ABSTRACT
Bacteriophage T7 terminator Tφ is a class I intrinsic terminator coding for an RNA hairpin structure immediately followed by oligo(U), which has been extensively studied in terms of its transcription termination mechanism, but little is known about its physiological or regulatory functions. In this study, using a T7 mutant phage, where a 31-bp segment of Tφ was deleted from the genome, we discovered that deletion of Tφ from T7 reduces the phage burst size but delays lysis timing, both of which are disadvantageous for the phage. The burst downsizing could directly result from Tφ deletion-caused upregulation of gene 17.5, coding for holin, among other Tφ downstream genes, because infection of gp17.5-overproducing Escherichia coli by wild-type T7 phage showed similar burst downsizing. However, the lysis delay was not associated with cellular levels of holin or lysozyme or with rates of phage adsorption. Instead, when allowed to evolve spontaneously in five independent adaptation experiments, the Tφ-lacking mutant phage, after 27 or 29 passages, recovered both burst size and lysis time reproducibly by deleting early genes 0.5, 0.6, and 0.7 of class I, among other mutations. Deletion of genes 0.5 to 0.7 from the Tφ-lacking mutant phage decreased expression of several Tφ downstream genes to levels similar to that of the wild-type phage. Accordingly, phage T7 lysis timing is associated with cellular levels of Tφ downstream gene products. This suggests the involvement of unknown factor(s) besides the known lysis proteins, lysozyme and holin, and that Tφ plays a role of optimizing burst size and lysis time during T7 infection.
IMPORTANCEE. coli
INTRODUCTION
Bacteriophage T7 is an obligate lytic Escherichia coli phage that has been extensively studied for more than 60 years (1). T7 RNA polymerase, the only RNA polymerase (gp1) in the T7 genome, is one of the best-characterized RNA polymerases, and its transcription mechanisms have been studied in detail (2–6). In the T7 genome, an intrinsic termination signal for T7 RNA polymerase, terminator Tφ, is positioned between genes 10 and 11 at the late, class III region (Fig. 1A). Although the transcription termination mechanism of T7 RNA polymerase at terminator Tφ has been rather well studied, there have been few reports thus far on the in vivo roles of this terminator.
FIG 1.
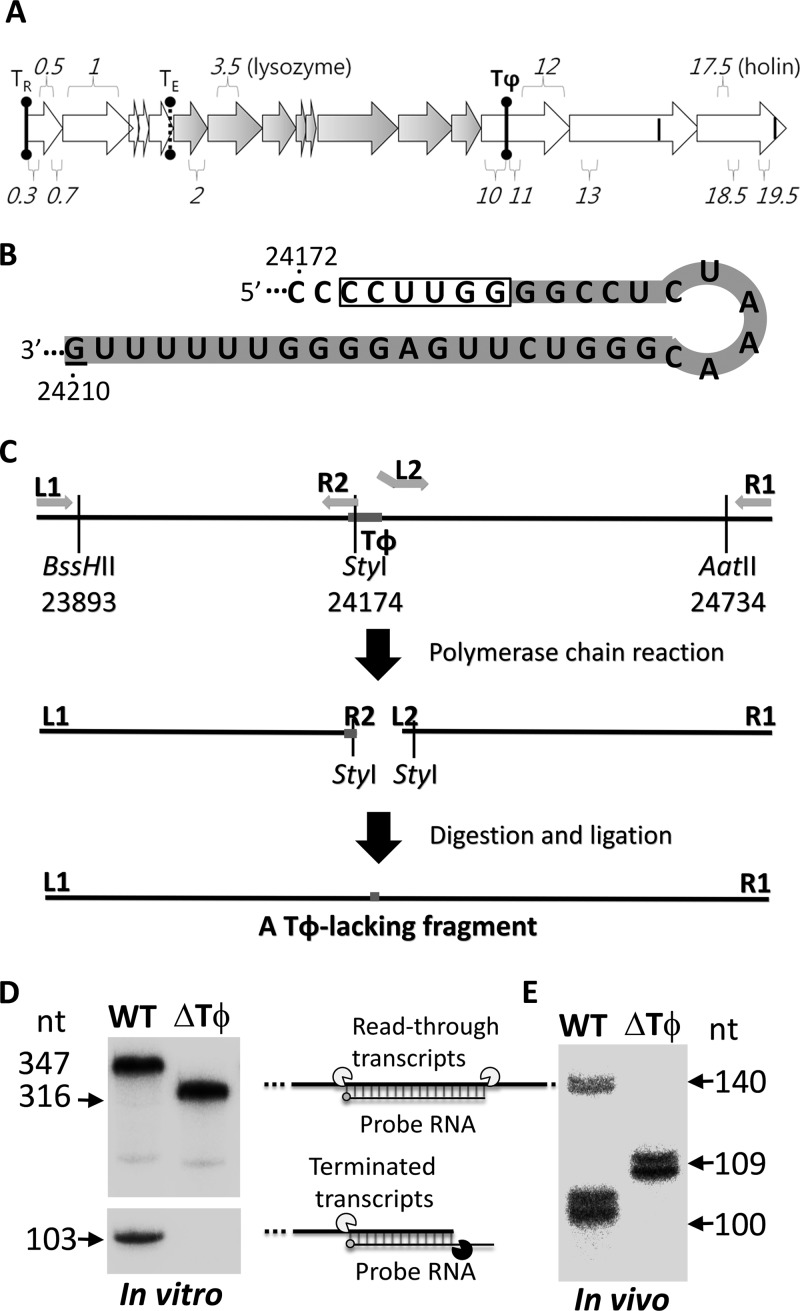
Terminator Tφ-lacking mutant of T7 phage. (A) Major transcripts from phage T7 genome. Arrows represent RNase III-processed transcripts of T7 class I (empty arrows on the left), II (gray arrows in the middle), and III (empty arrows on the right) genes, and each arrow starts at a promoter and ends at an RNase III cleavage site. Where multiple promoters are present between two RNase III cleavage sites, extra vertical bars are shown within arrows. Two terminators for T7 RNA polymerase (TR at the beginning of the genome and Tφ in the middle of the class III region) are shown with a solid-line dumbbell, while E. coli RNA polymerase terminator TE is shown with a dotted-line dumbbell. Among the 56 known or potential genes, only several genes mentioned in the text are shown. (B) RNA hairpin of the intrinsic terminator Tφ. Transcription terminates with the G residue at genomic position 24210. The shaded 31 residues (from positions 24180 to 24210) are deleted in the Tφ-lacking mutant. The StyI recognition sequence is boxed. (C) Strategy employed to create a Tφ-lacking fragment used in constructing a Tφ-lacking T7 mutant phage. (D) Confirmation of Tφ deletion using in vitro multiple-round transcription reactions. (E) Confirmation of Tφ deletion in E. coli after infection using RNase protection assays.
Terminator Tφ encodes a stable GC-rich RNA hairpin structure followed by a stretch of six U residues and the termination site G residue (Fig. 1B). Both the RNA secondary structure and primary sequence have been demonstrated to affect termination efficiency at Tφ. In vivo, read-through at terminator Tφ is necessary for the expression of T7 genes 11 and 12, as these two genes are located immediately downstream of Tφ and do not have their own promoters. It has also been reported that formation of a hairpin structure derived from the Tφ sequence at the 3′ end of the gene 10 transcript helps increase stability of the transcript mRNA and thereby raises its expression level more than 2-fold (7). Additionally, a frameshift in gene 10 mRNA translation, necessary for production of the minor capsid protein gp10B, also requires the Tφ-derived hairpin structure at the 3′ end of the mRNA, although failure of the frameshift shows little phenotypic effect on phage infection (8, 9). To our knowledge, there has been no report on the functions of terminator Tφ during T7 phage infection at phenotypic levels.
The replication cycle of phage T7 typically consists of three phases: (i) searching for and adsorption to a host, followed by (ii) transfer of its linear DNA genome into a host cell and expressing phage genes, and completion when (iii) host cells are lysed to release newly assembled phage progenies. All three phases have been well studied at molecular levels (1). The E. coli receptor for phage T7 is lipopolysaccharide on the surface, which binds to the C-terminal end of T7 tail fiber protein (gp17). Genomic DNA is then translocated into the host cell in a transcription-dependent manner, first by E. coli RNA polymerase and then by T7 RNA polymerase.
The lysis machinery of phage T7 comprises three components: holin (gp17.5), lysozyme (gp3.5), and possible spanins (gp18.5 and gp18.7). While deletion of the lysozyme greatly delayed lysis, there was no significant change in lysis time in the absence of holin (10). Spanins are needed for lysis only at high salt concentrations (11). During infection, the phage gene expression timings and levels are tightly regulated, mainly at transcriptional levels, by coupling of transcription with translocation of phage DNA. This suggests the importance of terminator Tφ in maintaining the balance of expression among T7 genes during phage infection.
In this study, we show that the presence of terminator Tφ is required to maintain normal burst size and lysis time of phage T7 during infection of E. coli. We observed that deletion of terminator Tφ from T7 phage led to a reduction in phage burst size and a delay in lysis time, which could not be explained by any known lysis pathway of T7 phage. Instead, the Tφ-lacking mutant was able to recover the wild-type lysis phenotype by deleting an early gene cluster. The results suggest for the first time the physiological roles of terminator Tφ during phage infection.
MATERIALS AND METHODS
Bacteria and phage preparation.
E. coli BL21 (Stratagene) was used as the host organism for T7 phage preparation and infection. T7 phage was purchased from the ATCC (BAA-1025-B2). A Tφ-lacking T7 phage mutant was constructed with a deletion of 31 bp of the Tφ terminator. Revertants 1 through 5 were evolved from the Tφ-lacking mutant phage. The Δ0.5-0.7 mutant of the T7 phage was constructed from revertant 1 by inserting the wild-type Tφ terminator sequence back into its genome. Phage lysate was prepared by adding either one drop of archival phage stock or a single plaque into a BL21 host culture with an absorbance at 595 nm (A595) of ∼0.5 and shaking until the culture was clarified. NaCl was added to a final concentration of 1 M, and the total lysate was centrifuged. The supernatant was then passed through a 0.45-μm syringe-driven filter unit (Millipore). Phage titer was measured using a soft-agar assay as previously described (12). E. coli DH5α (Invitrogen) was used to make Tφ-lacking phage mutants and for DNA cloning.
Assembly of T7 mutant genomes.
For the Tφ-lacking T7 mutant, a mutant phage of T7 lacking a 31-bp segment of Tφ was constructed and assembled (Fig. 1C) using a previously described strategy (13). In brief, a Tφ upstream part (amplicon L1-R2 from genomic positions 23823 to 24176) and a Tφ downstream part (amplicon L2-R1 from genomic positions 24211 to 24801) were juxtaposed using StyI digestion and ligation. The resulting Tφ-lacking fragment of 979 bp was then ligated into the T7 genome using two single-cut sites, BssHII (cut at genomic position 23893) and AatII (cut at 24734), to make a Tφ-lacking genome. The 40-kb Tφ-lacking T7 genomic DNA was then transfected into competent DH5α cells and plated on soft agar to allow plaque formation. Plaque PCR was then performed, and the PCR products were purified and sequenced to screen the correct mutant. The correct plaque was then used to make primary phage lysate. For making Δ0.5-0.7 T7 phage from revertant 1, the wild-type sequence of the Tφ terminator was inserted back in the genome using the same AatII and BssHII sites.
Measurement of transcription termination efficiency.
In vitro transcription termination efficiency was measured on DNA templates amplified from plasmid pKM01 (3) having either the wild-type terminator Tφ or a Tφ-lacking fragment cloned into the KpnI/HindIII site. The transcription reaction was carried out in a 20-μl reaction mixture containing 0.5 μg template DNA, 40 mM Tris-HCl, pH 7.9, 6 mM MgCl2, 100 mM KCl, 10 mM dithiothreitol, 0.5 mM ribonucleotide triphosphates, 20 μCi [α-32P]UTP (PerkinElmer), 4 U of RNasin (Promega), and 40 U of phage T7 RNA polymerase. The reaction mixture was incubated at room temperature for 1 h before being stopped by adding 10 μl of loading buffer (10 mM EDTA, 12 M urea, 0.025% bromophenol blue, and 0.025% xylene cyanol). To measure in vivo transcription termination efficiencies, an RNase protection assay was performed as previously described (3). Products were analyzed on 8 M urea–12% polyacrylamide gels. Band intensities were analyzed using TINA 2.0 software (DesignSoft).
Measurement of burst size.
Phage burst size was measured as previously described (14), with some modifications. Briefly, a BL21 culture at an A595 of 0.3 to 0.4 was infected with phages at a multiplicity of infection (MOI) of 0.02. After allowing phages to be attached to host cells for 2 min, the culture was centrifuged to discard all free phages and quickly diluted 10,000-fold into prewarmed media. Titers of samples at 6 and 8 min after infection were determined to obtain the number of infective centers. The culture was further diluted 100-fold, samples were extracted every 2 min from 15 to 25 min after infection and treated with chloroform, and the titers were determined. The burst size of individual cells was calculated by fitting the data from three independent experiments into a sigmoidal curve model using the least-squares fitting method.
Kinetic assay of lysis.
An E. coli BL21 culture at an A595 of 0.2 to 0.3 was infected with phages at an MOI of 3 to 4. Adsorption was allowed to proceed for 2 min at 37°C, and the culture was centrifuged to eliminate all free phages. The pellet was resuspended in a new medium and allowed to grow at 37°C in a shaking incubator. At every 2 min after infection, 200 μl of the culture was removed and placed in a 96-well plate kept on ice. Cell density was measured by A595 after all samples were taken. Lysis curve and average lysis time were estimated by fitting the data from at least two independent experiments into a sigmoidal curve using the least-squares fitting method.
Measurement of adsorption rates.
Freshly prepared phages were added to the BL21 culture at an A595 of ∼0.3 at an MOI of 0.001. After 5 min of infection at 37°C, 200 μl of culture was taken and quickly centrifuged to separate free phages from adsorbed phages. Another 200 μl of culture was taken at the same time and kept unspun. Both spun and unspun suspensions were then separately added to 2.5 ml of 0.7% LB agar and plated onto 1.5% LB agar to determine the total number of phages (Nt) from the unspun sample and the number of free phages (Nf) from the spun sample. The adsorption rate, α, was calculated using the equation α = −0.2[ln(Nf/Nt)].
Reverse Northern blot.
Total RNA was extracted from cell lysate samples using a RiboPure-bacteria kit (Ambion). Phage mRNA levels were measured semiquantitatively by reverse Northern blotting as previously described (15), except for the blotting step. For blotting, DNA probes first were amplified using PCR. The products were then denatured by boiling for 5 min in the presence of 0.35 M sodium hydroxide before being neutralized with 0.5 M sodium acetate. The denatured probe (100 μl) was then dot blotted onto a Zeta-Probe blotting membrane (Bio-Rad) by following the manufacturer's instructions. Radioactive intensity was measured and analyzed using TINA 2.0 software (DesignSoft).
Quantification of phage mRNA levels.
The postinfection cellular levels of phage gene mRNAs were measured quantitatively by reverse transcription followed by quantitative real-time PCR (RT-qPCR). The first-strand cDNA was synthesized from 1 μg of total RNA using biotinylated random 15-mer primers and reverse transcriptase ImpromII (Promega) and purified using streptavidin-coated magnetic beads (Dynabeads M-280; Invitrogen). Quantitative PCR for targeted mRNAs was performed using a CFX Connect system (Bio-Rad). All statistical analyses were performed using two-tailed Student's t tests.
Overexpression of holin gene.
A 204-bp segment of T7 gene 17.5 (encoding holin) was amplified from T7 genome DNA using a forward primer (5′-ATCACATATGCTATCATTAGACTTTAACAA-3′) and a reverse primer (5′-ATCACTCGAGTCACTCCTTATTGGCTTTCTT-3′) in PCR consisting of 35 cycles of denaturation at 95°C, annealing at 57°C, and extension at 72°C, each for 30 s. The resulting amplicon was cloned into the NdeI/XhoI site of the pET21 vector, and BL21 was transformed with the resulting plasmid for overproduction of T7 holin during phage infection.
Passaging of phage for experimental evolution.
Passaging was done by adding phage to 4 ml of log-phase BL21 host (A595 of 0.2 to 0.5) at low MOI (<0.1). The culture was then shaken at 37°C for 30 to 60 min. A drop from the previous passage was then added to 4 ml of a new log-phase host culture, and the process was repeated. If a phage stock was kept overnight before the next passage, 10 μl of chloroform was added and the final lysate was kept at 4°C.
RESULTS
A T7 phage mutant lacking terminator Tφ.
We created a T7 phage mutant lacking the transcription terminator Tφ to understand the roles of Tφ in the phage T7 lytic cycle. As Tφ is a typical class I intrinsic terminator (4), both the hairpin structure and immediate downstream U stretch are required for functionality. We deleted a 31-bp DNA sequence (from genomic positions 24180 to 24210) encoding the terminator hairpin (including five residues in the upstream side of the stem, all six in the loop, and all 13 in the downstream side of the stem), the following six U residues, and the termination site G residue of Tφ from T7 genome (Fig. 1B). A Tφ-lacking fragment was constructed by combining a Tφ upstream part and a Tφ downstream part through an StyI site (Fig. 1C). The combined fragment was inserted into the unique BssHII and AatII sites of the T7 genome before being transfected into competent E. coli DH5α for assembly of the Tφ-lacking mutant phage.
The deletion of Tφ was confirmed by DNA sequencing and in vitro and in vivo transcription assays (Fig. 1D). For in vitro multiple-round transcription assays, a 238-bp Tφ-containing segment from genomic positions 24153 through 24390 or a corresponding 207-bp Tφ-lacking segment was cloned from the wild-type or mutant phage, respectively, into the KpnI/HindIII site of a φ10 promoter-containing plasmid, pKM01, so that 347- or 316-nucleotide (nt)-long runoff transcripts, respectively, could be produced by T7 RNA polymerase transcription using PCR amplicon templates. The wild-type template yielded both 103-nt terminated transcripts and 347-nt runoff transcripts, showing a termination efficiency of 77% (Fig. 1D). In contrast, transcription of the Tφ-lacking template did not produce any terminated transcripts other than 316-nt runoff transcripts.
This in vitro confirmation agreed with measurements of in vivo transcripts using an RNase protection assay (Fig. 1E). For preparation of RNA probes, a Tφ-containing template (genomic positions from 24111 to 24250) and a corresponding Tφ-lacking template were amplified using PCR of the wild-type and mutant phages, respectively. The reverse primer had a φ10 promoter in the 5′ overhang for phage T7 RNA polymerase transcription of the amplicons to produce radioactively labeled 140- and 109-nt-long RNA probes, respectively, complementary to the T7 transcripts produced after infection.
The total RNA transcripts were extracted from E. coli BL21 infected with the wild-type or mutant phage and mixed with respective radiolabeled probes before being treated with a mixture of single-strand-specific RNases A and T1. The 140-nt RNA probe was hybridized to both terminated and read-through transcripts to yield protection of 100 and 140 nt, respectively, among total transcripts from the wild-type phage. In contrast, only the read-through transcripts were detected (109 nt protection) among those from the mutant phage. Accordingly, the mutant phage was demonstrated to lack the terminator Tφ physically and functionally.
Burst shrinkage and lysis delay by Tφ deletion.
With respect to the life of a lytic phage, two important indexes are burst size and lysis timing. Burst size is the number of progeny particles a phage produces after one cycle of infection, and lysis timing shows how fast or slow a phage lyses its bacterial hosts, in other words, how short or long the latent period is. These two parameters are normally regulated in a trade-off manner to maximize phage fitness: small bursts are compensated for by fast lysis, while delayed lysis usually yields enlarged bursts (16–20).
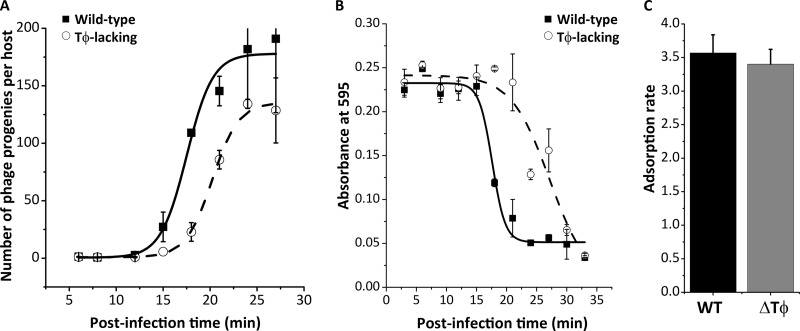
When measured under conditions where the host E. coli BL21 was in an early log phase, the burst sizes of wild-type and Tφ-lacking mutant phages were 179 and 130 particles per host, respectively (Fig. 2A and Table 1). The Tφ-lacking mutant achieved only 73% of the burst size of the wild-type phage (P = 0.01). The burst shrinkage was, however, accompanied by a significant delay rather than acceleration of lysis of the Tφ-lacking mutant (P = 0.0003). Lysis was delayed by approximately 10 min in the mutant compared to the wild-type phage, with the calculated mean lysis time shifted from 16.8 to 26.9 min, when BL21 was infected at 37°C (Fig. 2B and Table 1). Thus, deletion of Tφ from the T7 genome led to a delay in lysis time but shrinkage in burst size, both of which are disadvantageous for the phage adaptation to environments.
FIG 2.
Phenotypic characterization of the Tφ-lacking mutant phage. (A) One-step growth curve for burst size measurement of the wild-type (filled squares, solid line) and Tφ-lacking (open circles, dash line) phages. (B) Lysis curves for estimation of lysis time from the wild-type and Tφ-lacking mutant phage infections. (C) Adsorption rates measured for the wild-type (black column) and Tφ-lacking (gray column) phages. Vertical error bars represent standard deviations.
TABLE 1.
Phenotypic characterization of wild-type and mutant T7 phagesa
| Phage T7 type | Burst size (phage cell−1) | Lysis time (min) | Absorption rate (phage−1 ml−1 min−1) |
|---|---|---|---|
| Wild type | 179 ± 19 (reference) | 16.8 ± 0.7 (reference) | 3.6 ± 0.3 (reference) |
| Tφ-lacking | 130 ± 06 (P = 0.01) | 26.9 ± 0.3 (P = 0.0003) | 3.4 ± 0.2 (P = 0.5) |
| Revertant 1 | 170 ± 14 (P = 0.4) | 13.9 ± 0.6 (P = 0.004) | Not determined |
| Δ0.5-0.7 | 184 ± 13 (P = 0.2) | 12.6 ± 0.2 (P = 0.003) | Not determined |
P values were calculated against the wild type.
Phage adsorption rate unchanged by Tφ deletion.
Optimal lysis time is positively correlated with adsorption rate, and phage strains with higher adsorption rates show faster lysis than those with lower adsorption rates (21). To determine if the adsorption rate provides any contribution to the delay in lysis time observed with the Tφ-lacking mutant, we measured adsorption rates of the wild-type and mutant phages in infection of exponentially growing BL21 at 37°C (Fig. 2C and Table 1). No significant difference in adsorption rate was observed (P = 0.45) between the wild type (3.6 phage−1 ml−1 min−1) and Tφ-lacking mutant (3.4 phage−1 ml−1 min−1). These results are consistent with a previous finding that adsorption of T7 requires only the phage tail fiber protein (gp17) interacting with E. coli surface lipopolysaccharide (1), because deletion of the terminator Tφ would not change the tail protein.
Global elevation of Tφ downstream gene expression by Tφ deletion.
Because the terminator Tφ is located just upstream of gene 11 in the middle of the T7 genome (Fig. 1A), deletion of Tφ was expected to lead to elevated expression of Tφ downstream genes starting from gene 11, in the middle of the genome, through gene 19.5, at the end of the genome. To test this hypothesis, reverse Northern blotting was performed using 15 DNA probes of 200 to 500 bp designed specifically for T7 genes of classes I (early), II (middle), and III (late) to semiquantitatively measure their mRNA levels in the total RNA extracts from E. coli BL21 at 10 min after infection with the wild-type or mutant phage (Fig. 3A).
FIG 3.
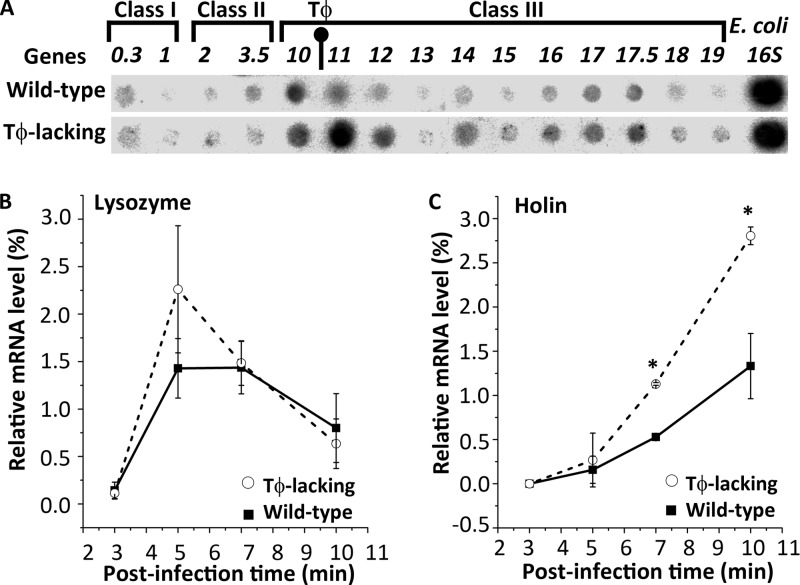
Transcription profiling of T7 genes during infection of the wild-type and Tφ-lacking phage. (A) The mRNA levels of several T7 genes in the wild-type and Tφ-lacking phage infections were measured using reverse Northern blotting. E. coli 16S rRNA was used as a normalization control. The layout of spots follows the sequence of T7 genes in the genome shown at the top, and terminator Tφ is located between genes 10 and 11. (B and C) The mRNA levels of two T7 lysis proteins, lysozyme (B) and holin (C), in the wild-type (filled squares) and Tφ-lacking phage infections (empty circles) were quantified in a time course manner using RT-qPCR.
Ten of the tested Tφ downstream genes, 11, 12, 13, 14, 15, 16, 17, 17.5, 18, and 19, were upregulated in the Tφ-lacking mutant infection compared to the wild-type phage infection. The extent of upregulation ranged from 1.3- to 6.8-fold after normalization against the levels of E. coli 16S rRNA, an internal control. Maximum elevation was observed for genes 11 and 12; this finding is conceivable because these two promoter-lacking genes rely entirely on read-through at Tφ for their expression, whereas the other genes can be transcribed from their own promoters in addition to the read-through at Tφ.
In contrast, four genes of the early and middle classes located upstream of Tφ in the T7 genome showed either no expression change (genes 0.3 and 3.5) or mild upregulation (genes 1 and 2) in the Tφ-lacking mutant infection. Expression of gene 10, located just upstream of Tφ, also appeared unchanged in the mutant phage infection. Thus, many T7 genes were upregulated to various degrees, but most Tφ downstream genes, especially the Tφ-dependent genes 11 and 12, were highly upregulated in the Tφ-lacking mutant phage infection compared to the wild-type phage infection.
mRNA levels of T7 lysis proteins, lysozyme, and holin.
Lysis is the destruction of the host cell wall and membrane. It releases phage progenies; therefore, it is essential for phage propagation. The lysis machinery in most phages with large double-stranded DNA consists of two components under normal salt concentrations: a lysin, a muralytic enzyme that degrades the cell wall, and a holin, a transmembrane protein that creates lesions in the host inner membrane through which lysin can gain access to the cell wall (22). In phage T7, these proteins are lysozyme encoded by a middle gene, 3.5, and holin encoded by a late gene, 17.5.
The mRNA levels of these two lysis proteins were quantitatively measured in a time course manner after infection started using RT-qPCR (Fig. 3B). The lysozyme mRNA level was not significantly different between the wild-type and Tφ-lacking mutant phages throughout the infection course (P > 0.05 at all time points tested). Accordingly, lysozyme levels are not associated with delayed lysis or downsized burst observed in the Tφ-lacking mutant infection.
On the other hand, the holin mRNA level was significantly elevated in the Tφ-lacking mutant infection, reaching a 2.1-fold increase at 7 (P = 0.002) and 10 min (P = 0.003) after infection (Fig. 3C). To assess if the holin upregulation was the cause of lysis delay and burst downsizing in the Tφ-lacking mutant infection, we overexpressed the holin gene in the E. coli BL21 host during the wild-type phage infection and measured lysis time and burst size.
Burst shrinkage but lysis acceleration by holin overproduction.
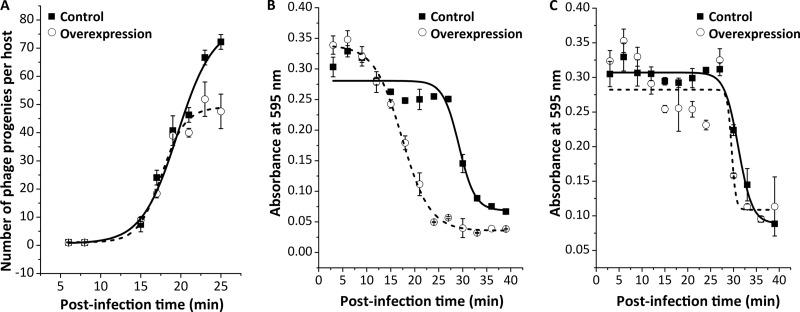
BL21 was transformed to take up a pET21 plasmid carrying the holin gene and infected with the wild-type phage, while holin gene overexpression was induced by adding 1.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) or noninduced as a control. The burst size was 50 versus 80 progenies/host under induced and noninduced conditions, respectively (Fig. 4A); therefore, it was significantly reduced (by 37%) by overproduction of holin (P = 0.02). Accordingly, the burst shrinkage observed in the Tφ-lacking mutant infection could result from upregulation of holin gene expression, a consequence of Tφ deletion.
FIG 4.
Effects of holin overproduction on burst size and lysis time of T7 phage infection. (A and B) The T7 phage burst size (A) and lysis time (B) were measured with infection of the BL21 host carrying a pET21 vector containing the holin gene under 1.5 mM IPTG-induced (empty circles, dash line) and noninduced (filled squares, solid line) conditions. (C) A control experiment measuring T7 lysis time with infection of the BL21 host carrying an empty pET21 vector under the same conditions.
However, the holin overproduction significantly accelerated rather than delayed lysis of the wild-type phage (P = 0.003), as the calculated mean lysis time was 16.5 and 28.2 min under the induced and noninduced conditions, respectively (Fig. 4B). The lysis acceleration was a bona fide effect of holin overproduction, because there was no difference in lysis time when BL21 was transformed with an empty pET21 vector: 25.7 and 27.0 min under induced and noninduced conditions, respectively (Fig. 4C). Accordingly, the lysis delay observed in the Tφ-lacking mutant infection cannot result from the upregulation of holin gene expression, which leads to an opposite consequence in the wild-type phage infection.
Furthermore, all of these results together suggest that overexpression of multiple Tφ downstream genes, including the holin gene, had a greater effect than merely counteracting the lysis-accelerating effect of holin gene overexpression to result in a further delayed lysis. Accordingly, a yet-unknown factor(s) other than lysozyme and holin is involved in causing the lysis delay in the Tφ-lacking mutant infection.
Lysis time and burst size recovered by deletion of T7 early genes.
Because the known lysis pathway of T7 phage involving lysozyme and holin could not explain the lysis delay, we took the classical genetics approach of experimental evolution. The Tφ-lacking mutant phage with a delayed lysis property was passed consecutively through a series of log-phase hosts until some spontaneous mutations rendered lysis as fast as that of the wild-type phage. Five independent adaptations were performed to reveal common suppressor mutations.
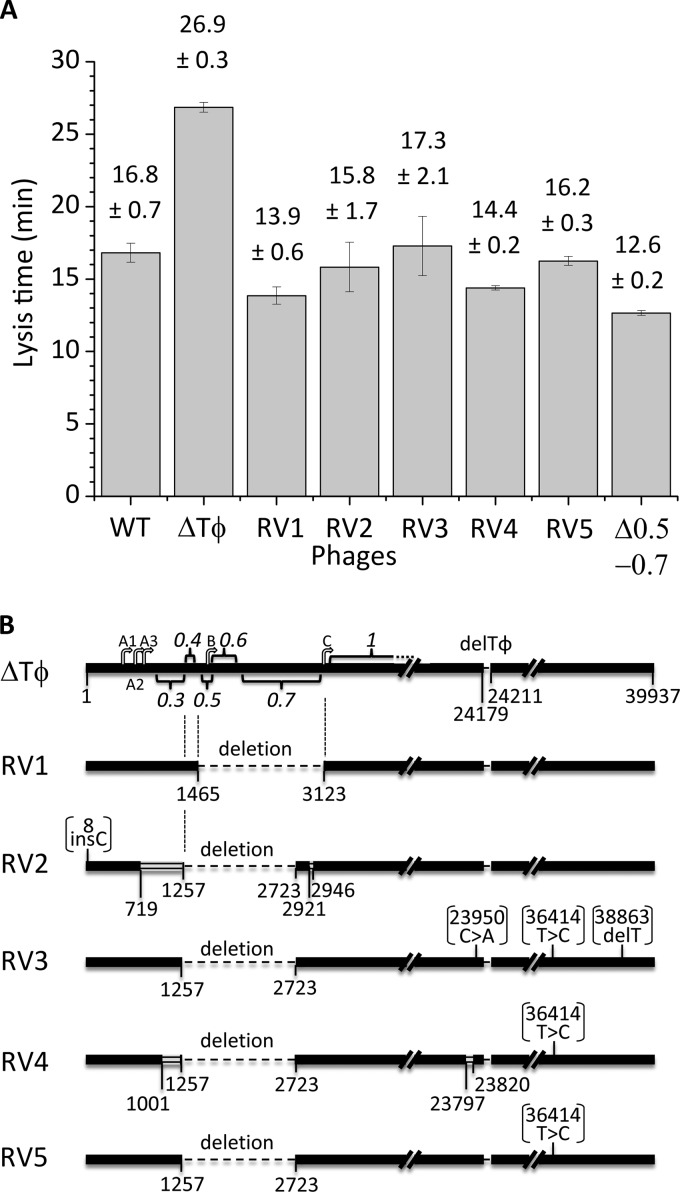
In the first adaptation, a fast-lysing revertant arose at passage 29, here referred to as revertant 1. The other four revertants, revertants 2 to 5, arose at passage 27 of their respective adaptations. All five revertants lysed their hosts significantly faster than the ancestral Tφ-lacking mutant with a lysis time of 26.9 min (P < 0.05) (Fig. 5A). Revertants 1 (13.9 min) and 4 (14.4 min) lysed even faster than the wild-type phage, with a lysis time of 16.8 min (P = 0.004 and 0.016, respectively), whereas revertants 2 (15.8 min), 3 (17.3 min), and 5 (16.2 min) lysed as fast as the wild type (P > 0.05).
FIG 5.
Experimental adaptation of the Tφ-lacking mutant. (A) Lysis time was measured for the wild-type (WT) and Tφ-lacking (ΔTφ) phages, the five fast-lysing revertants (RV1 through RV5), and the early-gene-lacking control (Δ0.5-0.7). (B) Genome maps of the ancestral Tφ-lacking mutant (ΔTφ) and the five revertants (RV1 though RV5). Relative genomic positions of some E. coli promoters (A1, A2, A3, B, and C) and T7 genes (0.3, 0.4, 0.5, 0.6. 0.7, and 1) are shown for ΔTφ on the top. Dashed lines indicate deleted regions. Double bars indicate heterologous regions. Point mutations shown in parentheses are either synonymous or intergenic.
To identify the mutations that recovered fast lysis, we sequenced the entire genomes of all five revertants (Fig. 5B). Revertant 1 carried a deletion from genomic positions 1466 to 3123 (harboring genes 0.5, 0.6, and 0.7). Revertants 2, 3, 4, and 5 commonly carried a larger deletion from positions 1255 to 2722 (genes 0.3 through 0.7) and a synonymous substitution at position 36414 (T to C within gene 17.5). Additionally, revertant 2 carried an addition of C at position 8 and revertant 3 carried a substitution at 23950 (C to A), an addition of G at 35299, and a deletion of T at 38863. Revertant 4 additionally carried a substitution at 297 (T to A), a deletion of G at 23800, an addition of A at 23818, and a substitution at 23820 (G to T), but revertant 5 carried no additional changes.
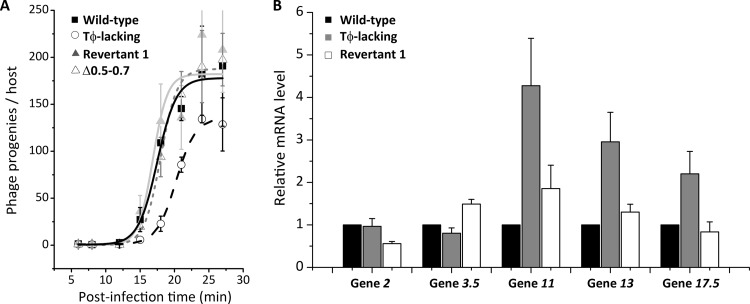
Thus, the mutations commonly shared by all five revertants were deletion of three nonessential genes, 0.5, 0.6, and 0.7, spanning 1,257 bp from positions 1466 to 2722, suggesting that deletion of these three early genes was sufficient to suppress the lysis delay effect of Tφ deletion. Because the deletion in revertant 1 affected only three genes while the other revertants lacked more genes, we further characterized revertant 1 as a representative revertant. Revertant 1 also recovered the wild-type burst size, reaching 170 progenies/host (P = 0.4) (Fig. 6A and Table 1), substantially larger than the Tφ-lacking mutant (130 phages/host). These results together showed that deletion of three nonessential early genes, 0.5, 0.6, and 0.7, could effectively suppress both lysis delay and burst-downsizing effects of Tφ deletion.
FIG 6.
Characterization of revertant 1. (A) Burst size measurement in wild-type T7 (filled square), Tφ-lacking mutant (empty circle), revertant 1 (filled triangle), and Δ0.5-0.7 (empty triangle). (B) The mRNA levels of five T7 genes, 2, 3.5, 11, 13, and 17.5, were quantified at 10 min after infection in wild-type T7 (black), Tφ-lacking mutant (gray), and revertant 1 (white) using RT-qPCR.
To assess the effect of deletion of these early genes, 0.5 to 0.7, alone in the presence rather than the absence of Tφ terminator, we inserted the wild-type Tφ sequence back into revertant 1, resulting in a mutant designated Δ0.5-0.7. The burst size of this control mutant was 184 phage progenies/host (Table 1), not significantly different from that of the wild type (P = 0.2). In contrast, the lysis time was 12.6 min (Table 1), significantly faster than that of the wild type (P = 0.003). Thus, the effect of deletion of genes 0.5 to 0.7 alone was to accelerate lysis while hardly affecting burst size.
Burst size associated with holin gene expression level.
Deletion of Tφ led to upregulation of the holin gene 17.5 (Fig. 3B), which in turn led to premature breakage of E. coli host and reduced burst size. Since revertant 1 recovered the burst size, we examined whether the holin expression level was downregulated in revertant 1 infection compared to the Tφ-lacking mutant infection. We measured mRNA levels of holin at 10 min after infections using RT-qPCR (Fig. 6B). The holin mRNA level was increased 2-fold in the Tφ-lacking mutant infection compared to the wild-type phage infection but decreased in revertant 1 infection to a level comparable to that of the wild-type phage infection. Consistent with a finding that the burst size was associated with the holin expression level (Fig. 3B), these results could explain how the burst size was recovered in revertant 1.
Lysis time associated with Tφ downstream gene expression levels.
Although lysis time was not associated with the holin gene expression level (Fig. 4B), most other Tφ downstream genes were upregulated by Tφ deletion (Fig. 3A). Thus, mRNA levels of two other Tφ downstream genes, 11 and 13, were quantitatively measured (Fig. 6B). Their mRNA levels were increased in the Tφ-lacking mutant infection compared to the wild-type phage infection but decreased again in revertant 1 infection compared to the Tφ-lacking mutant infection. In contrast, mRNA levels of two Tφ upstream genes, 2 and 3.5 (as controls), were similar in the wild-type, Tφ-lacking mutant and revertant 1 infections. These results suggest that lysis time is associated with mRNA levels of Tφ downstream genes (other than the holin gene) rather than Tφ upstream genes (including the lysozyme gene).
DISCUSSION
This study is the first to address the physiological role of the phage T7 terminator Tφ during infection of E. coli. We report that direct effects of Tφ deletion from the phage T7 genome are a delay in lysis time accompanied by a shrinkage rather than an enlargement of burst size at the phenotypic level (Fig. 2) and global elevation in mRNA levels of most, if not all, Tφ downstream genes at the molecular level (Fig. 3). All of these molecular and phenotypic effects of Tφ deletion could be suppressed by deletion of early genes, especially genes 0.5, 0.6, and 0.7 (Fig. 5 and 6). Burst size appeared to be associated with cellular levels of T7 holin, a Tφ downstream gene product, but lysis time was associated with Tφ downstream gene product(s) other than holin; this suggests that phage T7 lysis involves an unknown factor(s) other than the known lysis proteins of phage T7.
Holin creates sudden membrane lesions in a concentration-dependent manner on the phage lysis pathway (20, 23–25). This saltatory manner of holin action is important to ensure that production of phage progenies unabatedly continues until the moment of lysis. A straightforward interpretation of the observed shrinkage of burst in the Tφ-lacking mutant infection (Fig. 2A) and recovery of burst size in the revertant 1 infection (Fig. 6A) would be that Tφ deletion results in fast accumulation of holin, which directly leads to two consequences: first, early saltatory inner cell membrane leakage that allows lysozyme to pass through and degrade the cell wall, causing fast lysis; and second, premature cessation of progeny production, leading to burst downsizing. We observed both phenomena when holin-overproducing E. coli cells were infected with the wild-type T7 phage (Fig. 4A), providing further evidence supporting holin association with burst size. This is the first report that the presence of the phage T7 terminator Tφ is required for maintaining optimal burst size during T7 infection of E. coli.
There are many factors, both external and internal, that regulate phage burst size in its infection cycle, as the lysis function is strictly regulated in phages (26). Internally, burst size (b) is determined by the length of the eclipse period (e), the rate of maturation (m), and lysis time (l), approximately following the equation b = m (l − e) (19). The correlation between burst size and lysis time would not always be this linear, however, because burst size would reach saturation if the phage has exhausted all of the resources in the host cell. In general, nevertheless, burst size increases when lysis time increases, as lysis is delayed for larger bursts in order to maintain its maximal adaptiveness. Thus, burst size and lysis time are in a simple trade-off relation. In fact, holin overproduction, which led to downsized burst, resulted in accelerated lysis in the wild-type phage infection (Fig. 4B).
Surprisingly, however, in the Tφ-lacking mutant infection, the burst was smaller even though lysis was substantially delayed compared to the wild-type phage infection (Fig. 2). Thus, with Tφ deletion both burst size and lysis time are far from optimal values, and the trade-off relationship between the two parameters no longer holds. Lysis requires degradation of not only the inner cell membrane by holin but also the cell wall and the outer cell membrane by lysozyme (and spanins, under certain conditions). The rate-limiting step in the retarded lysis of the Tφ-lacking mutant lies in either the cell wall or the outer membrane degradation step, although the mechanism remains elusive.
It is known that release of T7 lysozyme is spontaneous upon holin-induced membrane lesion. T7 lysozyme is known to interact only with T7 RNA polymerase and with itself (27), which is important for its transcription-inhibitory function but not for its degradation of the host cell wall. Lysis was not delayed but rather accelerated by holin overproduction in the wild-type phage infection (Fig. 4B), suggesting that overexpression of multiple Tφ downstream genes did more than counteract the effect of holin overexpression on lysis time. Accordingly, it is suggested that some unknown factors other than holin, lysozyme, and spanins are involved in the lysis pathway of T7 phage. Heineman et al. (14) also suggested that T7 contains additional holins, as their optimality failed to predict the lysis time of T7 phage.
Previous studies have shown the power of fast evolution in a given phage group to overcome disadvantages caused by mutation or deletion of some beneficial gene functions (28, 29). Similarly, in our search for unknown factors that can overcome delay in lysis and shrinkage in burst size caused by Tφ deletion, suppressor mutations of Tφ deletion obtained through experimental evolutions were identified in this study (Fig. 5 and 6). According to the optimality model (30), a given trait can reach its optimal value through simple natural selection when placed under a few selective forces. The slow-lysing Tφ-lacking mutant phage successfully adapted fast lysis in all five independent evolution experiments.
Deletion of three early nonessential genes, 0.5, 0.6, and 0.7, from the Tφ-lacking genome was sufficient to suppress both of the two disadvantageous phenotypes, namely, delayed lysis and downsized burst of Tφ-lacking mutant phage, suggesting that one or more of these gene products can reverse the action. The deletion of this early gene cluster could have two effects on T7 infection. First, the T7 genome is shortened to reduce the time taken for genome penetration into host cells, which would accelerate lysis (31). Second, the three gene products, gp0.5, gp0.6, and gp0.7, are not produced; therefore, their function(s) and effect(s) on infection would be absent. Whereas the functions of gp0.5 and gp0.6 are not known yet, gp0.7 is known to have a protein kinase activity that can ultimately affect the expression of phage genes.
A serine-threonine protein kinase encoded by the T7 gene 0.7 phosphorylates many host proteins of the transcription (such as E. coli RNA polymerase β and β′ subunits), RNA processing (such as RNases III and E), and translation machineries (such as elongation factors G and P and ribosomal proteins S1 and S6). This phosphorylation was reported to help inactivate host E. coli transcription and stabilize T7 mRNAs for increased translation (32, 33). If so, deletion of gene 0.7 should reduce T7 mRNA levels, among other effects. In fact, deletion of gene 0.7 and others from the Tφ-lacking mutant genome reduced the holin mRNA level in revertant 1 infection to a level comparable to that of wild-type phage infection (Fig. 6B). In other words, burst downsizing is caused by deletion of Tφ, leading to elevation of holin mRNA level (in the Tφ-lacking mutant infection), and reversed by deletion of gene 0.7, leading to reduction of the holin mRNA level (in revertant 1 infection). Accordingly, these data support that the burst size is associated with holin levels.
In contrast, lysis time appears to be associated with a yet-unknown T7 factor. The factor would be a Tφ downstream gene product(s), because lysis was delayed when deletion of Tφ elevated the mRNA levels of Tφ downstream genes (Fig. 3A) but recovered when further deletion of genes 0.5 to 0.7 reduced mRNA levels of genes 11 and 13 (Fig. 6B) and presumably other Tφ downstream genes as well. However, holin, a product of a Tφ downstream gene (17.5), is not associated with lysis time, because the effect of holin overproduction was opposite that in the wild-type phage infection (accelerating lysis) (Fig. 4B) and the Tφ-lacking mutant infection (delaying lysis) (Fig. 2B).
Among the 15 genes located downstream of Tφ in the T7 genome, nine genes, 11, 12, 13, 14, 15, 16, 17, 18, and 19, are essential, and their gene products all act on the assembly of progeny phages (1). Three other genes, 17.5, 18.5, and 18.7, are regarded as nonessential, but their functions are known. Gene 17.5 encodes holin (of type II), which makes holes or channels in the inner membrane so that the lysozyme (gp3.5) can reach the cell wall and degrade it. The overlapping genes, 18.5 and 18.7, apparently encode spanins, which span the periplasm and are involved in disrupting the outer cell membrane under high salt concentrations, and they are homologous to the overlapping genes Rz and Rz1 of phage λ (11). The remaining three genes, 19.2, 19.3, and 19.5, are hypothetical at present but conserved among close relatives of phage T7. If these hypothetical genes are ever expressed as proteins or noncoding RNA, they can be candidates for the unknown lysis time-associated factor, among many others, although loss of gene 19.5 has previously caused a reduced burst (1).
In summary, we have observed for the first time the roles of terminator Tφ in maintaining burst size and lysis time of phage T7 during infection of E. coli. Phage T7 burst size is associated with T7 holin, as the burst is downsized by elevation of holin, which can be observed when the phage lacks Tφ. T7 lysis time is not associated with holin or lysozyme but with an unknown Tφ downstream gene product(s), which can be upregulated by Tφ deletion, leading to delayed but not accelerated lysis. It will be worthwhile to search for a new T7 lysis time-associated factor.
ACKNOWLEDGMENTS
We thank Sooncheol Lee and Kook Sun Ha for helpful discussions and preparations of phage T7 RNA polymerase and pKM01 plasmid.
This work was supported in part by a grant from the National Research Foundation of Korea (2012-0008847).
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Molineux I. 2006. The T7 group, p 277–301 In Calendar R. (ed), The bacteriophages. The Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 2.Durniak KJ, Bailey S, Steitz TA. 2008. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science 322:553–557. 10.1126/science.1163433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S, Nguyen HM, Kang C. 2010. Tiny abortive initiation transcripts exert antitermination activity on an RNA hairpin-dependent intrinsic terminator. Nucleic Acids Res. 38:6045–6053. 10.1093/nar/gkq450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macdonald LE, Durbin RK, Dunn JJ, McAllister WT. 1994. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J. Mol. Biol. 238:145–158. 10.1006/jmbi.1994.1277 [DOI] [PubMed] [Google Scholar]
- 5.Shen H, Kang C. 2001. Two site contact of elongating transcripts to phage T7 RNA polymerase at C-terminal regions. J. Biol. Chem. 276:4080–4084. 10.1074/jbc.M008616200 [DOI] [PubMed] [Google Scholar]
- 6.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. 2002. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature 420:43–50. 10.1038/nature01129 [DOI] [PubMed] [Google Scholar]
- 7.Mertens N, Remaut E, Fiers W. 1996. Increased stability of phage T7g10 mRNA is mediated by either a 5′- or a 3′-terminal stem-loop structure. Biol. Chem. 377:811–817 [DOI] [PubMed] [Google Scholar]
- 8.Condron BG, Atkins JF, Gesteland RF. 1991. Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol. 173:6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condron BG, Gesteland RF, Atkins JF. 1991. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 19:5607–5612. 10.1093/nar/19.20.5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heineman RH, Molineux IJ, Bull JJ. 2005. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J. Mol. Evol. 61:181–191. 10.1007/s00239-004-0304-4 [DOI] [PubMed] [Google Scholar]
- 11.Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373:1098–1112. 10.1016/j.jmb.2007.08.045 [DOI] [PubMed] [Google Scholar]
- 12.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501:69–76. 10.1007/978-1-60327-164-6_7 [DOI] [PubMed] [Google Scholar]
- 13.Chan LY, Kosuri S, Endy D. 2005. Refactoring bacteriophage T7. Mol. Syst. Biol. 1:2005.0018. 10.1038/msb4100025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heineman RH, Bull JJ, Molineux IJ. 2009. Layers of evolvability in a bacteriophage life history trait. Mol. Biol. Evol. 26:1289–1298. 10.1093/molbev/msp037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Zhang R, Liang P. 1997. Differential screening of differential display cDNA products by reverse northern. Methods Mol. Biol. 85:87–93 [DOI] [PubMed] [Google Scholar]
- 16.Abedon ST, Herschler TD, Stopar D. 2001. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 67:4233–4241. 10.1128/AEM.67.9.4233-4241.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abedon ST, Hyman P, Thomas C. 2003. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl. Environ. Microbiol. 69:7499–7506. 10.1128/AEM.69.12.7499-7506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heineman RH, Bull JJ. 2007. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution 61:1695–1709. 10.1111/j.1558-5646.2007.00132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang IN. 2006. Lysis timing and bacteriophage fitness. Genetics 172:17–26. 10.1534/genetics.105.045922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang IN, Dykhuizen DE, Slobodkin LB. 1996. The evolution of phage lysis timing. Evol. Ecol. 10:545–558. 10.1007/BF01237884 [DOI] [Google Scholar]
- 21.Shao Y, Wang IN. 2008. Bacteriophage adsorption rate and optimal lysis time. Genetics 180:471–482. 10.1534/genetics.108.090100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan GL, Rutenberg AD. 2007. Clocking out: modeling phage-induced lysis of Escherichia coli. J. Bacteriol. 189:4749–4755. 10.1128/JB.00392-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vukov N, Scherer S, Hibbert E, Loessner MJ. 2000. Functional analysis of heterologous holin proteins in a lambdaDeltaS genetic background. FEMS Microbiol. Lett. 184:179–186. 10.1016/S0378-1097(00)00041-0 [DOI] [PubMed] [Google Scholar]
- 25.Wang IN, Deaton J, Young R. 2003. Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J. Bacteriol. 185:779–787. 10.1128/JB.185.3.779-787.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkhout B, van Strien SBA, van Boom J, van Westrenen J, van Duin J. 1987. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J. Mol. Biol. 195:517–524. 10.1016/0022-2836(87)90180-X [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Studier FW. 1997. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J. Mol. Biol. 269:10–27. 10.1006/jmbi.1997.1016 [DOI] [PubMed] [Google Scholar]
- 28.Berkhout B, Klaver B, Das AT. 1997. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 25:940–947. 10.1093/nar/25.5.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsthoorn RC, Licis N, van Duin J. 1994. Leeway and constraints in the forced evolution of a regulatory RNA helix. EMBO J. 13:2660–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orzack SHES. 1994. Optimality models and the test of adaptationism. Am. Nat. 143:361–380. 10.1086/285608 [DOI] [Google Scholar]
- 31.Grigoriev A. 2001. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 29:3513–3519. 10.1093/nar/29.17.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchand I, Nicholson AW, Dreyfus M. 2001. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol. Microbiol. 42:767–776. 10.1046/j.1365-2958.2001.02668.x [DOI] [PubMed] [Google Scholar]
- 33.Robertson ES, Nicholson AW. 1992. Phosphorylation of Escherichia coli translation initiation factors by the bacteriophage T7 protein kinase. Biochemistry 31:4822–4827. 10.1021/bi00135a012 [DOI] [PubMed] [Google Scholar]