Abstract
How hantaviruses assemble and exit infected cells remains largely unknown. Here, we show that the expression of Andes (ANDV) and Puumala (PUUV) hantavirus Gn and Gc envelope glycoproteins lead to their self-assembly into virus-like particles (VLPs) which were released to cell supernatants. The viral nucleoprotein was not required for particle formation. Further, a Gc endodomain deletion mutant did not abrogate VLP formation. The VLPs were pleomorphic, exposed protrusions and reacted with patient sera.
TEXT
Hantaviruses are rodent-borne, enveloped, negative-strand, tripartite RNA viruses that form a separate genus in the Bunyaviridae family. Their transmission to humans can produce severe diseases, such as hemorrhagic fever with renal syndrome caused by Puumala hantavirus (PUUV) in Northern Europe and hantavirus pulmonary syndrome caused by Andes hantavirus (ANDV) in Argentina and Chile (1–3).
Hantavirus virions are roughly spherical and highly heterogenic, varying from 120 to 160 nm in size. They expose glycoprotein spikes locally ordered into tetramers and also contain naked membrane patches on their surfaces (4, 5). The viral envelope membrane is acquired from infected cells during virus budding and encloses the three single-stranded RNA (ssRNA) segments that encode four structural proteins: the Gn and Gc glycoproteins, nucleocapsid protein (N), and RNA-dependent RNA polymerase. As other bunyaviruses, hantaviruses do not have a matrix protein that mediates assembly and budding; hence, a role for the cytoplasmic tails of glycoproteins has been proposed (6–8). Previous studies of orthobunyavirus mutant glycoproteins showed that the endodomains of both glycoproteins are required for virus-like particle (VLP) and virus assembly (9). Further, studies on the Uukuniemi phlebovirus Gn tail showed that the Gn endodomain plays a crucial role in genome packaging into virus particles (10). Bunyavirus glycoproteins originate from a single glycoprotein precursor (GPC) through cotranslational cleavage in the endoplasmic reticulum (11–14). Hantaviruses are believed to bud at internal membranes, most probably derived from the Golgi apparatus, and exit cells via exocytosis; alternatively, they may bud directly from the plasma membrane (11, 15, 16).
To study viral assembly and budding processes, individual or isolated viral components are expressed in cells to test their release into the culture medium as VLPs corresponding to membrane-containing viral structures (17). Previously, it has been reported that hantavirus VLPs are produced when Gn, Gc, and N proteins are coexpressed (18, 19). However, not all of these proteins may be necessary for VLP production. This notion is supported by the observation that animals elicit high neutralizing antibody responses after DNA vaccination solely using a hantavirus Gn and Gc coding plasmid (20–23), which may be indicative for VLP formation in vivo, in the absence of the N protein. In addition, for other members of the Bunyaviridae, such as phleboviruses, it has been reported that the glycoproteins are the only viral components required for the formation of VLPs (24, 25). To test whether the hantavirus N protein is required for the assembly and budding of hantavirus-like particles, in the present work, hantavirus VLP formation was assessed by plasmid-driven expression of hantavirus Gn and Gc glycoproteins. To this end, 293FT cells (Invitrogen) grown in 10-cm dishes were transfected by the calcium phosphate protocol (26) using 8 μg of pI.18/ANDV-GPC plasmid coding for ANDV-GPC under the control of the cytomegalovirus promoter (27). As a positive control for hantavirus VLP formation, 293FT cells were cotransfected with pI.18/ANDV-GPC and pCMV-Bios/N (28) coding for ANDV-N under the cytomegalovirus promoter. The expression of ANDV Gn, Gc, and N was analyzed by Western blotting of cell lysates at 48 h posttransfection (Fig. 1A). To this end, the monoclonal antibodies (MAbs) anti-Gn 6B9/F5 (29), anti-Gc 2H4/F6 (30), and anti-N 7B3/F7 (31) and secondary antibody anti-mouse IgG peroxidase conjugate (Sigma) were used. In the lysates of cells transfected with one or both plasmids, ANDV Gn monomers were detected at ∼70 kDa and ANDV Gc monomers at ∼55 kDa (Fig. 1A, left). Higher-molecular-mass bands of Gn and Gc were also observed as reported previously (27, 29). N could be detected in cells cotransfected with pCMV-Bios/N with a size of ∼50 kDa. To analyze whether the viral proteins were also present in cell supernatants, they were concentrated by ultracentrifugation at 100,000 × g, as described previously (27). As seen in Fig. 1A (right), Gn and Gc were detected in concentrated supernatants of cells expressing both envelope proteins alone or in combination with the viral N protein, confirming our hypothesis that N was not required for the release of glycoproteins into the cell supernatant. To test whether the release of ANDV glycoproteins was a particular property of ANDV or if it was a more general feature of hantaviruses, plasmid-driven expression of PUUV glycoproteins was next tested. To this end, 293FT cells were transfected with 8 μg of the plasmid pWRG/PUU-M(s2) coding for PUUV-GPC under the cytomegalovirus promoter (32). When cell lysates and the concentrated supernatant of pWRG/PUU-M(s2)-transfected cells were analyzed by Western blotting, PUUV Gc was detected in both fractions with its expected molecular mass (Fig. 1B), indicating that, as for ANDV, the glycoproteins of PUUV were also released into the cell supernatant in the absence of additional viral molecules. To test whether the presence of viral glycoproteins in cell supernatants was related to virus-like structures, concentrated supernatants of cells transfected with plasmids coding for ANDV-GPC or PUUV-GPC were next analyzed by electron microscopy (EM) using phosphotungstic acid at a pH of ∼7.4 for negative staining, as described previously (33). Electron micrographs of concentrated supernatants of ANDV or PUUV glycoprotein-expressing cells showed virus-like structures that were variable in size and shape (Fig. 2A to D and E to H, respectively). No apparent morphological differences could be detected between ANDV VLPs produced with or without the N protein (data not shown). It has been well described that EM of hantaviruses reveals a characteristic grid-like pattern (34–36). Negative-staining EM of ANDV and PUUV VLPs fixed with 0.5% glutaraldehyde allowed to discern a grid-like pattern (Fig. 2C and G, respectively). Further, isolated surface projections could be distinguished (Fig. 2G, arrows), resembling a Y-shape similar to the described molecular structure of Hantaan virus and Tula virus spikes that are composed of a tetrameric, globular head domain connected to the membrane by a thinner, central stalk region (4, 5).
FIG 1.
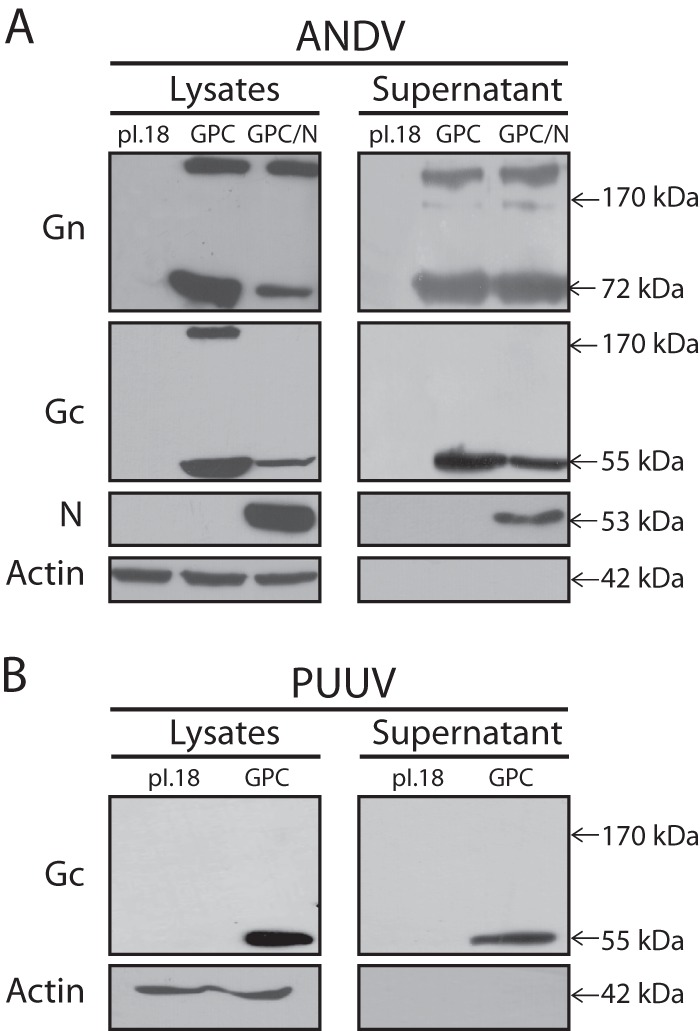
Detection of viral proteins in cell lysates and supernatants. Western blots of lysates and concentrated supernatant of 293FT cells transfected with different plasmids. (A) Transfection of empty plasmid, pI.18/ANDV-GPC, or cotransfection of pI.18/ANDV-GPC and pCMV-Bios/ANDV-N. (B) Transfection of empty plasmid or pWRG/PUU-M(s2). ANDV Gn and N proteins were detected with anti-Gn 6B9/F5 and anti-N 7B3/F7, respectively. ANDV and PUUV Gc was detected with MAb anti-Gc 2H4/F6. No MAb against PUUV Gn was available.
FIG 2.
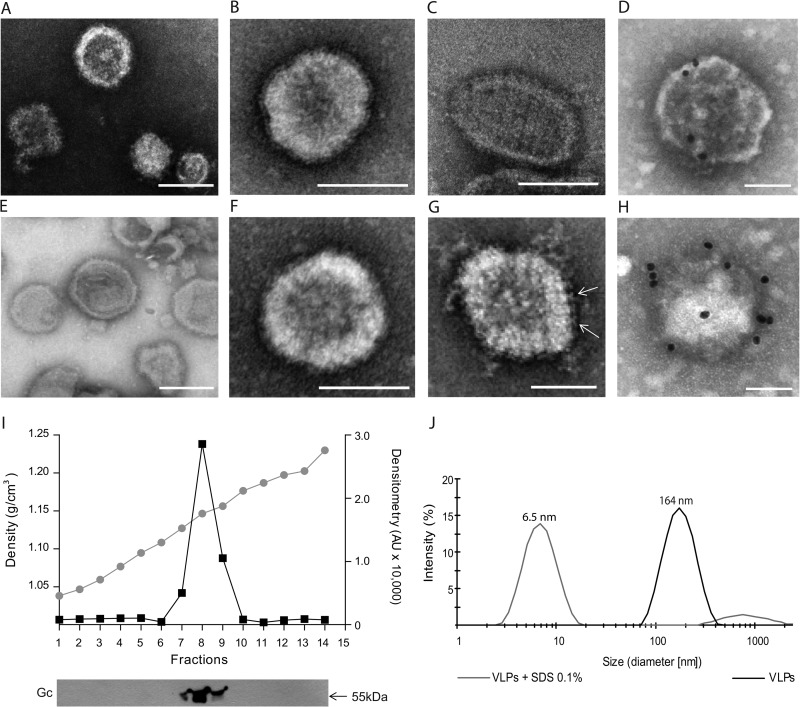
Characterization of hantavirus VLPs. Negative-stain EM of concentrated supernatants from cells transfected with pI.18/ANDV-GPC (A to D) or pWRG/PUU-M(s2) (E to H). Negative-stain EM using phosphotungstic acid of unfixed ANDV VLPs (B), PUUV VLPs (E, F), glutaraldehyde-fixed ANDV VLPs (A, C), and PUUV VLPs (G). Immunogold EM of ANDV VLPs (D) and PUUV VLPs (H) stained with uranyl acetate using patient sera (1:100) and protein A conjugated to gold beads (20 nm). (I) Sucrose gradient sedimentation of ANDV VLPs and detection of VLPs through Western blotting using anti-ANDV Gc MAb 2H4/F6. Buoyant density was determined by refractometry, and relative intensity of Gc bands was quantified as arbitrary units using ImageJ (40). (J) Dynamic light scattering of ANDV VLPs at pH 7.4 and ANDV VLPs treated for 30 min with SDS 0.1%. Bars, 100 nm.
The observed virus structures were further characterized by immunogold EM. For this purpose, VLPs were adsorbed to Formvar/carbon-coated copper grids and blocked with 5% bovine serum albumin. Subsequently, the immobilized VLPs were incubated with hantavirus patient sera (1:100) derived from Chilean patients infected with ANDV or ANDV-related species or with sera from European patients infected with PUUV or PUUV-related species. After 3 washes with Tris-buffered saline (TBS), primary antibody binding was detected with protein A conjugated to gold beads (Sigma). As can be seen in Fig. 2D and H, ANDV and PUUV VLPs were recognized by the respective specific patient sera. Normal patient sera were used as negative controls (n = 3; data not shown). To further characterize the VLPs in terms of density, sucrose gradient sedimentation was performed by ultracentrifugation for 16 h at 38,000 rpm using an SW55 rotor. The refractive index of each fraction was analyzed at 20°C, and the presence of VLPs was examined by Western blotting using MAb anti-Gc 2H4/F6. VLPs derived from ANDV glycoprotein expression peaked in fraction 8, corresponding to a buoyant density of 1.15 g/ml (Fig. 2I) that coincides with the density range of 1.15 to 1.18 g/ml, which has been reported for infectious hantaviruses and other bunyaviruses (34, 37). The size range of particles generated by glycoprotein expression was next determined by dynamic light scattering (Zetasizer Nano ZS; Malvern Instruments). The size of over 90% of VLPs varied within the range of 90 to 255 nm (Fig. 2J). When VLPs were incubated with 0.1% SDS, their size diminished below 20 nm, confirming their membranous composition (Fig. 2J). Taken together, these data indicate that the release of glycoproteins into cell supernatants in the form of virus-like structures does not require the participation of the viral N protein. Further, the hantavirus glycoproteins are the only viral components required for the assembly and release of VLPs.
The hantavirus VLPs were further characterized in terms of their antigenicity using the sera of hantavirus patients. For this purpose, ANDV or PUUV VLPs contained in concentrated supernatants were immobilized on enzyme-linked immunosorbent assay (ELISA) plates by incubation for 1 h at room temperature (RT). Subsequently, wells were blocked with 4% casein-sucrose for 2 h at RT, and human sera were then added at a dilution of 1:250. After 1.5 h of incubation at RT, the wells were washed 5 times with 0.05% Tween 20–phosphate-buffered saline (PBS). Next, wells were incubated for 1 h with anti-human immunoglobulins G, A, and M conjugated to peroxidase and finally revealed with a tetramethylbenzidine peroxidase substrate (KPL). The reaction was stopped within 10 min by the addition of 1 M phosphoric acid, and the absorbance was read at 450 nm. As seen in Fig. 3, ANDV VLPs reacted with sera derived from Chilean patients. Weak cross-reactivity with ANDV VLPs was detected with the sera of patients from North America infected with Sin Nombre virus or Sin Nombre virus-related species. No reactivity against ANDV VLPs was observed with sera from PUUV-infected patients and with negative-control sera. When PUUV VLPs were incubated with patient sera, reactivity was detected with PUUV-infected patient sera, and a weak cross-reactivity was observed with the sera of patients from America. No reactivity of PUUV VLPs was observed with the negative-control sera. In summary, these data confirm that the ANDV and PUUV VLPs contain glycoproteins on their surfaces that expose epitopes that are recognized by patient sera reactive against the respective native hantavirus.
FIG 3.
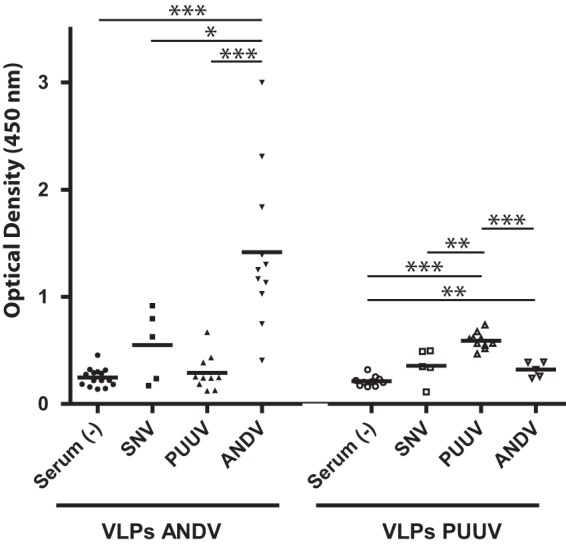
Antigenicity of hantavirus VLPs. ELISA plates were activated with concentrated ANDV or PUUV VLPs, and their reactivity was tested with sera derived from patients infected with different hantavirus species (ANDV, PUUV, Sin Nombre virus [SNV]). A Student t test was used for statistical evaluation: ***, P < 0.00025; **, P < 0.0025; *, P < 0.025.
To test the requirement of the Gc endodomain for VLP assembly, a Gc endodomain deletion mutant (GPCΔGcCT) was generated, based on the prediction of the Gc transmembrane region (38, 39). DNA mutagenesis was performed by Pfx polymerase (Invitrogen)-driven PCR amplification of the ANDV-GPC coding region using the forward primer 5′-TAGATCTATTATGGAAGGGTGGTATCTGGTTGC-3′ and the reverse primer 5′-CTCGAGCTAGCACAGAACACTGAACATTATGATAGAGAG-3′. Subsequently, the PCR product was subcloned into the pI.18 expression vector. To test expression and folding of the GPCΔGcCT mutant, 293FT cells were surface biotinylated 48 h posttransfection, using a cell surface protein isolation kit (Pierce) as described previously (29). Previous to biotinylation, the cell supernatant was collected for particle concentration, and the cell lysates were separated into two fractions: the unbiotinylated fraction containing intracellular proteins and the biotinylated fraction containing surface proteins. When these fractions were analyzed by Western blotting, no apparent difference among the wild-type (WT) and mutant proteins was detected in terms of Gc surface localization or the presence of glycoproteins in the concentrated supernatant (Fig. 4A). Further, negative-stain EM of the concentrated supernatant revealed virus structures similar to those of VLPs formed by the WT glycoproteins (Fig. 4B). These data indicate that in contrast to the orthobunyavirus Gc endodomain, the ANDV Gc endodomain is not required for the assembly of virus structures. This difference may be explained in part by the different length of these regions, conserving nine residues among hantaviruses (39) compared to 26 residues among orthobunyaviruses (9).
FIG 4.
Deletion analysis of ANDV Gc endodomain mutant. (A) Western blot analysis using anti-Gc and anti-actin MAbs of different fractions corresponding to the nonbiotinylated fraction (intracellular proteins), the biotinylated fraction (surface proteins), or the concentrated supernatant of 293FT cells that were transfected with pI.18/ANDV-GPC, pI.18/ANDV-GPCΔGcCT, or empty vector and biotinylated 48 h posttransfection. (B) Negative-stain EM analysis of concentrated supernatants derived from 293FT cells transfected with pI.18/ANDV-GPCΔGcCT. Bar, 100 nm.
In the absence of a reverse-genetics system for hantaviruses, the VLP formation system is an important tool for the characterization of the maturation and exit functions as well as cell entry of hantavirus glycoproteins. Further mutagenesis studies are currently assessing glycoprotein properties required for their self-assembly and cell exit. It remains to be determined whether self-assembly is a general feature of glycoproteins of the Hantavirus genus and the Bunyaviridae.
ACKNOWLEDGMENTS
We thank Jay Hooper (USAMRIID) for providing the plasmid pWRG/PUU-M(s2). Furthermore, we acknowledge Brian Hjelle (University of New Mexico, USA) and Hector Galeno (Instituto de Salud Púbica, Chile) for providing patient sera. Our thanks also extend to Alejandro Muñizaga (Advanced Microscopy Unit, Pontificia Universidad Católica, Chile) for his advice on immunogold EM.
This work was financed by CONICYT through grant FONDECYT 1100756 and basal funding PFB-16. R.A. was supported by a CONICYT doctoral fellowship.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Macneil A, Nichol ST, Spiropoulou CF. 2011. Hantavirus pulmonary syndrome. Virus Res. 162:138–147. 10.1016/j.virusres.2011.09.017 [DOI] [PubMed] [Google Scholar]
- 2.Krautkramer E, Zeier M, Plyusnin A. 2013. Hantavirus infection: an emerging infectious disease causing acute renal failure. Kidney Int. 83:23–27. 10.1038/ki.2012.360 [DOI] [PubMed] [Google Scholar]
- 3.Schmaljohn C, Hjelle B. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95–104. 10.3201/eid0302.970202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huiskonen JT, Hepojoki J, Laurinmaki P, Vaheri A, Lankinen H, Butcher SJ, Grunewald K. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84:4889–4897. 10.1128/JVI.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battisti AJ, Chu YK, Chipman PR, Kaufmann B, Jonsson CB, Rossmann MG. 2011. Structural studies of Hantaan virus. J. Virol. 85:835–841. 10.1128/JVI.01847-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Alminaite A, Vaheri A, Plyusnin A. 2010. Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 151:205–212. 10.1016/j.virusres.2010.05.008 [DOI] [PubMed] [Google Scholar]
- 7.Hepojoki J, Strandin T, Wang H, Vapalahti O, Vaheri A, Lankinen H. 2010. Cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J. Gen. Virol. 91:2341–2350. 10.1099/vir.0.021006-0 [DOI] [PubMed] [Google Scholar]
- 8.Strandin T, Hepojoki J, Vaheri A. 2013. Cytoplasmic tails of bunyavirus Gn glycoproteins—could they act as matrix protein surrogates? Virology 437:73–80. 10.1016/j.virol.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Kohl A, Li P, Elliott RM. 2007. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J. Virol. 81:10151–10160. 10.1128/JVI.00573-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overby AK, Pettersson RF, Neve EP. 2007. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 81:3198–3205. 10.1128/JVI.02655-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettersson R, Melin L. 1996. Synthesis, assembly and intracellular transport of Bunyaviridae membrane proteins. In Elliott RM. (ed), The Bunyaviridae. Plenum Press, New York, NY [Google Scholar]
- 12.Spiropoulou CF, Goldsmith CS, Shoemaker TR, Peters CJ, Compans RW. 2003. Sin Nombre virus glycoprotein trafficking. Virology 308:48–63. 10.1016/S0042-6822(02)00092-2 [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Elliott RM. 2004. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J. Virol. 78:5414–5422. 10.1128/JVI.78.10.5414-5422.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lober C, Anheier B, Lindow S, Klenk HD, Feldmann H. 2001. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology 289:224–229. 10.1006/viro.2001.1171 [DOI] [PubMed] [Google Scholar]
- 15.Goldsmith CS, Elliott LH, Peters CJ, Zaki SR. 1995. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch. Virol. 140:2107–2122. 10.1007/BF01323234 [DOI] [PubMed] [Google Scholar]
- 16.Xu F, Yang Z, Wang L, Lee YL, Yang CC, Xiao SY, Xiao H, Wen L. 2007. Morphological characterization of hantavirus HV114 by electron microscopy. Intervirology 50:166–172. 10.1159/000098959 [DOI] [PubMed] [Google Scholar]
- 17.Lyles DS. 2013. Assembly and budding of negative-strand RNA viruses. Adv. Virus Res. 85:57–90. 10.1016/B978-0-12-408116-1.00003-3 [DOI] [PubMed] [Google Scholar]
- 18.Betenbaugh M, Yu M, Kuehl K, White J, Pennock D, Spik K, Schmaljohn C. 1995. Nucleocapsid- and virus-like particles assemble in cells infected with recombinant baculoviruses or vaccinia viruses expressing the M and the S segments of Hantaan virus. Virus Res. 38:111–124. 10.1016/0168-1702(95)00053-S [DOI] [PubMed] [Google Scholar]
- 19.Li C, Liu F, Liang M, Zhang Q, Wang X, Wang T, Li J, Li D. 2010. Hantavirus-like particles generated in CHO cells induce specific immune responses in C57BL/6 mice. Vaccine 28:4294–4300 [DOI] [PubMed] [Google Scholar]
- 20.Hooper JW, Kamrud KI, Elgh F, Custer D, Schmaljohn CS. 1999. DNA vaccination with hantavirus M segment elicits neutralizing antibodies and protects against Seoul virus infection. Virology 255:269–278. 10.1006/viro.1998.9586 [DOI] [PubMed] [Google Scholar]
- 21.Hooper JW, Custer DM, Thompson E. 2003. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306:181–195. 10.1016/S0042-6822(02)00038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper JW, Custer DM, Smith J, Wahl-Jensen V. 2006. Hantaan/Andes virus DNA vaccine elicits a broadly cross-reactive neutralizing antibody response in nonhuman primates. Virology 347:208–216. 10.1016/j.virol.2005.11.035 [DOI] [PubMed] [Google Scholar]
- 23.Boudreau EF, Josleyn M, Ullman D, Fisher D, Dalrymple L, Sellers-Myers K, Loudon P, Rusnak J, Rivard R, Schmaljohn C, Hooper JW. 2012. A phase 1 clinical trial of Hantaan virus and Puumala virus M-segment DNA vaccines for hemorrhagic fever with renal syndrome. Vaccine 30:1951–1958 [DOI] [PubMed] [Google Scholar]
- 24.Overby AK, Popov V, Neve EP, Pettersson RF. 2006. Generation and analysis of infectious virus-like particles of Uukuniemi virus (bunyaviridae): a useful system for studying bunyaviral packaging and budding. J. Virol. 80:10428–10435. 10.1128/JVI.01362-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandell RB, Koukuntla R, Mogler LJ, Carzoli AK, Freiberg AN, Holbrook MR, Martin BK, Staplin WR, Vahanian NN, Link CJ, Flick R. 2010. A replication-incompetent Rift Valley fever vaccine: chimeric virus-like particles protect mice and rats against lethal challenge. Virology 397:187–198. 10.1016/j.virol.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham FL, van der Eb AJ. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54:536–539. 10.1016/0042-6822(73)90163-3 [DOI] [PubMed] [Google Scholar]
- 27.Cifuentes-Munoz N, Darlix JL, Tischler ND. 2010. Development of a lentiviral vector system to study the role of the Andes virus glycoproteins. Virus Res. 153:29–35. 10.1016/j.virusres.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 28.Gupta S, Braun M, Tischler ND, Stoltz M, Sundstrom KB, Bjorkstrom NK, Ljunggren HG, Klingstrom J. 2013. Hantavirus-infection confers resistance to cytotoxic lymphocyte-mediated apoptosis. PLoS Pathog. 9:e1003272. 10.1371/journal.ppat.1003272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cifuentes-Muñoz N, Barriga GP, Valenzuela PDT, Tischler ND. 2011. Aromatic and polar residues spanning the candidate fusion peptide of Andes virus are essential for membrane fusion and infection. J. Gen. Virol. 92:552–563. 10.1099/vir.0.027235-0 [DOI] [PubMed] [Google Scholar]
- 30.Godoy P, Marsac D, Stefas E, Ferrer P, Tischler ND, Pino K, Ramdohr P, Vial P, Valenzuela PD, Ferres M, Veas F, Lopez-Lastra M. 2009. Andes virus antigens are shed in urine of patients with acute hantavirus cardiopulmonary syndrome. J. Virol. 83:5046–5055. 10.1128/JVI.02409-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischler ND, Rosemblatt M, Valenzuela PD. 2008. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus Res. 135:1–9. 10.1016/j.virusres.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 32.Brocato RL, Josleyn MJ, Wahl-Jensen V, Schmaljohn CS, Hooper JW. 2013. Construction and nonclinical testing of a Puumala virus synthetic M gene-based DNA vaccine. Clin. Vaccine Immunol. 20:218–226. 10.1128/CVI.00546-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Bonsdorff CH, Pettersson R. 1975. Surface structure of Uukuniemi virus. J. Virol. 16:1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White JD, Shirey FG, French GR, Huggins JW, Brand OM, Lee HW. 1982. Hantaan virus, aetiological agent of Korean haemorrhagic fever, has Bunyaviridae-like morphology. Lancet i:768–771 [DOI] [PubMed] [Google Scholar]
- 35.Lee HW, Cho HJ. 1981. Electron microscope appearance of Hantaan virus, the causative agent of Korean haemorrhagic fever. Lancet i:1070–1072 [DOI] [PubMed] [Google Scholar]
- 36.Martin ML, Lindsey-Regnery H, Sasso DR, McCormick JB, Palmer E. 1985. Distinction between Bunyaviridae genera by surface structure and comparison with Hantaan virus using negative stain electron microscopy. Arch. Virol. 86:17–28. 10.1007/BF01314110 [DOI] [PubMed] [Google Scholar]
- 37.Obijeski JF, Murphy FA. 1977. Bunyaviridae: recent biochemical developments. J. Gen. Virol. 37:1–14. 10.1099/0022-1317-37-1-1 [DOI] [PubMed] [Google Scholar]
- 38.Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. 374:166 [Google Scholar]
- 39.Tischler ND, Fernandez J, Muller I, Martinez R, Galeno H, Villagra E, Mora J, Ramirez E, Rosemblatt M, Valenzuela PD. 2003. Complete sequence of the genome of the human isolate of Andes virus CHI-7913: comparative sequence and protein structure analysis. Biol. Res. 36:201–210 [DOI] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]