Abstract
The 2009 pandemic H1N1 influenza virus (pdm/09) is typically mildly virulent in mice. In a previous study, we identified four novel swine isolates of pdm/09 viruses that exhibited high lethality in mice. Comparing the consensus sequences of the PB2 subunit of human isolates of pdm/09 viruses with those of the four swine isolate viruses revealed one consensus mutation: T588I. In this study, we determined that 588T is an amino acid mutation conserved in pdm/09 viruses that was exceedingly rare in previous human influenza isolates. To investigate whether the PB2 with the T5581 mutation (PB2-T558I) has an effect on the increased pathogenicity, we rescued a variant containing PB2-588I (Mex_PB2-588I) in the pdm/09 virus, A/Mexico/4486/2009(H1N1), referred to as Mex_WT (where WT is wild type), and characterized the variant in vitro and in vivo. The results indicated that the mutation significantly enhanced polymerase activity in mammalian cells, and the variant exhibited increased growth properties and induced significant weight loss in a mouse model compared to the wild type. We determined that the mutation exacerbated PB2 inhibition of mitochondrial antiviral signaling protein (MAVS)-mediated beta interferon (IFN-β) expression, and PB2-588I was observed to bind to MAVS more efficiently than PB2-588T. The variant induced lower levels of host IFN-β expression than the WT strain during infection. These findings indicate that the pdm/09 influenza virus has increased pathogenicity upon the acquisition of the PB2-T588I mutation and highlight the need for the continued surveillance of the genetic variation of molecular markers in influenza viruses because of their potential effects on pathogenicity and threats to human health.
INTRODUCTION
Influenza A viruses can infect a variety of animal species; the 2009 pandemic H1N1 influenza virus (pdm/09) was reported to be transmitted to other animal species (1–4). The emergence of pdm/09 influenza virus in swine herds in China and Alberta, Canada, demonstrated that the transmission of the pdm/09 virus from humans to pigs occurred in multiple countries. Swine have α-2,3 and α-2,6 receptors and are considered to be “mixing vessels” or intermediate hosts for generating novel reassortants or pandemic influenza viruses. The 2009 pandemic influenza virus is a swine-origin triple reassortant with human, avian, and swine influenza virus genes (5), and enhanced pathogenicity was observed after transmission to swine (6). Virulence and the interspecies transmission of influenza viruses are a concern for human health. The molecular mechanisms of severe virulence have been explored after mouse adaptation or the introduction of mutated molecular markers that are correlated with virulence, such as PB2-E627K/E677G/D701N/T271A/E158G, PB1-F2–N66S, PA-T97I/F35L, hemagglutinin (HA)-D222G/Q226R (7–11), and others. PB2-E627K/D701N could help H5N1 avian influenza virus to replicate efficiently in mammalian cells and break the interspecies barrier (12, 13). When PB2-E627K or -D701N was introduced into the 2009 pandemic H1N1 influenza virus, no significant changes in virulence were observed, especially with respect to the pathogenicity in mice (14–16).
The PB2 subunit of the RNA polymerase (RNP) complex is a major virulence and host range determinant of influenza viruses, but the exact molecular mechanisms of its function in host range are unknown. Mutations in RNP could increase polymerase activity and improve viral replication (7–13), but not all mutations alter virus virulence (14–16). It was recently reported that PB2 and PB1-F2 could affect virulence by interacting with mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, VISA, or Cardif) and inhibiting beta interferon (IFN-β) expression (17, 18). MAVS mediates the activation of the NF-κB (nuclear factor κB) and IFN regulatory factor 3 (IRF-3) transcription factors in response to viral infections, and it functions downstream of the viral RNA sensors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (Mda-5) and plays an important role in the induction of IFN-β expression (19). Type I interferons are known to be critical factors in the innate immune defense against influenza virus infections (20). The significance of the PB2 inhibition of IFN-β expression during influenza virus infection has not been elucidated in detail.
In this study, we demonstrate that the pdm/09 virus isolated from swine possesses unknown molecular determinants of pathogenicity. We generated a virus containing the PB2-T588I mutation that a different in pathogenicity in mice. The mutation increases viral polymerase activity and plays a role in the regulation of host innate immune responses, suggesting that both viral and host factors are responsible for the increased pathogenicity.
MATERIALS AND METHODS
Cell culture.
Porcine kidney (PK15) cells and Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's minimal essential medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco, Auckland, New Zealand). Human embryonic kidney (293T) cells were maintained in RPMI 1640 medium (Invitrogen) with 10% FBS, and human type II alveolar epithelial (A549) cells were propagated in Ham's F12K medium with 10% FBS.
Plasmid and site-directed mutagenesis.
The PHW-Mex PB2, PB1, PA, NP, HA, neuraminidase (NA), M, and NS plasmids were constructed by synthesizing the gene sequences from the pdm/09 virus, A/Mexico/4486/2009 H1N1, and cloning each of the gene segments into the pHW2000 vector as described previously (21). A QuickChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used to generate the T588I mutation in the PB2 gene. MAVS, RIG-I, MDA-5, and TBK-1 expression plasmids and three luciferase (Luc) reporter plasmids containing either the IFN-β promoter (IFN-Luc), NF-κB promoter (NF-κB-Luc), or IRF-3 promoter (IRF-3-Luc) were kindly provided by Zhengfan Jiang (University of Beijing); the luciferase reporter plasmid pPoll-NP-Luc (pNP-Luc) was kindly provided by Hualan Chen (Harbin Veterinary Research Institute). A Renilla control plasmid (pGL4.75 hRluc/CMV, where CMV is cytomegalovirus) (Promega) was used to control for the cell number and the transfection efficiency.
Virus rescue and growth curves.
The recombinant viruses Mex_WT (where WT is wild type) and Mex_PB2-T588I were generated by an eight-plasmid reverse genetics system as described previously (21). The growth kinetics of the WT and the variant were determined by inoculating MDCK, PK15, or A549 cells at a multiplicity of infection (MOI) of 0.001 50% tissue culture infective dose (TCID50) per cell. One hour after inoculation, the cells were washed twice with phosphate-buffered saline (PBS) and fresh medium containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK), and trypsin (Sigma, St. Louis, MO, USA) was added (0.2 μg/ml TPCK in A549 cells). The supernatants were sampled at 12, 24, 36, and 48 h postinfection (hpi). The virus titers were determined by calculating the log10 TCID50/ml in MDCK cells.
Mouse experiment.
The animal studies were performed according to the protocols approved by the Hubei Provincial Animal Care and Use Committee (approval number SYXK 2008-0004).
To determine the morbidity and mortality, groups of six 5-week-old female BALB/c mice were intranasally inoculated with 50 μl of 104 TCID50s/ml, 105 TCID50s/ml, or 106 TCID50s/ml of the WT or the variant viruses. The body weights and survival of the six mice in each group were monitored daily for 14 days postinfection (dpi). The 50% mouse lethal dose (MLD50) values were determined by inoculating groups of five 4-week-old female BALB/c mice intranasally with 50 μl of 10-fold serial dilutions of the viruses, and the percentage body weight change of each mouse was calculated by comparing the group average weight with the initial weight of each mouse on day 0. The mice that lost >25% of their original body weights were humanely euthanized. To assess the levels of IFN-β, the pathological changes in the lungs of the infected mice, and the viral growth in the mouse organs, 12 mice per group were intranasally inoculated with 50 μl of the recombinant viruses at a dose of 105 TCID50s/ml. Three mice per group were euthanized at 1, 3, 5, and 7 dpi. The right lungs of the mice were removed to determine the virus titers and the IFN-β levels. The left lungs were fixed in formalin, and the fixed tissues were embedded in paraffin and stained with hematoxylin and eosin (H&E) for histopathological analysis.
Luciferase assay of polymerase activity and IFN-β expression.
The transfection of 293T cells in 12-well plates was performed using 0.4 μg of pNP-Luc, PHW-PB2, PHW-PB1, PHW-PA, and PHW-NP and 0.01 μg of pGL4.75 (hRluc/CMV) using 5 μl of Lipofectamine 2000 (Invitrogen). The 293T cells in 12-well plates were transfected with 0.5 μg of IFN-β-Luc, NF-κB-Luc, or IRF-3-Luc, 0.01 μg of pGL4.75 (hRluc/CMV), and 0.5 μg of MAVS with three copies of a Flag tag (3Flag-MAVS) with 2 μg of the influenza virus PB2-expressing plasmids (PCMV-myc-PB2, where PCMV is porcine CMV) using 6 μl of Lipofectamine 2000. The cells were incubated for 24 h and then lysed in 200 μl of passive lysis buffer (PLB; Promega). Luciferase and Renilla activities were assessed using a Dual-Luciferase Assay Kit (Promega).
Immunoprecipitation and Western blotting.
The 293T cells were plated onto a 15-cm dish and transfected with 20 μg of the expression plasmids using 60 μl of Lipofectamine 2000. The plates were incubated at 37°C. At 24 h posttransfection, the cells were collected, washed in ice-cold PBS, and lysed in 1 ml of Tris lysis buffer (Cell Signaling) on ice for 45 min. The clarified cell lysate (400 μl) was used for the immunoprecipitations with 5 μl of rabbit anti-Flag (Sigma). The beads were washed with Tris lysis buffer and resuspended in 50 μl of SDS-PAGE protein loading dye. The cell lysates and immunoprecipitates were analyzed by SDS-PAGE, and the proteins were transferred to nitrocellulose. The membranes were incubated with mouse anti-Flag or anti-Myc (Cell Signaling) primary antibodies, followed by a goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling). The signals were detected using an Immobilon Western Chemiluminescent HRP Substrate kit (Thermo Fisher) and ChemBis (Eastwin).
IFN-β assay.
To measure the levels of IFN-β expression in vitro and in vivo during influenza virus infection, A549 cells were infected with Mex_WT or Mex_PB2-588I at an MOI of 0.1 TCID50 per cell. The total cellular RNA was isolated using the TRIzol reagent (Invitrogen) at 12 and 24 h postinfection (hpi). The mRNA was reverse transcribed into cDNA using avian myeloblastosis virus (AMV; TaKaRa) and a T18 primer, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Relative quantitative PCR (qPCR) was performed to measure the IFN-β mRNA using a SYBR green PCR Kit (Roche) and ABI ViiA7 equipment. The data were analyzed using ViiA7 software (Applied Biosystems). The level of IFN-β in the lungs of the mice infected with 105 TCID50s/ml of the viruses was measured using a VeriKine Mouse IFN-β enzyme-linked immunosorbent assay (ELISA) kit (Biolegend) according to the manufacturer's instructions. An IFN-β standard curve was created and used to calculate the IFN-β concentrations in the lung samples. The infections were performed in triplicate using two independently rescued viruses of the same genetic backgrounds.
Statistical analyses.
All experiments used statistical analyses. The statistical significance of the data was determined using Student's t test with GraphPad Prism software (San Diego, CA). All the assays were run in triplicate and are representative of at least three separate experiments. The error bars represent the standard deviation; a P value of <0.01 was considered to be significant.
RESULTS
A single T-to-I mutation at position 588 in PB2 significantly enhances polymerase activity in mammalian cells.
In a previous study, we found four novel swine isolates of pdm/09 viruses that showed increased virulence; these viruses shared one consensus point mutation, PB2-T588I (22). To investigate whether the variation present at this residue occurs in human isolates or swine isolates of pdm/09 viruses, we performed a comprehensive sequence analysis with regard to position 588 of PB2 (Table 1). The percentages of all the available sequences of the human isolates and the swine isolates of pdm/09 viruses encoding PB2-588I were 3.38% and 28.57%, respectively. All of the swine isolates of the pdm/09 viruses with the PB2-T588I change were isolated in China (22–24). A comparison of the consensus protein sequences of the PB2 genes of pdm/09 with those of human influenza viruses collected between 2000 and 2008 demonstrated that 588T was an amino acid variant conserved in the pdm/09 viruses that was exceedingly rare in previous human isolates; 588I is more prevalent in human isolate viruses and other viruses that caused pandemic influenza viruses in humans before 2009, including H2N2 (1957), H3N2 (1968), and H1N1 (1977) (25).
TABLE 1.
Amino acid frequencies at position 588 in PB2 of human and swine pdm/09 viruses and 2000–2008 human H1N1 influenza viruses
| PB2 amino acid at position 588 by virusa | |||||
|---|---|---|---|---|---|
| pdm/09 human H1N1 (n = 2,984) |
2000–2008 Human H1N1 (n = 2,854) |
pdm/09 swine H1N1 (n = 56) |
|||
| Residue | Frequency (% [n]) | Residue | Frequency (% [n]) | Residue | Frequency (% [n]) |
| T | 96.0 (2,867) | I | 94.5 (2697) | I | 28.57 (16) |
| I | 3.38 (101) | T | 0.25 (7) | T | 69.64 (39) |
| A | 0.2 (6) | A | 4.91 (140) | A | 1.79 (1) |
| S | 0.13 (4) | S | 0.11 (3) | ||
| P | 0.03 (1) | L | 0.21 (6) | ||
| X | 0.17 (5) | X | 0.03 (1) | ||
The amino acid frequencies were determined for unique PB2 full-length sequences collected using the indicated criteria from the NCBI Influenza Virus Resource database (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database). n, number of sequences analyzed; X, amino acid deletion.
To investigate whether the mutation PB2-T588I affects the enzymatic activity of RNP, we performed a polymerase activity assay in 293T cells at different temperatures. The temperatures 33°C and 37°C were used to mimic the temperatures in the upper respiratory tract and the lower respiratory tract of humans, respectively (Fig. 1). The polymerase activities of the variant and the WT exhibited a temperature-dependent pattern and were higher at the higher temperature but much lower at 33°C; these results were similar to those described previously (26). The PB2-T588I mutation increased the polymerase activity at both temperatures, and the increases were approximately 2.3- and 1.9-fold compared with the WT level (P < 0.001, n = 3). The PB2-T588I mutation did not affect the PB2 expression levels.
FIG 1.
Viral RNA polymerase activity. Polymerase activity assays were performed in human 293T cells at 33°C and 37°C by transfecting vectors expressing the polymerase subunits and a pNP-Luc luciferase reporter. After cells were cultured at 33°C and 37°C for 24 h, luciferase production was measured. The data were normalized to the activity observed for the polymerase subunits and are presented on a percentage scale; the results are presented as the means ± standard deviations of three independent experiments. **, P < 0.001, as determined by a t test.
In vitro characterization of the recombinant viruses in human, porcine, and canine cells.
To investigate the growth kinetics of the WT and the variant viruses, we compared the multicycle growth of the two viruses in MDCK, A549, and PK15 cells. The variant titer peaked at 106.8 TCID50s/ml for MDCK cells, 103.6 TCID50s/ml for A549 cells, and 107.1 TCID50s/ml for PK15 cells, and the WT titer peaked at 105.3 TCID50s/ml for MDCK cells, 102.6 TCID50s/ml for A549 cells, and 105.6 TCID50s/ml for PK15 cells. The variant exhibited a significantly increased growth ability in the three types of cells, with virus titers more than 10-fold higher than in the WT at 36 hpi, and the trend continued at the later times points (Fig. 2) (P < 0.001, n = 3). These results indicate that the variant exhibits advantageous growth properties in all three cell types compared to the WT virus in vitro.
FIG 2.
Growth kinetics of recombinant viruses in cells. A549, MDCK, and PK-15 cells were infected with the WT or variant viruses at an MOI of 0.001. At the indicated hours postinfection (hpi), the virus titers in the supernatant were determined with the MDCK cells. The reported values are presented as the means ± standard deviations of three independent experiments. **, P < 0.001, as determined by a t test.
Recombinant virus containing PB2-T588I exhibited enhanced virulence in mice.
We further evaluated the growth and pathogenicity of the two recombinant viruses in a mouse model. The variant had an MLD50 of 104.5 TCID50s/ml, and the WT resulted in a nonlethal infection even when inoculated at the highest dose of 106 TCID50s/ml (Fig. 3B). The mice infected with the variant (106 and 105 TCID50s/ml) showed a greater than 25% weight loss. The WT-infected mice experienced no substantial body weight loss (Fig. 3A). Even the mice infected with the variant at a dose of 104 TCID50s/ml exhibited more weight loss than the mice infected with the WT at a dose of 106 TCID50s/ml (Fig. 3A). The mice who showed losses of >25% of their initial body weights were euthanized during the infection.
FIG 3.
Pathogenicity of the WT and the variant in mice. Six mice per group were intranasally inoculated with 104 TCID50s/ml, 105 TCID50s/ml, and 106 TCID50s/ml (each in 50 μl) of the WT or the variant. (A) Mouse body weights were monitored daily for 14 days. The values represent the average scores of the overall body weight loss compared with the initial body weight ± standard deviations. (B) The percent survival values were calculated by observing the infected mice. (C) The mice were intranasally inoculated with 105 TCID50s/ml of the WT or the variant virus. Tissues were collected from the mice (n = 3) on the indicated days postinfection, and the virus titers were determined in MDCK cells. *, P < 0.01, **, P < 0.01, as determined by a t test.
We investigated the virus titers in different body tissues of the mice infected with viruses at a dose of 105 TCID50s/ml by cell culture methods (Fig. 3C). In the WT-infected mice, the virus titer in the lungs was approximately 104 TCID50s/ml at 1 and 3 dpi, and the titer decreased rather quickly to 102.2 TCID50s/ml at 5 dpi. In the variant-infected mice, the virus titer in the lungs was approximately 105 TCID50s/ml, which was 10-fold higher than that of the WT at 1 and 3 dpi (P < 0.01 and P < 0.001, respectively; n = 3). In sharp contrast to the titer in WT-infected mice, the virus titer of the variant-infected mice remained at a high level at 5 dpi (103.9 TCID50s/ml), approximately 100-fold higher than that of the WT-infected mice (P < 0.001, n = 3). No virus was detected in the lungs of the infected mice at 7 dpi or in the extrapulmonary body tissues. These results indicate that early viral replication and higher virus titers contribute to the increased pathogenicity in mice. We performed histological analyses (Fig. 4). At 5 dpi, the negative-control (mock infection) mice displayed large air spaces and thin alveolar walls, which are indicative of a normal phenotype. The WT-infected mice show slight infiltration of inflammatory cells and alveolar wall thickening, and the variant-infected mice show large amounts of alveolar wall capillary hyperemia and acute pneumonia with focal lymphocyte infiltration. The histological analysis shows that the variant causes more severe pathological changes in the mice.
FIG 4.
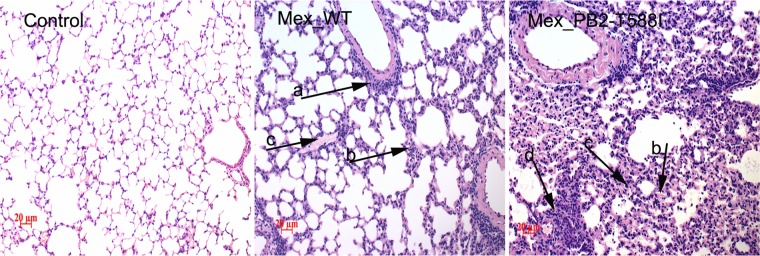
Histopathology analyses. The lungs of the infected mice were fixed with formalin, embedded in paraffin, and stained with hematoxylin and eosin. The images were obtained at a magnification of ×20. Arrow a, infiltration of inflammatory cells; arrow b, alveolar wall thickening; arrow c, alveolar wall capillary hyperemia; arrow d, focal lymphocyte infiltration.
The mutation T588I exacerbated the PB2 inhibition of MAVS-mediated IFN-β expression and conferred increased binding efficiency of PB2 to MAVS.
In addition to the NS1 protein, the PB2 protein of the influenza virus is an inhibitor of IFN-β (17, 27, 28). To assess if PB2 could inhibit IFN-β expression, an IFN-β promoter-driven luciferase reporter gene assay was performed. RIG-I, MDA-5, MAVS, and TBK-1 are known activators of the IFN-β promoter (19, 29–31). As expected from previous reports, the PB2 protein significantly decreased IFN-β expression when RIG-I (P < 0.001, n = 3), MDA-5 (P < 0.001, n = 3), or MAVS (P < 0.01, n = 3) was used as an activator (Fig. 5A, B, and C). No inhibitory effect was observed when TBK-1 was used as an activator (Fig. 5D) (P > 0.05, n = 3), which suggested that the PB2 protein could inhibit the activation of downstream MAVS, affecting the production of IFN-β (17, 27).
FIG 5.
PB2-588I exacerbates the inhibition of IFN-β. (A to D) The transfection of 293T cells was performed with a luciferase reporter plasmid under the control of an IFN-β promoter and a Renilla control plasmid, PB2 expression plasmids, and plasmids expressing either RIG-I, MDA-5, MAVS, or TBK-1. After 24 h, the cells were lysed, and the luciferase and Renilla activities were measured. The Renilla-adjusted luciferase activity (RLU) in the presence of overexpressed RIG-I, MDA-5, MAVS, or TBK-1 but in the absence of the PB2 protein (empty vector) was set to 100%. The activity in the presence of the PB2 protein was expressed as a percentage of that of the empty vector control. (E and F) The transfection of the 293T cells was performed with a luciferase reporter plasmid under the control of an NF-κB or IRF-3 promoter, a MAVS expression plasmid, and a plasmid expressing the PB2 protein or an empty vector. (G) The transfection of the 293T cells was performed with a luciferase reporter plasmid under the control of an IFN-β promoter, a MAVS expression plasmid, and a plasmid expressing either the wild-type PB2 or the PB2-T588I mutant protein. (H) The lysates from the 293T cells transfected with Flag-MAVS and PCMV-Myc-PB2-588T or PCMV-Myc-PB2-588I and immunoprecipitates were analyzed by Western blotting using anti-Flag and anti-Myc antibodies. (I) Densitometry analysis of the Western blot results of the binding efficiency of coimmunoprecipitate PB2-588I and PB2-588T with Flag-MAVS was done using ImageJ (NIH); the value of PB2-588T was set as the standard, and values shown are ratios of their bands to the standard. The bars represent the standard errors of the means, based on three experiments. *, P < 0.01; **, P < 0.001 (as determined by a two-tailed Student's t test). IP, immunoprecipitation; a-, anti.
The regulation of IFN-β synthesis requires the following transcription factors: ATF-2/c-Jun (AP-1), NF-κB, and IRF-3. MAVS mediates the activation of NF-κB and IRF-3 in response to viral infections (19). To determine whether the PB2 protein inhibits this activation, we analyzed the MAVS-mediated activation of NF-κB and IRF-3. PB2 strongly inhibited the activation of both NF-κB and IRF-3(Fig. 5E and F) (P < 0.001, n = 3). These results are consistent with previous reports, suggesting that the PB2 protein could inhibit MAVS-mediated IFN-β expression through NF-κB- and IRF-3-dependent signaling pathways (17, 27).
To assess whether the PB2-T588I mutation has an effect on the inhibition of IFN-β, we compared the inhibition of the PB2-588T protein to that of the PB2-588I protein. The IFN-β level in the cells transfected with PB2-588I was 41% of the level in the cells transfected with PB2-588T (Fig. 5G) (P < 0.001, n = 3). These data suggest that the PB2 protein could participate in the regulation of host innate immune responses by decreasing IFN-β expression. The T588I mutation significantly exacerbated the inhibition.
We investigated whether the PB2-T588I mutation affected the binding of PB2 to MAVS (Fig. 5H). We performed an immunoprecipitation assay using 293T cells cotransfected with a Flag-MAVS plasmid and a Myc-PB2 expression plasmid. We found that both PB2-588T and PB2-588I coimmunoprecipitated with Flag-MAVS and that the PB2-588I mutation conferred increased binding efficiency. These results show that the PB2 protein interacts with MAVS and that the PB2-T588I mutation leads to a stronger binding capability of PB2 to MAVS and exacerbates the PB2 protein inhibition of MAVS-mediated IFN-β synthesis.
The recombinant virus containing PB2-588I induced low levels of IFN-β expression.
We investigated whether the T588I mutation had an effect on the expression of IFN-β in response to a viral infection. A549 cells were infected with the variant or the WT virus; the IFN-β mRNA in the cells was quantified by quantitative reverse transcription-PCR (RT-qPCR) at 12 and 24 hpi. The WT and the variant induced significant levels of IFN-β mRNA compared with the mock-infected cells (Fig. 6A), and our data showed that the level of the IFN-β mRNA in the cells infected with the WT was significantly higher than the level in the cells infected with the variant at 12 and 24 hpi (25-fold and 8-fold, respectively) (P < 0.001, n = 3). These results are consistent with exacerbated inhibition of IFN-β expression in the transfected cells expressing the PB2-588I protein (Fig. 5G), suggesting that the mutation decreased IFN-β expression during a viral infection in vitro.
FIG 6.
The Mex_PB2-588I induced lower levels of IFN-β expression than the Mex_WT virus. (A) A549 cells were infected with either the Mex_WT or the Mex_PB2-588I virus at an MOI of 0.1 TCID50/ml. At 12 and 24 h postinfection, total RNA was isolated, and RT-qPCR was performed to measure IFN-β mRNA levels. (B) Twelve mice per group were infected with either the Mex_WT or the Mex_PB2-588I virus at 105 TCID50s/ml. At 1, 3, 5, and 7 dpi, IFN-β was measured using a mouse IFN-β ELISA kit. Columns represent mean observed concentrations of cytokines from three mice. The error bars represent the standard errors of the means. *, P < 0.01; **, P < 0.001 (as determined by a two-tailed Student's t test).
We analyzed the IFN-β level in the lungs of mice infected with the variant or the WT (Fig. 6B). On days 1, 3, 5, and 7 postinfection, the IFN-β expression levels in the mice infected with the WT virus were higher than the levels in the mice infected with the variant virus (P < 0.01 and P < 0.001, n = 3), suggesting that the variant had higher viral titers but attenuated IFN-β expression during infection in vivo. The data suggest that the variant is associated with an inhibition of host antiviral response, resulting in increased virulence.
DISCUSSION
The PB2 subunit of the influenza virus RNA polymerase was found to determine the host range and to affect the virulence of the influenza virus (17, 27, 32). The identified “signature” amino acids (PB2-E627K, PB2-E667G, PB2-D701N, and PB1-F2–N66S) in the polymerase genes altered the virulence of the H5N1 virus in mammalian hosts and in a mouse model. The introduction of these amino acids into the pdm/09 H1N1 virus did not alter the virulence in a mouse model (16, 33). In this study, we focused on the contribution of the PB2-T588I mutation to the 2009 pandemic H1N1 influenza virus.
The PB2 protein was hypothesized to be important for host restriction, with amino acid polymorphisms at positions 627, 271, 590/591, 701, and others. Position 588 was previously identified as a signature amino acid that contributed to the host-specific activity of both the A/PR/8/34 and the A/turkey/England/50-92/91(H5N1) viruses (32). Isoleucine (I), threonine (T), and alanine (A) residues at PB2 588 were commonly observed in human, swine, and avian viruses (34). PB2-588I was observed in an avian influenza virus that affected humans and in other viruses that caused outbreaks and pandemics in humans before 2009, but the residue is a T in the pdm/09 viruses (35). Taken together, these data provide strong evidence that position 588 in the PB2 protein is a biomarker and that 588T is an amino acid mutation conserved in the pdm/09 virus that is exceedingly rare in previous human influenza virus isolates.
The variant virus exhibited more efficient growth kinetics in three types of cells than the WT virus, suggesting that it has improved growth capability in mammalian hosts. Consistent with the cell culture results, the variant virus showed increased virulence in a mouse model, causing significant weight loss and mortality. These findings suggest the existence of an underlying molecular mechanism by which the mutation could affect virulence.
The PB2-590/591 (with a serine at position 590 and an arginine at position 591 [SR]) polymorphism, which had been shown to cause an increase in polymerase activity and viral replication in human and porcine cells, rescued PB2 with the restrictive glutamic acid at amino acid position 627 (7). The 588 and 590/591 residues located within 627 domains (32) and the 588I mutation increased the polymerase activity and viral replication in human and porcine cells in our study.
Previous studies have shown that the NS1 protein of the 1918 virus pandemic strain mediated the delay of type I interferon induction, which contributed to the virulence of this strain (36). The decreased ability of the divergent NS1 protein to antagonize the human type I IFN system explained why some H5N1 strains replicated well in human hosts (37). The NS1 mutations in an H3N2 subtype of the virus significantly enhanced IFN-β antagonism and reduced IFN-β production in mammalian cells (38). The PB1-F2 protein inhibited the induction of type I interferon by binding to MAVS, and PB1-F2 delayed the initial immune response but increased the pulmonary inflammatory response during the late stages of H5N1 infection (39). PB1-F2–66S was observed to bind to MAVS more efficiently than PB1-F2–66N, and the 66S in the PB1-F2 protein increased the virulence of the 1918 and H5N1 pandemic viruses (18). In this study, we determined that in addition to the NS1 and PB1-F2 proteins, the PB2 protein also participates in regulating the host innate immune responses as previously reported.
Type I interferon is known to be important in host antiviral responses, and RIG-I and MAVS mediate the cellular IFN-β-inducing pathway (29, 40). The polymerase binds to MAVS and inhibits IFN-β production; the PB2 subunit plays a critical role in this inhibition (17, 27). Our data, together with previous reports, indicate that the PB2 protein inhibits MAVS-mediated IFN-β expression through IRF-3- and NF-κB-dependent signaling pathways (19, 41–43). The PB2-T588I mutation exacerbated the inhibition and enhanced the interaction of PB2 and MAVS (Fig. 5H). The increased binding of the PB2-588I to MAVS may be due to a higher binding affinity than PB2-588T, but we have no molecular understanding of this phenomenon. These results indicate that the PB2 protein inhibited IFN-β production, and the mutation significantly enhanced IFN-β antagonism in addition to its contribution to viral polymerase activity.
We found that the variant virus induced lower levels of IFN-β than the WT virus during cell infection. We analyzed the IFN-β levels in the lungs of mice infected with the variant or the WT virus and determined that the variant inhibits IFN-β expression compared with the WT during mouse infections. These data suggest that the PB2-T588I mutation in the context of the influenza virus infection regulates the host antiviral innate immune pathways in vitro and in vivo. The significant weight loss observed in the infected mice was caused by the better growth kinetics of the variant virus and possibly by the fact that the variant decreased the host antiviral response.
The virulence of an influenza virus depends on a great deal of factors, such as cell tropism, viral infectivity, and host immunity. Our data, together with previous studies, clearly show that an increased polymerase activity is not the sole reason for alterations in viral replication or virulence (16). The synergism of the increased polymerase activity with the enhanced IFN-β antagonism contributes to the high virulence. The finding that the PB2 protein regulates host IFN-β expression suggests that influenza viruses have developed multiple strategies to evade host antiviral responses.
In summary, we determined that the amino acid 588I in the PB2 protein of the 2009 pandemic H1N1 influenza virus contributes to viral replication in mammalian cells and pathogenicity in a mouse model. The PB2-T588I mutation exacerbates the inhibition of IFN-β expression and enhances the binding of PB2 to MAVS, thereby antagonizing the host innate immune response. Both viral and cellular factors determine the virulence of influenza viruses, and an understanding of the virus-host interactions will facilitate the development of control strategies for influenza viruses.
ACKNOWLEDGMENT
This work was supported by the National Transgenic Major Program (2009ZX08009-141B).
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Berhane Y, Ojkic D, Neufeld J, Leith M, Hisanaga T, Kehler H, Ferencz A, Wojcinski H, Cottam-Birt C, Suderman M, Handel K, Alexandersen S, Pasick J. 2010. Molecular characterization of pandemic H1N1 influenza viruses isolated from turkeys and pathogenicity of a human pH1N1 isolate in turkeys. Avian Dis. 54:1275–1285. 10.1637/9422-061410-Reg.1 [DOI] [PubMed] [Google Scholar]
- 2.Dundon WG, De Benedictis P, Viale E, Capua I. 2010. Serologic evidence of pandemic (H1N1) 2009 infection in dogs, Italy. Emerg. Infect. Dis. 16:2019–2021. 10.3201/eid1612.100514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasma T, Joseph T. 2010. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg. Infect. Dis. 16:706–708. 10.3201/eid1604.091636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sponseller BA, Strait E, Jergens A, Trujillo J, Harmon K, Koster L, Jenkins-Moore M, Killian M, Swenson S, Bender H, Waller K, Miles K, Pearce T, Yoon KJ, Nara P. 2010. Influenza A pandemic (H1N1) 2009 virus infection in domestic cat. Emerg. Infect. Dis. 16:534–537. 10.3201/eid1603.091737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbs AJ, Armstrong JS, Downie JC. 2009. From where did the 2009 “swine-origin” influenza A virus (H1N1) emerge? Virol. J. 6:207. 10.1186/1743-422X-6-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Zou W, Jia G, Ke J, Zhu J, Lin X, Zhou H, Jin M. 2013. The 2009 pandemic (H1N1) viruses isolated from pigs show enhanced pathogenicity in mice. Vet. Res. 44:41. 10.1186/1297-9716-44-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312–21316. 10.1073/pnas.0911915106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song MS, Pascua PN, Lee JH, Baek YH, Park KJ, Kwon HI, Park SJ, Kim CJ, Kim H, Webby RJ, Webster RG, Choi YK. 2011. Virulence and genetic compatibility of polymerase reassortant viruses derived from the pandemic (H1N1) 2009 influenza virus and circulating influenza A viruses. J. Virol. 85:6275–6286. 10.1128/JVI.02125-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Li Y, Halpin R, Hine E, Spiro DJ, Wentworth DE. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza a viruses in mice. J. Virol. 85:357–365. 10.1128/JVI.01694-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJ, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, García-Sastre A, Perez DR, Fouchier RA. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J. Virol. 84:11802–11813. 10.1128/JVI.01136-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Childs RA, Matrosovich T, Wharton S, Palma AS, Chai W, Daniels R, Gregory V, Uhlendorff J, Kiso M, Klenk HD, Hay A, Feizi T, Matrosovich M. 2010. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J. Virol. 84:12069–12074. 10.1128/JVI.01639-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, Guo D. 2009. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 144:123–129. 10.1016/j.virusres.2009.04.008 [DOI] [PubMed] [Google Scholar]
- 13.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252. 10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagger BW, Memoli MJ, Sheng ZM, Qi L, Hrabal RJ, Allen GL, Dugan VG, Wang R, Digard P, Kash JC, Taubenberger JK. 2010. The PB2-E627K mutation attenuates viruses containing the 2009 H1N1 influenza pandemic polymerase. mBio 1(1):e00067–10. 10.1128/mBio.00067-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Wang J, Wang P, Song W, Zheng Z, Chen R, Guo K, Zhang T, Peiris JS, Chen H, Guan Y. 2010. Substitution of lysine at 627 position in PB2 protein does not change virulence of the 2009 pandemic H1N1 virus in mice. Virology 401:1–5. 10.1016/j.virol.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herfst S, Chutinimitkul S, Ye J, de Wit E, Munster VJ, Schrauwen EJ, Bestebroer TM, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Osterhaus AD, Perez DR, Fouchier RA. 2010. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J. Virol. 84:3752–3758. 10.1128/JVI.02634-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graef KM, Vreede FT, Lau YF, McCall AW, Carr SM, Subbarao K, Fodor E. 2010. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 84:8433–8445. 10.1128/JVI.00879-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga ZT, Grant A, Manicassamy B, Palese P. 2012. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J. Virol. 86:8359–8366. 10.1128/JVI.01122-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 20.García-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in détente. Science 312:879–882. 10.1126/science.1125676 [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113. 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, Wang C, Yang Y, Guo X, Kang C, Chen H, Jin M. 2011. Pandemic (H1N1) 2009 virus in swine herds, People's Republic of China. Emerg. Infect. Dis. 17:1757–1759. 10.3201/eid1709.101916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, Fan Q, Zhong L, Li Y, Liu W, Liu X, Gao S, Peng D. 2012. Isolation and phylogenetic analysis of pandemic H1N1/09 influenza virus from swine in Jiangsu province of China. Res. Vet. Sci. 93:125–132. 10.1016/j.rvsc.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Zhang J, Qiao C, Yang H, Zhang Y, Xin X, Chen H. 2013. Co-circulation of pandemic 2009 H1N1, classical swine H1N1 and avian-like swine H1N1 influenza viruses in pigs in China. Infect. Genet. Evol. 13:331–338. 10.1016/j.meegid.2012.09.021 [DOI] [PubMed] [Google Scholar]
- 25.Pan C, Cheung B, Tan S, Li C, Li L, Liu S, Jiang S. 2010. Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS One 5:e9549. 10.1371/journal.pone.0009549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W, Zhu Y, Qin K, Yu Z, Gao R, Yu H, Zhou J, Shu Y. 2012. Mutations in polymerase genes enhanced the virulence of 2009 pandemic H1N1 influenza virus in mice. PLoS One 7:e33383. 10.1371/journal.pone.0033383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwai A, Shiozaki T, Kawai T, Akira S, Kawaoka Y, Takada A, Kida H, Miyazaki T. 2010. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon beta promoter stimulator 1. J. Biol. Chem. 285:32064–32074. 10.1074/jbc.M110.112458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liniger M, Moulin HR, Sakoda Y, Ruggli N, Summerfield A. 2012. Highly pathogenic avian influenza virus H5N1 controls type I IFN induction in chicken macrophage HD-11 cells: a polygenic trait that involves NS1 and the polymerase complex. Virol. J. 9:7. 10.1186/1743-422X-9-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 30.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. 2002. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637–642. 10.1073/pnas.022637199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. 2006. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 80:9676–9686. 10.1128/JVI.00508-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foeglein Á, Loucaides EM, Mura M, Wise HM, Barclay WS, Digard P. 2011. Influence of PB2 host-range determinants on the intranuclear mobility of the influenza A virus polymerase. J. Gen. Virol. 92:1650–1661. 10.1099/vir.0.031492-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, García-Sastre A, Tumpey TM, Palese P. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442–4450. 10.1128/JVI.02717-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, Shih SR. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 12:1353–1360. 10.3201/eid1209.060276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. 2013. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect. 15:432–439. 10.1016/j.micinf.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 36.Meunier I, von Messling V. 2011. NS1-mediated delay of type I interferon induction contributes to influenza A virulence in ferrets. J. Gen. Virol. 92:1635–1644. 10.1099/vir.0.032193-0 [DOI] [PubMed] [Google Scholar]
- 37.Matthaei M, Budt M, Wolff T. 2013. Highly pathogenic H5N1 influenza A virus strains provoke heterogeneous IFN-α/β responses that distinctively affect viral propagation in human cells. PLoS One 8:e56659. 10.1371/journal.pone.0056659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forbes NE, Ping J, Dankar SK, Jia JJ, Selman M, Keleta L, Zhou Y, Brown EG. 2012. Multifunctional adaptive NS1 mutations are selected upon human influenza virus evolution in the mouse. PLoS One 7:e31839. 10.1371/journal.pone.0031839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leymarie O, Jouvion G, Hervé PL, Chevalier C, Lorin V, Lecardonnel J, Da Costa B, Delmas B, Escriou N, Le Goffic R. 2013. Kinetic characterization of PB1-F2-mediated immunopathology during highly pathogenic avian H5N1 influenza virus infection. PLoS One 8:e57894. 10.1371/journal.pone.0057894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269. 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 42.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 43.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]