism 6 (GraphPad Software). For human serum samples, the mean level of IgG bound to each VLP was compared using ordinary one-way analysis of variance (ANOVA), with Tukey's multiple comparison (MC) posttest correction. The mean level of rabbit anti-rAU08 polyclonal serum IgG binding to each VLP was compared using two-way ANOVA. Bonferroni's MC posttest was used to compare the serum binding affinity of each VLP to that of VLP rAU08 (the immunizing antigen). The mean level of binding of Mimotope peptides to rabbit anti-rAU08 polyclonal serum was compared using two-way ANOVA, with Tukey's MC posttest correction. A significant difference in VLP/peptide binding capacity for all tests was classified by a P value less than or equal to 0.05.
RESULTS
GII.3 VLP binding specificity.
The binding characteristics of the panel of GII.3 VLPs were first compared using archival human serum samples collected from three pediatric patients. All of the GII.3 VLPs bound to each of the serum samples, with no significant difference between the reactivity of each VLP, regardless of serum sample (Fig. 1) (one-way ANOVA, Tukey's MC test, where P ≤ 0.05). Preliminary data from previous unpublished observations showed that serum samples 1 and 2 were reactive with the rWR and rFV non-GII.3 VLPs; however, limited serum prevented further investigation of this reactivity (Kirkwood, unpublished).
FIG 1.
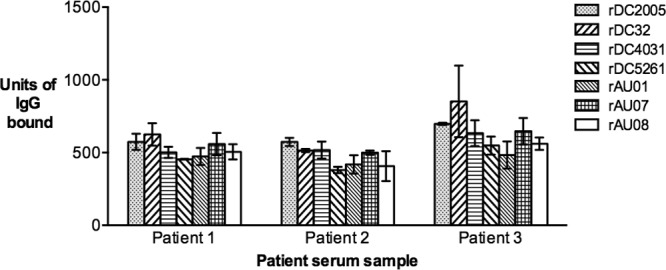
Binding of time-ordered GII.3 VLPs to human sera. The reactivity of serum IgG in human serum samples with the panel of time-ordered GII.3 VLPs was measured by ELISA and the results presented as an arbitrary concentration of bound IgG (y axis). The patient from whom each sample was collected is indicated (x axis). The bars represent the mean level of IgG bound to each VLP from two independent experiments. Error bars represent standard deviations.
The IgG binding profile of the anti-rAU08 polyclonal serum was determined for the GII.3 VLP panel, as well as for two non-GII.3 VLPs, GII.5 (rWR) and GII.6 (rFV). As shown in Fig. 2, there was no significant difference in the binding capacity of the homologous VLP (rAU08) compared to the other six GII.3 VLPs, regardless of time when the strain was circulating (two-way ANOVA, Bonferroni's MC test where P ≤ 0.05). The non-GII.3 VLPs, GII.5 and GII.6, bound anti-rAU08 polyclonal serum at a significantly lower level than the GII.3 VLP panel (two-way ANOVA, Bonferroni's MC test where P ≤ 0.05).
FIG 2.
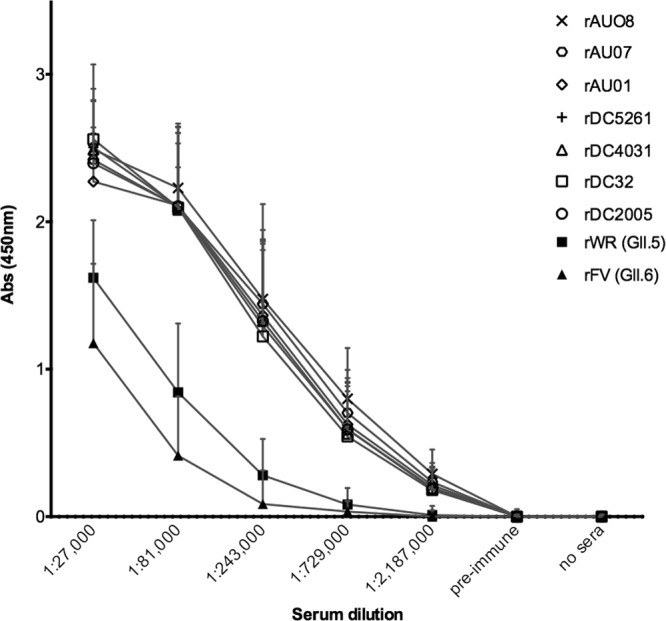
Binding of time-ordered GII.3 VLPs to anti-rAU08-specific polyclonal serum. The reactivity of rabbit anti-rAU08-specific polyclonal serum IgG with the panel of time-ordered GII.3 VLPs and two non-GII.3 VLPs (rWR and rFV) was measured by ELISA. Absorbance is shown on the y axis, and the polyclonal dilution series is shown on the x axis. Symbols represent the mean absorbance level of two independent experiments for each VLP at each serum dilution, and the error bars represent standard deviations.
Identification of immunoreactive regions on the capsid protein of norovirus AU08.
To localize immunoreactive regions (epitope-containing regions) of the GII.3 capsid protein, the most contemporary GII.3 VLP, rAU08, was digested using the endoproteinase Glu-C. Digestion of the rAU08 GII.3 VLP resulted in short peptides, of 5 to 41 residues in length, and a sequence coverage of ∼65 to 78% of the entire protein (Fig. 3A).
FIG 3.
Digestion and immunoprecipitation products as sequenced by liquid chromatography-MS/MS. An example of a typical mass spectrometry result with detected peptides aligned with the primary amino acid sequence of the capsid protein of strain AU08 (N terminus to C terminus) is shown. Amino acids making up the protein sequence are indicated by one-letter identifiers, and amino acid position is indicated above the sequence every 10 residues. The large gray bars below the amino acid sequence indicate that a peptide was detected that matches the expected mass of a peptide with that sequence composition. The small dark gray boxes correspond to the identified residue masses as determined through MS/MS sequencing. A greater number of dark gray boxes indicates greater result certainty. (A) Peptides detected following digestion of the VLP, prior to immunoprecipitation. (B) Peptides detected following immunoprecipitation of digested VLP with rabbit anti-rAU08 serum. (C) Peptides detected following immunoprecipitation of digested VLP with rabbit preimmune serum.
The peptides generated by Glu-C digestion of rAU08 VLP were subjected to immunoprecipitation using the rabbit anti-rAU08 polyclonal serum IgG. The precipitated peptides were sequenced by MS, and six peptides (IR-1 to IR-6) were identified as containing immunoreactive sequence based on results of three independent experiments (Table 2 and Fig. 3B). Two or three nonspecific peptides were consistently detected following immunoprecipitation with preimmune serum, and these peptides were discounted (Fig. 3C). The six immunoreactive peptides ranged in size from 17 to 38 amino acid residues, with an average length of 25 residues, and all matched a cleavage product as predicted for Glu-C (Table 2). Alignment of the six immunoreactive peptide sequences with the primary amino acid sequence of VP1 showed that two peptides each mapped to the S, P1, and P2 domains. Due to the denaturation steps required in the protease digestion protocol, it is likely that the majority of epitopes in these regions were linear.
TABLE 2.
Immunoreactive regions on the 2008 GII.3 capsid protein
| Name | Amino acid positiona (domainb) and sequencec | No. of fixed amino acid mutationsd |
|---|---|---|
| IR-1 | aa 26–63 (S), AMALEPVAGAAIAAPLTGQQNIIDPWIMNNFVQAPGGE | 1 (E30D, Lin B/D) |
| IR-2 | aa 64–80 (S), FTVSPRNSPGEVLLNLE | 0 |
| IR-3 | aa 237–255 (P1), MSNSRFPVPIDSLHTSPTE | 0 |
| IR-4 | aa 362–379 (P2), AKVDTTSGRFTPKLGSLE | 3 (V364I, Lin E; S368A, Lin D; A372T, Lin B/C/D/E) |
| IR-5 | aa 380–405 (P2), ISTESDDFDQNKPTRFTPVGIGVDHE | 7 (S381T/I, Lin B/C; S385D/G, Lin D/E; S389P/T/Q, Lin B/C/D/E; Q391K, Lin E; P392S, Lin B/C; R394K, Lin B/C; N404H, Lin D/E) |
| IR-6 | aa 497–526 (P1), AKLHKLGFITIAKNGDSPITVPPNGYFRFE | 0 |
Amino acid position corresponds to sequence numbering of strain AU08.
Capsid protein structural domain. P2 is the most exposed.
Amino acid sequence of strain AU08.
The number of amino acid sites in the immunoreactive region containing a mutation that has become conserved over time with reference to the oldest GII.3 strain (DC2005, isolated in 1975). The mutation and the GII.3 lineages (Lin) within which a mutation has been conserved are listed in parentheses. Lineage A, strains from 1975 to 1983; lineage B, strains from the 1980s; lineage C, strains from the 1990s; lineage D, strains from 2001 to 2010; lineage E, strains from 2006 to 2010 (47).
Sequence conservation of GII.3 antibody-binding regions in the capsid protein.
To discern the level of conservation in the identified immunoreactive regions over time, 74 GII.3 capsid protein sequences from 1975 to 2010 were aligned. Immunoreactive regions in the S and P1 domains of GII.3 capsid sequences were highly conserved, with occasional amino acid changes seen in single strains (Fig. 4). Immunoreactive regions IR-2 (S; aa 64 to 80), IR-3 (P1; aa 237 to 255), and IR-6 (P1; aa 497 to 525) contained no conserved substitutions among GII.3 strains. IR-1 (S; aa 26 to 63) contained one amino acid substitution (E30D) that became conserved in two GII.3 lineages (Fig. 4 and Table 2). The S and P1 domain epitopes combined had a lower proportion of variable amino acid sites (21%) than the S and P1 domains in their entirety (27%). Conversely, the two immunoreactive regions within the P2 domain, IR-4 (P2; aa 362 to 379) and IR-5 (P2; aa 380 to 405), were highly variable (Table 2 and Fig. 4). IR-4 and IR-5 had a combined proportion of 48% variable sites, which is more variable than the P2 domain in its entirety (43%).
FIG 4.
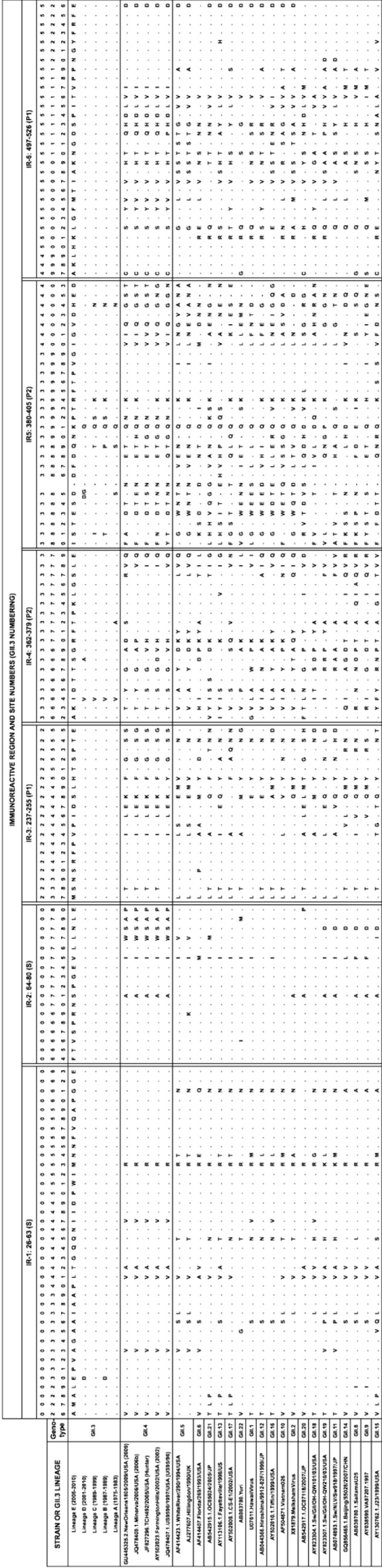
Amino acid alignment of immunoreactive regions. The immunoreactive regions of the capsid consensus sequence of each GII.3 lineage was aligned with the capsid sequence of FV (GII.6), WR (GII.5), five representative GII.4 epidemic strains, and a representative strain for each GII genotype. The immunoreactive region and individual site numbers (GII.3 rAU08 numbering) are indicated at the top, and the GII.3 lineage or strain and genotype are listed on the left. The capsid domain for each immunoreactive region is also indicated in parentheses: S, P1, and P2. Residues are indicated by 1-letter amino acid identifiers, and dots indicate identical residues to the reference sequence (GII.3 lineage E consensus sequence). A dash represents gaps in the alignment. Note that the AU08 strain varies from the lineage E consensus sequence at sites 364 (V) and 505 (I).
To explore the conservation of GII.3 immunoreactive regions within other GII genotypes, the VP1 consensus sequence of each GII.3 lineage was aligned with 25 VP1 protein sequences, representing the remaining GII genotypes (Fig. 4). All GII.3 immunoreactive regions were variable across the GII genogroup. The two immunoreactive regions in the S domain exhibited a high level of similarity (mean similarity: IR-1, 81%, and IR-2, 85%) between GII.3 and other genotypes. IR-2 (S; aa 64 to 80) was the most conserved, exhibiting 100% amino acid identity between GII.3, GII.10, GII.12, and GII.18 representative sequences while exhibiting a minimum identity of 65% between GII.3 and GII.4 sequences. The two immunoreactive regions within the P1 domain (IR-3 and IR-6) had average levels of conservation of 61% and 62%, while those in the P2 domain (IR-4 and IR-5) displayed 50% and 43% conservation between different genotypes. IR-5 (P2; aa 380 to 405) exhibited the greatest variability, where GII.3 shared a maximum amino acid identity of 58% (with GII.19) and a minimum identity of just 27% (with GII.21) (Fig. 4).
Confirmation and minimization of immunoreactive regions in the P2 domain using Mimotopes.
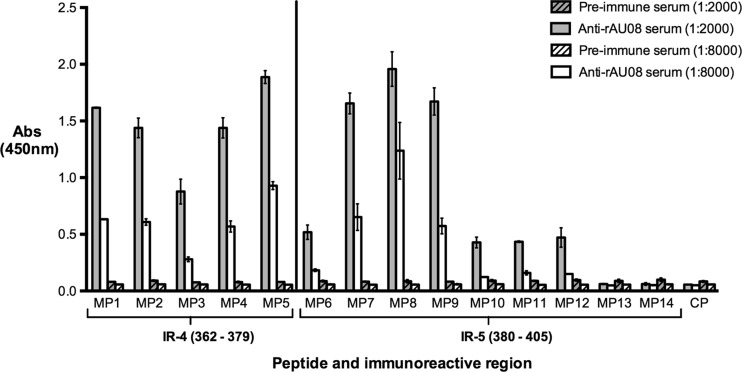
The two immunoreactive regions within the P2 domain (IR-4 and IR-5) were selected for further minimization using Mimotope technology. Fourteen 10-mer biotin-linked overlapping Mimotope peptides (MP1 to MP14) spanning the immunoreactive regions were screened for binding affinity to anti-rAU08 polyclonal serum. As seen in Fig. 5, at the lowest serum dilution, peptides MP1 to MP12 bound to the anti-rAU08 serum at a significantly higher level than the preimmune serum, while at the higher serum dilution, MP6 and MP10 to MP12 lost reactivity to the anti-rAU08 polyclonal antisera. Peptides MP13 and MP14 did not bind to anti-rAU08 serum at either dilution.
FIG 5.
Binding of Mimotope peptides to anti-rAU08-specific polyclonal serum. The reactivity of rabbit anti-rAU08-specific polyclonal serum IgG (at two dilutions) with Mimotope peptides (MP1 to MP14) was measured by ELISA. Absorbance is shown on the y axis, while the peptide name (Table 1) and the immunoreactive region that the peptides span are indicated on the x axis. The bars represent the mean absorbance level of two independent experiments, and the error bars represent standard deviations.
The Mimotope peptides (MP6 to MP14) spanning immunoreactive region IR-5 (P2; aa 380 to 405) revealed a binding trend that enabled identification of a minimal binding region. Peptides MP6 and MP10 to MP14 demonstrated low or no significant affinity with the polyclonal antibody, while peptides MP7, MP8, and MP9 demonstrated high antibody affinity. MP7 and MP9 showed comparable binding affinities, yet both had significantly lower affinities than MP8 (Fig. 5). Thus, the minimal binding region of immunoreactive region IR-5 was located to 384SDDFDQNKPT393 (Table 1). Residues at either end of the minimal binding region (384SD385 and 392PT393) significantly enhanced binding; however, they were not crucial for binding (Table 1). The IR-5 minimized epitope is highly variable, with 80% variable sites.
The Mimotope peptides (MP1 to MP5) spanning immunoreactive region IR-4 (P2; aa 362 to 379) all bound to anti-rAU08 IgG (Fig. 5). Peptide MP5 bound to anti-rAU08 serum at significantly higher levels than MP1 to MP4, while MP3 had a significantly lower affinity to anti-rAU08 (Table 1). The minimal binding residues of this immunoreactive region could not be defined through this assay alone and are further discussed in the homology modeling section below.
Mapping of immunoreactive regions onto a GII.3 structural homology model.
To locate the immunoreactive regions on the capsid tertiary structure, a homology model of the GII.3 strain AU08 capsid protein dimer was constructed, and immunoreactive regions were highlighted (Fig. 6). Several residues from both P2 domain immunoreactive regions are closely associated with the HBGA binding sites, and site 386(D) of IR-5 (P2; aa 380 to 405) is itself an HBGA binding site (25). IR-4 (P2; aa 362 to 379) is exposed on the surface of the capsid dimer in two distinct regions: on the upper surface of the P2 domain adjacent to the HBGA binding sites, and around the side of the P2 domain, beneath a protruding loop (Fig. 6). Since the surface-exposed region on the top of the P2 domain only constitutes amino acid 363K and the side chain of 365D, it is highly unlikely that it constitutes an epitope. Furthermore, these two residues are not part of the Mimotope peptide that had the highest affinity with the polyclonal serum. Thus, it likely that the residues that are exposed on the surface below the protruding loop (aa 366 to 372 and 375 to 377) form this epitope. Since peptide MP5 (aa 370 to 379) demonstrated the highest binding affinity to the polyclonal serum, surface-exposed residues 370RFT372 and 375LGS377 probably form a high-affinity epitope. Internal residues 378LE379 and possibly 373PK374 are important for epitope formation, since the loss of 378LE379 leads to significant reduction in binding (Table 1). Since peptides MP1 (aa 362 to 371) and MP2 (aa 364 to 373) also display a significant binding affinity to the anti-rAU08 polyclonal despite lacking half of the residues thought to form the high-affinity epitope, surface-exposed residues 366TTSG369 may also combine with residues 370RF371 to form a second, less reactive epitope. The loss of internal residues 364VD365 leads to a significant reduction in binding affinity, suggesting an important role for these residues in structurally forming the less reactive epitope (Table 1).
FIG 6.
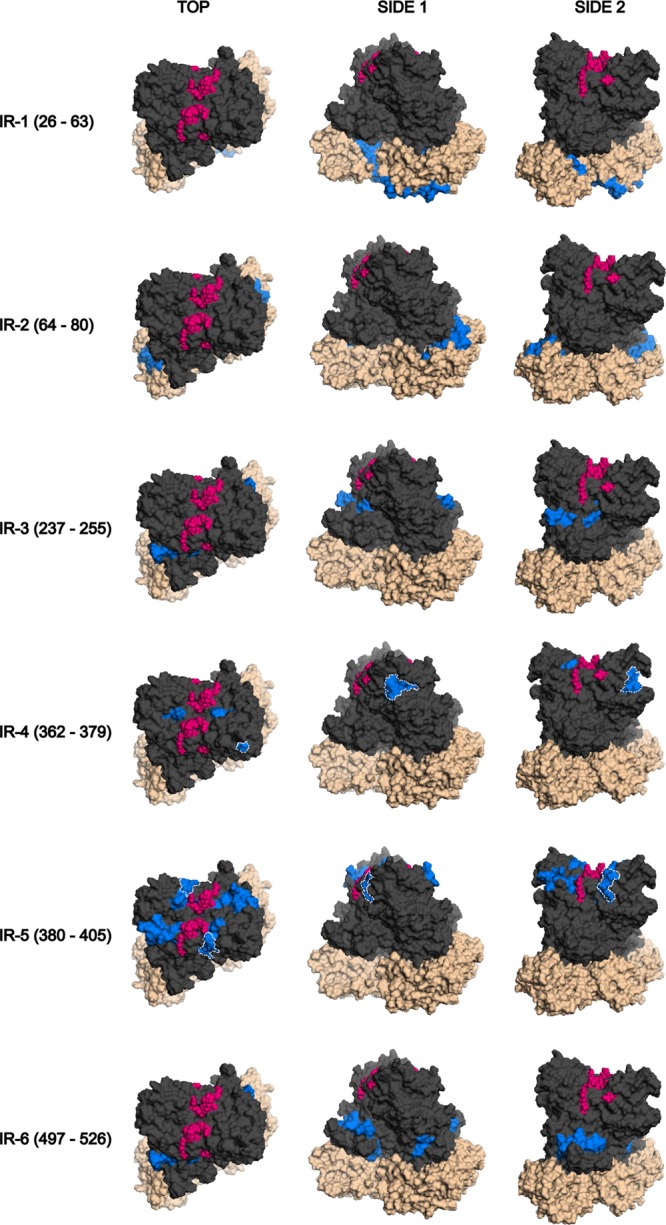
Immunoreactive regions mapped on the GII.3 capsid dimer. The six immunoreactive regions that were mapped in this study are displayed on the GII.3 (rAU08) capsid dimer homology model from top to bottom in the order that they appear in the primary sequence. The immunoreactive region is listed to the left of the models. Three angles for each immunoreactive region are displayed, the top and two side views, as indicated above the models. The protruding domain is displayed in dark gray, while the shell domain is denoted by beige coloring. The HBGA binding sites are indicated in pink, and the immunoreactive region is indicated in blue. The minimized epitopes of IR-4 (aa 362 to 379) and IR-5 (aa 380 to 405) are outlined by white dotted lines. Two overlapping epitopes constitute IR-4, which were inferred through a combination of Mimotope ELISA results and homology modeling. The minimized epitope of IR-5 was clearly determined through Mimotope ELISA results.
The two immunoreactive regions of the P1 domain are adjacent to each other in the tertiary structure, despite their distance in primary sequence (Fig. 6). The P1 immunoreactive regions form part of an outward protrusion from the dimer and are visible from the top of the dimer. The S domain immunoreactive region IR-2 (S; aa 64 to 80) also protrudes slightly outwards and is visible from the top of the dimer (Fig. 6). Residues of IR-1 (S; aa 26 to 63) are predominantly located on the base of the dimer. Both of the S domain immunoreactive regions are most likely accessible only during viral particle assembly or upon dissociation into dimers or partial viral particles.
DISCUSSION
Genotype GII.3 norovirus strains are a major cause of sporadic gastroenteritis, particularly in children (47). Norovirus GII.3 strains evolve at a rate comparable to that of GII.4 strains on a genomic level, yet not at the protein level, perhaps indicating limited change in immune targeted epitopes on the GII.3 capsid, restricting infection to the pediatric population (10). This study aimed to identify and characterize antibody-binding epitopes on the GII.3 capsid protein and to investigate temporal immunological variation.
To establish the differences in antibody-binding properties of GII.3 strains circulating in different years, we produced seven GII.3 VLPs, representing strains collected from 1975 to 2008, together with a strain-specific anti-GII.3 rabbit polyclonal targeting the most contemporary VLP (rAU08). The IgG binding profiles of the panel of GII.3 VLPs were compared using three human serum samples (collected from 1978 to 1980) and the strain-specific anti-rAU08 polyclonal serum, with no significant difference observed in the serum binding capacity of each GII.3 VLP (Fig. 1 and 2). This suggests that antibodies elicited following a presumed norovirus GII.3 infection during the 1970s or 1980s recognize the epitopes present on GII.3 noroviruses circulating at a similar time, as well as those circulating decades later. Equally, antibodies produced against the contemporary 2008 strain were reactive with GII.3 strains from previous years. Thus, it is likely that GII.3 antibody-binding epitopes are highly conserved over time, and strong intragenotype cross-reactivity is evident. Since intact VLPs were used for the ELISAs, we can assume that these cross-reactive epitopes are, for the most part, conformational. In addition, our epitope mapping experiments confirmed that GII.3 immunoreactive regions containing linear epitopes are also highly conserved overall, with the exception of those in the P2 domain (Fig. 4). The high level of intragenotype cross-reactivity and conservation of epitopes suggests that GII.3 strains may be limited to the pediatric population due to herd immunity.
Our findings are in contrast to results from Hansman and colleagues, who found various levels of cross-reactivity between five different GII.3 VLPs (isolated in the late 1990s or at an unknown date) and their respective antiserum (52). They reported that while some GII.3 VLPs were cross-reactive with specific heterologous GII.3 antisera, other GII.3 VLPs bound heterologous antisera with a 2- to 8-fold decrease in binding strength (52). However, three of the GII.3 strains studied by Hansman and colleagues were unusual strains. Two were uncommon recombinant strains with a GII.a RdRp from the late 1990s, and the third does not cluster within any GII.3 lineage (isolation date unknown) (47, 52). The VLPs in our study represented strains with either a pre-GII.3 ancestral RdRp (nontypeable) (47) or the highly common GII.3 or GII.b RdRp (47). Therefore, while our results show a consistency in GII.3 VLP-antibody binding, there may be some novel or uncommon strains that do not follow this pattern.
Previous studies have also shown a very broad intergenotype cross-reactivity of rabbit anti-GII.3 polyclonal serum or anti-GII.3 MAbs, which were able to bind to GI and GII VLPs. However, as seen in this study, these heterologous affinities were much lower than with the homologous GII.3 antigen (31, 44, 52, 53). Additionally, the sequence homology of the GII.3 immunoreactive regions mapped in this study varied greatly across genotypes (Fig. 4). This suggests a possible intergenotype cross-protective role for a GII.3-elicited immune response; however, protection would be limited, restricted perhaps only to certain genotypes.
It is likely that the intragenotype and intergenotype cross-reactive antibodies are elicited, for the most part, by the highly conserved S and P1 domain epitopes. Antibodies targeting the S and P1 domain epitopes may be elicited to disrupt replication, as seen for other viruses (54, 55). There is evidence to suggest this in the case of the epitope(s) within region IR-1 (S; aa 26 to 63), which contains the 48IDPWI52 motif that is thought to interact with the VP2 protein in the formation of the capsid structure (56). VP2 is thought to play a role in stabilization of the capsid structure and encapsidation of the genome (56); therefore, antibodies targeting this region may prevent assembly of stable capsids containing genomic material. Additionally, IR-1 appears to be highly immunogenic and cross-reactive, as it overlaps with epitopes 48IDPWI52, 44QQNIIDPWIMN54, and 51WIRNNF56 as previously mapped for MAbs TV20, 1B4/IF6, and N2C3, generated against GII.3 (strain TV24), GII.3 (strain NV36), and GII.4 (strain NVgz01) norovirus capsids, respectively (31, 36, 57). These overlapping epitopes are highly conserved across genogroups, particularly the recently mapped epitope 48IDPWI52, which is completely conserved across all genotypes of the Norovirus genus (57). Furthermore, MAbs that bound these S domain epitopes were found to be highly cross-reactive with norovirus strains of several genogroups (31, 36, 57). Thus, antibodies targeting the highly conserved S and P1 domain epitopes are often cross-reactive, and while they are unlikely to neutralize viruses, they may function to disrupt norovirus replication and limit disease progression.
The two immunoreactive regions mapped to the P2 domain in this study overlap a larger epitope (aa 361 to 403) previously mapped for three anti-GII.3 MAbs, further supporting the validity of these immunoreactive regions and our epitope mapping method (58). Our epitope mapping method allowed identification of large immunoreactive regions containing at least one epitope for sequence characterization. However, while most antigen-antibody contact surfaces are large (15 to 20 amino acids) and all residues may affect affinity, only 5 or 6 residues are energetically important in forming an epitope and are likely to be involved in productive antibody binding (59). Due to their great variability, the two P2 domain immunoreactive regions were selected for further minimization to characterize the key “productive binding” residues forming the epitope.
Based on combined evaluation of the Mimotope assay and homology modeling results, immunoreactive region IR-4 (P2; aa 362 to 379) likely contains two overlapping epitopes formed by residues 364 to 371 or 370 to 379, both located below a protruding loop (Fig. 6). Sites 368 and 372 were previously identified as evolutionarily important residues, and conserved changes at these sites may be due to immune pressure (47). However, our ELISA data demonstrate that these changes do not affect overall serum reactivity, since the variety of GII.3 VLPs on the panel represented all previous permutations at these sites and serum was completely cross-reactive. The GII.3 capsid protein evolves by a cyclic nature of reversion back to previously used residues, which may contribute to cross-reactivity among temporally distant norovirus strains, despite immune pressured change (10).
The minimal binding region of IR-5 (P2; aa 380 to 405) was located to residues 384SDDFDQNKPT393. Residue pairs at either end of the minimal binding region (384SD385 and 392PT393) are not imperative but significantly enhance binding. This minimized epitope has a substantially higher proportion of variable sites (80%) than the P2 region in its entirety (43%), suggesting immune-driven change in this epitope. Specifically, residues 385, 389, 391, and 392 in this epitope are highly variable, evolutionarily important residues, and substitutions at these sites are likely to be immune driven (47). In particular, residues 385 and 389 have previously been shown to be evolving under positive selection (10, 47). Furthermore, residue 385 aligns with residue 372 of the GII.4 mapped blockade epitope “A,” suggesting that this epitope may play a similar blockade role (30). This suggestion is endorsed by the epitope location, as epitope residues surround (sites 384, 385, and 387) or constitute (site 386) the HBGA binding sites (Fig. 6) (60). The high variability in this epitope suggests minimal contribution to GII.3 intragenotype and intergenotype cross-reactivity; however, changes at these sites do not reduce intragenotype overall serum reactivity, as demonstrated by universal serum binding to GII.3 VLPs possessing various permutations at evolutionarily important sites. Additionally, a previous study showed that anti-GII.3 convalescent-phase serum was able to block attachment of GII.4 VLPs to an HBGA carbohydrate, albeit to a lesser extent than the homologous convalescent-phase serum (45). Therefore, anti-GII.3 blockade antibodies may have the ability to bind to nonhomologous antigens to some degree.
In conclusion, this study characterized the intragenotype immune cross-reactivity of GII.3-targeted antibodies, while also utilizing a novel approach to identify and characterize major linear antibody-binding epitopes on the norovirus GII.3 capsid protein. Our data demonstrate that linear and conformational epitopes of the norovirus GII.3 capsid are mostly highly conserved and that a significant degree of cross-reactivity is seen within this genotype. The high cross-reactivity and limited evolution in GII.3 epitopes suggest that herd immunity against GII.3 may account for reduced GII.3 prevalence in adults. While further studies are required to confirm that this cross-reactive immunity actually prevents disease, our study suggests that a GII.3 strain would be a valuable inclusion in a pediatric targeted multivalent vaccine. Continued characterization of major norovirus antibody-binding epitopes is important for effective vaccine design, as targeting key antibody-binding residues in vaccine reformulation may prevent the emergence of antigenic variants in an immune population.
ACKNOWLEDGMENTS
This study was supported by the Victorian Government's Operational Infrastructure Support Program and the Intramural Research Program of the NIH, NIAID.
J. E. Mahar was supported by an Australian Postgraduate Award, and C. D. Kirkwood was supported by a Research Fellowship from the National Health and Medical Research Council of Australia (607347).
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231. 10.3201/eid1408.071114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi JN, Graham DY, Wang KN, Estes MK. 1990. Norwalk virus genome cloning and characterization. Science 250:1580–1583. 10.1126/science.2177224 [DOI] [PubMed] [Google Scholar]
- 4.Green KY, Ando T, Balayan MS, Berke T, Clarke IN, Estes MK, Matson DO, Nakata S, Neill JD, Studdert MJ, Thiel HJ. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl 2):S322–S330. 10.1086/315591 [DOI] [PubMed] [Google Scholar]
- 5.Katayama K. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225–239. 10.1006/viro.2002.1568 [DOI] [PubMed] [Google Scholar]
- 6.Jiang X, Wang M, Wang K, Estes MK. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51–61. 10.1006/viro.1993.1345 [DOI] [PubMed] [Google Scholar]
- 7.Bertolotti-Ciarlet A, Crawford SE, Hutson AM, Estes MK. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 77:11603–11615. 10.1128/JVI.77.21.11603-11615.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mesquita JR, Barclay L, Nascimento MS, Vinje J. 2010. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 16:980–982. 10.3201/eid1606.091861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. 2013. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 158:2059–2068. 10.1007/s00705-013-1708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boon D, Mahar JE, Abente EJ, Kirkwood CD, Purcell RH, Kapikian AZ, Green KY, Bok K. 2011. Comparative evolution of GII.3 and GII.4 norovirus over a 31-year period. J. Virol. 85:8656–8666. 10.1128/JVI.00472-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright PJ, Gunesekere IC, Doultree JC, Marshall JA. 1998. Small round-structured (Norwalk-like) viruses and classical human caliciviruses in southeastern Australia, 1980–1996. J. Med. Virol. 55:312–320. [DOI] [PubMed] [Google Scholar]
- 12.Guo L, Song J, Xu X, Ren L, Li J, Zhou H, Wang M, Qu J, Wang J, Hung T. 2009. Genetic analysis of norovirus in children affected with acute gastroenteritis in Beijing, 2004–2007. J. Clin. Virol. 44:94–98. 10.1016/j.jcv.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 13.Sdiri-Loulizi K, Ambert-Balay K, Gharbi-Khelifi H, Sakly N, Hassine M, Chouchane S, Guediche MN, Pothier P, Aouni M. 2009. Molecular epidemiology of norovirus gastroenteritis investigated using samples collected from children in Tunisia during a four-year period: detection of the norovirus variant GGII.4 Hunter as early as January 2003. J. Clin. Microbiol. 47:421–429. 10.1128/JCM.01852-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker SP, Cubitt WD, Jiang X. 1995. Enzyme immunoassay using baculovirus-expressed human calicivirus (Mexico) for the measurement of IgG responses and determining its seroprevalence in London, UK. J. Med. Virol. 46:194–200. 10.1002/jmv.1890460305 [DOI] [PubMed] [Google Scholar]
- 15.Smit TK, Steele AD, Peenze I, Jiang X, Estes MK. 1997. Study of Norwalk virus and Mexico virus infections at Ga-Rankuwa Hospital, Ga-Rankuwa, South Africa. J. Clin. Microbiol. 35:2381–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levett PN, Gu M, Luan B, Fearon M, Stubberfield J, Jamieson F, Petric M. 1996. Longitudinal study of molecular epidemiology of small round-structured viruses in a pediatric population. J. Clin. Microbiol. 34:1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chhabra P, Dhongade RK, Kalrao VR, Bavdekar AR, Chitambar SD. 2009. Epidemiological, clinical, and molecular features of norovirus infections in western India. J. Med. Virol. 81:922–932. 10.1002/jmv.21458 [DOI] [PubMed] [Google Scholar]
- 18.Phan TG, Kaneshi K, Ueda Y, Nakaya S, Nishimura S, Yamamoto A, Sugita K, Takanashi S, Okitsu S, Ushijima H. 2007. Genetic heterogeneity, evolution, and recombination in noroviruses. J. Med. Virol. 79:1388–1400. 10.1002/jmv.20924 [DOI] [PubMed] [Google Scholar]
- 19.Phan TG, Kuroiwa T, Kaneshi K, Ueda Y, Nakaya S, Nishimura S, Yamamoto A, Sugita K, Nishimura T, Yagyu F, Okitsu S, Muller WE, Maneekarn N, Ushijima H. 2006. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 78:971–978. 10.1002/jmv.20649 [DOI] [PubMed] [Google Scholar]
- 20.Mahar JE, Kirkwood CD. 2011. Characterization of norovirus strains in Australian children from 2006 to 2008: prevalence of recombinant strains. J. Med. Virol. 83:2213–2219. 10.1002/jmv.22215 [DOI] [PubMed] [Google Scholar]
- 21.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290. 10.1126/science.286.5438.287 [DOI] [PubMed] [Google Scholar]
- 22.Debbink K, Donaldson EF, Lindesmith LC, Baric RS. 2012. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J. Virol. 86:1214–1226. 10.1128/JVI.06189-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindesmith LC, Debbink K, Swanstrom J, Vinje J, Costantini V, Baric RS, Donaldson EF. 2012. Monoclonal antibody-based antigenic mapping of norovirus GII. 4-2002. J. Virol. 86:873–883. 10.1128/JVI.06200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansman GS, Biertumpfel C, Georgiev I, Mclellan JS, Chen L, Zhou T, Katayama K, Kwong PD. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J. Virol. 85:6687–6701. 10.1128/JVI.00246-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957. 10.1128/JVI.00219-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan M, Xia M, Cao S, Huang P, Farkas T, Meller J, Hegde RS, Li X, Rao Z, Jiang X. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324–334. 10.1016/j.virol.2008.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale AD, Tanaka TN, Kitamoto N, Ciarlet M, Jiang X, Takeda N, Brown DW, Estes MK. 2000. Identification of an epitope common to genogroup 1 “Norwalk-like viruses.” J. Clin. Microbiol. 38:1656–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker TD, Kitamoto N, Tanaka T, Hutson AM, Estes MK. 2005. Identification of genogroup I and genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 79:7402–7409. 10.1128/JVI.79.12.7402-7409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parra GI, Abente EJ, Sandoval-Jaime C, Sosnovtsev SV, Bok K, Green KY. 2012. Multiple antigenic sites are involved in blocking the interaction of GII.4 norovirus capsid with ABH histo-blood group antigens. J. Virol. 86:7414–7426. 10.1128/JVI.06729-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen DJ, Noad R, Samuel D, Gray JJ, Roy P, Iturriza-Gomara M. 2009. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol. J. 6:150. 10.1186/1743-422X-6-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoda T, Suzuki Y, Terano Y, Yamazaki K, Sakon N, Kuzuguchi T, Oda H, Tsukamoto T. 2003. Precise characterization of norovirus (Norwalk-like virus)-specific monoclonal antibodies with broad reactivity. J. Clin. Microbiol. 41:2367–2371. 10.1128/JCM.41.6.2367-2371.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batten CA, Clarke IN, Kempster SL, Oliver SL, Bridger JC, Lambden PR. 2006. Characterization of a cross-reactive linear epitope in human genogroup I and bovine genogroup III norovirus capsid proteins. Virology 356:179–187. 10.1016/j.virol.2006.07.034 [DOI] [PubMed] [Google Scholar]
- 33.Oliver SL, Batten CA, Deng Y, Elschner M, Otto P, Charpilienne A, Clarke IN, Bridger JC, Lambden PR. 2006. Genotype 1 and genotype 2 bovine noroviruses are antigenically distinct but share a cross-reactive epitope with human noroviruses. J. Clin. Microbiol. 44:992–998. 10.1128/JCM.44.3.992-998.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lochridge VP. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799–2806. 10.1099/vir.0.81134-0 [DOI] [PubMed] [Google Scholar]
- 35.Hansman GS, Taylor DW, Mclellan JS, Smith TJ, Georgiev I, Tame JRH, Park SY, Yamazaki M, Gondaira F, Miki M, Katayama K, Murata K, Kwong PD. 2012. Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle. J. Virol. 86:3635–3646. 10.1128/JVI.06868-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Zhou R, Wang Y, Sheng H, Tian X, Li H, Qiu H. 2009. Identification and characterization of a native epitope common to norovirus strains GII/4, GII/7 and GII/8. Virus Res. 140:188–193. 10.1016/j.virusres.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 37.Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. 2012. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 8:e1002705. 10.1371/journal.ppat.1002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, Johnston RE, Weber DJ, Baric RS. 2010. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J. Virol. 84:1800–1815. 10.1128/JVI.02179-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. 2011. Norovirus vaccine against experimental human Norwalk Virus illness. N. Engl. J. Med. 365:2178–2187. 10.1056/NEJMoa1101245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez K, Wahid R, Richardson C, Bargatze RF, El-Kamary SS, Sztein MB, Pasetti MF. 2012. Intranasal vaccination with an adjuvanted Norwalk virus-like particle vaccine elicits antigen-specific B memory responses in human adult volunteers. Clin. Immunol. 144:98–108. 10.1016/j.clim.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 41.Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan M, Jiang X. 2012. Norovirus P particle: a subviral nanoparticle for vaccine development against norovirus, rotavirus and influenza virus. Nanomedicine (Lond.) 7:889–897. 10.2217/nnm.12.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parra GI, Bok K, Taylor R, Haynes J, Sosnovtsev SV, Richardson C, Green KY. 2012. Immunogenicity and specificity of norovirus consensus GII.4 virus-like particles in monovalent and bivalent vaccine formulations. Vaccine 30:3580–3586. 10.1016/j.vaccine.2012.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okame M, Shiota T, Hansman G, Takagi M, Yagyu F, Takanashi S, Phan TG, Shimizu Y, Kohno H, Okitsu S, Ushijima H. 2007. Anti-norovirus polyclonal antibody and its potential for development of an antigen-ELISA. J. Med. Virol. 79:1180–1186. 10.1002/jmv.20906 [DOI] [PubMed] [Google Scholar]
- 45.Rockx B, Baric RS, De Grijs I, Duizer E, Koopmans MP. 2005. Characterization of the homo- and heterotypic immune responses after natural norovirus infection. J. Med. Virol. 77:439–446. 10.1002/jmv.20473 [DOI] [PubMed] [Google Scholar]
- 46.Bok K, Abente EJ, Realpe-Quintero M, Mitra T, Sosnovtsev SV, Kapikian AZ, Green KY. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901. 10.1128/JVI.00864-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahar JE, Bok K, Green KY, Kirkwood CD. 2013. The importance of intergenic recombination in norovirus GII.3 evolution. J. Virol. 87:3687–3698. 10.1128/JVI.03056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroneman A, Vennema H, Deforche K, Avoort HVD, Peñaranda S, Oberste MS, Vinjé J, Koopmans M. 2011. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51:121–125. 10.1016/j.jcv.2011.03.006 [DOI] [PubMed] [Google Scholar]
- 49.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrödinger LLC. 2010. The PyMOL molecular graphics system, version 1.3. Schrödinger, LLC, New York, NY [Google Scholar]
- 52.Hansman GS, Natori K, Shirato-Horikoshi H, Ogawa S, Oka T, Katayama K, Tanaka T, Miyoshi T, Sakae K, Kobayashi S, Shinohara M, Uchida K, Sakurai N, Shinozaki K, Okada M, Seto Y, Kamata K, Nagata N, Tanaka K, Miyamura T, Takeda N. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87:909–919. 10.1099/vir.0.81532-0 [DOI] [PubMed] [Google Scholar]
- 53.Belliot G, Noel JS, Li JF, Seto Y, Humphrey CD, Ando T, Glass RI, Monroe SS. 2001. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses.” J. Clin. Microbiol. 39:4288–4295. 10.1128/JCM.39.12.4288-4295.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donker NC, Foley M, Tamvakis DC, Bishop R, Kirkwood CD. 2011. Identification of an antibody-binding epitope on the rotavirus A non-structural protein NSP2 using phage display analysis. J. Gen. Virol. 92:2374–2382. 10.1099/vir.0.032599-0 [DOI] [PubMed] [Google Scholar]
- 55.Wright A, Yan H, Lamm ME, Huang YT. 2006. Immunoglobulin A antibodies against internal HIV-1 proteins neutralize HIV-1 replication inside epithelial cells. Virology 356:165–170. 10.1016/j.virol.2006.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vongpunsawad S, Prasad BV, Estes MK. 2013. Norwalk virus minor capsid protein VP2 associates within the VP1 shell domain. J. Virol. 87:4818–4825. 10.1128/JVI.03508-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parra GI, Azure J, Fischer R, Bok K, Sandoval-Jaime C, Sosnovtsev SV, Sander P, Green KY. 2013. Identification of a broadly cross-reactive epitope in the inner shell of the norovirus capsid. PLoS One 8:e67592. 10.1371/journal.pone.0067592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoda T, Terano Y, Suzuki Y, Yamazaki K, Oishi I, Utagawa E, Shimada A, Matsuura S, Nakajima M, Shibata T. 2000. Characterization of monoclonal antibodies generated against Norwalk virus GII capsid protein expressed in Escherichia coli. Microbiol. Immunol. 44:905–914. 10.1111/j.1348-0421.2000.tb02582.x [DOI] [PubMed] [Google Scholar]
- 59.Novotny J. 1991. Protein antigenicity: a thermodynamic approach. Mol. Immunol. 28:201–207. 10.1016/0161-5890(91)90062-O [DOI] [PubMed] [Google Scholar]
- 60.Tan M, Xia M, Chen Y, Bu W, Hegde RS, Meller J, Li X, Jiang X. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. 10.1371/journal.pone.0005058 [DOI] [PMC free article] [PubMed] [Google Scholar]