Abstract
The structural flexibility found in human immunodeficiency virus (HIV) envelope glycoproteins creates a complex relationship between antigenicity and sensitivity to antiviral antibodies. The study of this issue in the context of viral particles is particularly problematic as conventional virus capture approaches can perturb antigenicity profiles. Here, we employed a unique analytical system based on fluorescence correlation spectroscopy (FCS), which measures antibody-virion binding with all reactants continuously in solution. Panels of nine anti-envelope monoclonal antibodies (MAbs) and five virus types were used to connect antibody binding profiles with neutralizing activities. Anti-gp120 MAbs against the 2G12 or b12 epitope, which marks functional envelope structures, neutralized viruses expressing CCR5-tropic envelopes and exhibited efficient virion binding in solution. MAbs against CD4-induced (CD4i) epitopes considered hidden on functional envelope structures poorly bound these viruses and were not neutralizing. Anti-gp41 MAb 2F5 was neutralizing despite limited virion binding. Similar antigenicity patterns occurred on CXCR4-tropic viruses, except that anti-CD4i MAbs 17b and 19e were neutralizing despite little or no virion binding. Notably, anti-gp120 MAb PG9 and anti-gp41 MAb F240 bound to both CCR5-tropic and CXCR4-tropic viruses without exerting neutralizing activity. Differences in the virus production system altered the binding efficiencies of some antibodies but did not enhance antigenicity of aberrant gp120 structures. Of all viruses tested, only JRFL pseudoviruses showed a direct relationship between MAb binding efficiency and neutralizing potency. Collectively, these data indicate that the antigenic profiles of free HIV particles generally favor the exposure of functional over aberrant gp120 structures. However, the efficiency of virion-antibody interactions in solution inconsistently predicts neutralizing activity in vitro.
INTRODUCTION
Human immunodeficiency virus (HIV) is an integrating retrovirus that establishes a permanent chronic infection. Accordingly, the development of preventive measures to block HIV replication prior to host cell entry remains a major goal. It is widely held that a successful HIV vaccine will have to provide sterilizing protection against infection mediated in part by antibodies that recognize conserved domains on HIV envelope proteins (gp120 and gp41) and block viral replication through humoral effector mechanisms such as direct virus neutralization.
gp120 and gp41 form noncovalent complexes that in turn assemble into trimeric spikes on the surfaces of virions; these trimers provide the major target for antiviral agents and antibodies. Areas of broadest vulnerability across virus strains include the CD4 receptor binding site on gp120 (1–6), a high-mannose cluster in gp120 (7–11) found primarily within the subtype B category of viruses (12, 13), and a conserved domain termed the membrane-proximal extracellular region (MPER) located at the base of gp41 (14–18). Certain cognate monoclonal antibodies (MAbs) to these domains exhibit potent cross-reactive neutralizing activity (19, 20). More recently, it has been shown that a series of conserved neutralizing antibody targets are formed by the quaternary structure of the envelope spike in concert with a unique carbohydrate component (21–23). An additional set of conserved epitopes is displayed after gp120 binds to CD4 (24–30) and assumes a constrained structure (18, 31–34). A subset of these CD4-induced (CD4i) epitopes forms a coreceptor binding site that interacts with chemokine receptors (CCR5 or CXCR4) in order to trigger viral entry; additional CD4i epitopes are located elsewhere on gp120 (35). One of the latter (A32) was recently defined as a major target for antibody-dependent cellular cytotoxicity (ADCC) activity mediated by antibodies from HIV-infected persons (36–38).
At the same time, certain types of HIV particles (e.g., pseudoviruses and virus-like particles) are thought to harbor a significant fraction of “misfolded” or “nonfunctional” trimers that fail to present neutralizing domains (39, 40) but present other epitopes in an aberrant context. It has been proposed that antibodies are neutralizing only if they specifically bind functional trimers on virion surfaces and ignore misfolded structures (40, 41). Thus, it has been postulated that the binding of broadly neutralizing MAb b12 or 2G12 to virions identifies intact, functional trimers on virions whereas the binding of MAbs such as those against CD4i epitopes indicates the presence of misfolded, nonfunctional structures such as envelope monomers (40, 41). Conversely, misfolded structures are believed to heavily skew immune responses toward nonneutralizing antibodies (40, 41).
Given the potential heterogeneity of HIV envelope structures, there is ongoing interest in determining the antigenic nature of HIV virions and how this equates with sensitivity to neutralization by cognate antibodies versus binding to nonneutralizing antibodies. To date, analyses of envelope antigenicity on HIV particles have relied heavily on various types of assays in which epitope exposure is measured as a function of virion capture by immobilized MAbs. The captured virions are typically detected by indirect means such as infectivity assays (40, 42–46). This methodology has produced discordant findings regarding the neutralizing versus capture activities of anti-envelope MAbs (43). For example, neutralizing anti-MPER MAbs capture virions only weakly (47), whereas poorly neutralizing antibodies capture particles relatively well (40). The latter effects have been attributed to a preponderance of misfolded trimers on virion surfaces, potentially introduced in part by the capture method, and/or to avidity effects that can produce artificially strong virion binding to immobilized antibodies (40, 45). A means to directly analyze antibody-virion interactions with all reactants in solution could present a more unalloyed picture of surface epitope exposure under key in vivo and/or in vitro conditions.
Fluorescence correlation spectroscopy (FCS) is a method that allows real-time analyses of protein-protein interactions with all reactants continuously in solution (48, 49). FCS has been used to study the associations between HIV-1 integrase and fluorescently labeled oligonucleotides and other high-molecular-mass protein-DNA complexes (50). It has been used to study binding interaction between HIV-1 integrase and LEDGF/P75 (51) but has not yet been applied to protein interactions with HIV surface glycoproteins. A favorable feature of FCS is that fluorescently labeled target proteins are continuously replenished by diffusion into a small observation volume. This allows observation for extended periods of time and, importantly, does not require immobilization of reactants on solid substrates. Random diffusion of the fluorophores results in time-dependent fluorescence intensity fluctuations in the observed volume, which quantify diffusion rates that are inversely proportional to the size of the labeled species. Thus, FCS registers the binding of fluorescently tagged antibodies to free virions as a function of changes in diffusion rate, given that virion-bound antibody exhibits an 8-fold-lower diffusion rate than does free antibody. The fraction of rapidly diffusing species (free antibody) that become more slowly diffusing (bound antibody) reflects the efficiency of virus-antibody binding as governed by the relative antigenicity of a cognate epitope on a virion surface.
In this study, we characterized and employed an FCS-based system to evaluate epitope exposure on HIV particles in solution as evinced by a panel of characterized anti-HIV envelope MAbs. FCS binding patterns were then compared with neutralization activities on matched target virions. This approach indicates that epitopes marking aberrant envelope structures are poorly exposed on CCR5- or CXCR4-tropic virions in solution whereas epitopes marking functional epitopes are efficiently presented. At the same time, the susceptibility of HIV to antibody-mediated neutralization is not generally predicted by the efficiency of antibody-virion binding in solution.
MATERIALS AND METHODS
Production of pseudoviruses and infectious molecular clones.
To produce the HIV-1 NL4-3 infectious molecular clone, HEK293T cells were transfected with a full-length pNL4-3 HIV genome expression plasmid obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID (1), using FuGENE (Promega, Madison, WI) at a reagent/DNA ratio of 3:1. To produce the infectious molecular clone of transmitted/founder HIV-1AD17 virus (52), HEK293T cells were transfected with the AD17 plasmid (kindly provided by B. Hahn, University of Pennsylvania) at a FuGENE-to-DNA ratio of 3:1. The HIV-1JRFL, HIV-1HXB2, and HIV-1BaL pseudoviruses were produced by cotransfection of HEK293T cells with an Env-deficient HIV-1 backbone plasmid, pNL4-3-ΔE-EGFP (53), along with Env expression plasmid (54, 55) pCAGGS-JRFL or pCAGGS-HXB2 (kindly provided by J. Binley, Torrey Pines Institute of Molecular Studies, San Diego, CA) or pHIV-1-BaL 0.1 (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID). Control particles lacking Env (delE) were produced by transfection with pNL4-3-ΔE-EGFP backbone alone.
HIV-1AD17 viruses and HIV-1BaL and HIV-1NL4-3 infectious molecular clones were also produced in SupT1-R5 cells. The relevant plasmids were first used to transfect HeLa/Tat cells using FuGENE at a reagent/DNA ratio of 3:1. The next day, SupT1-R5 cells were added to the transfected HeLa/Tat cells and cocultured overnight. The infected SupT1-R5 cells in suspension were then separated from the adherent HeLa/Tat cells and expanded to produce viruses over 3 days, at which time virus-containing supernatants were harvested.
Supernatants were concentrated approximately 10-fold using PEG-it virus precipitation solution (System Biosciences, Mountain View, CA) overnight at 4°C. The antigen content of all pseudovirus preparations was quantified using p24 and gp120 antigen capture enzyme-linked immunosorbent assays (ELISAs). Infectivity was established using standardized procedures (12) and quantified as a function of 50% tissue culture infective dose (TCID50) in TZM-bl cells.
To assess the amounts of free viral antigen in the final preparations, concentrated virions were incubated at 37°C for 3 h in the presence or absence of soluble CD4 (sCD4) (100 μg/ml), and then pelleted. gp120 and p24 levels in the supernatants and pellets were assayed by antigen capture ELISA. Less than 10% of the total gp120 and 10% of the total p24 antigen were routinely detected in the supernatant fractions of all preparations in the presence and absence of sCD4 treatment.
Antibodies.
MAbs b12, 2G12, and 2F5 were purchased from Polymun Scientific (Vienna, Austria). MAbs PG9, 17b, A32, C11, 19e, and F240 were expressed from plasmid clones in HEK293T cells using an IgG1 backbone for heavy-chain variable regions and either a κ- or λ-chain expression vector for light-chain variable regions. MAbs were purified from culture supernatants by protein A chromatography. The humanized monoclonal anti-respiratory syncytial virus antibody Synagis (MedImmune LLC, Gaithersburg, MD) and nonspecific human plasma IgG1 (Calbiochem, La Jolla, CA) were used as negative controls. All MAbs were fluorescently labeled and purified with an Alexa Fluor 647 monoclonal antibody labeling kit (Invitrogen, Molecular Probes, Eugene, OR). Briefly, the Alexa Fluor 647 reactive dye has a succinimidyl ester moiety that reacts efficiently with primary amines of antibody to form stable dye-protein conjugates. Each labeling reaction was performed with 100 μg of a monoclonal antibody. The labeled antibody was separated from unreacted dye by centrifugation through a spin column at 1,100 × g for 5 min. Recovered antibodies were dialyzed against phosphate-buffered saline as necessary. Dye-to-protein ratios were determined by measuring absorbance at 280 nm (protein) versus 650 nm (dye). Ratios varied from 3 to 5 depending on the antibody. Labeled antibodies were quantified by a UV-visible (UV-vis) spectrometer (Nanodrop 2000; Thermo-Scientific, Wilmington, DE).
FCS measurements.
All FCS experiments used virus preparations diluted to 10 μg/ml p24 equivalent. gp120-to-p24 antigen ratios were typically 1:10 to 1:50; for the transmitted/founder HIV-1AD17 viruses, the ratios were 1:100 and 1:200 with viruses grown in SupT1-R5 and HEK293T cells, respectively. The 10-μg/ml p24 equivalent value typically corresponded to TCID50/ml values in the range of 200,000 to 650,000 (BaL), 200,000 to 500,000 (JRFL), 100,000 to 300,000 (HXB2), 200,000 to 1,000,000 (NL4-3), 100,000 to 650,000 (HIVAD17 in SupT1-R5 cells), and 600,000 to 1,000,000 (HIVAD17 in 293T cells) as determined by TCID50 assay (12). The reaction buffer for FCS measurements contained Dulbecco modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, antibiotics, 0.1 mg of G418 (Gibco-BRL)/ml, and 0.05 mg of hygromycin B/ml (complete medium). Pseudoviruses with 1-μg p24 equivalent concentrations in 100 μl reaction buffer were first treated for 90 min at 37°C with 1.5 μl of nonspecific (Calbiochem) IgG1 (to produce a final IgG1 concentration of 100 μg/ml in reaction mixture) to block nonspecific binding. This was followed by addition of 1 μl Alexa Fluor 647-conjugated MAbs to produce final reaction concentrations of MAbs in the range of 4.5 to 6.6 μg/ml. MAbs were allowed to interact with virions for 90 min at 37°C. For the study of CD4-induced conformational changes, viruses were incubated for 90 min at 37°C with 1.5 μl of sCD4 (Biogen, Cambridge, MA) to achieve a final reaction concentration of 100 μg/ml, along with nonspecific IgG1 as described above. An aliquot (11 μl) of a given reaction mixture was loaded onto a glass coverslip sample chamber and sealed for spectroscopic measurements. FCS measurements were performed using a confocal microscope (MicroTime 200; PicoQuant, Berlin, Germany). The FCS system has single-molecule detection sensitivity. The excitation laser (λex, ∼635 nm) was reflected by a dichroic mirror to a high-numerical-aperture (NA) oil objective (100×; NA, 1.3) and focused onto the solution sample. The fluorescence was collected by avalanche photodiodes through a dichroic beam splitter and a long-pass (655-nm; Chroma) filter, thus eliminating the scattered excitation light and collecting the fluorescence from the Alexa Fluor 647 probes in the region of interest. FCS measurements were performed in a constant detection volume (1 fl). The lower and upper limits of detection of the FCS system are 10 pM and 100 nM, respectively (56). PicoQuant Symphotime software was used to generate the autocorrelation curves and in analyzing the data. The autocorrelation function of the fluorescence intensities is given by the product of the intensity at time t, I(t), with the intensity after a delay time τ, I(t + τ), averaged over a large number of measurements. The time t refers to the actual time as the intensities are observed. We have collected the data for each sample for 60 s. The delay time τ is the difference in real time between measurement of I(t) and I(t + τ), typically in the range from 10−2 to 102 ms. If the intensity fluctuations are slow compared to τ, then I(t) and I(t + τ) will be similar in magnitude. That is, if I(t) is larger than the average intensity ⟨I⟩, then I(t + τ) is likely to be larger than ⟨I⟩. If the intensity fluctuations are fast relative to τ, then the value of I(t) and I(t + τ) will not be related. The most commonly used autocorrelation function is given by
| (1) |
where G(τ) is the autocorrelation function of fluorescence fluctuations. The data are typically the number of photon counts in a given time interval, typically about a microsecond, which are due to a small number of fluorophores. The intensity is dependent on the number of photons detected from each fluorophore in a given period of time. The autocorrelation function for a diffusional model is given by
| (2) |
where G(τ) is the autocorrelation function of fluorescence fluctuations, G(0) is the amplitude when the delay time τ is 0, and D is the diffusion coefficient. The diffusion coefficient for the ith species traversing a three-dimensional (3D) Gaussian volume with radius ω0 and half-axial height z0 is given by
| (3) |
The autocorrelation of multiple diffusing species is a linear combination of the autocorrelations for each species separately. To fit with two species with the same brightness of detected photon per time interval, the diffusion model equation becomes
| (4) |
The values of N1/(N1 + N2) and N2/(N1 + N2) are taken to represent the percentage of diffusing free MAbs and bound MAbs to virions, respectively. The diffusion coefficients of fluorescently labeled MAbs dramatically decrease as a consequence of virion binding. Autocorrelation analyses of the fluorescence intensity fluctuations (equations 2 to 4) reveal the fraction of labeled MAb that acquires a lower diffusion rate in the observation volume when virions are present. The predicted diffusion coefficient (D1) of a fluorescently labeled immunoglobulin molecule (60 μm2/s) (57) is significantly higher than the coefficient (D2) predicted for immunoglobulin bound to a 100-nm viral particle (5 μm2/s) modeled as a sphere with a 50-nm radius of gyration (r), using the Stokes-Einstein relation, D = kBT/(6πηr), where kB is the Boltzmann constant, T is absolute temperature, and η is the solvent viscosity (0.01 g/cm · s for water at 293 K). These predicted values matched coefficients measured during routine calibrations with free immunoglobulins (61 ± 4 μm2/s) or with fluorescently labeled calibration beads (Invitrogen) known to be 100 nm in diameter (7 ± 2 μm2/s).
The percentage of total MAb (at given test concentrations) that shifts into the more slowly diffusing species which is the virion-bound fraction (N2/N1 + N2) reflects the relative magnitude of cognate epitope exposure in the target population of virions. Epitope exposure is taken as “positive” if MAb binding on a target virus is above what is seen with delta Env particles and negative-control antibodies. The final concentrations of labeled antibodies tested were based on the upper limit of fluorophore concentration approximating 100 nM (56). Accordingly, labeled MAbs at approximately 5-μg/ml (33 nM) final concentrations were used for FCS, given dye/protein rations of 3 to 5. The precise amounts of MAbs used in each experiment are indicated elsewhere in the text.
The FCS system was also characterized for the detection of immune complexes that might be created if gp120 was shed from the virions during the observation period. Protein complexes were generated experimentally by incubating 100 μg/ml of BaL gp120 with 6 μg/ml of b12 or 2G12. FCS analyses determined that these complexes exhibit diffusion coefficients in the range of 39 to 43 μm2/s, distinct from predicted diffusion coefficients for free antibody and virion-bound antibody. No species with such diffusion coefficients were detected in any of our experiments.
Neutralization assays.
Neutralizing activity was determined using TZM-bl target cells as previously described (12). This assay measures Tat-driven luciferase expression (relative luminescence units [RLU]) following a single round of virus infection. To enable matched comparisons with FCS measures, neutralization assays were performed with Alexa Fluor 647-labeled antibodies and workable amounts of viruses approaching what was used in the FCS reaction chambers. Briefly, TZM-bl cells (8,300/well) were cultured overnight in 96-well microtiter plates at 37°C. Amounts of the HIVAD17 infectious virus; HIV-1NL4-3 and HIV-1BaL molecular clones; and HIV-1JRFL, HIV-1BaL, or HIV-1HXB2 pseudovirus equal to 10 μg/ml p24 (final TCID50 values of 25,000 for HIVAD17, 30,000 for NL4-3, 22,000 for JRFL, 20,000 for BaL, and 5,200 for HXB2) were incubated with 3-fold serially diluted concentrations of test antibodies starting at 50 μg/ml for 1 h at 37°C and then added to the cells. Assay controls included replicate wells of TZM-bl cells alone or TZM-bl cells with virus in the absence of antibody. After 48 h at 37°C in a CO2 incubator, Bright-Glo reagent (Promega, Madison, WI) was added to the cells and RLU were measured using a Victor 3 luminometer (PerkinElmer, Waltham, MA). Values obtained in experimental wells were corrected for background signal by subtracting RLU measurements in the absence of virus. Percent infection or percent inhibition was calculated by dividing the background-corrected RLU for each experimental well by the corrected RLU for control wells containing only cells and virus. All assays were carried out in triplicate.
RESULTS
Our approach was to examine antibody-virion interactions in solution using a selection of virions representing different coreceptor tropisms, different sensitivities to neutralization, and direct relevance to antiviral systems typically used to assess humoral anti-HIV immunity and antiviral activities. Toward this end, we assessed pseudovirions expressing the CCR5-tropic JRFL envelope (a “tier 2” difficult-to-neutralize envelope); pseudovirions expressing the CCR5-tropic BaL envelope (a “tier 1” easily neutralized envelope); pseudovirions expressing the HXB2 CXCR4-tropic envelope (a “tier 1” easily neutralized envelope); and an infectious clone, NL4-3, which is highly related (∼98% envelope sequence homology) to the HXB2 envelope sequence. The latter allowed us to compare highly related envelope sequences on infection-competent viruses with those on replication-defective pseudovirions.
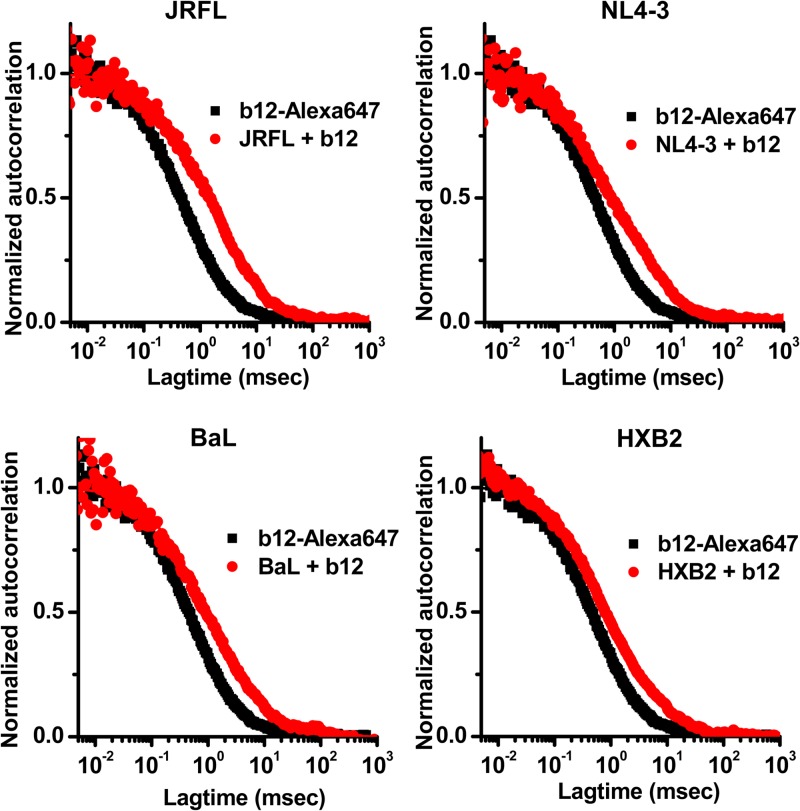
FCS analyses of epitope presentation on virion-associated HIV envelope were carried out with human anti-HIV envelope monoclonal antibodies (MAbs). Initial tests of the system were conducted with MAb b12, which recognizes a conserved neutralizing epitope in the CD4 binding site on gp120 (4, 58) that is preferentially exposed on functional envelope spikes (40). As expected, autocorrelation plots of Alexa Fluor 647-labeled MAb b12 (Fig. 1) in the absence of virions could be fitted to a single-species diffusion model, consistent with a diffusion coefficient of 65 μm2/s for free immunoglobulin (57). However, in the presence of NL4-3 virus or JRFL, BaL, or HXB2 pseudovirus, autocorrelation curves consistently fit a two-species diffusion model in which one species exhibited a high diffusion coefficient (e.g., 65 μm2/s) and the other showed a lower diffusion coefficient of approximately 8 μm2/s (Fig. 1). The latter value agrees with what is predicted for an antibody bound to a 100-nm retroviral particle (5 μm2/s; see Materials and Methods). In the case of JRFL pseudoviruses, the amount of total antibody exhibiting the lower diffusion rate was 56%; with BaL pseudoviruses, it was 44%; with HXB2 pseudoviruses, it was 40%; and with NL4-3 viruses, it was 48% (Fig. 1).
FIG 1.
Correlation spectroscopy curves for the interaction of Alexa 647-labeled MAb b12 with various types of virus particles. Autocorrelation curves obtained with antibody (6.6-μg/ml final concentration) alone are shown in black; those obtained with antibody in the presence of the indicated viruses are shown in red. The experiment was repeated three times with similar results.
Varying the number of virions in solution confirmed that the slowly diffusing MAb species signals the formation of antibody-virion complexes. A fixed concentration (6.6-μg/ml final concentration) of MAb b12 was incubated with serial amounts (0.1 μg/ml to 50 μg/ml p24 equivalents) of JRFL or BaL pseudovirus or the NL4-3 infectious virus. As shown in Fig. 2, dose-dependent MAb binding was detected above 1-μg/ml p24 equivalent concentrations of each virus. No significant MAb binding was detected between the 0.1- and 1-μg/ml p24 antigen equivalents. At the highest achievable concentrations of virus (50 μg/ml p24 equivalents), roughly 50 to 70% of MAbs adopted the low diffusion rate indicative of virion binding.
FIG 2.
The fraction of antibody adopting a lower diffusion rate in FCS is virion dose dependent. The indicated viruses were titrated as a function of p24 antigen equivalents (see Materials and Methods) and incubated with a fixed quantity of Alexa Fluor 647-labeled MAb b12 (6.6 μg/ml). The relative fraction of MAb b12 that exhibits a lower diffusion coefficient (∼8 μm2/s) as a result of virion binding in solution (see Materials and Methods) is shown. All experiments were repeated three times, and average values are shown. Error bars indicate standard deviations.
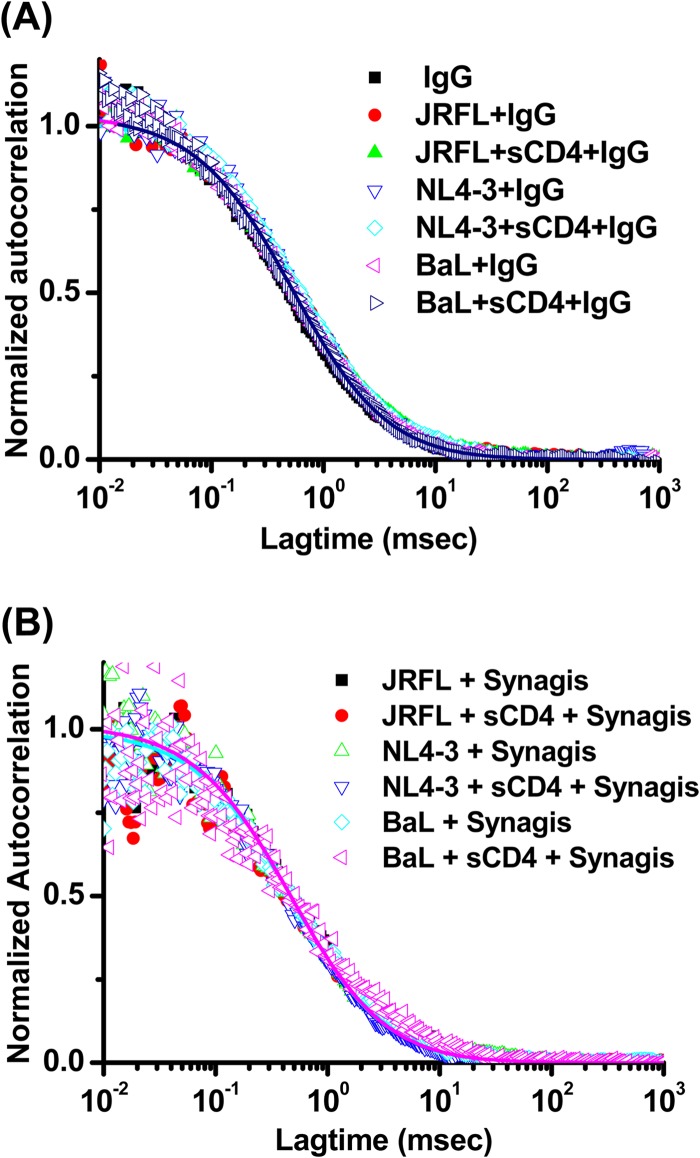
To establish the signal-to-noise dynamics of the system, we assessed the binding of Alexa Fluor 647-labeled, nonspecific human IgG1 or irrelevant Synagis MAb (6-μg/ml final concentrations) to the viruses analyzed above (Fig. 3). When tested in the absence of virus, both reagents fit single-species diffusion models with 65-μm2/s diffusion coefficients, similar to MAb b12. However, in the presence of viruses the autocorrelation plots of the antibodies were unchanged; no detectable fraction of antibody exhibited a lower diffusion coefficient. Treatment of virions with sCD4 (100 μg/ml) to trigger structural rearrangements in envelope produced the same results. These data indicated that nonspecific virion interactions in solution are beneath the level of detection in the FCS system.
FIG 3.
Absence of nonspecific antibody interactions with virions in FCS. Fluorescence correlation plots with JRFL or BaL pseudovirus and NL4-3 virus in the presence or absence of 100 μg/ml sCD4 are shown for Alexa Fluor 647-labeled nonspecific human IgG1 (A) or for Alexa Fluor 647-labeled Synagis (6 μg/ml) (B). The experiment was repeated with similar results.
A second set of experiments was carried out to assess the specificity of apparent MAb-virion binding signatures in solution. Since MAb b12 binds the CD4 binding site, soluble CD4 (sCD4) was expected to competitively reduce the fraction of MAb that adopted the lower diffusion rate, if the latter was due to epitope-specific interactions. Figure 4 shows analyses of Alexa Fluor 647-labeled MAb b12 (6.6-μg/ml final concentration) with JRFL, NL4-3, or BaL particles in the presence of sCD4 concentrations from 0 μg/ml to 100 μg/ml. In the absence of sCD4, the autocorrelation curves again fit a two-species diffusion model in which the more slowly diffusing species (∼8 μm2/s) was taken as virion-bound antibody. This species represented 56%, 48%, and 44% of the total antibody for JRFL, NL4-3, and BaL envelope particles, respectively, in accordance with the experiments shown in Fig. 1. At an sCD4 concentration of 100 μg/ml, the autocorrelation plots obtained in the presence of all three virus types reflected only a single species with diffusion coefficients matching free antibody (65 μm2/s). This profile suggested that the binding of MAb b12 to virions was extensively blocked by sCD4 in accordance with its epitope specificity. Interestingly, the dose-effect curves seemed biphasic. There was a sharp decrease in bound MAb from 0 to 20 μg/ml sCD4, but from 20 to 100 μg/ml sCD4, the decline was more gradual. This biphasic curve suggests that the test viruses express two populations of gp120 that bind b12 and/or CD4 in a different manner. This possibility could not be reliably elaborated with the FCS configuration developed here but warrants more detailed analyses with ancillary methods.
FIG 4.
Effects of soluble CD4 on the binding of anti-gp120 MAbs to virus particles in solution. (A) Alexa-647 labeled MAb b12 (6.6 μg/ml) was tested for binding to virions treated for 90 min at 37°C with sCD4 concentrations ranging between 0 and 100 μg/ml. (B) Alexa 647-labeled MAb 17b (5.5 μg/ml) was tested under the same conditions. The relative fraction of MAb that adopts a lower diffusion coefficient (∼8 μm2/s) as a result of virion binding (see Materials and Methods) is shown. All experiments were repeated three times, and average values are shown. Error bars indicate standard deviations.
Converse experiments were conducted with Alexa 647-labeled MAb 17b (5.5-μg/ml final concentration), which recognizes a CD4i epitope on gp120 that is stabilized by CD4 binding. Tests were conducted with the JRFL, NL4-3, and BaL envelope particles in the absence or presence of serial sCD4 concentrations ranging from 0 to 100 μg/ml (Fig. 4B). In the absence of sCD4, a small fraction of MAb 17b (10 to 18% depending on the virus) adopted the lower diffusion coefficient indicative of virion binding. However, this fraction increased with sCD4 concentration; at 100 μg/ml sCD4, roughly 54, 55, and 39% of MAb 17b reflected virion attachment with JRFL, NL4-3, and BaL envelope particles, respectively. Such dose-effect trends are entirely consistent with previous studies of the 17b epitope using various HIV envelope antigens and infectivity systems (26, 59). Taken together, the data obtained with MAbs 17b and b12 indicated that the FCS assay system accurately reflects the expected exposure patterns of functional receptor binding sites on virus particles in solution.
FCS with a broader panel of Alexa 647-labeled anti-envelope MAbs was used to examine the antigenicity patterns of conserved epitopes on various viruses. These experiments included the subtype B, CCR5-tropic transmitted/founder HIV variant AD17 (52) produced from HEK293T cells transfected with an infectious molecular clone (see Materials and Methods). Test antibodies included MAb 2G12, against a carbohydrate cluster on gp120 (10); MAb PG9, a potent broadly neutralizing antibody against a conserved epitope involving glycans and the variable loops of gp120 (21, 60–62); MAb A32, against a conformational CD4i epitope comprising the gp120 C1 domain that is hidden on intact trimers (37); MAb 19e, against a hybrid CD4i epitope (63); MAb C11, against a conserved gp120 epitope that is poorly exposed on intact trimers (64); MAb 2F5, against the membrane-proximal external region (MPER) of gp41 (17); and F240, against a cluster 1 epitope in gp41 (65, 66).
This MAb panel comprised divergent anti-HIV activities, with MAbs 2G12, PG9, 2F5, and b12 characterized as typically neutralizing; F240 characterized as poorly neutralizing to nonneutralizing (65, 67, 68); and MAbs C11, A32, and 19e characterized as entirely nonneutralizing (63, 69). Thus, the binding of MAb b12, 2G12, PG9, or 2F5 was expected to mark the presence of functional envelope structures; the binding of MAb F240, A32, or C11 was expected to signal the presence of aberrant epitope exposures within misfolded structures on virion surfaces. Similarly, the binding of MAb 17b or 19e in the absence of CD4 should signal aberrant structures. Synagis and nonspecific human IgG1 served as negative controls, which exhibited no binding to any of the test viruses (Fig. 5).
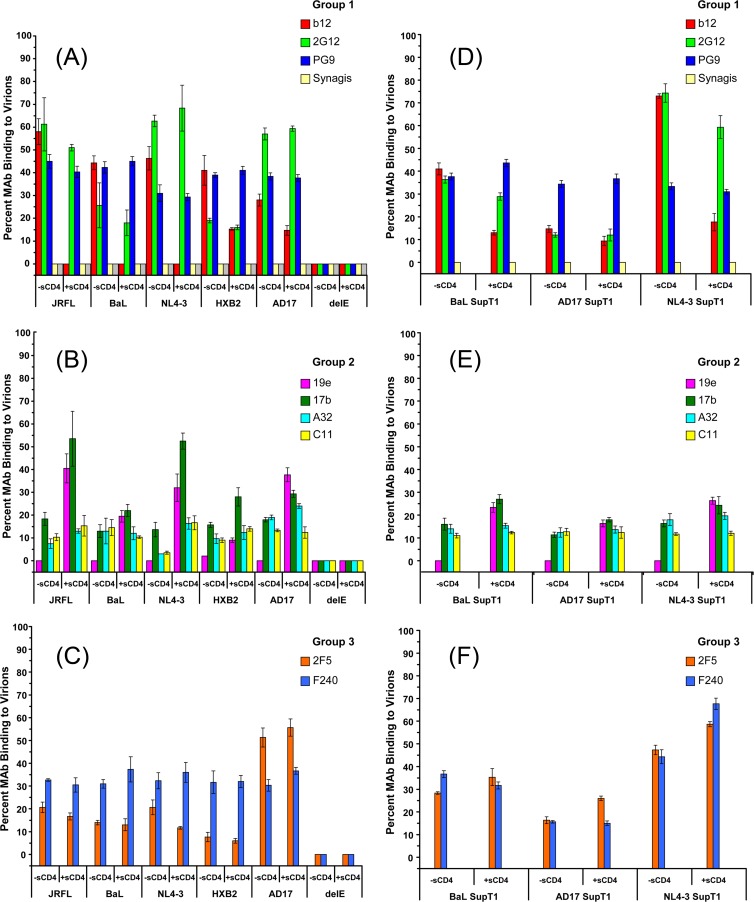
FIG 5.
Anti-envelope MAb binding to viral particles in solution. Viruses were produced in either HEK293T cells (A to C) or SupT1 cells (D to F) and tested at 10-μg/ml p24 equivalent concentrations with or without 100 μg/ml sCD4. The indicated groups of MAbs were tested at 4.5- to 6.6-μg/ml final concentrations (see Materials and Methods). Binding data for MAbs b12 and 17b from Fig. 4 with JRFL, BaL, and NL4-3 viruses are included in panels A and B, respectively. The relative fraction of MAb that adopts a lower diffusion coefficient (∼8 μm2/s) as a result of virion binding (see Materials and Methods) is shown. All experiments were repeated at least three times, and average values are shown. Error bars indicate standard deviations.
We verified that the test MAbs exhibited little or no envelope-independent nonspecific interactions with virion surfaces in solution. This was assessed with “delta env” viruses (delE), produced in the absence of an envelope expression construct (see Materials and Methods). Test antibodies were labeled with Alexa Fluor 647 and tested at 4.5- to 6.6-μg/ml final concentrations against delE virions in the presence and absence of 100 μg/ml sCD4. In each case, the FCS data fit only a single-species diffusion model with the signature diffusion coefficient (65 μm2/s) of free antibody. Specifically, an undetectable fraction of each MAb adopted a lower diffusion rate indicative of virion binding (Fig. 5).
MAb binding patterns using pseudoviruses or molecular clones produced in HEK293T cells are shown in Fig. 5, grouped according to whether they bind functional gp120 epitopes (group 1), bind epitopes expected to be hidden on intact trimers in the absence of CD4 (group 2), or recognize gp41 epitopes (group 3). Among the group 1 MAbs, 2G12 adopted the lower diffusion coefficient signaling virion binding in all cases, ranging from over 60% with JRFL pseudoviruses to roughly 20% with HXB2 pseudoviruses. These measures were not appreciably altered by the presence of sCD4. Notably, the MAb 2G12 binding fraction was lower on HXB2 virions than on the NL4-3 infectious virus despite the strong envelope sequence homology shared by the two types of particles (Fig. 5A). As noted above (Fig. 4), MAb b12 binding was also observed with all virus types in FCS, with the AD17 virus showing the lowest b12 antigenicity. MAb binding was reduced in the presence of 10 μg/ml sCD4 in all cases (Fig. 4 and 5A). Notably, the sCD4 inhibitory effect was lower on the AD17 infectious virus and on HXB2 pseudovirions than on the other virus types grown in HEK293T cells. As with MAbs 2G12 and b12, significant fractions of PG9 exhibited virion binding (Fig. 5A), ranging from 30% to 50% depending on the virus. The fractions of virion-bound antibody were not significantly altered in the presence of sCD4.
Only minor fractions (≤20%) of group 2 antibodies exhibited virion-bound diffusion coefficients (Fig. 5B). The binding signals for MAbs C11 and A32 were not substantially increased in the presence of sCD4 as predicted by previous studies (70). MAb 19e showed undetectable virion binding in the absence of sCD4 (Fig. 5B) but measurable binding in the presence of sCD4 (Fig. 5B). The CD4-induced binding effect was least pronounced on HXB2 pseudovirions. In general, these patterns agree with previous evidence that this MAb is specific for a hybrid gp120-CD4 epitope (63). Minor but detectable fractions of MAb 17b exhibited virion-bound diffusion coefficients in the absence of sCD4 (Fig. 5B). As expected, the fraction of virion-bound antibody increased in the presence of sCD4. This increase varied among viruses and was lowest with HXB2.
Between the group 3 MAbs, 2F5 exhibited minor binding to JRFL, BaL, HXB2, and NL4-3 virions but substantial binding to AD17 (Fig. 5C). In no case was the fraction of MAb 2F5 substantially altered by sCD4 treatment. A consistent fraction (30% and 40%) of MAb F240 bound to all viruses in a manner that was not altered by the presence of sCD4.
The differences in MAb binding between the HXB2 pseudovirus and the NL4-3 infectious virus (Fig. 5) suggested that the antigenicity of virus particles in solution could vary according to production conditions. To explore this question further, we generated viral particles using another method capable of yielding enough material to facilitate the FCS analyses. Infectious molecular clones of BaL, NL4-3, and AD17 (designated BaL SupT1, NL4-3 SupT1, and AD17 SupT1, respectively) were generated in the SupT1-R5 suspension cell line (see Materials and Methods) and tested against the groups of MAbs. As shown in Fig. 5D, the efficiency of group 1 MAb binding to the BaL SupT1 and NL4-3 SupT1 viruses was similar to what was observed with the corresponding HEK293T-derived viruses (Fig. 5A). MAbs 2G12 and b12, but not PG9, showed substantially less efficient binding to the AD17 SupT1 virus than to its counterpart produced in HEK293T cells. The binding of group 2 MAbs to the BaL SupT1 virus (Fig. 5E) was similar to what was observed with the corresponding pseudovirus (Fig. 5B). The efficiency of MAb 19e binding was increased by sCD4 treatment, indicating that a hybrid CD4i epitope was formed on the virion surfaces. However, even after sCD4 treatment the efficiency of binding of SupT1-R5-derived viruses to MAb 19e was less than what was seen with the HEK293T-derived counterparts. Similarly, the ability of sCD4 to enhance the efficiency of MAb 17b binding to SupT1-R5-derived viruses (Fig. 5E) was less than what was observed with the HEK293T-derived viruses. The SupT1-R5-derived viruses also produced somewhat different group 3 MAb binding profiles (Fig. 5F) than those produced using HEK293T cells (Fig. 5C). Group 3 MAb binding was generally lower on AD17 SupT1 viruses than on viruses produced in HEK293T cells. Conversely, MAb 2F5 and MAb F240 bound BaL SupT1 and NL4-3 SupT1 viruses, respectively, somewhat more efficiently than their HEK293T cell-derived counterparts. It was also noteworthy that MAb F240 binding to NL4-3 SupT1 was significantly increased in the presence of sCD4 (P < 0.01; t test), whereas such conditions did not induce significant changes in F240 binding to the BaL SupT1 and AD17 SupT1 viruses or to NL4-3 viruses grown in HEK293T cells (P > 0.05).
It was important to determine that the FCS binding signals comprised interactions with infectious virions. This was accomplished by testing the MAbs for their ability to neutralize the same viruses used in FCS, following the rationale that a binding antibody must be interacting with infectious particles if it inhibits infection. Accordingly, we performed neutralization assays that were deliberately aligned with the FCS studies, using matched virus preparations and Alexa 647-labeled antibodies. Given their generally superior antigenicity, the HEK293T cell-derived pseudoviruses and the NL4-3 virus were used for these experiments. Neutralizing activity (Fig. 6) was assessed in a standardized in vitro assay using TZM-bl target cells (12). To enable precise cross-comparisons, the concentration ranges of labeled antibodies and amounts of virus tested paralleled what was used in FCS (e.g., Fig. 5).
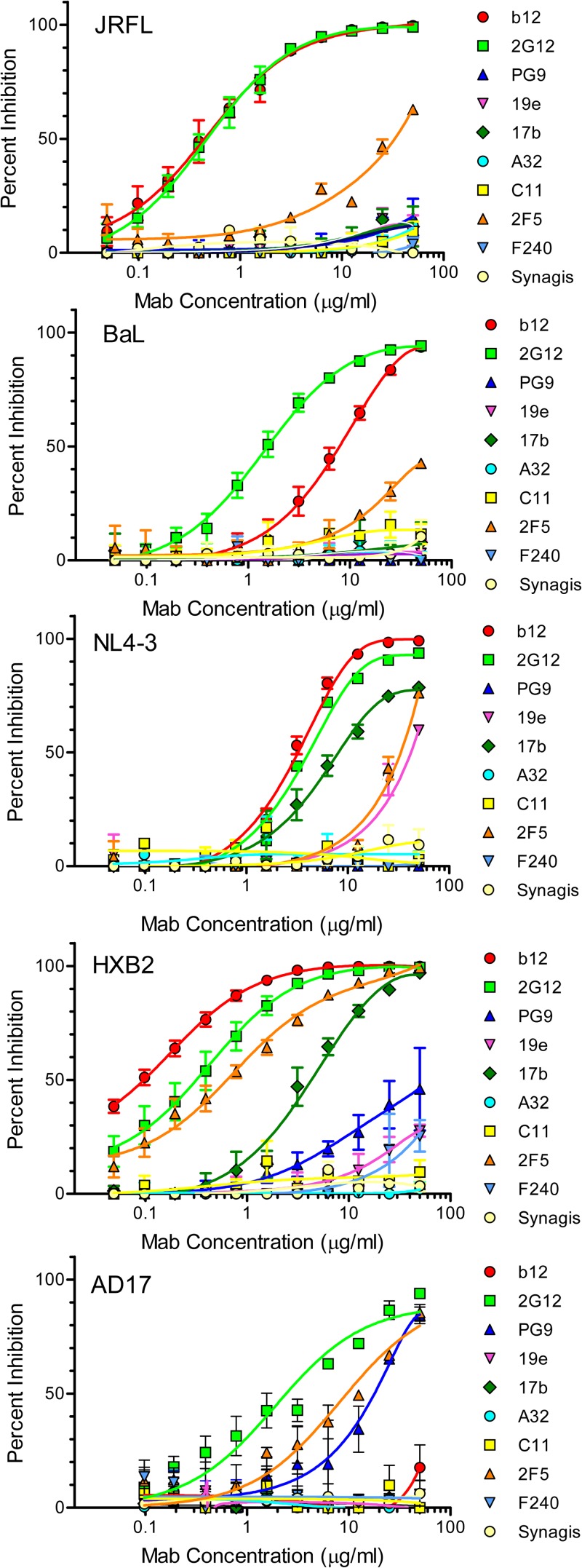
FIG 6.
Neutralizing activity of anti-envelope MAbs on JRFL, BaL, or HXB2 pseudovirus or AD17 or NL4-3 virus in vitro. Assays were carried out with the indicated viruses and concentrations of Alexa Fluor-labeled MAbs using TZM-bl cells as target cells (see Materials and Methods). All assays were performed in triplicate. Average values are shown; error bars indicate standard deviations.
Among the group 1 MAbs, 2G12 neutralized all viruses, MAb b12 neutralized all viruses except AD17, and MAb PG9 neutralized only the AD17 virus and HXB2 pseudovirus (Fig. 6). The lack of MAb PG9 neutralizing activity against the JRFL and BaL pseudoviruses agrees with previous tests with viruses expressing these envelopes (21). Among the group 2 MAbs, C11 and A32 did not neutralize any of the test viruses (Fig. 6) in accordance with their inefficient virion binding apparent in the FCS analyses (Fig. 5). MAb 17b neutralized the CXCR4-tropic HXB2 pseudovirus and NL4-3 infectious virus but not the CCR5-tropic pseudoviruses (Fig. 6). This neutralization bias did not track with unusually strong MAb binding to unliganded NL4-3 or HXB2 virions (i.e., those tested in the absence of sCD4) compared to the other virus types. For example, a comparison of unliganded viruses showed that the percentages of virion-bound MAb were equivalent (less than 20%) with the BaL pseudovirus and the NL4-3 virus. However, only the latter virus was neutralized (Fig. 6). Labeled MAb 19e also exhibited neutralizing activity against HXB2 and NL4-3, albeit less potent than that of MAb 17b (Fig. 6). Between the group 3 MAbs, 2F5 neutralized all viruses in the panel with various degrees of potency (Fig. 6), whereas F240 neutralized only the HXB2 pseudovirus to a minor extent.
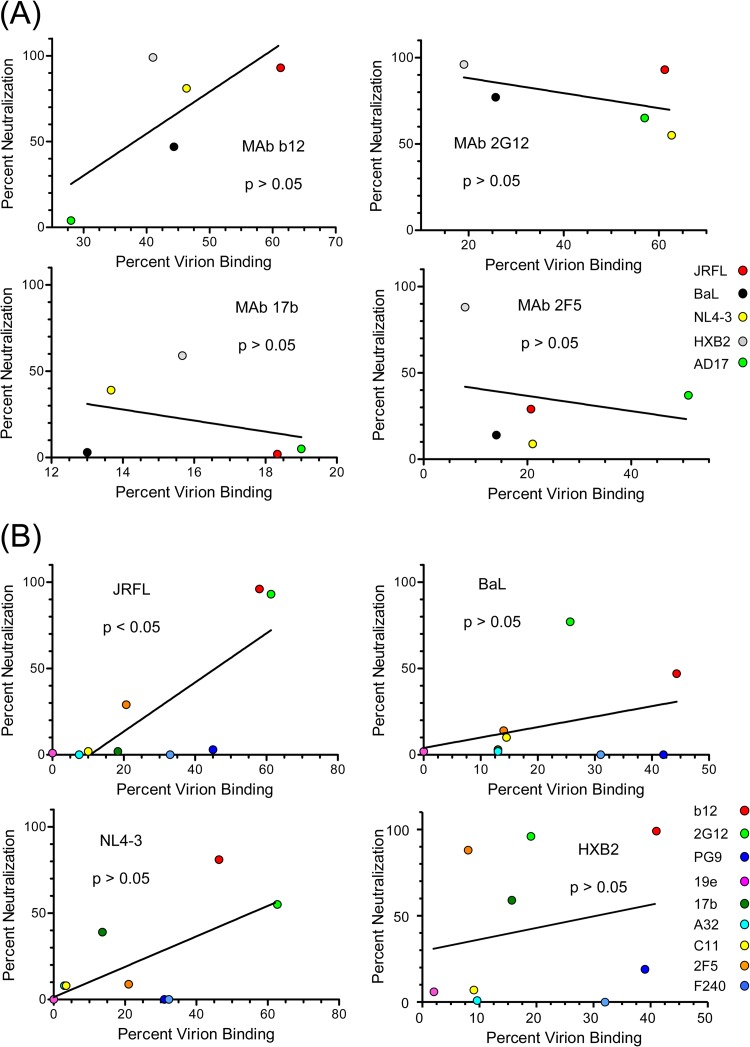
The relationship between the efficiency of MAb binding to soluble virions and neutralizing activity was evaluated in two ways. First, MAbs that neutralized more than one test virus were used to compare FCS binding measures (percent antibody bound) with corresponding neutralizing potencies (percent neutralization observed with the antibody concentration used in FCS). As shown in Fig. 7A, none of the MAbs produced a clear relationship between neutralizing potency and the capacity to bind virions in solution. Second, viruses that were neutralized by multiple MAbs were used to compare FCS binding measures (percent antibody bound) with neutralization sensitivity (percent neutralization observed with the antibody concentration used in FCS). As shown in Fig. 7B, the neutralizing effects of test MAbs against JRFL were directly and significantly related to the fraction of antibody that bound to virions in solution. Although comparable trends were seen with the other viruses, they were not statistically significant.
FIG 7.
Comparisons of neutralizing activity with binding efficiency in FCS. Data from Fig. 5 for the indicated MAbs were compared with neutralizing effects at the concentrations used for FCS. The latter values were derived from the dose-effect curves in Fig. 6. Relationships were tested by linear regression analyses; a P value of <0.05 was taken as significant. (A) For each MAb, percent binding to the indicated viruses was compared with neutralizing potency. (B) For each virus, MAb binding in FCS was compared with neutralization effect. Antibodies are color coded as in Fig. 6.
DISCUSSION
It is widely held that HIV virions express a mixture of functional and misfolded/nonfunctional envelope structures (39–41) that determine their overall antigenicity and susceptibility to antiviral antibodies. However, it is not entirely clear how these antigenic profiles link to neutralization sensitivity, mechanisms of neutralization, or binding to antibodies that might mediate other (“nonneutralizing”) forms of humoral immunity. Answers to these questions can provide important guidance for designing antiviral strategies such as vaccines to raise protective antibody responses. Accordingly, there have been intensive efforts to address this issue by probing soluble versions of HIV envelope proteins, cell-cell fusion systems (70–73), or captured virions (40, 42–46) with anti-envelope antibodies thought to mark key antiviral targets.
In general, such efforts face several caveats. Analyses of soluble glycoproteins may not reflect the impact of cell surface proteolysis (74), interactions with lipid bilayers, and/or matrix contacts (75–77) on trimeric envelope structure. Similarly, cell-cell fusion systems may not replicate the envelope-matrix interactions that occur on budding virions (78) or represent the spectrum of envelope structures that occurs on free virions. Data from virus capture assays may be biased by mechanical forces (e.g., via centrifugation) applied to virions, avidity effects, and/or indirect detection of antibody-virion complexes (40, 45). Solution-based antibody binding was previously used to explore virion-associated envelope antigens (79); however, the method involved washing and fixation steps followed by negative-stain electron microscopy to image virion-bound antibody. Thus, data from this approach are derived from visual surveys of a limited range of selected particles.
Here, we employed a unique FCS-based method that probes the antigenicity of infectious virion populations with all reactants continuously in solution. This approach not only mitigates the aforementioned caveats but also estimates in vivo and/or in vitro conditions where contacts between soluble HIV particles and cognate antibodies occur during infection.
In initial tests with MAb b12, FCS readily distinguished bound and unbound antibody in the observation volume as a function of relatively lower versus higher diffusion rates, respectively (Fig. 1). This reflects the fact that FCS obviates the need for manipulations to remove unbound antibody. The system also distinguishes specific and nonspecific antibody interactions as evinced by findings that the MAb b12 binding signal (i.e., the slowly diffusing antibody species) was inhibited by sCD4 on all types of viruses tested (Fig. 4 and 5), MAbs did not produce a binding signal on envelope-deleted particles (Fig. 5), and non-HIV-specific antibodies did not adopt lower diffusion rates in the presence of HIV-1 virions (Fig. 3 and 5). Having established that FCS is serviceable for studying HIV, we employed it for more comprehensive analyses of viral antigenicity.
Based on available data (40), MAb b12 was used as a benchmark probe for functional structures on viral particles. This MAb exhibited appreciable solution binding across the viruses expressed in HEK293T cells (Fig. 5). The MAb also inhibited infection in matched neutralization assays (Fig. 6), establishing that the virions bound by MAb b12 in FCS include ones that are infectious. MAb 2G12, which also recognizes functional structures, was also consistently neutralizing but exhibited more variable binding efficiencies across viruses. Among viruses, MAb b12 bound the HXB2 pseudovirus and the AD17 infectious virus with lowest efficiency in solution. In both cases, sCD4 caused comparatively weak inhibition of binding (Fig. 5). Notably, efficient MAb b12 binding and strong sCD4 blocking were observed with the NL4-3 infectious virus, which expresses a nearly identical envelope sequence. MAb 2G12 also bound the NL4-3 virus more efficiently than the HXB2 pseudovirus. Taken together, the data indicate that b12 and 2G12 interactions in solution are envelope dependent but not specifically determined by envelope sequence, coreceptor tropism, or viral phenotype.
The group 2 anti-gp120 MAbs served to evaluate the antigenicity of functional envelope trimers that expose CD4i epitopes, including the coreceptor binding site, in response to CD4-gp120 interactions. The fraction of MAbs 17b or 19e that bound to particles in solution variably increased in the presence of sCD4 (Fig. 4 and 5) in an envelope-dependent manner. The least extensive changes in CD4i antigenicity were evident with the BaL and HXB2 pseudoviruses (Fig. 5) and the AD17 infectious virus, suggesting that certain virus types express a lower percentage of envelopes capable of CD4-induced structural conversions in solution. The marginal increase in MAb 19e binding to sCD4-treated HXB2 pseudoviruses is noteworthy since MAb 19e relies on CD4 to form part of its cognate epitope (63). Thus, sCD4 binding efficiency may be especially low on HXB2 virions. Nevertheless, the FCS patterns indicate that functional structures are generally antigenic across different virus types.
The group 2 MAbs C11, A32, and 17b further served as a means to probe unliganded virions for the expression of misfolded/nonfunctional envelope structures such as nontrimeric gp120-gp41 complexes, aberrantly constrained gp120 in the absence of CD4, or virion-bound monomeric gp120 (39–41). In the absence of sCD4, a consistently small percentage of these MAbs (20% or less) bound to any virus produced in HEK293T cells (Fig. 5). Notably, cryo-electron tomography of AT-2-inactivated BaL virions indicated that MAb 17b can bind to a certain fraction of envelope trimers in the absence of sCD4 (80). Such binding could explain the limited FCS binding signals that we observed (Fig. 5).
None of the antibodies tested here exhibited a clear relationship between neutralizing activity and binding to virion populations in solution (Fig. 7A). As noted above, neutralizing activity served as an indicator of binding to infectious virions. Additionally, in tests of unliganded (i.e., absent sCD4), HEK293T cell-derived viruses, only the JRFL pseudovirus exhibited a statistically significant direct relationship between neutralization sensitivity and antibody binding in solution (Fig. 7B). One obvious explanation for this disconnect was that certain MAbs were sometimes neutralizing despite inefficient solution binding to the unliganded virions. For example, MAb 17b selectively neutralized NL4-3 and HXB2 (Fig. 6 and 7) even though it bound to these and other unliganded virus types with similarly poor efficiencies (Fig. 5 and 7). These data suggest that MAbs such as 17b may inhibit infection by targeting a postattachment structure, in which cognate epitope exposure is transiently enhanced, before putative steric restrictions of the coreceptor binding site occur (81). However, the binding data do not clarify why the CXCR4-tropic viruses seemed to be more sensitive to such effects. For example, MAb 17b showed equivalent binding to NL4-3 (which it neutralized) and to JRFL (which it did not neutralize) in the presence of soluble CD4 (Fig. 5B). Detailed analyses of surface-bound virions may be needed to resolve the mechanisms of neutralization by antibodies such as MAb 17b.
In accordance, MAb 19e, which is specific for a hybrid gp120-CD4 epitope, also exhibited neutralizing activity against the CXCR4-tropic viruses despite undetectable binding in FCS analyses without sCD4. Other neutralizing anti-gp120 antibodies that are exquisitely dependent on gp120-CD4 complex formation, including ones that we have recently described (38), may act through similar mechanisms. However, it should be noted that although unconjugated MAb 19e neutralizes certain HIV-2 variants treated with sCD4 (82), it typically does not neutralize HIV-1 strains (83), including the viruses tested here (data not shown). We speculate that dye conjugation altered the binding characteristics of MAb 19e to facilitate the modest neutralizing activity that we observed with the Alexa Fluor-labeled MAb (Fig. 6). The anti-gp41 MAb 2F5 was another example where virus neutralization occurred despite sparse virion binding in FCS (Fig. 5, 6, and 7). The only virus type that was efficiently bound by MAb 2F5 was AD17 grown in HEK293T cells, indicating that this particular envelope may have unique structural aspects.
The relationship between virion binding in solution and neutralizing activity was complicated further by MAbs such as PG9 and F240. Anti-gp41 MAb F240 was distinct in that it exhibited little or no neutralizing activity despite generally efficient binding across viruses. This agrees with previous studies showing that MAb F240 is nonneutralizing yet capable of capturing viral particles (68). Notably, such binding may have some significance for antiviral immunity, as it was shown that passive immunization of macaques with MAb F240 provided partial protection from simian-human immunodeficiency virus (SHIV) challenge (68). Virion binding by MAb F240 has been attributed to the recognition of gp41 “stumps” (41), which have been distinguished from functional envelope structures by their susceptibility to certain proteases (84). We attempted to address this question using protease treatment methods (84) that were reported to selectively degrade nonfunctional structures from viruslike particles. However, application of the method to the virions used here produced equivocal data; for example, the binding of MAbs b12 and 2G12 was also abrogated. It is possible that the protease sensitivities of envelope structures could vary according to virion production and/or binding assay conditions. We cannot exclude the possibility that MAb F240 marks the presence of a misfolded form of gp41 common to many viruses. At the same time, evidence that sCD4 significantly enhances MAb F240 binding (Fig. 5F) to NL4-3 SupT1 viruses suggests that epitope exposure may not be exclusive to gp41 stump-like structures, at least on certain virus types. In any case, it is possible that the potential antiviral properties of MAb F240 could be substantially influenced by virus type and/or production method.
MAb PG9 was neutralizing against the HXB2 and AD17 viruses but not the JRFL virus, in accordance with previous studies (85), yet bound to all virus types with similar efficiencies in FCS. The broad neutralizing activity reported for MAb PG9 (85) clearly establishes that it binds infectious virions in the context of in vitro infectivity assays. Since FCS showed binding in the absence of neutralization, MAb PG9 may also bind noninfectious virions and/or nonfunctional envelope structures on particles in solution. The available information (60, 61, 85) indicates that PG9 may bind envelope trimers in a range of orientations, some of which may explain our observations.
Notably, the polyreactivity linked with a variety of anti-HIV envelope MAbs did not apparently perturb our observations. Among the MAbs in our panel, MAb 2F5 was the most likely candidate in this regard. It is well established that anti-MPER antibodies are variably autoreactive with certain cellular proteins and phospholipids (86, 87) and may establish membrane lipid interactions to enable neutralizing activity (88–91). However, in the system used here MAb 2F5 did not exhibit detectable binding to delE particles in solution (Fig. 5), indicating little or no binding to non-HIV antigens. A recent study that compared capture assay formats suggested that anti-MPER MAbs bind misfolded envelope structures trapped on virions and neutralize via interactions with fusion intermediates formed postattachment (45). Our findings are consistent with a postattachment neutralization mechanism, although the FCS analyses failed to show sCD4-induced changes in MAb 2F5-virion binding in contrast with previous observations (79, 92). At the same time, the reported impact of CD4 on MPER epitope exposure has been variable (15, 92–95). Our data cannot exclude the possibility that MPER epitopes are induced by envelope interactions with cell surface CD4 (20), which differs from sCD4 in several respects (96, 97).
A practical difficulty presented by HIV derived from cell cultures is that envelope antigenicity can be influenced by virus production method. We explored this issue here to a limited extent by comparative examination of NL4-3, BaL, and AD17 virions produced in SupT1 cells. One expectation was that the alternative method might increase the antigenicity of putative misfolded envelope structures. However, this was not the case. The overall patterns of gp120 antigenicity in the absence of sCD4 were roughly the same on viruses produced in SupT1 and HEK293T cells (Fig. 5), although the absolute MAb binding efficiencies varied to a minor degree. The most apparent difference was that sCD4 induction of group 2 MAb binding was lower on the SupT1 viruses even though all test viruses contained equivalent antigen content, and regardless of production method, gp41 epitopes showed more variability in antigenicity depending on virus production method. Compared to their counterparts produced in HEK293T cells, the SupT1-derived NL4-3 viruses exhibited higher 2F5 and F240 antigenicity, which moderately increased in the presence of sCD4. Conversely, the antigenicity of these epitopes on SupT1-derived AD17 virus was lower. Thus, the comparative analyses indicate that it may be difficult to predict how the antigenic profiles of virions in solution will change according to virus production method, although we recognize that more exhaustive tests with FCS might clarify the issue.
Considered in aggregate, our data provide a view of HIV-antibody interactions with all reactants continuously in solution, a condition that is relevant to certain in vitro systems and/or aspects of natural HIV infection. Under these conditions, the antigenic profiles of HIV particles tend to favor efficient binding to functional over misfolded structures. However, since binding occurs without neutralization and neutralization occurs through multiple mechanisms, antigenicity profiles do not always predict the neutralization sensitivity. Cases where antibody-virion interactions occur in the absence of conventional neutralizing activity further introduce the possibility for other antiviral mechanisms, such as Fc receptor-dependent processes, to come into play during infection. In this regard, FCS may provide a useful tool for interpreting the design and outcome of passive immunization and vaccine studies and for characterizing vaccines based on viral particles.
ACKNOWLEDGMENTS
The present work was supported by National Institutes of Health grant K25AI087968 (K.R.) and the Bill and Melinda Gates Foundation grant OPP1033109 (G.K.L.).
We thank Yongjun Guan for preparation of A32, C11, PG9, and F240 antibody clones; Christine Obreicht for producing purified antibodies; and Jacqueline Martin for producing viruses.
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Burton DR, Stanfield RL, Wilson IA. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U. S. A. 102:14943–14948. 10.1073/pnas.0505126102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236. 10.1038/ni0304-233 [DOI] [PubMed] [Google Scholar]
- 3.Burton DR, Montefiori DC. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl A):S87–S98 [PubMed] [Google Scholar]
- 4.Burton DR, Pyati J, Koduri R, Thornton GB, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Moore JP, Ho DD, Barbas CF., III 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. 10.1126/science.7973652 [DOI] [PubMed] [Google Scholar]
- 5.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155–1159. 10.1126/science.1061692 [DOI] [PubMed] [Google Scholar]
- 6.Poignard P, Saphire EO, Parren PW, Burton DR. 2001. Gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253–274. 10.1146/annurev.immunol.19.1.253 [DOI] [PubMed] [Google Scholar]
- 7.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scanlan CN, Pantophlet R, Wormald MR, Saphire EO, Calarese D, Stanfield R, Wilson IA, Katinger H, Dwek RA, Burton DR, Rudd PM. 2003. The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv. Exp. Med. Biol. 535:205–218. 10.1007/978-1-4615-0065-0_13 [DOI] [PubMed] [Google Scholar]
- 9.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071. 10.1126/science.1083182 [DOI] [PubMed] [Google Scholar]
- 10.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 102:13372–13377. 10.1073/pnas.0505763102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt LS, Earl PL, Vogt J, Eller LA, Chandran D, Liu J, Robinson HL, Moss B. 2008. Correlation of immunogenicities and in vitro expression levels of recombinant modified vaccinia virus Ankara HIV vaccines. Vaccine 26:486–493. 10.1016/j.vaccine.2007.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790. 10.1128/JVI.01730-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray ES, Meyers T, Gray G, Montefiori DC, Morris L. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3:e255. 10.1371/journal.pmed.0030255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724–10737. 10.1128/JVI.78.19.10724-10737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892–10905. 10.1128/JVI.75.22.10892-10905.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757–1765. 10.1089/08892220152741450 [DOI] [PubMed] [Google Scholar]
- 17.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884 [DOI] [PubMed] [Google Scholar]
- 19.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252. 10.1128/JVI.78.23.13232-13252.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks ET, Moore PL, Richman D, Robinson J, Crooks JA, Franti M, Schulke N, Binley JM. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101–113 [PMC free article] [PubMed] [Google Scholar]
- 21.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. 2010. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:11483–11488. 10.1073/pnas.1004600107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232–5237. 10.1128/JVI.79.8.5232-5237.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. 10.1038/384179a0 [DOI] [PubMed] [Google Scholar]
- 25.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–186. 10.1038/384184a0 [DOI] [PubMed] [Google Scholar]
- 26.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore JP, Sodroski J. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. 10.1038/312763a0 [DOI] [PubMed] [Google Scholar]
- 28.Pal R, DeVico A, Rittenhouse S, Sarngadharan MG. 1993. Conformational perturbation of the envelope glycoprotein gp120 of human immunodeficiency virus type 1 by soluble CD4 and the lectin succinyl Con A. Virology 194:833–837. 10.1006/viro.1993.1326 [DOI] [PubMed] [Google Scholar]
- 29.Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway RS, Jr, Leary J, Bugelski PJ. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 88:2189–2193. 10.1073/pnas.88.6.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1038/31405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. 10.1038/nature03327 [DOI] [PubMed] [Google Scholar]
- 33.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. 10.1038/31514 [DOI] [PubMed] [Google Scholar]
- 34.Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244–1247. 10.1038/nm1196-1244 [DOI] [PubMed] [Google Scholar]
- 35.DeVico AL. 2007. CD4-induced epitopes in the HIV envelope glycoprotein, gp120. Curr. HIV Res. 5:561–571. 10.2174/157016207782418560 [DOI] [PubMed] [Google Scholar]
- 36.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036. 10.1128/JVI.00171-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao HX, Alam SM, Mascola JR, Robinson J, Ma B, Montefiori DC, Rhein M, Sutherland LL, Scearce R, Haynes BF. 2004. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 78:5270–5278. 10.1128/JVI.78.10.5270-5278.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. U. S. A. 110:E69–E78. 10.1073/pnas.1217609110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338:154–172. 10.1016/j.virol.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 40.Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353–365. 10.1128/JVI.77.1.353-365.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528. 10.1128/JVI.80.5.2515-2528.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavacini L, Posner M. 2004. Native HIV type 1 virion surface structures: relationships between antibody binding and neutralization or lessons from the viral capture assay. AIDS Res. Hum. Retroviruses 20:435–441. 10.1089/088922204323048186 [DOI] [PubMed] [Google Scholar]
- 43.Burrer R, Haessig-Einius S, Aubertin AM, Moog C. 2005. Neutralizing as well as non-neutralizing polyclonal immunoglobulin (Ig)G from infected patients capture HIV-1 via antibodies directed against the principal immunodominant domain of gp41. Virology 333:102–113. 10.1016/j.virol.2004.12.034 [DOI] [PubMed] [Google Scholar]
- 44.Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384–9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leaman DP, Kinkead H, Zwick MB. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J. Virol. 84:3382–3395. 10.1128/JVI.02363-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P, Yates NL, Shen X, Bonsignori M, Moody MA, Liao HX, Fong Y, Alam SM, Overman RG, Denny T, Ferrari G, Ochsenbauer C, Kappes JC, Polonis VR, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Montefiori DC, Gilbert P, Michael NL, Kim JH, Haynes BF, Tomaras GD. 2013. Infectious virion capture by HIV-1 gp120-specific IgG from RV144 vaccinees. J. Virol. 87:7828–7836. 10.1128/JVI.02737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson JD, Brunel FM, Jensen R, Crooks ET, Cardoso RM, Wang M, Hessell A, Wilson IA, Binley JM, Dawson PE, Burton DR, Zwick MB. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J. Virol. 81:4033–4043. 10.1128/JVI.02588-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hess ST, Huang S, Heikal AA, Webb WW. 2002. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochemistry 41:697–705. 10.1021/bi0118512 [DOI] [PubMed] [Google Scholar]
- 49.Hess ST, Webb WW. 2002. Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophys. J. 83:2300–2317. 10.1016/S0006-3495(02)73990-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vercammen J, Maertens G, Gerard M, De Clercq E, Debyser Z, Engelborghs Y. 2002. DNA-induced polymerization of HIV-1 integrase analyzed with fluorescence fluctuation spectroscopy. J. Biol. Chem. 277:38045–38052. 10.1074/jbc.M205842200 [DOI] [PubMed] [Google Scholar]
- 51.Maertens G, Vercammen J, Debyser Z, Engelborghs Y. 2005. Measuring protein-protein interactions inside living cells using single color fluorescence correlation spectroscopy. Application to human immunodeficiency virus type 1 integrase and LEDGF/p75. FASEB J. 19:1039–1041. 10.1096/fj.04-3373fje [DOI] [PubMed] [Google Scholar]
- 52.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. 10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, Li Q, Pham P, Cofrancesco J, Persaud D, Siliciano RF. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718–1729. 10.1128/JVI.78.4.1718-1729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Binley JM, Cayanan CS, Wiley C, Schulke N, Olson WC, Burton DR. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678–5684. 10.1128/JVI.77.10.5678-5684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, Wyatt R. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 80:1414–1426. 10.1128/JVI.80.3.1414-1426.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhury S, Ray K, Lakowicz JR. 2012. Silver nanostructures for fluorescence correlation spectroscopy: reduced volumes and increased signal intensities. J. Phys. Chem. Lett. 3:2915–2919. 10.1021/jz301229m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Starr TE, Thompson NL. 2002. Local diffusion and concentration of IgG near planar membranes: measurement by total internal reflection with fluorescence correlation spectroscopy. J. Phys. Chem. B 106:2365–2371. 10.1021/jp012689f [DOI] [Google Scholar]
- 58.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. 10.1038/nature10696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 110:4351–4356. 10.1073/pnas.1217537110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521. 10.1128/JVI.00552-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis GK, Fouts TR, Ibrahim S, Taylor BM, Salkar R, Guan Y, Kamin-Lewis R, Robinson JE, Devico AL. 2011. Identification and characterization of an immunogenic hybrid epitope formed by both HIV gp120 and human CD4 proteins. J. Virol. 85:13097–13104. 10.1128/JVI.05072-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore JP, Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, Posner MR. 1998. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses 14:1271–1280. 10.1089/aid.1998.14.1271 [DOI] [PubMed] [Google Scholar]
- 66.Cavacini LA, Duval M, Robinson J, Posner MR. 2002. Interactions of human antibodies, epitope exposure, antibody binding and neutralization of primary isolate HIV-1 virions. AIDS 16:2409–2417. 10.1097/00002030-200212060-00005 [DOI] [PubMed] [Google Scholar]
- 67.Miranda LR, Duval M, Doherty H, Seaman MS, Posner MR, Cavacini LA. 2007. The neutralization properties of a HIV-specific antibody are markedly altered by glycosylation events outside the antigen-binding domain. J. Immunol. 178:7132–7138 [DOI] [PubMed] [Google Scholar]
- 68.Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, Dunlop DC, Poignard P, Doyle LA, Cavacini L, Veazey RS, Moore JP. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl. Acad. Sci. U. S. A. 108:11181–11186. 10.1073/pnas.1103012108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036. 10.1128/JVI.00171-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finnegan CM, Berg W, Lewis GK, DeVico AL. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096–11105. 10.1128/JVI.75.22.11096-11105.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finnegan CM, Berg W, Lewis GK, DeVico AL. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76:12123–12134. 10.1128/JVI.76.23.12123-12134.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss CD, Barnett SW, Cacalano N, Killeen N, Littman DR, White JM. 1996. Studies of HIV-1 envelope glycoprotein-mediated fusion using a simple fluorescence assay. AIDS 10:241–246. 10.1097/00002030-199603000-00001 [DOI] [PubMed] [Google Scholar]
- 73.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413–423. 10.1083/jcb.151.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J. Virol. 87:1884–1889. 10.1128/JVI.02765-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postler TS, Desrosiers RC. 2013. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J. Virol. 87:2–15. 10.1128/JVI.02053-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78:3429–3435. 10.1128/JVI.78.7.3429-3435.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forster MJ, Mulloy B, Nermut MV. 2000. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J. Mol. Biol. 298:841–857. 10.1006/jmbi.2000.3715 [DOI] [PubMed] [Google Scholar]
- 78.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. 10.1126/science.1226359 [DOI] [PubMed] [Google Scholar]
- 79.Rathinakumar R, Dutta M, Zhu P, Johnson WE, Roux KH. 2012. Binding of anti-membrane-proximal gp41 monoclonal antibodies to CD4-liganded and -unliganded human immunodeficiency virus type 1 and simian immunodeficiency virus virions. J. Virol. 86:1820–1831. 10.1128/JVI.05489-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565. 10.1128/JVI.77.19.10557-10565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419. 10.1084/jem.20042510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor BM, Foulke JS, Flinko R, Heredia A, Devico A, Reitz M. 2008. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J. Virol. 82:5460–5471. 10.1128/JVI.01049-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crooks ET, Tong T, Osawa K, Binley JM. 2011. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J. Virol. 85:5825–5839. 10.1128/JVI.00154-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators. Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908. 10.1126/science.1111781 [DOI] [PubMed] [Google Scholar]
- 87.Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, Spicer L, Vandeberg JL, Haynes BF, Kelsoe G. 2013. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J. Exp. Med. 210:241–256. 10.1084/jem.20121977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467:591–595. 10.1038/nature09385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, Wyatt R, Zwick MB, Nabel GJ, Mascola JR, Kwong PD. 2010. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J. Virol. 84:2955–2962. 10.1128/JVI.02257-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. 2010. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U. S. A. 107:1529–1534. 10.1073/pnas.0909680107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song L, Sun ZY, Coleman KE, Zwick MB, Gach JS, Wang JH, Reinherz EL, Wagner G, Kim M. 2009. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc. Natl. Acad. Sci. U. S. A. 106:9057–9062. 10.1073/pnas.0901474106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peachman KK, Wieczorek L, Polonis VR, Alving CR, Rao M. 2010. The effect of sCD4 on the binding and accessibility of HIV-1 gp41 MPER epitopes to human monoclonal antibodies. Virology 408:213–223. 10.1016/j.virol.2010.09.029 [DOI] [PubMed] [Google Scholar]
- 93.de Rosny E, Vassell R, Jiang S, Kunert R, Weiss CD. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 78:2627–2631. 10.1128/JVI.78.5.2627-2631.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sattentau QJ, Zolla-Pazner S, Poignard P. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713–717. 10.1016/S0042-6822(95)80094-8 [DOI] [PubMed] [Google Scholar]
- 95.Montero M, van Houten NE, Wang X, Scott JK. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72:54–84. 10.1128/MMBR.00020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matthias LJ, Azimi I, Tabrett CA, Hogg PJ. 2010. Reduced monomeric CD4 is the preferred receptor for HIV. J. Biol. Chem. 285:40793–40799. 10.1074/jbc.M110.190579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ashish, Juncadella IJ, Garg R, Boone CD, Anguita J, Krueger JK. 2008. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J. Biol. Chem. 283:2761–2772. 10.1074/jbc.M708325200 [DOI] [PubMed] [Google Scholar]