ABSTRACT
Bovine ephemeral fever virus (BEFV) is an arthropod-borne rhabdovirus that is classified as the type species of the genus Ephemerovirus. In addition to the five canonical rhabdovirus structural proteins (N, P, M, G, and L), the large and complex BEFV genome contains several open reading frames (ORFs) between the G and L genes (α1, α2/α3, β, and γ) encoding proteins of unknown function. We show that the 10.5-kDa BEFV α1 protein is expressed in infected cells and, consistent with previous predictions based on its structure, has the properties of a viroporin. Expression of a BEFV α1-maltose binding protein (MBP) fusion protein in Escherichia coli was observed to inhibit cell growth and increase membrane permeability to hygromycin B. Increased membrane permeability was also observed in BEFV-infected mammalian cells (but not cells infected with an α1-deficient BEFV strain) and in cells expressing a BEFV α1-green fluorescent protein (GFP) fusion protein, which was shown by confocal microscopy to localize to the Golgi complex. Furthermore, the predicted C-terminal cytoplasmic domain of α1, which contains a strong nuclear localization signal (NLS), was translocated to the nucleus when expressed independently, and in an affinity chromatography assay employing a GFP trap, the full-length α1 was observed to interact specifically with importin β1 and importin 7 but not with importin α3. These data suggest that, in addition to its function as a viroporin, BEFV α1 may modulate components of nuclear trafficking pathways, but the specific role thereof remains unclear.
IMPORTANCE
INTRODUCTION
Bovine ephemeral fever virus (BEFV) is an arthropod-borne rhabdovirus that is classified as the type species of the genus Ephemerovirus (1). BEFV causes an acute febrile disease of cattle and water buffalo in a zone extending from Africa to the Middle East, Asia, and Australia. The 14,900-nucleotide negative-sense single-stranded RNA (ssRNA) BEFV genome contains 10 long open reading frames (ORFs), arranged in the order 3′-N-P-M-GNS-α1-α2-β-γ-L-5′, encoding the five common rhabdovirus structural proteins (N, P, M, G, and L) and several additional proteins of unknown function. The BEFV GNS gene encodes a large nonstructural glycoprotein of unknown function that is expressed on the cell surface but does not occur in virions and appears to have arisen by duplication of the G gene (2, 3). The other small BEFV accessory genes (α1, α2, β, and γ) also encode proteins of unknown function that have not previously been detected in infected cells or in virions. BEFV α1 is predicted to be a 10.5-kDa protein with structural characteristics that suggest that it may function as a viroporin with an N-terminal domain containing clusters of aromatic residues, a putative hydrophobic transmembrane domain, and a highly basic C-terminal domain (4, 5). Similar uncharacterized viroporin-like proteins are encoded in the genomes of all other ephemeroviruses (species Kimberley virus, Adelaide River virus, Obodhiang virus, and Kotonkan virus), as well as several other arthropod-borne rhabdoviruses, including the tibroviruses (species Tibrogargan virus and Coastal Plains virus) (5–8).
Viroporins are ion channel proteins that enhance membrane permeability to facilitate various aspects of virus replication. They are usually small nonglycosylated transmembrane proteins (50 to 120 amino acids [aa]) that generate hydrophilic membrane pores upon oligomerization (9, 10). Viroporins (or putative viroporins) have been described in a wide range of viruses, including positive-sense ssRNA viruses (Coronaviridae, Flaviviridae, Picornaviridae, Togaviridae), negative-sense ssRNA viruses (Orthomyxoviridae, Paramyxoviridae), double-stranded RNA viruses (Reoviriade), reverse-transcribing RNA viruses (Retroviridae), and double-stranded DNA viruses (Poxviridae, Polyomaviridae, Papillomaviridae, Phycodnaviridae). They have been classified according to their various membrane topologies, which may feature one or more transmembrane domains with the N terminus and C terminus oriented either toward the lumen or toward the cytosol (10). By disrupting the ion balance in cells, viroporins have been shown to facilitate virus uncoating in endosomes (11), virus assembly, budding, and release (12), glycoprotein trafficking, and the induction of caspase-dependent apoptosis (13). Viroporins may also have secondary functions through their interaction with host cell proteins via the cytoplasmic domain. The C-terminal cytoplasmic domain of human immunodeficiency virus type 1 (HIV-1) viroporin Vpu interacts with and is subsequently involved in the degradation of CD4 in the endoplasmic reticulum; the 54-aa C-terminal cytoplasmic domain of the influenza A virus (IAV) M2 protein and the N-terminal cytoplasmic domain of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein have each been shown to interact with caveolin-1, a cholesterol-binding protein involved in lipid rafts (14–16). Due to their critical roles in infection and pathogenesis, viroporins are attracting increasing interest as targets for antiviral therapies (10).
In this paper, we demonstrate that the BEFV α1 protein has the functional characteristics of a viroporin. We show that α1 expression arrests growth in Escherichia coli and increases membrane permeability in E. coli and in mammalian cells, in which it localizes in the Golgi complex. We also provide evidence that α1 interacts specifically with the nuclear transport receptors importin β1 and importin 7.
MATERIALS AND METHODS
Origins and growth of viruses.
BEFV strain CS1865 was used as a source of the α1 gene. The virus was isolated from a clinical case of bovine ephemeral fever at Kowanyama, Queensland, Australia (141°44′E, 15°28′S), in 1969 and was passed 11 times intravenously in cattle, three times in suckling mice, and once in C6-36 Aedes albopictus cells (T. D. St. George, personal communication). The virus was then passaged once in BHK-BSR cells for preparation of total RNA. BEFV strain CS919 is a cell culture-adapted virus which does not express α1 due to a corruption in the transcription termination signal of the upstream ORF (GNS). CS919 was isolated from the same source as the prototype strain BB7721 (Charters Towers, Queensland, Australia [146°15′E, 20°04′S]) in 1968, but it has a different passage history. Bluetongue virus (BTV) serotype 1 (strain CS156) was used as the source of the NS3 gene. The virus was isolated from cattle blood collected at Beatrice Hill, Northern Territory, Australia (131°20′E, 12°39′S), in 1979 (17). It was passaged 13 times in BHK-21 and Vero cells prior to one passage in BHK-BSR cells for preparation of total RNA. The virus infection and transfection studies reported here were conducted in BHK-BSR, HEK293T, or COS-7 cells cultured at 37°C in Eagle's minimum essential medium (EMEM) supplemented with 10 mM HEPES, 6.7 mM NaHCO3, 2 mM l-glutamine, 137 mM streptomycin, 80 U/ml penicillin, and 5% fetal calf serum (growth medium) or 2.5% fetal calf serum (maintenance medium).
Plasmid construction and transfection.
Total RNA was extracted from infected BHK-BSR cells using a Qiagen RNeasy kit according to the manufacturer's specifications. cDNA was synthesized using a SuperScript III first-strand synthesis system (Life Technologies) and 10 μM random hexamer primers (GeneWorks). The BEFV α1 ORF was amplified using specific primers and cloned into the BamHI and SalI sites of pGBKT7 (Clontech). The ORF was subsequently excised from pGBKT7 with NdeI and PstI and cloned into the corresponding restriction sites of pMALc5x (New England BioLabs). To prepare green fluorescent protein (GFP) fusion constructs, the α1 ORF was excised from pGBKT7 using BamHI and SalI and cloned into the BglII and SalI sites of pAcGFP1c2 (Clontech).
The BTV NS3 ORF was amplified using specific primers and cloned into the pGEM-T Easy vector (Promega). The ORF was subsequently excised with EcoRI and cloned into the corresponding site of pAcGFP1c2. The ORF was subcloned from pAcGFP1c2 into pMALc5x using EcoRI. The ORF expressing the Nipah virus (NiV) V protein was amplified from pCAGGS-NiV-V (18) using specific primers and cloned into the BglII and BamHI sites of pEGFPc1 (Clontech).
The histone H1° ORF was amplified using gene-specific primers modified to introduce sites for Gateway cloning (Life Technologies). The ORF was subsequently cloned into pDEST-RFP (19) using the Gateway system according to the manufacturer's specifications to yield pDEST-RFP-H1°.
All constructs were sequenced to verify their integrity. Plasmid transfections of mammalian cells were conducted using the Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer's specifications.
SDS-PAGE and immunoblotting.
Protein extracts were treated and analyzed using precast NuPAGE Novex bis-Tris SDS-polyacrylamide gels (Life Technologies) according to the manufacturer's instructions. The separated proteins were transferred from polyacrylamide gels to Hybond-LFP polyvinylidene difluoride membranes (Amersham) using an XCell II blot module (Life Technologies). Membranes were blocked with 5% skim milk in phosphate-buffered saline (PBS)–Tween (0.5%) for 1 h and then incubated for 1 h with primary antibody at the following dilutions: anti-BEFV α1 rabbit polyclonal antibody (GenScript), 1:2,000; anti-BEFV N monoclonal antibody 11A3 (20), 1:1,000; anti-maltose binding protein (MBP; New England BioLabs), 1:2,000; anti-importin β (Santa Cruz Biotechnology), 1:2,000; anti-importin 7 (Santa Cruz Biotechnology), 1:750; anti-importin α3 (Abcam), 1:500; and anti-GFP (Abcam), 1:1,000. After washing, membranes were incubated for 1 h with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2,000) or HRP-conjugated donkey anti-goat/sheep IgG (1:2000), washed again, immersed in Novex ECL chemiluminescent substrate reagent (Life Technologies) for 5 min, and exposed to Kodak BioMax Light film (Amersham). Raw image intensity was analyzed using either ImageJ software or the Gene Tools software package from SynGene.
Confocal microscopy.
Cells on glass coverslips were fixed with 4% paraformaldehyde in PBS for 40 min at room temperature and then washed and stored in PBS at 4°C. Cells were permeabilized with 0.1% Triton X-100 for 10 min, and nonspecific binding was blocked with 0.5% bovine serum albumin (BSA) in PBS (PBS-BSA) for 30 min. Cells were subsequently stained for 1 h with specific primary antibodies (described below), washed 3 times for 5 min each time in PBS, incubated for 1 h in species-specific secondary antibodies conjugated to either Alexa Fluor 568 or Alexa Fluor 488 (1:200 dilution; Life Technologies), washed 2 times for 5 min each time with PBS, and rinsed twice in sterile distilled H2O. Nucleic acids were labeled with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; Sigma) in distilled water, and the coverslips were then mounted on microscope slides in Vectashield mounting medium (Vector Laboratories), sealed with nail varnish, and imaged using a Leica Microsystems SP5 confocal microscope with a ×60 oil immersion objective lens.
BEFV G, BEFV α1, and BEFV N were visualized in infected cells using anti-BEFV G polyclonal antibody produced in rabbits infected with a recombinant vaccinia virus expressing BEFV G (rVV-G; 1:200) (2), anti-BEFV α1 rabbit polyclonal antibody (1:500; GenScript), and BEFV N monoclonal antibody 11A3 (1:500), respectively. To investigate the subcellular localization of BEFV α1, transfected cells were treated with leptomycin B (2.8 ng/ml) for 3 h, stained with either Golgi complex marker anti-GM130 (1:500; BD Biosciences) or endoplasmic reticulum (ER) marker mouse anti-protein disulfide isomerase (PDI; 1:1,000; Quantum Scientific), fixed, and imaged. To assess the colocalization of BEFV α1 with importins β1, α3, and 7, cells transfected with either pAcGFPc2 or pAcGFPc2-BEFVα1 were immunostained with anti-importin β1 (1:500 dilution; Abcam), anti-importin 7 (1:500 dilution; GeneTex), or anti-importin α3 (1:200 dilution; Abcam) antibody. Nuclear accumulation of importin-β-dependent cargoes (simian virus 40 [SV40] large T antigen [T-Ag] and histone H1) was assessed by staining transfected COS-7 cells at 24 h postinfection (hpi) with SV40 T-Ag (v-300) antibody (1:750 dilution; Santa Cruz Biotechnology) or by cotransfection to express red fluorescent protein (RFP)-H1° before imaging, as described above.
The fluorescence intensity in digitized confocal microscope images was measured using ImageJ software to calculate the ratio of nuclear to cytoplasmic fluorescence corrected for background fluorescence (Fn/c) for >30 images (22), and statistical significance was calculated using a two-tailed Mann-Whitney test with a 95% confidence interval.
Growth, induction, and membrane permeability assays in bacteria.
Single colonies of E. coli DH5α (Life Technologies) transformed with plasmids containing MBP fusion constructs inoculated into 5 ml LB containing 100 μg/ml ampicillin (LB-amp) were incubated overnight at 37°C, diluted 1/100 in 5 ml of LB-amp, and grown to an optical density at 600 nm (OD600) of 0.4 before induction with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside). For membrane permeability assays, the cultures were treated with 0 mM, 0.5 mM, or 1.0 mM hygromycin B (Hyg; Life Technologies) for 10 min prior to induction with 2 mM IPTG. The cells were harvested by centrifugation for 30 min after induction, resuspended in 2× NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) containing β-mercaptoethanol (Sigma), boiled for 5 min, and then analyzed by SDS-PAGE and Western blotting using an anti-MBP polyclonal antibody (New England BioLabs).
Membrane permeability assays in mammalian cells.
Plasma membrane permeability assays in mammalian cells were conducted as described previously (23–26), with some modifications. HEK293T cells in 24-well plates were transfected with pAcGFP1c2, pAcGFP1c2-BEFVα1, or pAcGFP1c2-BTVNS3. At 21 h posttransfection, the medium was removed, the cells were washed 3 times with methionine plus cysteine-free EMEM (MP Chemicals) containing 0.1% BSA, and 1 ml of the same medium was added to each well. Cells were pretreated with 1 mM hygromycin B (Hyg) in 1 ml of the same medium for 20 min or maintained in Hyg-free medium (controls), and then 150 μCi l-[35S]methionine and cysteine (Perkin-Elmer) was added to each well for 3 h at 37°C. The cultures were then rinsed 3 times in the same medium and harvested in 200 μl of Y-PER yeast protein extraction reagent (Thermo-Scientific) containing 1× protease inhibitor cocktail set V, EDTA free (Calbiochem). Cell preparations were vortexed for 20 min, centrifuged at 10,000 × g for 20 min at room temperature, and then added in 1:1 volumes to 2× NuPAGE LDS sample buffer (diluted in sterile H2O with β-mercaptoethanol [Sigma]). Samples were heated at 85°C for 15 min and then subjected to electrophoresis (5 μl of sample was loaded) using 4 to 12% bis-Tris SDS-polyacrylamide gels and 1× MES (morpholineethanesulfonic acid) running buffer (Life Technologies) according to the manufacturer's instructions. Gels were fixed (70% methanol, 10% acetic acid) for 30 min, soaked in Amplify reagent (Amersham) for 30 min, placed in gel drying fluid (40% methanol, 10% glycerol, 7.5% acetic acid) for 5 min, and then dried overnight on glass plates using gel drying film (Promega). Once dried, the gels were exposed to BioMax MR film (Kodak) at −80°C, as required.
Propidium iodide staining was conducted in BHK-BSR cells transfected with pAcGFP1c2, pAcGFP1c2-BEFVα1, or pAcGFP1c2-BTVNS3. At 24 h posttransfection, cells were washed 2 times for 5 min each time with PBS and then stained with 250 μl of 20 ng/μl propidium iodide in Opti-MEM medium (Life Technologies) for 30 min at 37°C. Glass coverslips were removed, washed once in PBS, mounted on microscope slides, and imaged as described above. To assess membrane permeability in infected cells, BHK-BSR cells were infected with 0.1 50% tissue culture infective dose (TCID50)/cell of either BEFV CS1856 (which expresses α1) or BEFV CS919 (which is deficient in α1). At 1 hpi, 19 hpi, and 25 hpi, cells were washed 2 times for 5 min each time with PBS and then stained with 250 μl of 20 ng/μl propidium iodide in Opti-MEM medium for 30 min at 37°C. Cells were washed 2 times for 5 min each time in ice-cold PBS, placed on ice, and then stained for 1 h with anti-G rabbit polyclonal antibody and imaged as described above.
Affinity chromatography and mass spectrometry.
Affinity chromatography was conducted using GFP-binding protein coupled to agarose beads (ChromoTek). BHK-BSR cells were seeded and transfected in 6-cm petri dishes with either pAcGFP1c2 or pAcGFP1c2-BEFVα1. After 24 h, the cells were washed in PBS and harvested in 200 μl ice-cold lysis buffer (10 mM Tris HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1× Sigma P2714 protease inhibitor), incubated for 30 min on ice with frequent pipetting, and centrifuged at 10,000 × g for 5 min at 4°C. The clarified supernatants were transferred to precooled tubes and adjusted to 1 ml with ice-cold dilution buffer (lysis buffer without NP-40). The GFP beads were washed 3 times with 500 μl ice-cold dilution buffer, 20 μl of the commercial bead suspension was added to each lysate, and the mixture was incubated for 1 h at 4°C with gentle agitation. The beads were then harvested by centrifugation at 2,000 × g and washed 3 times with dilution buffer, resuspended in 20 μl of 2× NuPAGE LDS sample buffer, and boiled for 5 min prior to SDS-PAGE. Following electrophoresis, the gels were washed 3 times for 10 min each time in water, stained overnight with colloidal Coomassie brilliant blue R250, and destained in water. Stained bands were excised, and the proteins were analyzed by mass spectrometry (MS) (27). Capillary high-pressure liquid chromatography (HPLC)-tandem MS (MS/MS) analysis of the in-gel tryptic digests was performed using a Shimadzu Prominence HPLC system (Shimadzu) interfaced with a linear ion trap (LTQ)-Orbitrap-Velos Pro hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The tryptic digests were acidified, loaded onto a 120-Å, 3-μm-particle-size, 300-μm by 10-mm C18-AQ Reprosil-Pur trap column (SGE Australia Pty. Ltd.) at 30 μl/min in 98% solvent A (0.1% [vol/vol] aqueous formic acid) and 2% solvent B (80% [vol/vol] acetonitrile, 20% [vol/vol] H2O containing 0.1% [vol/vol] formic acid) for 3 min at 40°C, and subsequently gradient eluted onto a preequilibrated self-packed analytical column (Maisch GmbH Reprosil-Pur C18-AQ, 120 Å, 150 μm by 200 mm) using a flow rate of 900 nl/min. The LTQ-Orbitrap-Velos Pro mass spectrometer was controlled using Xcalibur (version 2.0) software (Thermo Fisher Scientific) and operated in a data-dependent acquisition mode to automatically switch between Orbitrap MS and ion trap MS/MS. The survey full-scan mass spectra (m/z 300 to 1,700) were acquired in the Orbitrap mass spectrometer with the resolving power set to 60,000 (at 400 m/z) after accumulating ions to an automatic gain control target value of 1.0 × 106 charges in the LTQ mass spectrometer. MS/MS spectra were acquired in parallel on the 15 most intense ions from the survey scan in the LTQ mass spectrometer filled to an automatic gain control target value of 5.0 × 103.
The liquid chromatography-MS/MS data were processed by Proteome Discoverer (version 1.3) software (Thermo Fisher Scientific), and searches against the complete Swiss-Prot protein database were performed with an in-house Mascot server (Matrix Science). The Mascot search parameters were enzymatic cleavage set to tryptic (allowing a maximum of two missed cleavages) and carbamidomethylation of cysteine specified as a fixed modification. Deamidation of asparagine and glutamine, and methionine oxidation were specified as variable modifications. Fragment ion and parent ion mass tolerances were set to 0.8 Da and 20 ppm, respectively (27). Protein identifications obtained from the Mascot search results were processed using Scaffold (version 3.0) software (Proteome Software, Inc.), and protein and peptides (minimum of two peptides for confident identification of a protein) were filtered with minimum identification probabilities of 95% and 99.9%, respectively.
RESULTS
Structural characteristics of the BEFV α1 protein.
The BEFV α1 gene encodes a 10.5-kDa (88-aa) protein (Fig. 1). Membrane topology predictions (with the TOPPRED, HMMTOP, and Phobius programs) indicate that the α1 protein contains a single central transmembrane domain, an aromatic-residue-rich 36-aa N terminus oriented toward the ER lumen, and a highly basic 35-aa C terminus which is located in the cytosol. Although other architectures occur among known viroporins, this topology is characteristic of class IA viroporins, such as the influenza A virus (IAV) M2 protein, HIV-1 Vpu, and SARS-CoV E protein (10). The BEFV α1 N-terminal ectodomain contains four tryptophan residues and is predicted to contain a single phosphorylation site (NetPhos); tryptophan residues that also feature in the N-terminal ectodomains of other class IA viroporins and in IAV M2 have been shown to act as a gate to facilitate asymmetric conductance through the ion channel (28). The α1 C-terminal endodomain is predicted with high probability (with the Jpred3 program) to assume an α-helical conformation throughout much of its length and to contain a single phosphorylation site and a strong nuclear localization signal (NLS). The α1 proteins of other ephemeroviruses (species Berrimah virus [BRMV], Adelaide River virus [ARV], and Obodhiang virus [OBOV]) are predicted to have the same topology and similar features, including a C-terminal NLS (Fig. 1).
FIG 1.
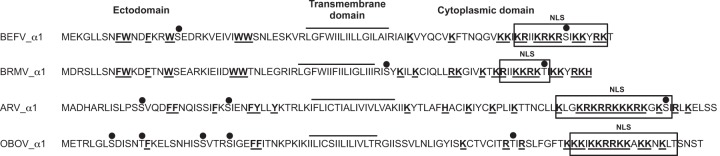
Structural features of the α1 proteins of ephemeroviruses BEFV, BRMV, ARV, and OBOV illustrating the predicted membrane topology, clusters of aromatic residues (W, F, Y) in the N-terminal ectodomain, basic residues (K, R) in the C-terminal endodomain (bold and underlined), predicted phosphorylation sites (●), and predicted NLSs (boxed).
Expression of α1 during BEFV infection of mammalian cells.
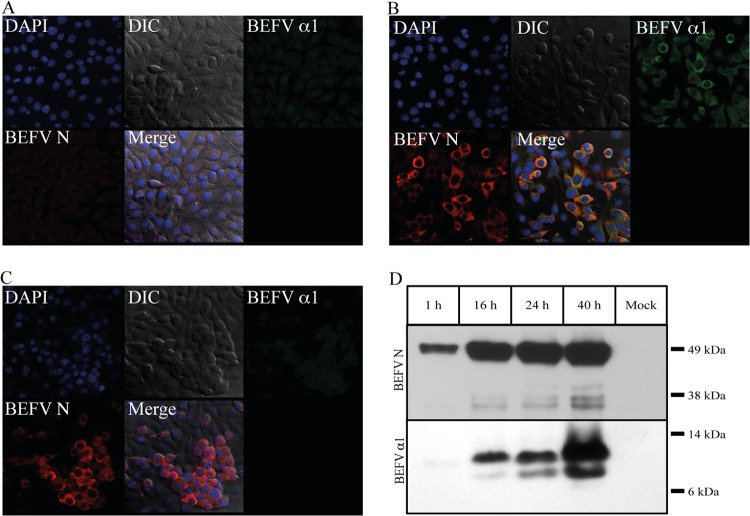
BHK-BSR cells were infected with BEFV CS1865 or BEFV CS919 at a multiplicity of infection of 0.1 TCID50/cell and stained at 24 hpi using BEFV α1 polyclonal antibody or BEFV N monoclonal antibody 11A3 (Fig. 2A and B). BEFV CS919 has a truncated transcription termination signal in the GNS gene immediately upstream of the α1 ORF and expresses only a GNS-α1 bicistronic mRNA (D. A. Joubert, L. Trinidad, and P. J. Walker, unpublished data). BEFV N monoclonal antibody 11A3 strongly stained the cytoplasm of cells infected with either strain, but BEFV α1 polyclonal antibody stained only cells infected with BEFV CS1865. For BEFV α1, the staining pattern was largely perinuclear, with little evidence of staining at the plasma membrane or in the nucleus. BEFV N protein appeared to be more evenly distributed throughout the cytoplasm. Immunoblots conducted on the BEFV CS1865-infected BHK-BSR cells indicated low-level expression of BEFV α1 from 1 hpi, with the band intensity increasing as infection progressed up to 40 hpi (Fig. 2C). An additional smaller band detected in cells infected with BEFV CS1865 appeared to be a degradation product. As expected, no BEFV α1 expression could be detected in cells infected with BEFV CS919 (result not shown).
FIG 2.
Detection of BEFV α1 and BEFV N in uninfected and BEFV-infected BHK-BSR cells. (A to C) Immunofluorescence images of uninfected (A), BEFV CS1865-infected (B), and BEFV CS919-infected (C) cells at 24 hpi. Frames illustrate differential interference contrast (DIC) images, cells stained with DAPI (blue) to highlight nuclei, cells stained with 1:500 dilutions of anti-BEFV α1 polyclonal rabbit serum (BEFV α1; green) or anti-BEFV N mouse monoclonal antibody 11A3 (BEFV N; red), and the merged fluorescence image (Merge). (D) Western blot analysis demonstrating the detection of BEFV N protein and BEFV α1 protein in BEFV-infected BHK-BSR cells at 1, 15, 24, and 40 hpi. Results for uninfected cells (mock) are also shown.
Effect of BEFV α1 induction on E. coli growth kinetics.
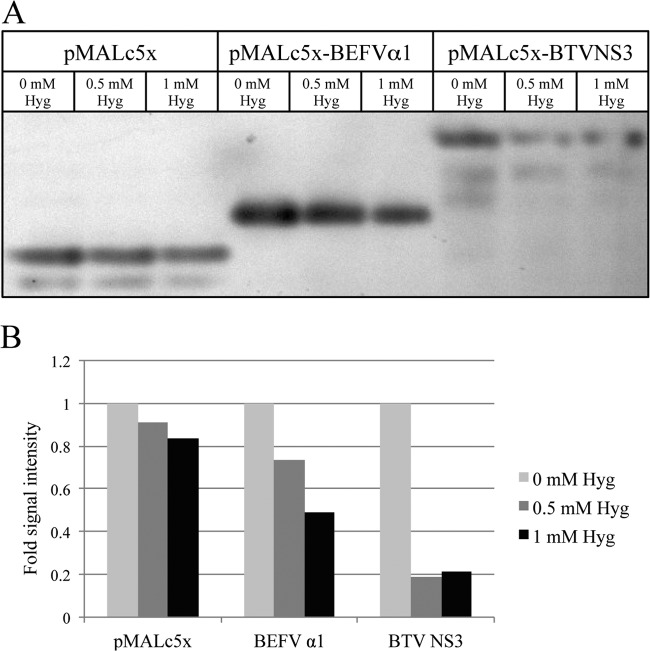
The BEFV α1 gene and the BTV NS3 gene (shown previously to encode a viroporin) (26) were cloned into vector pMALc5x downstream of the maltose binding protein, and the constructs (pMALc5x-BEFVα1 and pMALc5x-BTVNS3, respectively) were transformed into E. coli DH5α. The empty vector served as a negative control. The cultures were induced with IPTG when the OD600 reached 0.4. Immunoblot analysis of cultures at 30 min postinduction using anti-MBP antibody indicated that BEFV α1 and BTV NS3 were each induced efficiently (Fig. 3B and C). To determine the effect on E. coli growth kinetics, the OD600 of the cultures was assayed at 15-min intervals postinduction. As shown in Fig. 3A, all noninduced cultures and the IPTG-induced culture transformed with the empty vector exhibited very similar and typical E. coli growth kinetics. However, induction of BEFV α1 or BTV NS3 arrested E. coli growth, with almost complete growth arrest occurring in cultures transformed with the plasmid expressing the BEFV α1 fusion protein. Neither BEFV α1 nor BTV NS3 expression in E. coli DH5α (i.e., in the absence of T7 lysozyme) resulted in bacterial cell lysis.
FIG 3.
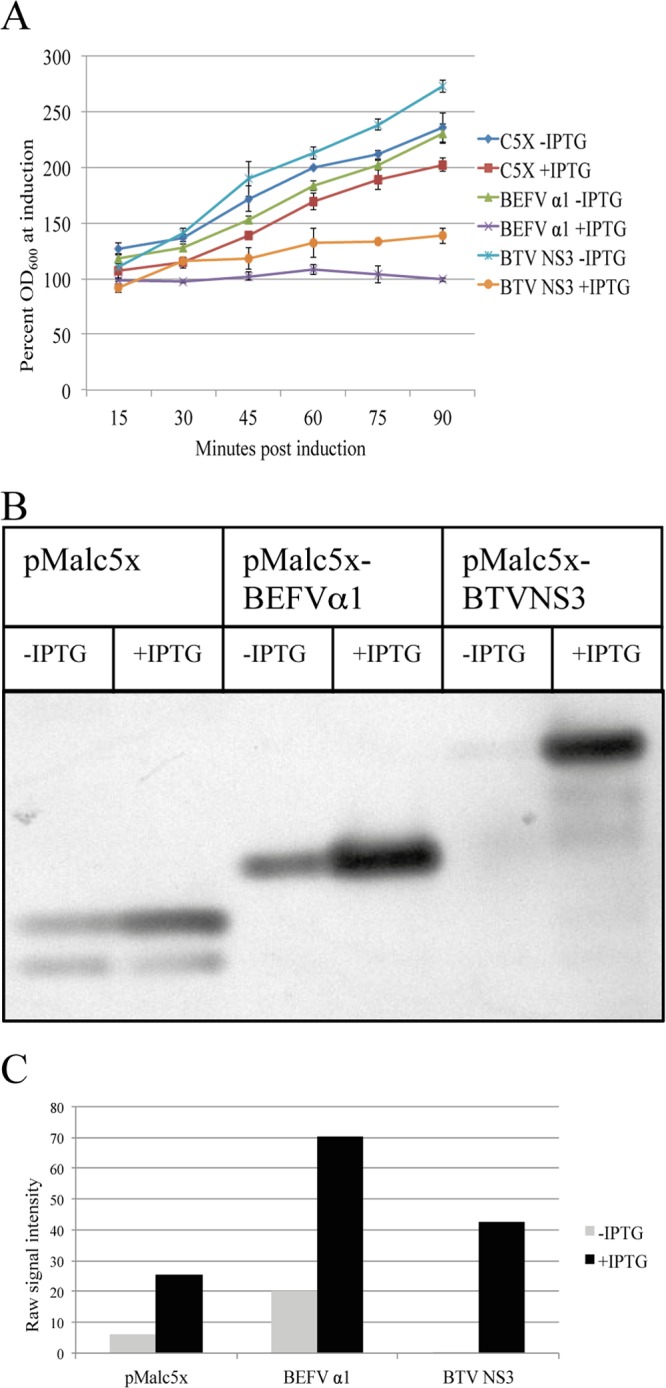
Expression of BEFV α1 arrests growth in E. coli. (A) Growth characteristics of E. coli transformed with pMALc5x, pMALc5x-BEFVα1, or pMALc5x-BTVNS3 at 15-min intervals postinduction with IPTG. OD600 measurements are expressed as the percentage of the OD600 at the time of induction. (B) Immunoblot using anti-MBP rabbit serum demonstrating expression of BEFV α1 and BTV NS3 fusion proteins at 30 min postinduction with IPTG. Sample volumes were normalized according to OD600 measurements to ensure equal protein loading. (C) Quantification of the signal intensity in immunoblots using the Gene Tools software package from SynGene. Experiments were conducted in duplicate, with the results of representative experiments shown.
Effect of BEFV α1 expression on cell membrane permeability.
Hygromycin B (Hyg) is a low-molecular-weight compound that inhibits translation in permeabilized cells and has been used previously to characterize viroporin activity (26). To determine the effect of BEFV α1 expression on E. coli membrane permeability, cells were transformed with pMALc5x, pMALc5x-BEFVα1, and pMALc5x-BTVNS3 and treated with Hyg 10 min prior to induction with IPTG. The effect of Hyg on protein synthesis at 30 min postinduction was determined by SDS-PAGE and immunoblotting using anti-MBP antibody (Fig. 4). Hyg treatment resulted in no inhibition of protein synthesis in E. coli expressing only MBP, but there was a marked dose-dependent reduction of protein synthesis in cells expressing the MBP-BEFVα1 or MBP-BTVNS3 fusion protein.
FIG 4.
Effect of BEFV α1 and BTV NS3 expression on membrane permeability in E. coli. (A) Immunoblot using anti-MBP rabbit serum of lysates of E. coli transformed with pMAL-c5x, pMALc5x-BEFVα1, or pMALc5x-BTVNS3 and induced with IPTG in the presence of increasing concentrations of Hyg; (B) quantification of the signal intensity in immunoblots using the Gene Tools software package from SynGene. Experiments were conducted in duplicate, with the results of representative experiments shown.
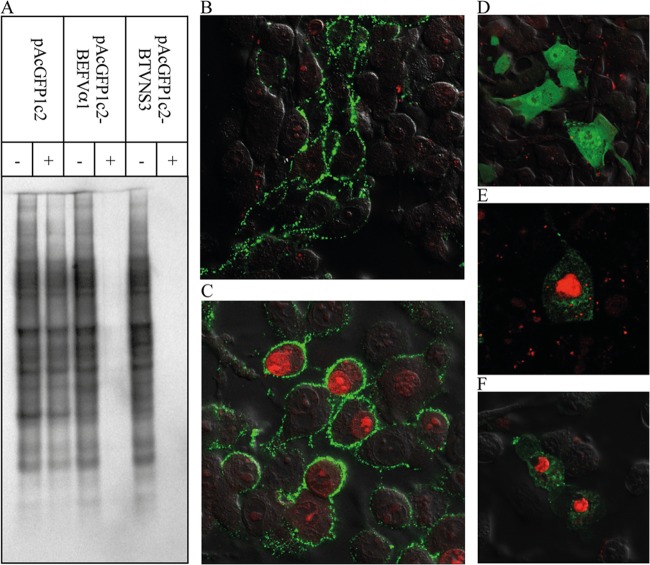
To determine the effect of BEFV α1 on mammalian cell permeability, HEK293T cells were transfected with plasmid pAcGFP1c2 (expressing the GFP reporter gene) or with pAcGFP1c2-BEFVα1 or pAcGFP1c2-BTVNS3 (expressing the respective GFP fusion proteins). At 21 h posttransfection, cysteine- and methionine-free medium was added, and at 24 h posttransfection, the cells were treated with Hyg in cysteine- and methionine-free medium for 20 min and then radiolabeled for 3 h with l-[35S]methionine and cysteine. The effect of Hyg treatment on protein synthesis was assessed by SDS-PAGE and autoradiography (Fig. 5A). HEK293T cells expressing only GFP showed very little or no inhibition of protein synthesis, whereas protein synthesis was almost completely inhibited in cells expressing BEFV α1 or BTV NS3.
FIG 5.
Effect of BEFV α1 and BTV NS3 expression on membrane permeability in mammalian cells. (A) Autoradiogram of lysates of HEK293T cells transfected with pAcGFP1c2, pAcGFP1c2-BEFVα1, or pAcGFP1c2-BTVNS3. The transfected cells were untreated (−) or treated with Hyg (+), and then radiolabeled with l-[35S]methionine and cysteine and analyzed by SDS-PAGE. (B and C) BHK-BSR cells infected with BEFV CS919 (B) or BEFV CS1865 (C). At 25 hpi, cells were stained with propidium iodide (red), followed by immunostaining with polyclonal antibodies against BEFV G protein (green) and imaging using confocal microscopy. (D to F) BHK-BSR cells transfected with pAcGFP1c2 (D), pAcGFP1c2-BEFVα1 (E), or pAcGFP1c2-BTVNS3 (F). At 24 hpi, cells were stained with propidium iodide (red) and imaged using confocal microscopy.
To assess further the effect of BEFV α1 on membrane permeability, BHK-BSR cells were either transfected as described above or infected with BEFV CS1865 (expressing α1) or BEFV CS919 (not expressing α1) and then stained with propidium iodide at 24 h posttransfection or 25 hpi. Control cells expressing only GFP showed minimal propidium iodide staining, whereas cells expressing BEFV α1 or BTV NS3 were clearly stained (results not shown). Similarly, cells infected with BEFV CS1865 could be stained with propidium iodide, whereas cells infected with BEFV CS919 showed very little staining (Fig. 5B and C).
Subcellular localization of BEFV α1.
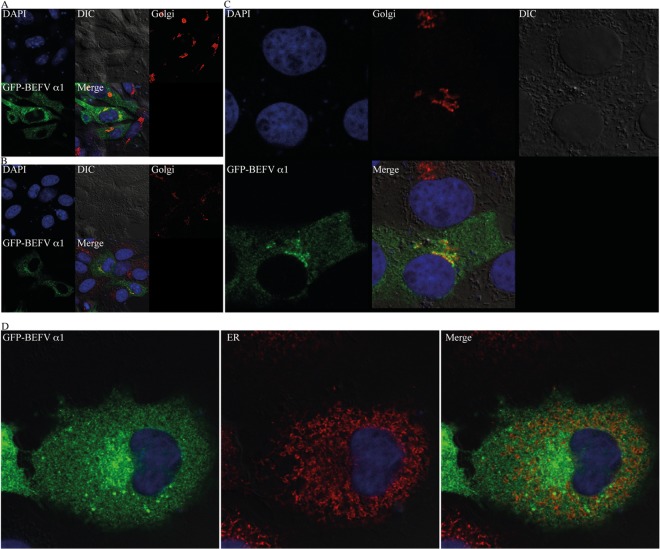
To identify the localization of BEFV α1 in infected cells, BHK-BSR cells or Vero cells were transfected with plasmid pAcGFP1c2-BEFVα1 expressing the GFP fusion protein. Transfected cells were fixed at 24 hpi, permeabilized with Triton X-100, stained with either GM130 (Golgi complex marker) or anti-PDI (ER marker), and visualized by confocal microscopy. BEFV α1 was observed to be cytoplasmic with evidence of strong colocalization with the Golgi complex marker, and no clear evidence for a strong association with the ER was found (Fig. 6). As the C-terminal cytoplasmic domain of BEFV α1 is predicted to contain a strong NLS, BHK-BSR cells were transfected with plasmid pAcGFP1c2-BEFVα1, and the nuclear export protein receptor CRM1 was blocked by treatment with leptomycin B at 24 to 27 h posttransfection to prevent nuclear export. The pAcGFP1c2 vector and pEGFPc1-NiV (expressing the Nipah virus V protein) were used as negative and positive controls, respectively. As shown in Fig. 7, leptomycin B resulted in no accumulation of BEFV α1-GFP in the nucleus, confirming that it has no detectable nuclear presence.
FIG 6.
Subcellular localization of BEFV α1 visualized by confocal microscopy in BHK-BSR cells (A and D) and Vero cells transfected with pAcGFP1c2-BEFVα1 (B and C). Frames illustrate cells stained with DAPI (blue) to highlight nuclei, differential interference contrast (DIC) images, cells stained with anti-GM130 polyclonal antibody (Golgi; red) or anti-PDI monoclonal antibody (ER; red), cells transfected with plasmid pAcGFP1c2-BEFVα1 (GFP-BEFVα1; green), and the merged fluorescence image (Merge).
FIG 7.
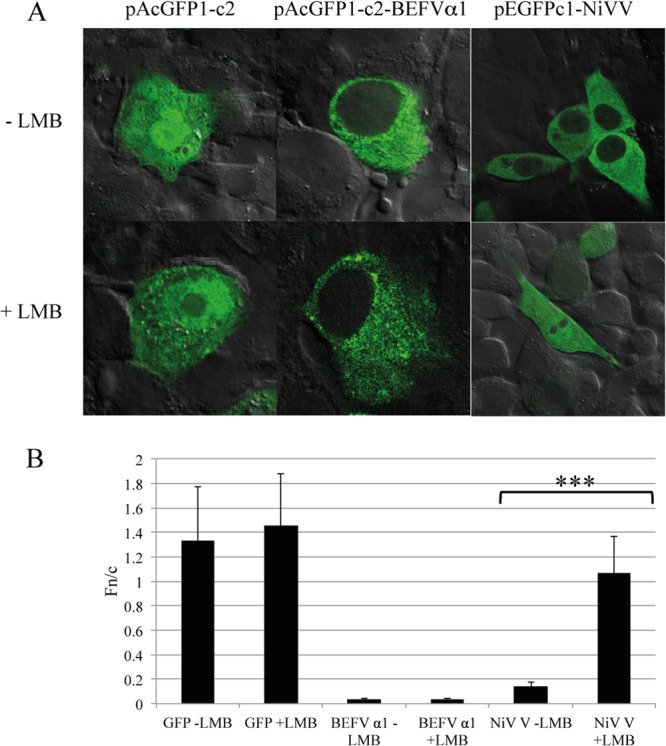
BEFV α1 is excluded from the nucleus of BHK-BSR cells. (A) Confocal microscopy of BHK-BSR cells transfected with pAcGFP1c2, pAcGFP1c2-BEFVα1, or pEGFP1c1-NiVV and treated for 3 h with either culture medium alone or culture medium supplemented with 2.8 ng/ml leptomycin B (LMB). (B) The ratio of nuclear fluorescence to cytoplasmic fluorescence (Fn/c), determined using ImageJ software from >30 imaged cells per transfection. Statistical analysis was performed using a Mann-Whitney test with a 95% confidence interval, and significant differences are indicated (***, P < 0.05).
To further assess the significance of the predicted NLS, the N-terminal ectodomain (M1-V35) and the C-terminal cytoplasmic domain (R53-T88) of BEFV α1 were cloned into plasmid pAcGFP1c2 to generate pAcGFP1c2-BEFVα1[N] and pAcGFP1c2-BEFVα1[C], respectively. BHK-BSR cells were transfected with these plasmids or pAcGFP1c2-BEFVα1 expressing the full-length BEFV α1 fusion protein and at 24 h posttransfection were examined by confocal microscopy as described above. As shown in Fig. 8, there was strong nuclear fluorescence in cells transfected with the C-terminal fragment. In contrast, both full-length BEFV α1 protein and the N-terminal fragment were detected mainly in the cytoplasm.
FIG 8.
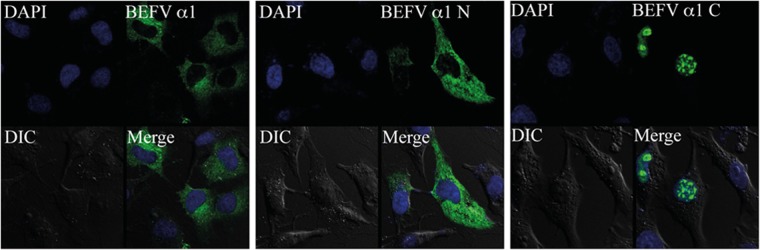
Subcellular localization of the N-terminal and C-terminal domains of BEFV α1. Confocal microscopy of BHK-BSR cells transfected with pAcGFP1c2-BEFVα1 (BEFV α1), pAcGFP1c2-BEFVα1[N] (BEFV α1 N), or pAcGFP1c2-BEFVα1[C] (BEFV α1 C). Frames illustrate cells stained with DAPI (blue) to highlight nuclei, differential interference contrast (DIC) images, GFP and GFP fusion proteins (BEFV α1, BEFV α1 N, and BEFV α1 C; green), and the merged fluorescence image (Merge).
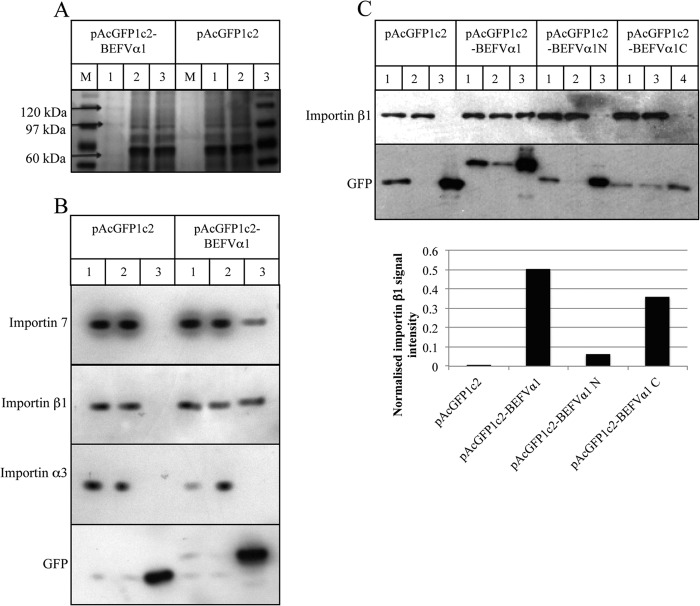
BEFV α1 specifically binds importins β1 and 7.
A GFP trap was employed to identify specific cytoplasmic interactions of BEFV α1 with cellular proteins. BHK-BSR cells were transfected with the vector pAcGFP1c2 or with pAcGFP1c2-BEFVα1. At 24 h posttransfection, clarified cell lysates were prepared and incubated with GFP-binding protein coupled to agarose beads. Proteins bound to the washed beads were eluted and resolved by SDS-PAGE. Three bands binding specifically to BEFV α1 (migrating at approximately 120 kDa, 97 kDa, and 60 kDa) were excised from the gels, and the components were identified by mass spectrometry (Fig. 9A). Two of the most abundant proteins in the binding fractions were identified as importin β1 (molecular mass, 97 kDa) and importin 7 (molecular mass, 120 kDa) (see Table S1 in the supplemental material), each of which has a role in the nuclear translocation of proteins through binding to NLS. Other karyopherins were also detected as minor components of the excised bands, including importin 5 (molecular mass, 124 kDa), importin 8 (molecular mass, 120 kDa), transportin 1 (molecular mass, 102 kDa), and importin α2 (molecular mass, 58 kDa). Proteins detected at low levels included serum proteins (e.g., albumin, plasminogen) and commonly detected contaminants (e.g., keratin, trypsin). The specificity of the interactions was confirmed in an independent GFP trap experiment in which the GFP-BEFV α1-binding fraction and the GFP-binding fraction were resolved by SDS-PAGE and examined by Western blotting using antibodies specific to importin β1, importin 7, and importin α3. As shown in Fig. 9B, importin β1 and importin 7, but not the related importin α3, were detected in the BEFV α1-binding fraction.
FIG 9.
BEFV α1 specifically interacts with importin 7 and importin β1 but not with importin α3. (A) SDS-polyacrylamide gel depicting bands selected and excised for mass spectrometry from GFP-binding fractions of lysates from BHK-BSR cells transfected with pAcGFP1c2 or pAcGFP1c2-BEFVα1. Lanes: M, molecular marker; 1, bound fraction; 2, unbound fraction; 3, input fraction. (B and C) Immunoblots and signal quantification of the GFP-binding fractions from lysates of BHK-BSR cells transfected with pAcGFP1c2, pAcGFP1c2-BEFVα1, pAcGFP1c2-BEFVα1[N], and pAcGFP1c2-BEFVα1[C]) using antibodies specific to importin 7, importin β1, importin α3, and GFP. Lanes: 1, input fraction; 2, unbound fraction; 3, bound fraction. The signal intensity in panel C was quantified using ImageJ software, and the results are shown at the bottom of panel C. All experiments were conducted in duplicate, with the results of representative experiments shown.
To identify the interactive domain, another GFP trap experiment was performed with clarified cell lysates prepared from HEK293T cells transfected with pAcGFPc2, pAcGFPc2-BEFVα1, pAcGFP1c2-BEFVα1[N] (expressing the 36-aa N-terminal domain), and pAcGFP1c2-BEFVα1[C] (expressing the 36-aa C-terminal domain). Western blotting with antibodies specific to importin β1 confirmed specific binding of pAcGFP1c2-BEFVα1[C] to importin β1. Quantification of the signal intensity indicated that pAcGFP1c2-BEFVα1[C] bound importin β1 much more strongly than pAcGFP1c2-BEFVα1[N], but both were less efficient at binding than pAcGFPc2-BEFVα1 (Fig. 9C).
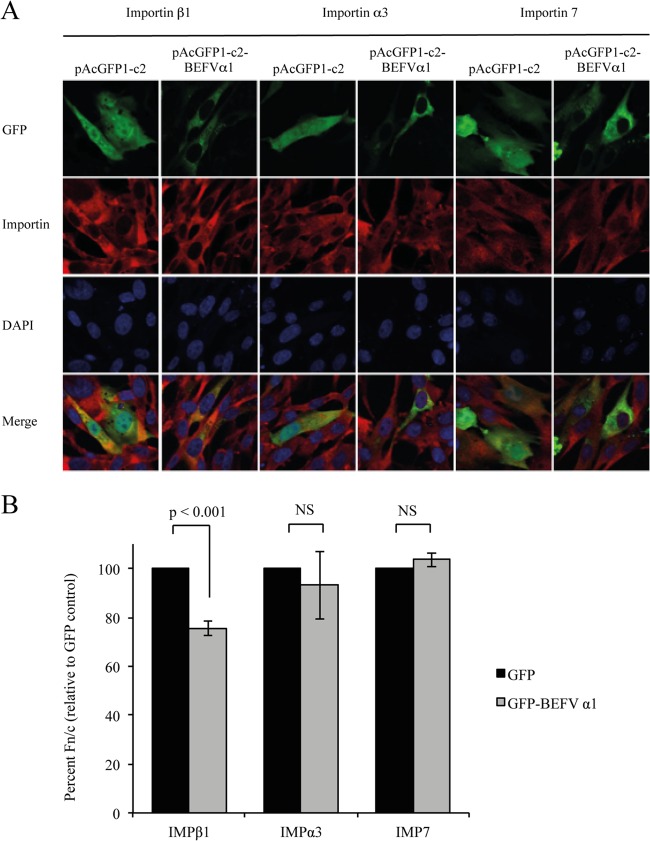
BEFV α1 inhibits the nuclear localization of importin β1.
To further assess the significance of the interaction between BEFV α1 and importins β1 and 7, the effect of BEFV α1 expression on importin β1, importin 7, and importin α3 localization was investigated. BHK-BSR cells were transfected with either pAcGFPc2 or pAcGFPc2-BEFVα1, fixed at 24 h posttransfection, immunostained with antibodies against importin β1, importin 7, or importin α3, and visualized by confocal microscopy (Fig. 10A). Expression of BEFV α1 reduced the ratio of nuclear fluorescence to cytoplasmic fluorescence (Fn/c) for cells stained for importin β1, indicating a significantly (P < 0.001) lower level of nuclear accumulation of importin β1 relative to that for cells transfected with pAcGFP2 alone (Fig. 10B). However, BEFV α1 expression had no significant effect on the localization of importin 7 or importin α3.
FIG 10.
BEFV α1 inhibits nuclear accumulation of importin β1 but not importin 7 or importin α3. (A) Immunofluorescence images of BHK-BSR cells transfected with pAcGFP1c2 or pAcGFP1c2-BEFVα1 and stained with antibodies against importin β1, importin 7, or importin α3. (B) The ratio of nuclear fluorescence to cytoplasmic fluorescence (Fn/c) from >30 imaged cells per transfection was determined using ImageJ software. Statistical analysis was performed using a Mann-Whitney test, as described in the legend to Fig. 7. IMP, importin; NS, not significant.
Lack of an effect of BEFV α1 expression on nuclear accumulation of SV40 T-antigen or histone H1.
To assess whether BEFV α1 expression affects the subcellular localization of nuclear import cargoes dependent on importin β1, COS-7 cells were cotransfected either with pAcGFP1c2 and pDEST-RFP-H1° or with pAcGFP1c2-BEFVα1 and pDEST-RFP-H1o and fixed at 24 h posttransfection. COS-7 cells expressing SV40 large T antigen were also transfected with either pAcGFP1c2 or pAcGFP1c2-BEFVα1, fixed at 24 h posttransfection, and immunostained with anti-T-antigen antibody. The cells were visualized by confocal microscopy, and Fn/c was determined for both histone H1 and T antigen. BEFV α1 had no significant effect on the nuclear accumulation of either histone H1 (Fig. 11) or SV40 large T antigen (results not shown).
FIG 11.
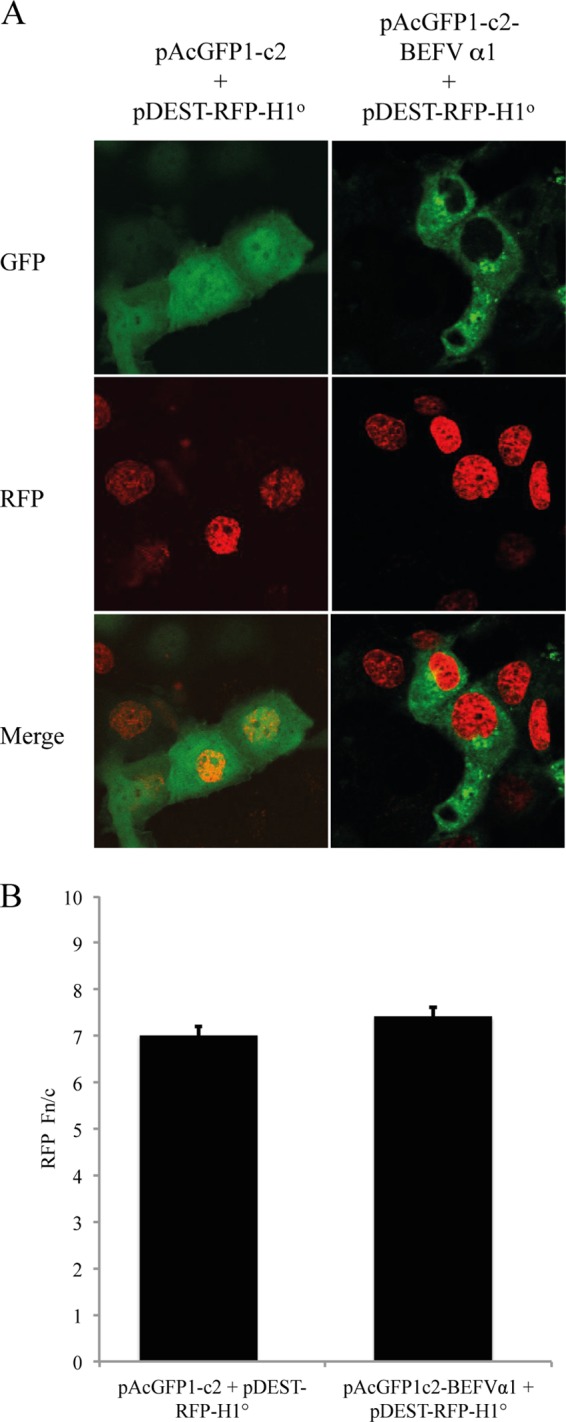
BEFV α1 does not inhibit nuclear accumulation of histone H1. (A) Immunofluorescence images of COS-7 cells transfected with pAcGFP1c2 and pDEST-RFP-H1° or pAcGFP1c2-BEFVα1 and pDEST-RFP-H1°. (B) The ratio of nuclear fluorescence to cytoplasmic fluorescence (Fn/c) from >30 imaged cells per transfection was determined using ImageJ software. Statistical analysis was performed using a Mann-Whitney test, as described in the legend to Fig. 7.
DISCUSSION
For many years, vesicular stomatitis virus (VSV) and rabies virus (RV) have served as important models for the study of negative-sense RNA viruses. However, with over 30 complete rhabdovirus genomes sequenced to date, it has become evident that the relatively simple genome organization and expression strategy of vesiculoviruses and lyssaviruses are far from representative of the Rhabdoviridae as a whole and that, in addition to the five major structural proteins (N, P, L, G, and L), all rhabdoviruses appear to encode one or more additional proteins, either as independent genes or as alternative or overlapping ORFs (5). Few of this growing number of putative rhabdoviruses proteins have yet been shown to be expressed during infection and characterized functionally.
Ephemerovirus genomes are among the largest and most complex of those of the rhabdoviruses sequenced to date, containing up to six additional ORFs in the region between the G and L genes. Here we demonstrate that the BEFV α1 protein is expressed during infection and, as predicted previously from the deduced amino acid sequence (4), functions as a viroporin. Recombinant BEFV α1 fusion proteins were shown to inhibit log-phase growth kinetics when expressed in E. coli and induce a dose-dependent increase in membrane permeability to Hyg in bacteria and mammalian cells. E. coli expression systems have been used extensively to assess viroporin-induced modifications to membrane permeability by inhibition of growth kinetics and induction of cell lysis by allowing access of coexpressed T7 lysozyme through the inner membrane to the peptidoglycan layer (29–35). IPTG-induced expression of BEFV α1 (or BTV NS3) as an MBP fusion protein in E. coli DH5α (lacking T7 lysozyme) resulted in an immediate arrest of log-phase cell growth but did not result in cell lysis. Similar results have been reported for the influenza virus M2 protein and rotavirus NSP4 when induced in the absence of T7 lysozyme, and this has been interpreted to indicate that the viroporin-induced inner membrane permeabilization is not the direct cause of bacterial cell lysis (30, 36). Sensitivity to Hyg has previously been employed to identify other recognized viroporins, including the BTV NS3 protein, IAV M2 protein, poliovirus 2B protein, murine hepatitis virus E protein, Sindbis virus 6K protein, hepatitis C virus p7 protein, and JC polyomavirus angoprotein (13, 26, 32, 37, 38). Hyg inhibits translation in bacterial and eukaryotic cells by interfering with aminoacyl-tRNA recognition and ribosomal translocation (39, 40), if access to the cytoplasm is facilitated by cell membrane permeabilization. In E. coli, the BEFV α1 protein was less effective than BTV NS3 in inducing sensitivity to Hyg under the conditions used. However, in mammalian cells, protein synthesis was totally abolished by Hyg treatment 24 h after transfection with plasmids expressing either BTV NS3 or BEFV α1.
Propidium iodide has also been used previously to evaluate membrane permeability (41). We have shown here for the first time that mammalian cells overexpressing BEFV α1 (or BTV NS3) are permeable to propidium iodide, while the dye is excluded from cells expressing GFP alone. Similarly, infection with BEFV strain CS1865 increased membrane permeability to propidium iodide, while the dye was excluded from cells infected with BEFV CS919, which does not express α1. Together, these data indicate that BEFV α1 has membrane permeabilization properties consistent with those described for other recognized viroporins.
Recombinant BEFV α1 protein also demonstrated a strong membrane association, colocalizing in transfected cells with cis-Golgi matrix marker GM130 (42). Although viroporins have been observed to be associated with various cellular membranes, a specific association with the Golgi complex has been reported for BTV NS3 protein, the poliovirus and coxsackievirus 2B proteins, and respiratory syncytial virus (RSV) SH protein (26, 43–46). The coxsackievirus 2B protein has been shown to perturb cellular Ca2+ homeostasis, resulting in an influx of extracellular Ca2+, decreased Ca2+ signaling between the endoplasmic reticulum and mitochondria, and the suppression of caspase activation and apoptotic cell death (47). The poliovirus 2B protein and the RSV SH have also been shown to inhibit apoptosis (48, 49), but there is also contrary evidence that the poliovirus 2B protein and several other viroporins induce apoptosis in some cell lines via the mitochondrial pathway (13). It has been suggested that viroporins may both suppress and induce apoptosis at different stages of infection (13). BEFV has been shown to induce apoptosis in a process that requires viral gene expression and occurs through activation of Fas- and mitochondrion-mediated pathways (50, 51), but it is not known at this time if α1 is involved.
The cytoplasmic tail domains of several viroporins have been reported to have secondary functions associated with virus assembly (52, 53), degradation of CD4 (15), and the binding of caveolin-1 (14, 16). Here, we show that the BEFV α1 C-terminal domain contains predicted nuclear localization signals and, when expressed in mammalian cells independently of the transmembrane domain, is translocated to the nucleus. We also show using a pulldown assay that the BEFV α1 protein binds to importins in transfected cells, with strong evidence of interaction with importin β1 and importin 7. Logically, this is likely to occur through the α1 C-terminal domain, and this was confirmed in pulldown assays. Furthermore, overexpression of BEFV α1 led to a significant reduction of importin β1 accumulation in the nucleus. It is not clear at this stage why there was no evidence of specific nuclear exclusion of importin 7, but it may have been due to a relatively higher affinity of BEFV α1 for importin β1.
Although they were not previously associated with viroporins, other viral proteins have been reported to bind and sequester karyopherins in the cytoplasm. Ebola virus VP24 blocks interferon signaling by binding to importin α1, importin α5, and importin α6, which are the NLS receptors for activated STAT-1, inhibiting its nuclear translocation (54, 55). A region of the capsid protein of the alphavirus Venezuelan equine encephalitis virus also contains nuclear import and nuclear export signals which mediate the formation of a tetrameric complex with importin α/β and the nuclear export receptor CRM1, obstructing translocation of cargo (56). Similarly, the SARS-CoV ORF6 protein is an ER/Golgi complex-associated transmembrane protein that tethers importin α2 and importin β2 to the membrane through its C-terminal tail, blocking the nuclear translocation of STAT-1 and interferon-stimulated gene expression (57). Although other karyopherins (including importin α2) were detected in the BEFV α1 binding fraction, the major interactions that we detected were with the karyopherin β family importins β1 and 7. The classical nuclear translocation pathway involves NLS recognition by importin α, which forms a complex with importin β to bind cargo. However, many proteins are known to bind directly to importin βs, including importin β1, which, in addition to their roles as nuclear transporters, are known to have diverse functions, including as microtubule motor adaptors, in controlling mitotic functions ranging from spindle assembly to nuclear envelope and nuclear pore assembly (58). Although the importin β1/importin 7 complex has been specifically implicated in the nuclear import and suppression of aggregation of highly basic cargoes such as histone H1 (58), we could find no evidence that BEFV α1 inhibited nuclear accumulation of histone H1 or, indeed, of the importin α/β cargo SV40 T antigen (59). This implies that importin binding by BEFV α1 does not impact nuclear transport, at least in the absence of other viral proteins/viral infection. Detailed examination of the role of BEFV α1 interaction with karyopherins during infection is a focus of future work in this laboratory.
Small hydrophobic proteins with structural similarity but little or no sequence identity to the BEFV α1 protein appear to occur commonly in animal rhabdoviruses (5). All other ephemeroviruses (Berrimah virus, Kimberley virus, Malakal virus, Adelaide River virus, Obodhiang virus, and Kotonkan virus) encode α1 proteins in the long G-L intergenic region, directly downstream of the nonstructural glycoprotein (GNS) gene (6, 7, 60). Tibroviruses (Tibrogargan virus and Coastal Plains virus), which lack a GNS gene, encode proteins of similar sizes and structures in the U3 ORF immediately downstream of the G gene (8), as do the Hart Park group rhabdoviruses (Wongabel virus and Flanders virus) in the U5 ORF, which overlaps the end of the G gene (5, 61). Other rhabdoviruses (Tupaia rhabdovirus, Durham virus, and Oak Vale virus) encode small hydrophobic proteins between the M and G genes (62–64), but they differ in structure from the α1 proteins, and it is yet to be established if they function as viroporins.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Rebecca Davies for advice on the GFP trap affinity chromatography and James Wynne and Julio Rodriguez for critical review of the manuscript prior to submission. We also acknowledge Dianne Green for technical assistance with immunofluorescence and confocal microscopy.
Access to proteomic infrastructure funding at the QIMR Berghofer Medical Research Institute Protein Discovery Centre was made possible through the Australian Government National Collaborative Infrastructure Scheme (NCRIS) and EIF Scheme provided via Bioplatforms Australia and the Queensland State Government. The work was also supported by National Health and Medical Research Council (Australia) project grant 1003244 (to G.W.M.) and Australian Research Council Discovery project DP110101749 (to G.W.M. and D.A.J.).
Footnotes
Published ahead of print 20 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01812-13.
REFERENCES
- 1.Dietzgen RG, Calisher CH, Kurath G, Kuzman IV, Rodriguez LL, Stone DM, Tesh RB, Tordo N, Walker PJ, Wetzel T, Whitfield AE. 2011. Rhabdoviridae, p 654–681 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier, San Diego, CA [Google Scholar]
- 2.Hertig C, Pye AD, Hyatt AD, Davis SS, McWilliam SM, Heine HG, Walker PJ, Boyle DB. 1996. Vaccinia virus-expressed bovine ephemeral fever virus G but not G(NS) glycoprotein induces neutralizing antibodies and protects against experimental infection. J. Gen. Virol. 77:631–640. 10.1099/0022-1317-77-4-631 [DOI] [PubMed] [Google Scholar]
- 3.Walker PJ, Byrne KA, Riding GA, Cowley JA, Wang Y, McWilliam S. 1992. The genome of bovine ephemeral fever rhabdovirus contains two related glycoprotein genes. Virology 191:49–61. 10.1016/0042-6822(92)90165-L [DOI] [PubMed] [Google Scholar]
- 4.McWilliam SM, Kongsuwan K, Cowley JA, Byrne KA, Walker PJ. 1997. Genome organization and transcription strategy in the complex GNS-L intergenic region of bovine ephemeral fever rhabdovirus. J. Gen. Virol. 78:1309–1317 [DOI] [PubMed] [Google Scholar]
- 5.Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. 2011. Rhabdovirus accessory genes. Virus Res. 162:110–125. 10.1016/j.virusres.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasdell KR, Voysey R, Bulach D, Joubert DA, Tesh RB, Boyle DB, Walker PJ. 2012. Kotonkan and Obodhiang viruses: African ephemeroviruses with large and complex genomes. Virology 425:143–153. 10.1016/j.virol.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 7.Blasdell KR, Voysey R, Bulach DM, Trinidad L, Tesh RB, Boyle DB, Walker PJ. 2012. Malakal virus from Africa and Kimberley virus from Australia are geographic variants of a widely distributed ephemerovirus. Virology 433:236–244. 10.1016/j.virol.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 8.Gubala A, Davis S, Weir R, Melville L, Cowled C, Boyle D. 2011. Tibrogargan and Coastal Plains rhabdoviruses: genomic characterisation, evolution of novel genes and seroprevalence in Australian livestock. J. Gen. Virol. 92:2160–2170. 10.1099/vir.0.026120-0 [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez ME, Carrasco L. 2003. Viroporins. FEBS Lett. 552:28–34. 10.1016/S0014-5793(03)00780-4 [DOI] [PubMed] [Google Scholar]
- 10.Nieva JL, Madan V, Carrasco L. 2012. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 10:563–574. 10.1038/nrmicro2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mould JA, Drury JE, Frings SM, Kaupp UB, Pekosz A, Lamb RA, Pinto LH. 2000. Permeation and activation of the M2 ion channel of influenza A virus. J. Biol. Chem. 275:31038–31050. 10.1074/jbc.M003663200 [DOI] [PubMed] [Google Scholar]
- 12.Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. 10.1371/journal.ppat.0030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madan V, Castello A, Carrasco L. 2008. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 10:437–451. 10.1111/j.1462-5822.2007.01057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padhan K, Tanwar C, Hussain A, Hui PY, Lee MY, Cheung CY, Peiris JS, Jameel S. 2007. Severe acute respiratory syndrome coronavirus ORF3a protein interacts with caveolin. J. Gen. Virol. 88:3067–3077. 10.1099/vir.0.82856-0 [DOI] [PubMed] [Google Scholar]
- 15.Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou P, Wu F, Lu L, Huang JH, Chen YH. 2009. The cytoplasmic domain of influenza M2 protein interacts with caveolin-1. Arch. Biochem. Biophys. 486:150–154. 10.1016/j.abb.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 17.St George TD, Cybinski DH, Della-Porta AJ, McPhee DA, Wark MC, Bainbridge MH. 1980. The isolation of two bluetongue viruses from healthy cattle in Australia. Aust. Vet. J. 56:562–563. 10.1111/j.1751-0813.1980.tb02598.x [DOI] [PubMed] [Google Scholar]
- 18.Shaw ML, Garcia-Sastre A, Palese P, Basler CF. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633–5641. 10.1128/JVI.78.11.5633-5641.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soboleva TA, Jans DA, Johnson-Saliba M, Baker RT. 2005. Nuclear-cytoplasmic shuttling of the oncogenic mouse UNP/USP4 deubiquitylating enzyme. J. Biol. Chem. 280:745–752. 10.1074/jbc.M401394200 [DOI] [PubMed] [Google Scholar]
- 20.Cybinski DH, Walker PJ, Byrne KA, Zakrzewski H. 1990. Mapping of antigenic sites on the bovine ephemeral fever virus glycoprotein using monoclonal antibodies. J. Gen. Virol. 71:2065–2072. 10.1099/0022-1317-71-9-2065 [DOI] [PubMed] [Google Scholar]
- 21. Reference deleted.
- 22.Moseley GW, Roth DM, DeJesus MA, Leyton DL, Filmer RP, Pouton CW, Jans DA. 2007. Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 18:3204–3213. 10.1091/mbc.E07-01-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldabe R, Barco A, Carrasco L. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134–23137. 10.1074/jbc.271.38.23134 [DOI] [PubMed] [Google Scholar]
- 24.Alonso MA, Carrasco L. 1982. Molecular basis of the permeabilization of mammalian cells by ionophores. Eur. J. Biochem. 127:567–569 [DOI] [PubMed] [Google Scholar]
- 25.Chang Y-S, Liao C-L, Tsao C-H, Chen M-C, Liu C-I, Chen L-K, Lin Y-L. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73:6257–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Z, Harty RN. 2004. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J. Biol. Chem. 279:43092–43097. 10.1074/jbc.M403663200 [DOI] [PubMed] [Google Scholar]
- 27.Hastie ML, Headlam MJ, Patel NB, Bukreyev AA, Buchholz UJ, Dave KA, Norris EL, Wright CL, Spann KM, Collins PL, Gorman JJ. 2012. The human respiratory syncytial virus nonstructural protein 1 regulates type I and type II interferon pathways. Mol. Cell. Proteomics 11:108–127. 10.1074/mcp.M111.015909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Qiu JX, Soto C, DeGrado WF. 2011. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr. Opin. Struct. Biol. 21:68–80. 10.1016/j.sbi.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez ME, Carrasco L. 1998. The human immunodeficiency virus type 1 Vpu protein enhances membrane permeability. Biochemistry 37:13710–13719. 10.1021/bi981527f [DOI] [PubMed] [Google Scholar]
- 30.Guinea R, Carrasco L. 1994. Influenza virus M2 protein modifies membrane permeability in E. coli cells. FEBS Lett. 343:242–246. 10.1016/0014-5793(94)80564-4 [DOI] [PubMed] [Google Scholar]
- 31.Hyser JM, Utama B, Crawford SE, Estes MK. 2012. Genetic divergence of rotavirus NSP4 results in distinct serogroup-specific viroporin activity and intracellular punctate structure morphologies. J. Virol. 86:4921–4934. 10.1128/JVI.06759-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madan V, Garcia MDJ, Sanz MA, Carrasco L. 2005. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 579:3607–3612. 10.1016/j.febslet.2005.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez M, Garcia-Barreno B, Melero JA, Carrasco L, Guinea R. 1997. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology 235:342–351. 10.1006/viro.1997.8696 [DOI] [PubMed] [Google Scholar]
- 34.Sanz MA, Perez L, Carrasco L. 1994. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 269:12106–12110 [PubMed] [Google Scholar]
- 35.Studier WF, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89. 10.1016/0076-6879(90)85008-C [DOI] [PubMed] [Google Scholar]
- 36.Hyser JM, Collinson-Pautz M, Utama B, Estes MK. 2010. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 1(5):e00265–10. 10.1128/mBio.00265-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madan V, Redondo N, Carrasco L. 2010. Cell permeabilization by poliovirus 2B viroporin triggers bystander permeabilization in neighbouring cells through a mechanism involving gap junctions. Cell. Microbiol. 12:1144–1157. 10.1111/j.1462-5822.2010.01460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H. 2010. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 6:e1000801. 10.1371/journal.ppat.1000801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borovinskaya MA, Shoji S, Fredrick K, Cate JHD. 2008. Structural basis of hygromycin B inhibition of protein biosynthesis. RNA 14:1590–1599. 10.1261/rna.1076908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabanas MJ, Vazquez D, Modolell J. 1978. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur. J. Biochem. 87:21–27. 10.1111/j.1432-1033.1978.tb12347.x [DOI] [PubMed] [Google Scholar]
- 41.Arroyo J, Boceta M, Gonzalez ME, Michel M, Carrasco L. 1995. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J. Virol. 69:4095–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715–1726. 10.1083/jcb.131.6.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Jong AS, Wessels E, Dijkman HBPM, Galama JMD, Melchers WJG, Willems PHGM, van Kuppeveld FJM. 2003. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 278:1012–1021. 10.1074/jbc.M207745200 [DOI] [PubMed] [Google Scholar]
- 44.Rixon HW, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153–1165. 10.1099/vir.0.19769-0 [DOI] [PubMed] [Google Scholar]
- 45.Sandoval IV, Carrasco L. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. 2013. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 68:66–75. 10.1136/thoraxjnl-2012-202182 [DOI] [PubMed] [Google Scholar]
- 47.Campanella M, de Jong AS, Lanke KW, Melchers WJ, Willems PH, Pinton P, Rizzuto R, van Kuppeveld FJ. 2004. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J. Biol. Chem. 279:18440–18450. 10.1074/jbc.M309494200 [DOI] [PubMed] [Google Scholar]
- 48.Fuentes S, Tran KC, Luthra P, Teng MN, He B. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81:8361–8366. 10.1128/JVI.02717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neznanov N, Kondratova A, Chumakov KM, Angres B, Zhumabayeva B, Agol VI, Gudkov AV. 2001. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75:10409–10420. 10.1128/JVI.75.21.10409-10420.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CJ, Shih WL, Yu FL, Liao MH, Liu HJ. 2004. Apoptosis induced by bovine ephemeral fever virus. J. Virol. Methods 122:165–170. 10.1016/j.jviromet.2004.08.016 [DOI] [PubMed] [Google Scholar]
- 51.Lin CH, Shih WL, Lin FL, Hsieh YC, Kuo YR, Liao MH, Liu HJ. 2009. Bovine ephemeral fever virus-induced apoptosis requires virus gene expression and activation of Fas and mitochondrial signaling pathway. Apoptosis 14:864–877. 10.1007/s10495-009-0371-5 [DOI] [PubMed] [Google Scholar]
- 52.Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070. 10.1128/JVI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCown MF, Pekosz A. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595–3605. 10.1128/JVI.79.6.3595-3605.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156–5167. 10.1128/JVI.02349-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469–13477. 10.1128/JVI.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atasheva S, Fish A, Fornerod M, Frolova EI. 2010. Venezuelan equine encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin α/β that obstructs nuclear pore complex function. J. Virol. 84:4158–4171. 10.1128/JVI.02554-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812–9824. 10.1128/JVI.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harel A, Forbes DJ. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319–330. 10.1016/j.molcel.2004.10.026, [DOI] [PubMed] [Google Scholar]
- 59.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282:5101–5105. 10.1074/jbc.R600026200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, McWilliam SM, Cowley JA, Walker PJ. 1994. Complex genome organization in the GNS-L intergenic region of Adelaide River rhabdovirus. Virology 203:63–72. 10.1006/viro.1994.1455 [DOI] [PubMed] [Google Scholar]
- 61.Gubala AJ, Proll DF, Barnard RT, Cowled CJ, Crameri SG, Hyatt AD, Boyle DB. 2008. Genomic characterisation of Wongabel virus reveals novel genes within the Rhabdoviridae. Virology 376:13–23. 10.1016/j.virol.2008.03.004 [DOI] [PubMed] [Google Scholar]
- 62.Allison AB, Palacios G, Travassos da Rosa A, Popov VL, Lu L, Xiao SY, DeToy K, Briese T, Lipkin WI, Keel MK, Stallknecht DE, Bishop GR, Tesh RB. 2011. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Res. 155:112–122. 10.1016/j.virusres.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quan PL, Williams DT, Johansen CA, Jain K, Petrosov A, Diviney SM, Tashmukhamedova A, Hutchison SK, Tesh RB, Mackenzie JS, Briese T, Lipkin WI. 2011. Genetic characterization of K13965, a strain of Oak Vale virus from Western Australia. Virus Res. 160:206–213. 10.1016/j.virusres.2011.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Springfeld C, Darai G, Cattaneo R. 2005. Characterization of the Tupaia rhabdovirus genome reveals a long open reading frame overlapping with P and a novel gene encoding a small hydrophobic protein. J. Virol. 79:6781–6790. 10.1128/JVI.79.11.6781-6790.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.