ABSTRACT
In certain sporadic, familial, and infectious prion diseases, the prion protein misfolds and aggregates in skeletal muscle in addition to the brain and spinal cord. In myocytes, prion aggregates accumulate intracellularly, yet little is known about clearance pathways. Here we investigated the clearance of prion aggregates in muscle of transgenic mice that develop prion disease de novo. In addition to neurodegeneration, aged mice developed a degenerative myopathy, with scattered myocytes containing ubiquitinated, intracellular prion inclusions that were adjacent to myocytes lacking inclusions. Myocytes also showed elevated levels of the endoplasmic reticulum chaperone Grp78/BiP, suggestive of impaired protein degradation and endoplasmic reticulum stress. Additionally, autophagy was induced, as indicated by increased levels of beclin-1 and LC3-II. In C2C12 myoblasts, inhibition of autophagosome maturation or lysosomal degradation led to enhanced prion aggregation, consistent with a role for autophagy in prion aggregate clearance. Taken together, these findings suggest that the induction of autophagy may be a central strategy for prion aggregate clearance in myocytes.
IMPORTANCE
INTRODUCTION
Infectious prions cause severe neurodegeneration that is ultimately fatal following a prolonged incubation period (1, 2). Prion diseases occur when the host-encoded monomeric prion protein, PrPC, misfolds and aggregates into a β-sheet-rich multimer, PrPSc, in an autocatalytic process that leads to widespread PrPSc deposition as well as neuronal death, spongiosis, astrogliosis, and microglial activation in the brain and spinal cord (3–7).
PrPSc accumulates in neural and other tissues, including skeletal muscle, lymph nodes, spleen, gastrointestinal tract, and adrenal glands, depending on the type of prion disease (8–11). The degenerative lesions, however, are largely restricted to the nervous system. Nevertheless, infectious prions or PrPSc aggregates arise in the muscles of patients with variant, iatrogenic, and sporadic Creutzfeldt-Jakob disease (CJD) (12, 13), as well as in cervids with chronic wasting disease (CWD) and in sheep and hamster models of scrapie (14–16). In addition, skeletal muscles of transgenic mice that overexpress PrPC develop myocyte degeneration and intracellular PrP aggregates (17, 18). Myocytes have also been shown to replicate infectious prions (19). Thus, skeletal muscle is a rare extracerebral tissue that develops prion-induced degeneration, aggregate formation, and prion infectivity.
The mature prion protein (approximately 209 amino acids) consists of an unstructured N terminus and a globular C-terminal domain composed of three α-helices and a short β-sheet (20). We previously demonstrated that transgenic mice expressing a “rigid-loop” structural variant of PrP (RL-PrP) due to two amino acid residue substitutions in the β2-α2 loop region (S170N, N174T) develop a transmissible prion disease characterized by hind limb paresis, muscle atrophy, and kyphosis. Intriguingly, PrPSc aggregates accumulated as extracellular plaques in the brain and as intracellular inclusions in muscle (21), suggesting that aggregate development or clearance mechanisms differ for neurons and myocytes.
Intracellular misfolded proteins are routed for degradation by the ubiquitin-proteasome system or by autophagy, where cargo is sequestered in autophagosomes and ultimately degraded in endolysosomal compartments (22–24). Inadequate clearance can lead to the accumulation of misfolded proteins in “aggresomes” or “inclusion bodies.” In patients with sporadic inclusion body myositis (sIBM), intracellular inclusion bodies in myocytes are composed of a mixture of misfolded, β-sheet-rich proteins (25, 26). In sIBM, endoplasmic reticulum (ER) stress and induction of the unfolded protein response (UPR) occur, as well as macroautophagy, yet cells ultimately fail to clear aggregates. Impaired autophagy due to mutations in p97/valosin-containing protein (VCP) has been proposed to cause inclusion body myopathy associated with Paget's disease of bone and frontotemporal dementia (IBMPFD) (27, 28).
The UPR is induced when secreted or membrane proteins misfold and accumulate in the ER (29). The UPR activates a transcriptional and translational program that increases the production of ER chaperones, enhances ER-associated protein degradation, and reduces the efficiency of ribosomal assembly on mRNAs, thereby attenuating protein translation (29). During a prion infection, the UPR is activated in the brain, decreasing protein translation through the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) and leading to impaired synaptic function (30). PrP aggregates have also been shown to directly interact with and inhibit the proteasome (31). In cells expressing mutant PrP, impairment of the proteasome results in the trafficking of misfolded PrP directly to the plasma membrane (32).
The pathways of PrP aggregate clearance in the diverse cell types affected during prion disease remain unclear. To better understand how PrP aggregates are cleared from the skeletal muscle in individuals with familial or sporadic prion disease, we investigated the role of the UPR, the ubiquitin-proteasome system, and autophagy in muscles of aged transgenic mice that express RL-PrP.
MATERIALS AND METHODS
Animals.
Aged tg1020 mice (which express RL-PrP) (310 to 536 days; n = 15), aged tga20 mice (which overexpress mouse PrP) (230 to 470 days; n = 14), and aged wild-type (WT) mice (173 to 524 days; n = 12) were utilized in these studies. The tg1020 and tga20 transgenic mice were developed using the “half-genomic” Prnp minigene containing the prion promoter and were on a Prnp−/− background (21, 33). Transgene-positive mice were identified from tail or ear samples by standard PCR (21). Mice were maintained under specific-pathogen-free conditions with unlimited access to food and water. All procedures involving animals were performed in such as way as to minimize suffering and were approved by the Institutional Animal Care and Use Committee at UC San Diego. Protocols were performed in strict accordance with good animal practices, as described in the Guide for the Care and Use of Laboratory Animals (34).
Plasmids and constructs.
Mouse Prnp cDNA containing the rigid-loop mutations (corresponding to S170N and N174T) were subcloned into the pcDNA3.1C vector (Invitrogen). Site-directed mutagenesis was performed to obtain WT-pcDNA3.1C and to introduce the 3F4 tag (residues 109M and 112M [human PrP numbering]) into the PrP sequence.
Cell culture, transfections, and drug treatments.
C2C12 myoblasts (ATCC) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 20% fetal bovine serum, penicillin, and streptomycin. For transient transfections, C2C12 cells were grown in a 6-well plate and were transfected with 1 μg plasmid by using the transfection reagent Lipofectamine 2000 (Invitrogen). Six hours posttransfection, the medium was changed to a complete medium containing dimethyl sulfoxide (DMSO) (Sigma), bafilomycin A (BafA) (160 nM; Calbiochem), or leupeptin (20 μM; Sigma), and cells were incubated for an additional 20 h at 37°C. MG132 (50 μM; Sigma) was added 16 h posttransfection, and incubation was performed for 6 h at 37°C.
Biochemical assays. (i) Tissue homogenization/cell lysis.
Muscle tissue was homogenized (5%, wt/vol) in ice-cold lysis buffer (1% Triton X-100, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) using a BeadBeater tissue homogenizer. C2C12 cells were lysed in a buffer consisting of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Complete; Roche).
(ii) Western blot analysis.
Fifty micrograms of total protein was resolved on 10% or 4-to-12% Bis-Tris gels and was transferred to a nitrocellulose membrane. Antibodies to Grp78/BiP (Abcam), ubiquitin (Dako), p62 (Sigma), beclin (Novus), LC3 (Novus), PrP 3F4 (Millipore), PrP POM1 (35) (a gift from Adriano Aguzzi), TDP-43 (Proteintech), transferrin receptor (Invitrogen), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling), and actin (Sigma) were used for protein detection along with the respective horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson Laboratories) and an enhanced chemiluminescence (ECL) substrate kit (Pierce). Signals were captured and quantified by using a Fujifilm LAS 4000 imager and MultiGauge software.
(iii) IP with anti-PrP antibody 15B3.
Fifty milligrams of muscle was homogenized in homogenization buffer (Prionics) supplemented with protease inhibitors (PI) (1 mM PMSF and Complete) and was ultracentrifuged at 150,000 × g for 1 h. The pellet was resuspended in immunoprecipitation (IP) buffer (Prionics) with PI. Samples were precleared with anti-IgM Dynabeads, followed by incubation with 15B3 (Prionics)-conjugated anti-IgM Dynabeads (1.5 μg of 15B3 per tube) at 25°C for 2 h. The beads were washed three times with IP buffer, and protein was eluted by heating the beads in LDS sample buffer (Invitrogen) containing β-mercaptoethanol at 95°C for 5 min. For IP analysis of transfected C2C12 cells, the ultracentrifugation step was omitted.
(iv) PK sensitivity assay.
Muscle homogenate was lysed in a Tris-based buffer (100 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2% sarcosyl) for 10 min at 37°C, and aliquots containing approximately 200 μg of protein were incubated either without proteinase K (PK) or in 1 to 10 μg/ml PK (final PK concentration, 1, 2, 3, 4, 5, or 10 μg/ml). The samples were incubated at 37°C for 30 min, and PK digestion was stopped by the addition of PMSF and protease inhibitors (Complete). Samples were transferred in triplicate to an enzyme-linked immunosorbent assay (ELISA) plate precoated with anti-PrP antibody POM2. PrP was detected with the anti-PrP biotinylated-POM1 antibody and streptavidin HRP-conjugated anti-mouse IgG as the secondary antibody. The signals were detected with the HRP substrate 1-Step Ultra tetramethylbenzidine (TMB) (Thermo Scientific). tg1020, tga20, and WT mice were analyzed in triplicate using 4 to 5 mice each.
(v) Deglycosylation assay.
Lysates from C2C12 cells expressing WT-PrP or RL-PrP were denatured in a denaturing buffer (New England BioLabs) for 10 min at 95°C, followed by incubation with endoglycosidase H (endo H) or peptide-N-glycosidase F (PNGase F) (New England BioLabs) at 37°C for 90 min.
Histology.
For PrP immunohistochemistry, paraffin-embedded tissue sections (2 μm) were treated with 98% formic acid for 6 min, followed by treatment with proteinase K (10 μg/ml) and autoclaving in citrate buffer (Dako). SAF84 (Cayman Chemicals) was used for labeling PrP. For beclin-1 (Abcam), LC-3 (Novus), CD68 (Novus), ubiquitin (Dako), and slow myosin heavy chain (Sigma) staining, the slides were autoclaved in Tris-EDTA (for beclin) or citrate (for LC-3, CD68, ubiquitin, and slow myosin) buffer prior to antibody incubation. Biotin-conjugated anti-mouse or anti-rabbit IgG (Jackson Laboratories) secondary antibodies were used along with peroxidase-conjugated streptavidin. Diaminobenzidine (DAB) was used for signal detection. TDP-43 (Proteintech) did not require pretreatment. For fast myosin heavy chain staining, an alkaline phosphatase-conjugated antibody (Sigma) was used without any pretreatment. The NBT (nitroblue tetrazolium)-BCIP (5-bromo-4-chloro-3-indolylphosphate) substrate was used to visualize the antibody.
Molecular biology analysis to detect ER stress.
Muscle tissues were lysed, and total RNA was collected (Qiagen). cDNA was generated from total RNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and was used as the template for PCR amplification across the fragment of the Xbp-1 cDNA bearing the intron target of IRE1. The primers used for mouse Xbp-1 were 5′-GGCCTTGTGGTTGAGAACCAGGAG-3′ and 5′-GAATGCCCAAAAGGATATCAGACTC-3′. PCR conditions were as follows: 95°C for 5 min; 95°C for 1 min; 68°C for 30 s; 72°C for 30 s; 72°C for 5 min with 35 cycles of amplification. PCR products were resolved on a 2% agarose–1× Tris-acetate-EDTA (TAE) gel.
For quantitative PCR, aliquots of cDNA were used as the template. The primers used included those for mouse GAPDH (5′-CTCAACTACATGGTCTACATGTTCCA-3′ and 5′-CCATTCTCGGCCTTGACTGT-3′), mouse ERdj4 (5′-CAGAATTAATCCTGGCCTCC-3′ and 5′-ACTATTGGCATCCGAGAGTG-3′), mouse BiP (5′-ACTCCGGCGTGAGGTAGAAA-3′ and 5′-AGAGCGGAACAGGTCCATGT-3′), and mouse Chop (5′-ACGGAAACAGAGTGGTCAGTGC-3′ and 5′-CAGGAGGTGATGCCCACTGTTC-3′). mRNA levels of GAPDH, a transcript whose levels are not altered by ER stress, served as an internal normalization standard. PCR conditions were as follows: 95°C for 5 min; 95°C for 10 s; 60°C for 10 s; 72°C for 10 s, with 50 cycles of amplification.
For the p62 quantitative PCR, primers and 6-carboxyfluorescein (FAM)-labeled p62 and 18S rRNA endogenous control probes (Life Technologies) were used.
RESULTS
Degenerative myopathy with PrP aggregates in tg1020 mice.
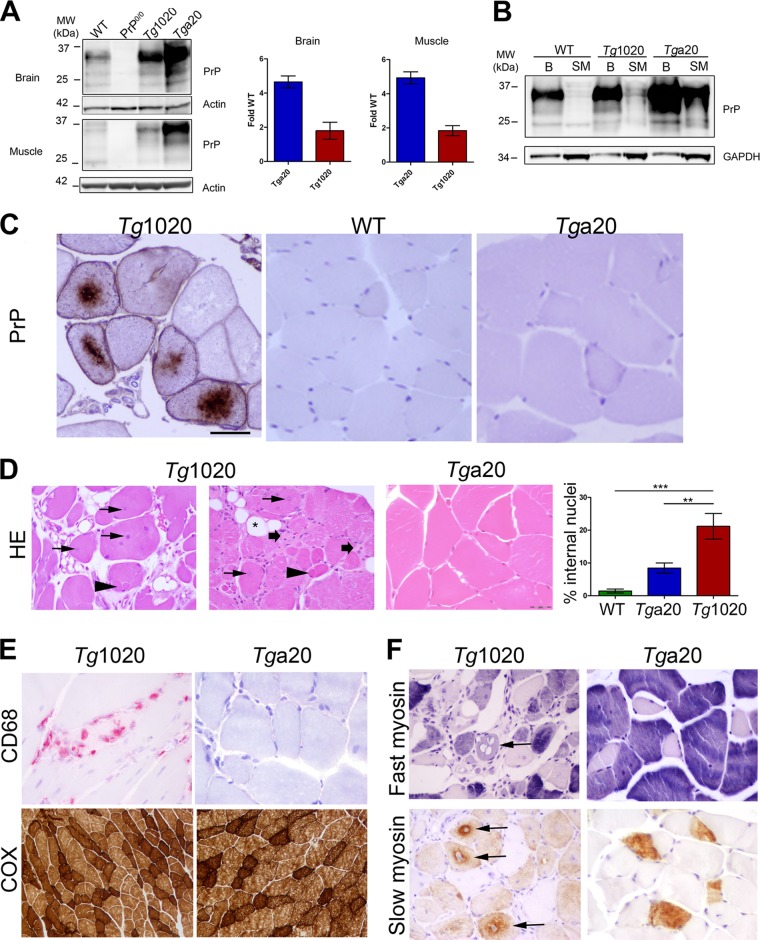
tg1020 mice express RL-PrPC at 2-fold WT levels, whereas tga20 control mice express mouse PrPC at 4- to 5-fold WT levels (Fig. 1A). The skeletal muscle expresses approximately 1/10 the PrPC level of the brain in WT, tg1020, and tga20 mice (Fig. 1B). Intracellular PrP aggregates developed in skeletal muscle from aged tg1020 mice (more than 300 days old), but not in that from aged WT or tga20 mice (Fig. 1C). PrP aggregates surrounded a central nucleus and occupied 10 to 40% of the sarcoplasm. Affected fibers were often clustered within the same fascicle and were interspersed with unaffected myocytes.
FIG 1.
Aged tg1020 mice develop a degenerative myopathy and accumulate central intracellular PrP aggregates in myocytes. (A). Western blot for PrP using equal amounts of protein from each mouse line. (B) Western blot of brain (B) and skeletal muscle (SM) shows higher PrP expression in the brain than in skeletal muscle, even with three times more protein loaded for the muscle samples (note higher GAPDH levels in muscles). POM1 was used for PrP detection. (C) Immunohistochemistry of skeletal muscle revealed coarse, dense perinuclear intracellular PrP inclusions in scattered myocytes of tg1020 mice but not in WT or tga20 mice. (D) Representative images of muscle show central nuclei (arrows), split fibers (arrowheads), endomysial fibrosis (broad arrows), and adipocyte replacement of myocytes (star) in tg1020 mice. HE, hematoxylin and eosin. Asterisks in the bar graph on the right indicate significant differences in the percentages of internal nuclei among the mice by Student's t test (**, P < 0.01; ***, P < 0.001). (E) Focal regions of macrophage infiltration (CD68) were occasionally observed in tg1020 muscle but not in tga20 muscle. A cytochrome oxidase stain (COX) reveals no clear difference in mitochondrial number or pattern between tg1020 and tga20 muscle. (F) Slow and fast myosin heavy chain immunostains show that both type I and type II fibers are degenerating (arrows) in tg1020 skeletal muscle.
The muscle of tg1020 mice showed widespread pathology, including abundant internal nuclei (approximately 23% of myocytes), split fibers, fibrosis, and myocyte replacement by adipocytes, consistent with a degenerative myopathy (Fig. 1D). Small numbers of macrophages infiltrated the interstitial space between myocytes (Fig. 1E). The mitochondria of tg1020 and tga20 myocytes were stained using cytochrome oxidase and did not differ significantly (Fig. 1E). Quantification of type I and II fibers by slow and fast myosin heavy chain antibody staining demonstrated that these fiber types were equally affected (Fig. 1F). Type II fibers predominated in all mice, as expected for perivertebral muscles (36).
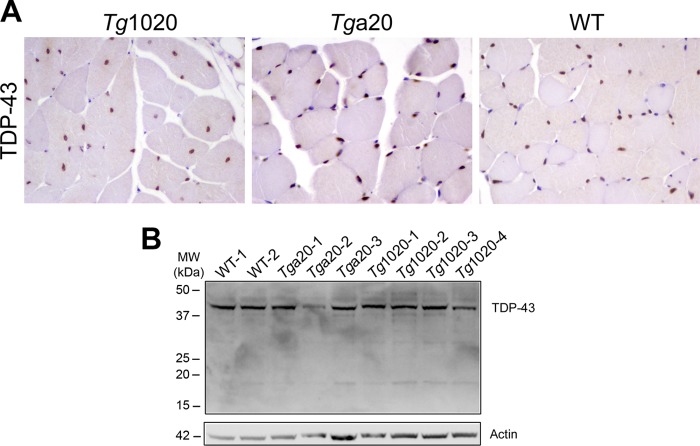
TDP-43 was not mislocalized in affected myocytes.
Mislocalization and modification of the nuclear protein TDP-43 have been reported in several neurodegenerative and neuromuscular disorders (37–39); however, TDP-43 staining was present exclusively within nuclei, and no mislocalization was evident in tg1020 myocytes (Fig. 2A). Additionally, no prominent cleaved fragments of TDP-43 were detected by Western blot analysis (Fig. 2B). Taken together, these results suggest that TDP-43 is traffics normally in RL-PrP-expressing myocytes, in agreement with a prior study that found no TDP-43 mislocalization in brain samples from deceased patients with prion disease (40).
FIG 2.
TDP-43 expression and localization in WT, tg1020, and tga20 skeletal muscle. (A) Immunostains for TDP-43 in skeletal muscle tissue show that TDP-43 is restricted to nuclei. (B) Western blotting of skeletal muscle using an anti-TDP-43 antibody shows no marked differences in TDP-43 levels among the mice.
Biochemical properties of PrPSc in muscle.
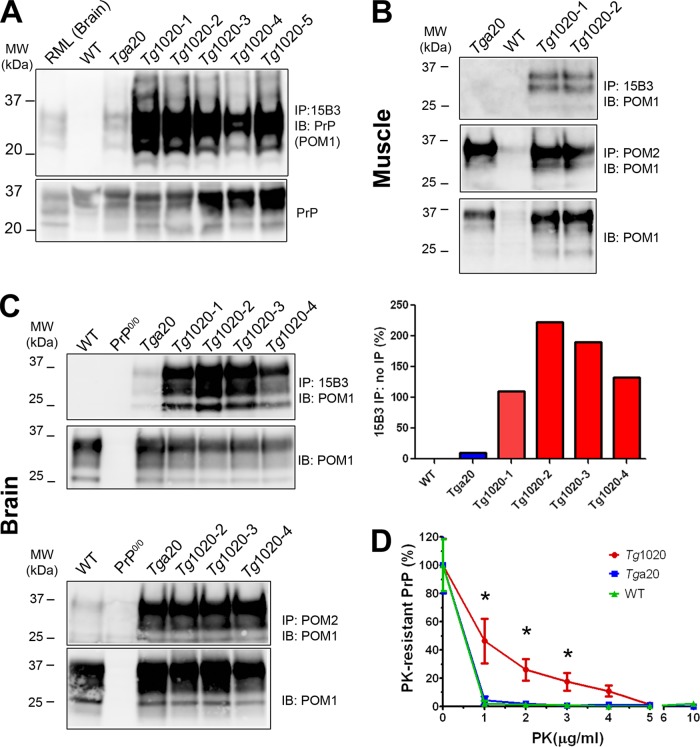
To measure the PrP aggregate load in skeletal muscle, we concentrated the insoluble PrP by ultracentrifugation and then immunoprecipitated native PrP aggregates using the PrP conformation-specific antibody, 15B3 (41). Abundant PrP aggregates were detected in the skeletal muscle of tg1020 mice, while none were found in WT mice (Fig. 3A). Low levels of PrP aggregates were detected in the skeletal muscle of tga20 mice (Fig. 3A), yet quantification revealed that the aggregate levels in the tg1020 mice were 2- to 4-fold higher.
FIG 3.
PrP aggregates accumulate in tg1020 muscle. (A) (Top) Immunoprecipitation using the PrP conformation-specific antibody 15B3 reveals abundant PrP aggregates in tg1020 skeletal muscle. (Bottom) The protein loaded for the IP contained similar amounts of PrP. Note that 4-fold-larger amounts of the WT sample were loaded. IB, immunoblot; POM1, anti-PrP antibody. (B and C) PrP in muscle and brain, respectively. Antibodies 15B3 and POM2 were used for nondenaturing and denaturing IP, respectively, of the insoluble PrP fraction obtained after ultracentrifugation. POM1 was used for detection by Western blotting. Lower panels show the PrP levels for each sample used for IP. (D) PrP in tg1020 muscle is more PK resistant than PrP in the muscle of tga20 or WT mice as analyzed by PrP ELISA (graphed values are normalized). Asterisks indicate significant differences (*, P ≤ 0.05) by Student's t test.
We next performed denaturing immunoprecipitation (IP) of PrP from the insoluble fractions of brain and muscle samples by using an antibody against a linear epitope (PrP antibody POM2, recognizing the octapeptide repeats in PrP residues 57 to 88 [35]). Similar levels of insoluble PrP were bound by POM2 in the tga20 and tg1020 homogenates (Fig. 3B and C, lower panel). In comparison, native IP using PrP conformation-specific antibody 15B3 recovered no PrP or only low levels of PrP in tga20 muscle and brain, respectively, whereas PrP levels in the brains of tg1020 mice were 10- to 20-fold higher, indicating that tg1020 mice harbored abundant PrP aggregates in a PrPSc-like conformation (Fig. 3B and C, upper panel).
Aggregated PrP may be sensitive or resistant to proteinase K. We next compared the proteinase K (PK) sensitivities of PrP from WT, tga20, and tg1020 muscles. Samples were incubated with increasing concentrations of PK, and PK-resistant PrP was measured by ELISA. PrP in the muscle of tg1020 mice was significantly more PK resistant at low PK concentrations (1 to 3 μg/ml) than PrP in the muscle of WT or tga20 mice (Fig. 3D).
ER stress response in tg1020 mice.
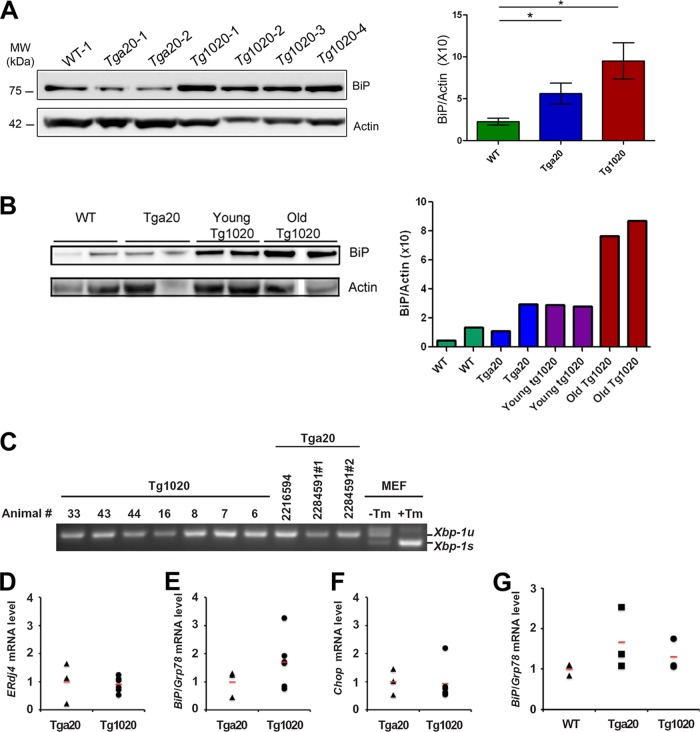
Since the expression of RL-PrP may alter normal protein homeostasis, we next determined whether the UPR was induced in the muscle of tg1020 mice. We measured levels of the ER chaperone and UPR marker Grp78/BiP (42) in the biceps femoris of aged mice. Grp78/BiP levels were significantly higher in tg1020 mice than in WT mice (BiP/actin ratio, 9.5 ± 2 for tg1020 mice and 2.2 ± 0.4 for WT mice [n, 10 to 16; P, 0.02 by an unpaired, two-tailed Student t test]) (Fig. 4A), indicating ER stress in the tg1020 mice. The tga20 mice showed a slight increase in Grp78/BiP levels (BiP/actin ratio, 5.6 ± 1), which were significantly different from the levels in WT mice (P, 0.03 by Student's t test), suggesting that PrP overexpression causes some ER stress. Young tg1020 mice (45 days old) showed slightly higher BiP levels than WT mice (Fig. 4B).
FIG 4.
Induction of the unfolded protein response and an analysis of select ER stress markers in tg1020 skeletal muscle. (A) Western blot analysis using an antibody for UPR marker Grp78/BiP shows an approximately 4-fold-higher level of Grp78/BiP in tg1020 muscle than in WT muscle. Asterisks indicate significant differences (*, P < 0.05) by Student's t test. (B) Young tg1020 mice had slightly higher levels of BiP than WT mice. Note: a tg1020 sample was removed from the blot due to very low actin levels. (C) Semiquantitative real-time PCR reveals no Xbp-1 mRNA splicing in tg1020 skeletal muscle compared to tga20 controls. Xbp-1u and Xbp-1s indicate the unspliced and spliced forms of Xbp-1 mRNA, respectively. Mouse embryonic fibroblasts (MEF) treated with tunicamycin (Tm) showed strong Xbp-1 mRNA splicing and served as a positive control. (D) Quantitative PCR analysis of mRNA levels of ERdj4, a downstream transcriptional target of XBP-1, showed no difference between tga20 and tg1020 animals. (E and F) Quantitative PCR analysis of BiP/Grp78 and Chop mRNA levels, respectively, revealed higher levels of BiP/Grp78 in the muscles of some tg1020 mice than in tga20 mice; however, the differences were not statistically significant. (G) Quantitative PCR analysis revealed no significant difference in BiP/Grp78 mRNA levels between the muscles of young tg1020 mice and those of young WT or tga20 mice. In panels D through F, 3 tga20 and 6 tg1020 animals were analyzed for quantitative PCR measurements, and the values for each animal were plotted individually.
To assess additional UPR-related events, we searched for Xbp-1 splicing, which is initiated by IRE1, and measured mRNA levels of ERdj4, an IRE1 downstream target, BiP/Grp78, an ATF6 downstream target, and Chop, a PERK downstream target, in the muscles of tga20 and tg1020 mice. We detected no evidence of Xbp-1 splice events and no increase in the mRNA levels of ERdj4 (Fig. 4C and D). Since there was variability in the levels of BiP/Grp7 and Chop mRNAs among the tg1020 mice, the differences between the levels in tg1020 and tga20 mice were not statistically significant. However, 3 of 6 tg1020 mice showed elevated levels of BiP/Grp78 mRNA, and 2 of 6 mice showed Chop mRNA levels more than 2-fold-higher than those of tga20 mice (Fig. 4E and F). The induction of BiP/Grp78 mRNA synthesis in some mice was consistent with the increase in BiP/Grp78 protein levels that was seen (Fig. 4A and E). BiP mRNA levels were not significantly elevated in young tg1020 mice (Fig. 4G). Together, these results are consistent with ER stress in some tg1020 mice.
Ubiquitinated inclusions in tg1020 muscle.
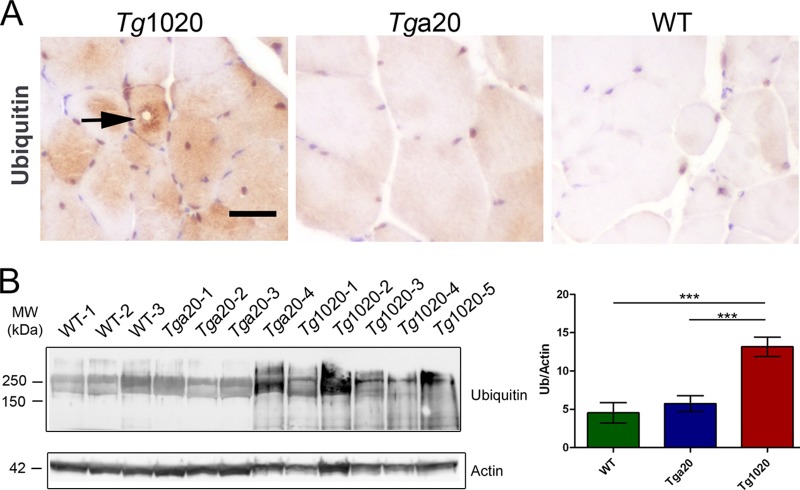
Ubiquitinated inclusions occur in several protein aggregation diseases, including myopathies (43–48). Ubiquitin-positive inclusions were found histologically in the skeletal muscles of tg1020 mice, but not in those of age-matched WT or tga20 mice (Fig. 5A). Higher levels of ubiquitinated proteins were also detected biochemically in tg1020 muscles than in tga20 or WT muscles (Fig. 5B) (ubiquitin/actin ratios, 13.1 ± 1 for tg1020 mice, 4.5 ± 1 for WT mice, and 5.7 ± 1 for tga20 mice; P, <0.001 for tg1020 versus WT or tga20 mice [n, 9 to 11]).
FIG 5.
Accumulation of ubiquitinated proteins in skeletal muscle of tg1020 mice (A) Immunohistochemistry shows ubiquitinated inclusions (arrow) in tg1020 muscle. (B) Assessment of total ubiquitinated protein by Western blotting shows an approximately 3-fold-higher level of ubiquitinated proteins in tg1020 muscle than in WT or Tga20 muscle. Asterisks indicate significant differences (***, P < 0.001) by Student's t test.
The ubiquitinated proteins in the tg1020 mice may consist of PrP or other proteins that were ubiquitinated due to disrupted protein homeostasis. To test for ubiquitination of PrP, we immunoprecipitated PrP aggregates using the conformation-specific antibody 15B3 under native conditions and antibody POM2 under denaturing conditions, followed by Western blot detection using the anti-ubiquitin antibody. We did not detect any ubiquitinated PrP (data not shown); however, ubiquitinated proteins can be difficult to detect. The lack of PrP ubiquitination in muscle suggests that PrP aggregates may be targeted to a nonproteasomal pathway for degradation. Alternatively, if the misfolded PrP is indeed ubiquitinated, then the turnover may be rapid and may preclude detection without the use of proteasome inhibitors in vivo. However, the higher levels of ubiquitinated proteins may result from a disruption of protein folding homeostasis by mutant PrP, leading to increased misfolding events and a buildup of ubiquitinated proteins, as was recently shown to occur in cells expressing a mutant huntingtin fragment (49).
Induction of autophagy in the tg1020 muscle.
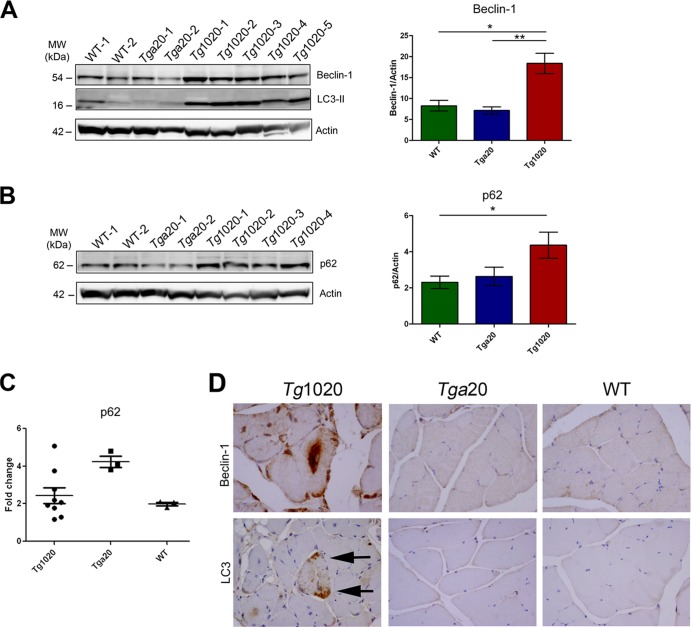
Beclin-1 regulates autophagy through the formation of an autophagy initiator complex that includes Vps34 and Vps15 (50). This complex contributes to the recruitment of membranes to form autophagosomes. Beclin-1-independent pathways of autophagy may also exist (51). To determine whether beclin-1 levels were altered in tg1020 mice, we measured beclin-1 levels in muscle homogenates by immunoblotting. Beclin-1 levels were elevated in tg1020 skeletal muscle, suggesting that autophagy had been induced (beclin-1/actin ratios, 18.3 ± 2 for tg1020 mice, 8.2 ± 1 for WT mice, and 7.1 ± 1 for tga20 mice [n, 8 to 17; P, 0.01 for WT versus tg1020 mice and 0.002 for tga20 versus tg1020 mice]) (Fig. 6A).
FIG 6.
Increased expression of autophagy-related proteins in tg1020 muscle. (A) Beclin-1 and LC3-II levels were higher in tg1020 muscle than in WT or tga20 muscle. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by Student's t test. (B) Muscle homogenates also showed increased levels of p62 in tg1020 muscle. (C) Quantitative real-time PCR analysis of mRNA in muscles revealed that p62 mRNA levels in tg1020 mice do not differ from those in WT or tga20 mice. (D) Beclin-1 immunostains revealed occasional large central aggregates in tg1020 myocytes, whereas LC3 showed punctate peripheral staining (arrows).
p62 levels were next assessed and were found to be elevated in tg1020 mice, which could indicate a block in the selective degradation of ubiquitinated aggregates (52) (p62/actin ratios, 4.3 ± 0.7 for tg1020 mice, 2.3 ± 0.3 for WT mice, and 2.6 ± 0.5 for tga20 mice [n, 10 to 15; P, 0.04 for WT versus tg1020 mice]) (Fig. 6B). To determine whether p62 was upregulated, we performed quantitative PCR. Here we found no differences in the p62 mRNA levels between tg1020 mice and WT or tga20 mice (Fig. 6C), although levels in some tg1020 mice were 2-fold higher than those in WT mice.
As a measure of the autophagosome number (53), LC3-II protein was examined in the muscle of tg1020 mice. LC3-II is the lipidated version of LC3-I and is involved in autophagosome formation (54). Seven of 11 (64%) tg1020 muscle samples showed conversion of LC3-I to LC3-II (Fig. 6A and data not shown), indicating either enhanced autophagosome synthesis or a block in LC3-II degradation (53).
Consistent with the biochemical findings, immunolabeling of skeletal muscle revealed dense foci of beclin-1 and LC3 in tg1020 mice (Fig. 6D). Taken together, these data suggest that autophagy is induced in response to PrP aggregates in the skeletal muscle.
PrP aggregates accumulate upon inhibition of autophagy and proteasomal degradation pathways in vitro.
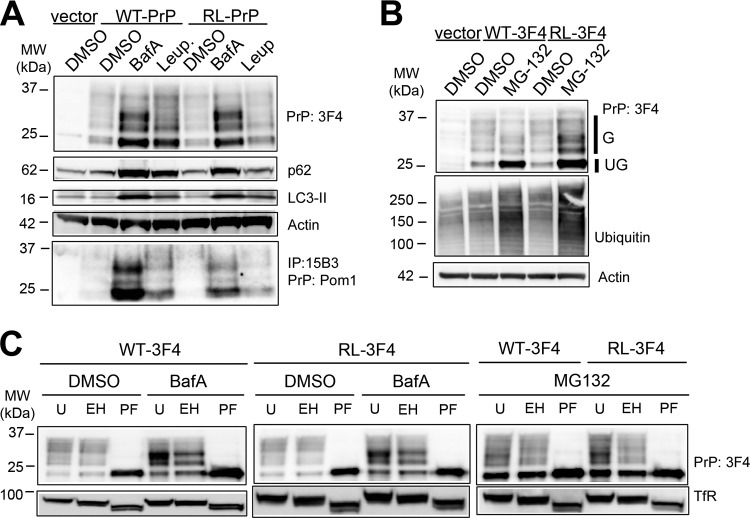
To investigate the clearance pathways of PrP in vitro, we expressed WT-PrP and RL-PrP with the 3F4 epitope tag, which is recognized by the 3F4 antibody (3F4 residues 109 to 112 [MKHM]) in mouse myoblasts (C2C12 cells). We tested the effects of two inhibitors of autophagy on the clearance of PrP: bafilomycin A (BafA), which blocks phagosome-lysosome fusion, and leupeptin, which inhibits lysosomal proteases and prevents the degradation of cargo. WT-PrP- or RL-PrP-expressing cells incubated with BafA showed 3-fold-higher levels of PrP than cells incubated with a vehicle (DMSO), whereas cells treated with leupeptin showed PrP levels 1.4- to 3-fold higher than those of vehicle-treated cells, as measured using a Fujifilm LAS 4000 imager and MultiGauge software (Fig. 7A). p62 and LC3-II levels were increased in BafA- or leupeptin-treated cells, indicating that the treatments were efficacious in inhibiting autophagy. PrP was driven by the cytomegalovirus promoter for constitutive protein expression, suggesting that the increases in PrP levels had developed due to a decrease in clearance.
FIG 7.
Inhibition of autophagy and proteasomal pathways leads to the accumulation of distinct PrP isoforms in C2C12 cells. (A) Lysates from C2C12 cells transfected either with an empty vector or with a vector encoding WT-PrP or RL-PrP show elevated levels of both unglycosylated and glycosylated PrP upon BafA or leupeptin treatment. Increased p62 and LC3-II levels show the efficacy of the BafA and leupeptin treatments in modulating autophagy. Immunoprecipitation with the PrP conformational antibody 15B3 shows that PrP aggregates accumulate in cells expressing WT-PrP and RL-PrP only upon autophagic inhibition using BafA or leupeptin. (B) Lysates from C2C12 cells show elevated levels of unglycosylated WT-PrP and RL-PrP upon MG132 treatment. Glycosylated and unglycosylated PrP signals are labeled G and UG, respectively. An increase in the level of total ubiquitinated proteins demonstrates the efficacy of MG132 treatment. Immunoblotting for actin shows that equal amounts of protein were loaded. (C) C2C12 cells were transfected with WT-PrP or RL-PrP and were treated with DMSO, BafA, or MG132. Lysates either were left untreated (U) or were treated with endo H (EH) or PNGase F (PF). Glycosylated WT-PrP and RL-PrP that accumulated upon BafA treatment were endo H resistant, yet upon MG132 treatment, they were endo H sensitive. Endo H and PNGase activities were demonstrated by shifts in the molecular weight of the transferrin receptor (TfR) corresponding to the losses of 1 and 2 glycans, respectively, compared to TfR in the untreated samples.
To test for the formation of PrP aggregates in myoblasts, we performed IP analysis using the PrP conformational antibody 15B3. Interestingly, 15B3-reactive WT-PrP and RL-PrP accumulated in BafA- and leupeptin-treated cells, but not in DMSO-treated cells (Fig. 7A, bottom). PrP overexpression may be a contributing factor underlying aggregate development in cells expressing WT-PrP, since PrP overexpression has been shown to lead to muscle and nerve degeneration (17).
We next tested the effect of the proteasomal inhibitor MG132 on WT-PrP and RL-PrP accumulation. Cells expressing WT-PrP or RL-PrP that were treated with DMSO or MG132 for 6 h showed elevated levels of ubiquitinated proteins (Fig. 7B). A previous report showed that treatment of CHO cells with proteasome inhibitors led to ubiquitination of PrP and PrP accumulation (55). Here, for both WT-PrP and RL-PrP, unglycosylated PrP levels increased by approximately 3-fold, whereas glycosylated PrP levels increased by 2-fold, in MG132-treated cells in 3 independent experiments (Fig. 7B). Interestingly, PrP that accumulated upon proteasomal inhibition was not detected following IP with the PrP conformational antibody 15B3 in 4 independent experiments (data not shown), suggesting that PrP was monomeric or was aggregated in a conformation not recognized by the antibody.
Inhibiting the proteasome with MG132 led to relatively higher increases in levels of unglycosylated PrP than glycosylated PrP. To determine the biochemical differences between glycosylated PrP that accumulated upon autophagy and those that accumulated upon proteasome inhibition, we performed endo H and PNGase F digestion of WT-PrP and RL-PrP samples obtained after DMSO, BafA, or MG132 treatment. DMSO-treated WT-PrP and RL-PrP were primarily endo H resistant (83%) and PNGase F sensitive (Fig. 7C). After BafA treatment to inhibit autophagy, WT-PrP and RL-PrP were only slightly less endo H resistant (70 to 73%), indicating that the glycosylated species were indeed present within the Golgi apparatus or in a post-Golgi compartment. In contrast, after MG132 treatment to inhibit the proteasome, glycosylated WT-PrP and RL-PrP differed in that RL-PrP was more endo H sensitive than WT-PrP (73% of WT-PrP and 52% of RL-PrP was resistant), indicating that the accumulated RL-PrP was likely in part ER derived (Fig. 7C).
DISCUSSION
Here we report that expression of mutant PrP in vivo recapitulates prion aggregation and features of degenerative myopathy in a patient with spontaneous prion disease (13). Notably, the myopathy in mice is characterized by internal nuclei, split fibers, central rimmed vacuoles, cell death and replacement by adipocytes, and the accumulation of intracellular aggregates. These lesions are similar to many other protein aggregate myopathies in patients and in mouse models, including gelsolin, amyloid β, and phosphorylated tau aggregate myopathies (46, 57–59). PrP inclusions were in type I and type II fibers, suggesting that aggregation was influenced neither by the oxidative capacity nor by the glycolytic ability of the cell. In contrast to sIBM, the prion protein myopathy was not an inflammatory disease, since the inflammatory cell influx was minimal and appeared to be limited to macrophages. Additionally, these studies offer evidence of autophagy as a prion clearance mechanism, an area which has been poorly understood in prion-infected individuals, particularly in non-central-nervous-system tissues, such as muscle.
Skeletal muscles from tg1020 mice exhibited evidence of ER stress, reflected by an increase in the levels of ER chaperones. ER stress may be directly or indirectly activated by RL-PrP and suggests that either (i) misfolded RL-PrP directly induces ER stress in the sarcoplasmic reticulum (SR) or (ii) RL-PrP disrupts general protein folding in the SR, leading to ER stress. Chronic stress from mutant PrP may overwhelm the capacity of the SR to properly fold proteins, leading to the initiation of proapoptotic signaling pathways and cell death (37), as noted by an increase in Chop levels. Induction of apoptosis through Chop would be expected to develop asynchronously in myocytes. Here we found that not all muscle samples showed detectable increases in Chop levels, potentially due to the heterogenous distribution of the affected fibers. Further studies would be needed to clarify the pathways of myocyte death in our model. Nevertheless, our findings were consistent with reports showing ER stress in prion-expressing cell models, brains of cattle with bovine spongiform encephalopathy, and brains of mice with experimental prion infection (30, 60–65). ER stress may indirectly enhance the formation of PrP aggregates, since induction of the UPR in persistently infected scrapie cells and in cell lines expressing mutant PrP showed increased levels of PrPSc (32, 66).
To maintain proteostasis, myocytes appear to respond to the misfolded PrP by activating autophagy, suggested by the elevated levels of beclin-1 and LC3-II in the skeletal muscle in vivo and in C2C12 cells. Induction of autophagy has been described in other protein-misfolding myopathies. Muscle biopsy specimens from sIBM patients show an upregulation of p62 (67). Additionally, in patients with IBMPFD, mutations in VCP/p97 protein disrupted autophagy due to a defect in autophagosome maturation and led to central inclusions in myocytes similar to those seen in tg1020 mice (28, 68). Furthermore, mutations in α-B crystallin led to protein aggregation and cardiomyopathy that were markedly exacerbated by inhibiting autophagy through heterozygous inactivation of beclin-1 (69), underscoring the importance of autophagy in aggregate clearance. Autophagy has also been shown previously to be activated in the brains of prion-infected animals (70) and in prion-infected cultured cells (71), and pharmacologic induction of autophagy has been shown to decrease PrPSc levels in vitro and in vivo (71–73). It will be of great interest to extend these studies with measurements of autophagic flux in prion models.
Although we could not detect evidence for proteasome degradation of mutant PrP in the muscles of mice, there were increased levels of unglycosylated and glycosylated PrP in cultured cells upon proteasomal inhibition. Glycosylated RL-PrP was particularly sensitive to endo H treatment, suggesting that a small fraction of RL-PrP is retained in the ER and is degraded in a proteasome-dependent manner. We failed to immunoprecipitate this species with a PrP aggregate-specific antibody, suggesting that it was monomeric or in a conformation different from the aggregated form that accumulates upon inhibition of autophagy.
Taken together, these findings highlight a role for ER stress and autophagy in the clearance of PrP from skeletal muscle in prion-infected individuals. Our data suggest that mutant PrP induces ER stress and that a small fraction of PrP is exported from the ER for degradation by the proteasome. Misfolded PrP also enters the secretory pathway, and post-Golgi PrP becomes sequestered in autophagosomes and ultimately degraded by autophagy. Additional studies may elucidate differences in aggregate clearance mechanisms in familial and transmissible prion diseases.
ACKNOWLEDGMENTS
We thank Diane Shelton for helpful discussions, Keno Wu and Mona Farahi for experimental help, and the animal caretakers for excellent animal care. We thank Adriano Aguzzi for generously providing anti-PrP antibodies (POM series).
These studies were supported by the National Institutes of Health (NS076896, NS069566, and U54AI0359 [to C.J.S.]; EY020846 [to J.H.L.]) and by a Harmonia grant (507/1-034-04/507-10-049 [to P.L.]).
Footnotes
Published ahead of print 4 December 2013
REFERENCES
- 1.Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136–144. 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. 1984. Prions: novel infectious pathogens. Adv. Virus Res. 29:1–56. 10.1016/S0065-3527(08)60404-2 [DOI] [PubMed] [Google Scholar]
- 3.Wadsworth JD, Collinge J. 2011. Molecular pathology of human prion disease. Acta Neuropathol. 121:69–77. 10.1007/s00401-010-0735-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguzzi A, Falsig J. 2012. Prion propagation, toxicity and degradation. Nat. Neurosci. 15:936–939. 10.1038/nn.3120 [DOI] [PubMed] [Google Scholar]
- 5.Deleault NR, Harris BT, Rees JR, Supattapone S. 2007. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U. S. A. 104:9741–9746. 10.1073/pnas.0702662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Wang X, Yuan CG, Ma J. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132–1135. 10.1126/science.1183748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. 2010. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119:177–187. 10.1007/s00401-009-0633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbe MM, Soldati GG, Zlinszky KK, Wunderlin SS, Ehrensperger FF. 2009. Immunohistochemical study of PrPSc distribution in neural and extraneural tissues of two cats with feline spongiform encephalopathy. BMC Vet. Res. 5:11. 10.1186/1746-6148-5-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sigurdson CJ, Spraker TR, Miller MW, Oesch B, Hoover EA. 2001. PrPCWD in the myenteric plexus, vagosympathetic trunk and endocrine glands of deer with chronic wasting disease. J. Gen. Virol. 82:2327–2334 http://vir.sgmjournals.org/content/82/10/2327.long [DOI] [PubMed] [Google Scholar]
- 10.Ironside JW. 2010. Variant Creutzfeldt-Jakob disease. Haemophilia 16(Suppl. 5):175–180. 10.1111/j.1365-2516.2010.02317.x [DOI] [PubMed] [Google Scholar]
- 11.Herzog C, Riviere J, Lescoutra-Etchegaray N, Charbonnier A, Leblanc V, Sales N, Deslys JP, Lasmezas CI. 2005. PrPTSE distribution in a primate model of variant, sporadic, and iatrogenic Creutzfeldt-Jakob disease. J. Virol. 79:14339–14345. 10.1128/JVI.79.22.14339-14345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glatzel M, Abela E, Maissen M, Aguzzi A. 2003. Extraneural pathologic prion protein in sporadic Creutzfeldt-Jakob disease. N. Engl. J. Med. 349:1812–1820. 10.1056/NEJMoa030351 [DOI] [PubMed] [Google Scholar]
- 13.Kovacs GG, Lindeck-Pozza E, Chimelli L, Araujo AQ, Gabbai AA, Strobel T, Glatzel M, Aguzzi A, Budka H. 2004. Creutzfeldt-Jakob disease and inclusion body myositis: abundant disease-associated prion protein in muscle. Ann. Neurol. 55:121–125. 10.1002/ana.10813 [DOI] [PubMed] [Google Scholar]
- 14.Thomzig A, Kratzel C, Lenz G, Kruger D, Beekes M. 2003. Widespread PrPSc accumulation in muscles of hamsters orally infected with scrapie. EMBO Rep. 4:1–4. 10.1038/sj.embor.embor717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreoletti O, Simon S, Lacroux C, Morel N, Tabouret G, Chabert A, Lugan S, Corbiere F, Ferre P, Foucras G, Laude H, Eychenne F, Grassi J, Schelcher F. 2004. PrPSc accumulation in myocytes from sheep incubating natural scrapie. Nat. Med. 10:591–593. 10.1038/nm1055 [DOI] [PubMed] [Google Scholar]
- 16.Angers RC, Browning SR, Seward TS, Sigurdson CJ, Miller MW, Hoover EA, Telling GC. 2006. Prions in skeletal muscles of deer with chronic wasting disease. Science 311:1117. 10.1126/science.1122864 [DOI] [PubMed] [Google Scholar]
- 17.Westaway D, DeArmond SJ, Cayetano-Canlas J, Groth D, Foster D, Yang SL, Torchia M, Carlson GA, Prusiner SB. 1994. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell 76:117–129. 10.1016/0092-8674(94)90177-5 [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Liang J, Zheng M, Li X, Wang M, Wang P, Vanegas D, Wu D, Chakraborty B, Hays AP, Chen K, Chen SG, Booth S, Cohen M, Gambetti P, Kong Q. 2007. Inducible overexpression of wild-type prion protein in the muscles leads to a primary myopathy in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 104:6800–6805. 10.1073/pnas.0608885104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosque PJ, Ryou C, Telling G, Peretz D, Legname G, DeArmond SJ, Prusiner SB. 2002. Prions in skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 99:3812–3817. 10.1073/pnas.052707499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. 1996. NMR structure of the mouse prion protein domain PrP (121–231). Nature 382:180–182. 10.1038/382180a0 [DOI] [PubMed] [Google Scholar]
- 21.Sigurdson CJ, Nilsson KP, Hornemann S, Heikenwalder M, Manco G, Schwarz P, Ott D, Rulicke T, Liberski PP, Julius C, Falsig J, Stitz L, Wüthrich K, Aguzzi A. 2009. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc. Natl. Acad. Sci. U. S. A. 106:304–309. 10.1073/pnas.0810680105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 23.Ciechanover A. 2012. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Biochim. Biophys. Acta 1824:3–13. 10.1016/j.bbapap.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 24.Wong E, Cuervo AM. 2010. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 13:805–811. 10.1038/nn.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Askanas V, Engel WK, Nogalska A. 2009. Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain Pathol. 19:493–506. 10.1111/j.1750-3639.2009.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Askanas V, Engel WK. 2008. Inclusion-body myositis: muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer's and Parkinson's disease brains. Acta Neuropathol. 116:583–595. 10.1007/s00401-008-0449-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6:217–227. 10.4161/auto.6.2.11014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. 2009. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 187:875–888. 10.1083/jcb.200908115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- 30.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, Barrett DA, Tsaytler P, Bertolotti A, Willis AE, Bushell M, Mallucci GR. 2012. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 485:507–511. 10.1038/nature11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deriziotis P, Andre R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, Nathan JA, Rosenzweig R, Krutauz D, Glickman MH, Collinge J, Goldberg AL, Tabrizi SJ. 2011. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 30:3065–3077. 10.1038/emboj.2011.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nunziante M, Ackermann K, Dietrich K, Wolf H, Gadtke L, Gilch S, Vorberg I, Groschup M, Schatzl HM. 2011. Proteasomal dysfunction and endoplasmic reticulum stress enhance trafficking of prion protein aggregates through the secretory pathway and increase accumulation of pathologic prion protein. J. Biol. Chem. 286:33942–33953. 10.1074/jbc.M111.272617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. 1996. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15:1255–1264 [PMC free article] [PubMed] [Google Scholar]
- 34.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 35.Polymenidou M, Moos R, Scott M, Sigurdson C, Shi YZ, Yajima B, Hafner-Bratkovic I, Jerala R, Hornemann S, Wüthrich K, Bellon A, Vey M, Garen G, James MN, Kav N, Aguzzi A. 2008. The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One 3:e3872. 10.1371/journal.pone.0003872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hesse B, Fischer MS, Schilling N. 2010. Distribution pattern of muscle fiber types in the perivertebral musculature of two different sized species of mice. Anat. Rec. (Hoboken) 293:446–463. 10.1002/ar.21090 [DOI] [PubMed] [Google Scholar]
- 37.Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, Hanson PI, Kimonis V, Pestronk A. 2008. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry 79:1186–1189. 10.1136/jnnp.2007.131334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 39.Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA. 2009. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40:19–31. 10.1002/mus.21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isaacs AM, Powell C, Webb TE, Linehan JM, Collinge J, Brandner S. 2008. Lack of TAR-DNA binding protein-43 (TDP-43) pathology in human prion diseases. Neuropathol Appl. Neurobiol 34:446–456. 10.1111/j.1365-2990.2008.00963.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, Ehrensperger F, Hornemann S, Glockshuber R, Riek R, Billeter M, Wuthrich K, Oesch B. 1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 390:74–77. 10.1038/36337 [DOI] [PubMed] [Google Scholar]
- 42.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462–464. 10.1038/332462a0 [DOI] [PubMed] [Google Scholar]
- 43.Weihl CC, Miller SE, Hanson PI, Pestronk A. 2007. Transgenic expression of inclusion body myopathy associated mutant p97/VCP causes weakness and ubiquitinated protein inclusions in mice. Hum. Mol. Genet. 16:919–928. 10.1093/hmg/ddm037 [DOI] [PubMed] [Google Scholar]
- 44.Lin JH, Walter P, Yen TS. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 3:399–425. 10.1146/annurev.pathmechdis.3.121806.151434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosen KM, Veereshwarayya V, Moussa CE, Fu Q, Goldberg MS, Schlossmacher MG, Shen J, Querfurth HW. 2006. Parkin protects against mitochondrial toxins and beta-amyloid accumulation in skeletal muscle cells. J. Biol. Chem. 281:12809–12816. 10.1074/jbc.M512649200 [DOI] [PubMed] [Google Scholar]
- 46.Page LJ, Suk JY, Bazhenova L, Fleming SM, Wood M, Jiang Y, Guo LT, Mizisin AP, Kisilevsky R, Shelton GD, Balch WE, Kelly JW. 2009. Secretion of amyloidogenic gelsolin progressively compromises protein homeostasis leading to the intracellular aggregation of proteins. Proc. Natl. Acad. Sci. U. S. A. 106:11125–11130. 10.1073/pnas.0811753106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90:537–548. 10.1016/S0092-8674(00)80513-9 [DOI] [PubMed] [Google Scholar]
- 48.Lin WL, Lewis J, Yen SH, Hutton M, Dickson DW. 2003. Filamentous tau in oligodendrocytes and astrocytes of transgenic mice expressing the human tau isoform with the P301L mutation. Am. J. Pathol. 162:213–218. 10.1016/S0002-9440(10)63812-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. 2012. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J. Cell Biol. 196:573–587. 10.1083/jcb.201110093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park C, Cuervo AM. 2013. Selective autophagy: talking with the UPS. Cell Biochem. Biophys. 67:3–13. 10.1007/s12013-013-9623-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang R, Zeh HJ, Lotze MT, Tang D. 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18:571–580. 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barth S, Glick D, Macleod KF. 2010. Autophagy: assays and artifacts. J. Pathol. 221:117–124. 10.1002/path.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. 2009. In search of an “autophagomometer” Autophagy 5:585–589. 10.4161/auto.5.5.8823 [DOI] [PubMed] [Google Scholar]
- 54.Florey O, Overholtzer M. 2012. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 22:374–380. 10.1016/j.tcb.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yedidia Y, Horonchik L, Tzaban S, Yanai A, Taraboulos A. 2001. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 20:5383–5391. 10.1093/emboj/20.19.5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rane NS, Yonkovich JL, Hegde RS. 2004. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 23:4550–4559. 10.1038/sj.emboj.7600462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitazawa M, Green KN, Caccamo A, LaFerla FM. 2006. Genetically augmenting Aβ42 levels in skeletal muscle exacerbates inclusion body myositis-like pathology and motor deficits in transgenic mice. Am. J. Pathol. 168:1986–1997. 10.2353/ajpath.2006.051232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delaunay A, Bromberg KD, Hayashi Y, Mirabella M, Burch D, Kirkwood B, Serra C, Malicdan MC, Mizisin AP, Morosetti R, Broccolini A, Guo LT, Jones SN, Lira SA, Puri PL, Shelton GD, Ronai Z. 2008. The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PLoS One 3:e1609. 10.1371/journal.pone.0001609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nogalska A, D'Agostino C, Engel WK, Askanas V. 2011. Novel demonstration of conformationally modified tau in sporadic inclusion-body myositis muscle fibers. Neurosci. Lett. 503:229–233. 10.1016/j.neulet.2011.08.042 [DOI] [PubMed] [Google Scholar]
- 60.Brown AR, Rebus S, McKimmie CS, Robertson K, Williams A, Fazakerley JK. 2005. Gene expression profiling of the preclinical scrapie-infected hippocampus. Biochem. Biophys. Res. Commun. 334:86–95. 10.1016/j.bbrc.2005.06.060 [DOI] [PubMed] [Google Scholar]
- 61.Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C, Glimcher LH. 2008. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 105:757–762. 10.1073/pnas.0711094105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hetz C, Russelakis-Carneiro M, Maundrell K, Castilla J, Soto C. 2003. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 22:5435–5445. 10.1093/emboj/cdg537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hetz C, Russelakis-Carneiro M, Walchli S, Carboni S, Vial-Knecht E, Maundrell K, Castilla J, Soto C. 2005. The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J. Neurosci. 25:2793–2802. 10.1523/JNEUROSCI.4090-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang Y, Xiang W, Terry L, Kretzschmar HA, Windl O. 2010. Transcriptional analysis implicates endoplasmic reticulum stress in bovine spongiform encephalopathy. PLoS One 5:e14207. 10.1371/journal.pone.0014207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM, Ortori CA, Willis AE, Fischer PM, Barrett DA, Mallucci GR. 2013. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 5:206ra138. 10.1126/scitranslmed.3006767 [DOI] [PubMed] [Google Scholar]
- 66.Hetz C, Castilla J, Soto C. 2007. Perturbation of endoplasmic reticulum homeostasis facilitates prion replication. J. Biol. Chem. 282:12725–12733. 10.1074/jbc.M611909200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nogalska A, Terracciano C, D'Agostino C, King Engel W, Askanas V. 2009. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 118:407–413. 10.1007/s00401-009-0564-6 [DOI] [PubMed] [Google Scholar]
- 68.Ju JS, Weihl CC. 2010. Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy. Hum. Mol. Genet. 19:R38–R45. 10.1093/hmg/ddq157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. 2008. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 105:9745–9750. 10.1073/pnas.0706802105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y, Tian C, Wang SB, Xie WL, Guo Y, Zhang J, Shi Q, Chen C, Dong XP. 2012. Activation of the macroautophagic system in scrapie-infected experimental animals and human genetic prion diseases. Autophagy 8:1604–1620. 10.4161/auto.21482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cortes CJ, Qin K, Cook J, Solanki A, Mastrianni JA. 2012. Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Straussler-Scheinker disease. J. Neurosci. 32:12396–12405. 10.1523/JNEUROSCI.6189-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, Ertmer A. 2009. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 5:361–369. 10.4161/auto.5.3.7662 [DOI] [PubMed] [Google Scholar]
- 73.Heiseke A, Aguib Y, Riemer C, Baier M, Schatzl HM. 2009. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J. Neurochem. 109:25–34. 10.1111/j.1471-4159.2009.05906.x [DOI] [PubMed] [Google Scholar]