Abstract
Mouse embryonic cells are unable to support the replication of Moloney murine leukemia virus (MLV). The integrated viral DNA is transcriptionally silenced, largely due to binding of host transcriptional repressors to the primer binding site (PBS) of the provirus. We have previously shown that a PBS DNA-binding repressor complex contains ZFP809 and TRIM28. Here, we identified ErbB3-binding protein 1 (EBP1) to be a novel component of the ZFP809-TRIM28 silencing complex and show that EBP1 depletion reduces PBS-mediated retroviral silencing.
TEXT
Murine leukemia viruses (MLVs) are unable to replicate in embryonic stem (ES) or embryonic carcinoma (EC) cells (1–3). In embryonic cells, reverse transcription and proviral integration proceed normally, but viral transcription is repressed, and hence no viral gene products can be detected. Several mechanisms are likely involved in the transcriptional silencing, including (i) the absence of enhancer proteins recognizing binding sites in the viral long terminal repeat (LTR) (1, 2), (ii) recruitment of trans-acting transcriptional repressors (3–5), and (iii) de novo DNA methylation (6). One critical site for transcriptional silencing, termed the repressor binding site (RBS), shows extensive overlap (17/18 bp) with the primer binding site (PBSpro) of MLV (7–9), which normally functions during the priming of reverse transcription by host proline tRNA. The RBS by itself is sufficient to induce potent transcriptional repression of reporter constructs in EC cells, irrespective of its orientation or position (10, 11). In addition, electrophoretic mobility shift assays (EMSAs) using RBS as the probe demonstrate the presence of stem cell-specific nuclear factors. A single base pair mutation in the RBS, termed the B2 mutation, is sufficient to abolish nuclear factor binding and thereby restore viral gene expression (10, 11). These findings have allowed the characterization of the stem cell-specific trans-acting transcriptional repressor complex as containing TRIM28 (also known as KAP-1 or Tif1-beta), a known transcriptional corepressor (12–14), and Krüppel-associated box (KRAB) zinc finger protein 809 (ZFP809) (15), a DNA binding transcription factor. The current model for PBS-mediated silencing consists of the binding of PBSpro by ZFP809 and the subsequent recruitment of TRIM28 to the provirus via the ZFP809 KRAB domain. This association permits TRIM28-dependent recruitment of additional chromatin modifiers, such as the histone H3K9 methyltransferase SETDB1 (16), heterochromatin-associated protein 1 (HP1) (12), and the nucleosome remodeling histone deacetylase (NuRD) complex (17). Together, the ensuing histone and DNA methylation of the proviral DNA ensures transcriptional silencing and genomic stability.
The ZFP809-TRIM28 complex detected by EMSA is of high molecular weight (13, 15, 18), and PBS-dependent silencing involves many effector proteins. It is thus fully plausible that ZFP809-TRIM28-dependent silencing requires additional unknown host factors. In an effort to identify novel components of the ZFP809-TRIM28 silencing complex, we generated stable HEK-293 cell lines expressing a Flag-tagged version of ZFP809 lacking its C terminus [Flag-ZFP809(1-353)]. As shown previously, compared to full-length ZFP809, this truncation mutant is more readily expressed and retains full capacity to restore PBS-mediated retroviral silencing in differentiated cells (15). Lysates from tagged ZFP809-expressing HEK-293 cells were subjected to immunoprecipitation with an anti-Flag antibody followed by mass spectrometric analysis of ZFP809-associated proteins. Immunoprecipitates using nonspecific IgG were analyzed similarly as the negative control. Seventeen unique ZFP809-interacting proteins were identified (see Table S1 in the supplemental material), with TRIM28 having the largest number of peptides, consistent with its major role in PBS-dependent retroviral silencing in embryonic cells (13–15, 19). In addition, peptides corresponding to proliferation-associated protein 2G4 (PA2G4), also known as ErbB3-binding protein 1 (EBP1), were isolated specifically from ZFP809-expressing cells. EBP1's known involvement in the transcriptional repression of E2F1 and androgen receptor-regulated genes (20–24) raises the possibility that EBP1, through its association with ZFP809, may exert similar repressor functions in the context of PBS-dependent retroviral silencing.
To confirm the authenticity of the EBP1-ZFP809 interaction, lysates from Flag-ZFP809-expressing HEK-293 cells were subjected to immunoprecipitation with either anti-Flag or control anti-hemagglutinin (anti-HA) antibodies, and the bound proteins were then analyzed by immunoblotting with either anti-EBP1 or anti-TRIM28 antibodies. Endogenous EBP1 was specifically detected in anti-Flag but not anti-HA immunoprecipitates (Fig. 1A). As expected, TRIM28 also associated with ZFP809. We observed the reciprocal interactions upon immunoprecipitation of endogenous EBP1 from Flag-ZFP809-expressing HEK-293 cells followed by immunoblotting with either Flag or TRIM28 antibodies (Fig. 1B). Furthermore, in F9 embryonic carcinoma cells, a cell line known to be nonpermissive to MLV infection, the association of endogenous EBP1 with TRIM28 immunoprecipitates could also be detected (Fig. 1C). The lack of specific ZFP809 antibodies prevented us from performing coimmunoprecipitation studies of endogenous ZFP809. Taken together, these results suggest that EBP1 may be a novel component of the ZFP809-TRIM28 transcriptional repression complex.
FIG 1.
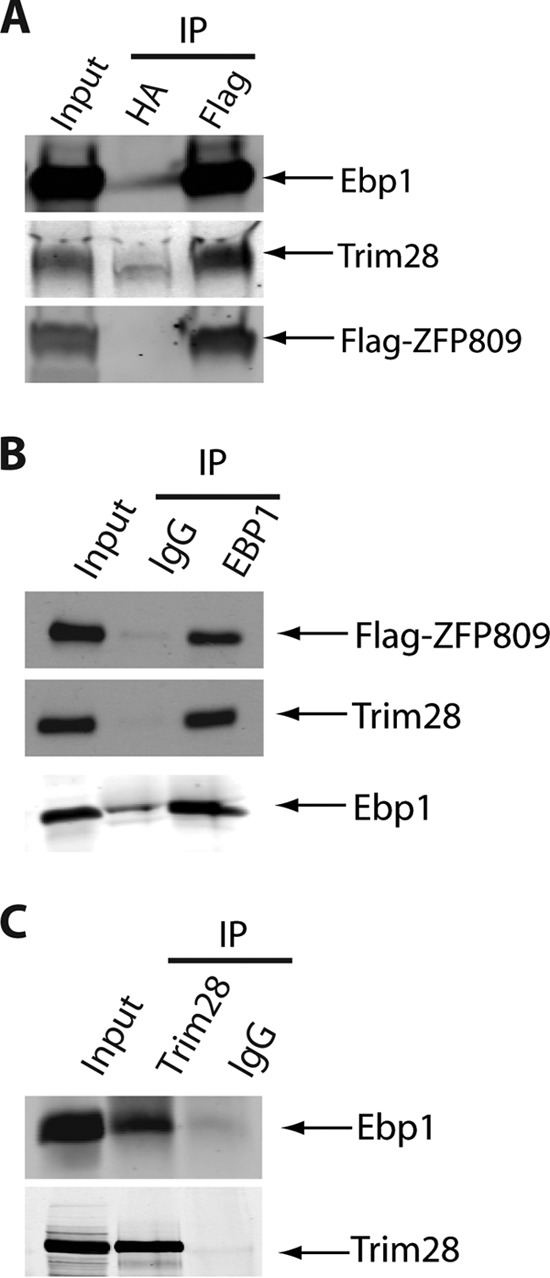
EBP1 forms a complex with ZFP809 and TRIM28 in vivo. (A) Total cell lysates were prepared as described before (15) from HEK-293 cells stably expressing Flag-tagged ZFP809, and proteins were immunoprecipitated with anti-Flag or control anti-HA antibody. Bound proteins were subjected to 10% SDS-PAGE and immunoblotted with anti-EBP1, anti-TRIM28, and anti-Flag antibodies as indicated on the right. The input lane represents 2% of the material used for the immunoprecipitations. (B) Total cell lysates from Flag-ZFP809-expressing HEK-293 cells were subjected to immunoprecipitation and immunoblotting as indicated. The input lane represents 2% of the material used for each immunoprecipitation. (C) Total cell lysates prepared from F9 embryonic carcinoma cells were subjected to immunoprecipitation and immunoblotting as indicated. The input lane represents 2% of the material used for the immunoprecipitations. Note that all of the data shown in Fig. 1 are representative of at least three independent experiments.
The observation that EBP1 forms an endogenous complex with both ZFP809 and TRIM28 raises the possibilities that (i) EBP1 directly interacts with ZFP809, (ii) EBP1 is recruited to ZFP809 via association with TRIM28, or (iii) EBP1 is capable of forming physical contacts with both ZFP809 and TRIM28. To test these possibilities, we expressed each protein fused to glutathione S-transferase (GST) in Escherichia coli, recovered the GST fusion proteins on glutathione-Sepharose beads, and performed in vitro binding assays. Beads containing GST-ZFP809, GST-TRIM28, or, as a negative control, GST-ZFP124, an unrelated zinc finger protein not implicated in PBS-mediated silencing (15), were incubated with purified His6-EBP1, and bound proteins were visualized by immunoblotting with an anti-His6 antibody. EBP1 bound specifically not only to GST-ZFP809 as expected but remarkably also to GST-TRIM28 even in the absence of ZFP809 (Fig. 2A). EBP1 showed no binding at all to ZFP124. The lack of binding to ZFP124 is striking, given that like ZFP809, it contains an N-terminal KRAB domain and a C-terminal cluster of zinc fingers. The yield of EBP1 bound by ZFP809 was higher than the yield bound by TRIM28. The yield bound by a mixture of ZFP809 and TRIM28 was similar to that bound by ZFP809 alone. To further map the regions in ZFP809 responsible for EBP1 interaction, we created two additional deletion mutants targeting its KRAB domain (ZFP809ΔKRAB) or its seven zinc fingers (ZFP809ΔZF). Initial attempts to express these mutants as GST fusion proteins in bacteria were unsuccessful, and we therefore chose to overexpress these mutants in mammalian cells and carried out His6-EBP1 pulldown assays using mammalian cell lysates. As shown in Fig. 2B, ZFP809(1-353) and ZFP809ΔKRAB mutants formed a complex with EBP1 in vitro, and such interaction was lost upon removal of the ZFP809 zinc fingers (ZFP809ΔZF). Given the amino acid differences in the zinc finger domains of ZFP809 and ZFP124, this observation is consistent with EBP1's preferential association with ZFP809 and not ZFP124 (Fig. 2A). It further suggests that the zinc finger domains of ZFP809 may function in both DNA-binding and EBP1 recruitment. To test whether the presence of nonspecific DNA in the cell lysate may affect binding, we repeated the experiment after DNase I treatment of the cell lysate. No difference in binding was observed (data not shown).
FIG 2.
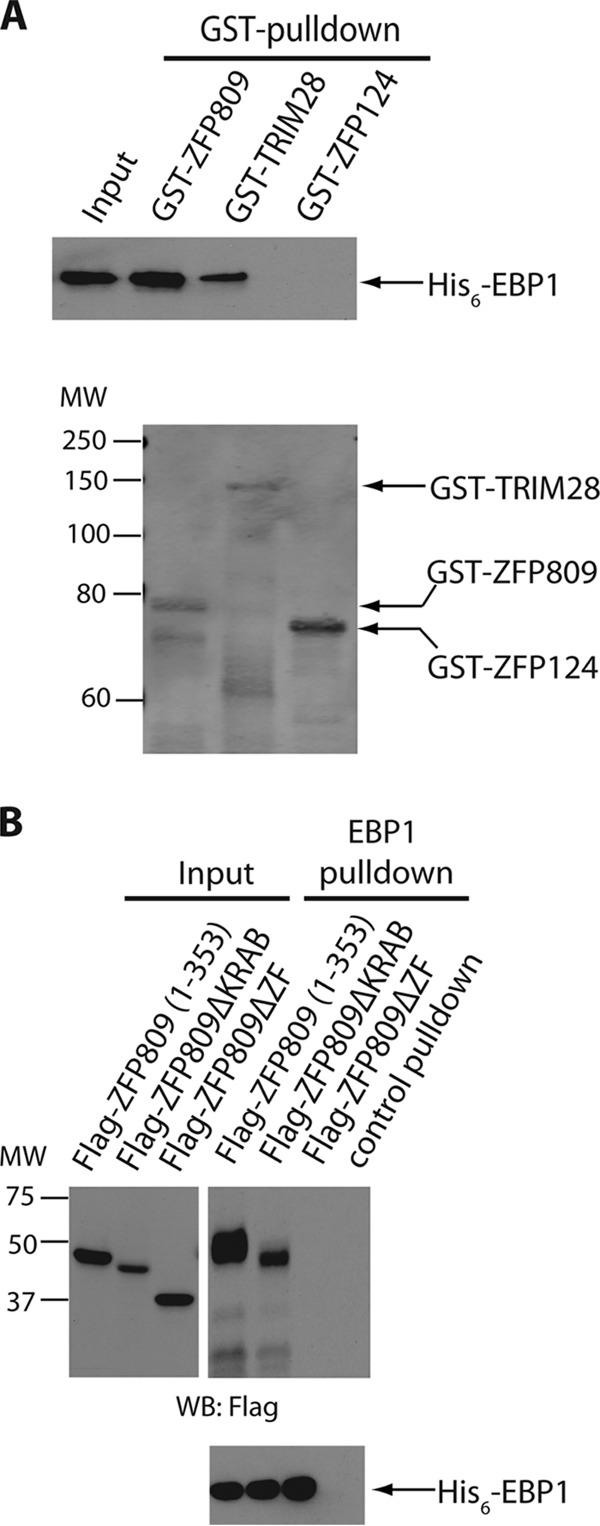
EBP1 interacts directly with ZFP809 and TRIM28 in vitro. (A) Top, purified His6-EBP1 was incubated with either GST-ZFP809, GST-TRIM28, or GST-ZFP124 (as a negative control) fusion proteins immobilized on glutathione-Sepharose beads. Bound proteins were eluted and subjected to 10% SDS-PAGE and immunoblotted with an anti-His6 antibody. The input lane represents 5% of total His6-EBP1 used for each pulldown reaction. Data shown are representative of three independent experiments. Bottom, the same blot as that shown in the top panel was stripped and reprobed with anti-GST antibody to show equal amounts of GST proteins used in each pulldown reaction. (B) Top, purified His6-EBP1 immobilized on Cobalt resin was incubated with HEK-293 total cell lysates expressing various Flag-tagged ZFP809 truncation mutants. As a negative control, Cobalt resin alone was incubated with Flag-ZFP809(1-353) lysates. Bound proteins were eluted and subjected to 10% SDS-PAGE and immunoblotted with an anti-Flag antibody. The input lane represents 2% of cell lysates used for each pulldown reaction. Data shown are representative of three independent experiments. Bottom, the same blot shown in the top panel was stripped and reprobed with anti-His6 antibody to show equal amounts of His6-EBP1 used for each pulldown reaction.
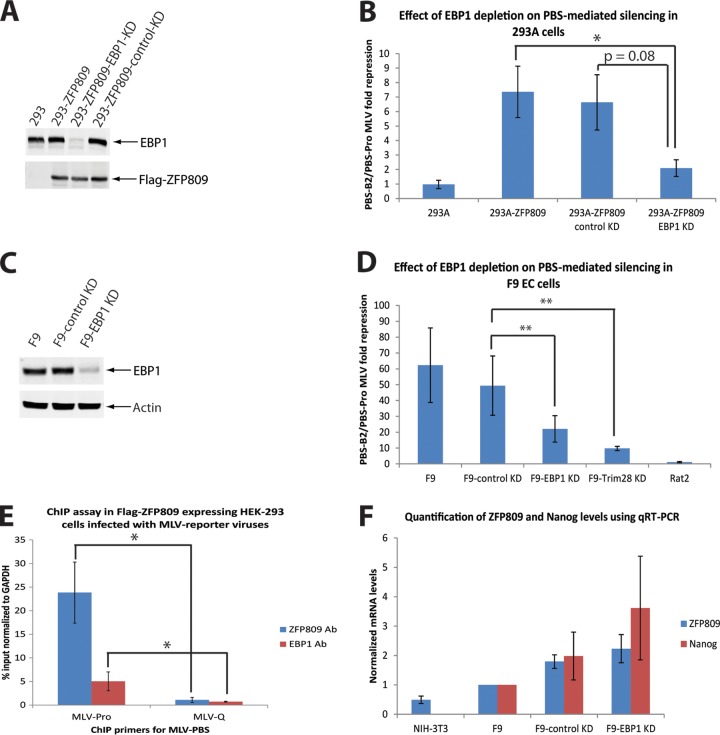
To investigate the functional relevance of the EBP1-ZFP809-TRIM28 interaction for PBS-mediated retroviral silencing, we utilized retroviral RNA interference (RNAi) methods to stably deplete endogenous EBP1 expression in Flag-ZFP809-expressing HEK-293 cells. As a negative control, cell lines containing scrambled short hairpin RNA (shRNA) were generated. As shown in Fig. 3A, efficient EBP1 knockdown was achieved. To quantify the level of PBS-mediated repression, we made use of transducing MLV reporter viruses expressing phleomycin D1 (Zeocin) resistance. We compared the titer of a wild-type MLV (MLV-WT) reporter with the wild-type PBS (hence a target for ZFP809-dependent silencing) to an MLV-B2 reporter with a mutant PBS escaping PBS-mediated silencing. The two viruses were used to infect the various cell lines outlined in Fig. 3B, and cells were then selected with Zeocin for 2 weeks before colony counting. The ratio of the number of B2 colonies to WT colonies represents the degree of PBS-mediated silencing. As expected, differentiated cell types, such as wild-type HEK-293 cells, lacked PBS-mediated silencing, as indicated by a B2/WT ratio close to 1 (Fig. 3B). In contrast, overexpression of ZFP809 in HEK-293 cells induced PBS-mediated silencing, as shown by a B2/WT ratio of 7. EBP1 depletion in ZFP809-expressing cells caused a 4-fold reduction in PBS-mediated restriction, indicating the functional importance of EBP1 in the ZFP809-TRIM28 complex. To further confirm our finding, we performed the analogous experiments in F9 EC cells (Fig. 3D). The ratio of MLV-B2 to WT reporter was close to 60-fold in F9 and F9-control KD cells. Such restriction was significantly reduced upon TRIM28 depletion, with a 6-fold loss in silencing. PBS-mediated silencing was reduced by 2- to 3-fold upon the depletion of endogenous EBP1 (Fig. 3D). Based on these observations, we next postulated that EBP1 may be capable of associating with the integrated provirus in vivo in a PBSpro-dependent manner. To test this, chromatin immunoprecipitation (ChIP) experiments were performed using Flag-ZFP809-expressing HEK-293 cells transduced with either an MLV-PBSpro virus or an MLV-PBSgln virus known to escape PBS-dependent silencing. As expected, ZFP809 is highly enriched (25-fold) on the PBSpro but not the PBSgln locus (Fig. 3E). Consistently, EBP1 showed a 5-fold enrichment on PBSpro, while no enrichment was seen on PBSgln (Fig. 3E). These results correlate with EBP1's repressive effect on PBSpro and are consistent with the model of ZFP809-mediated recruitment of EBP1 to the provirus in vivo. Finally, it has been demonstrated that cellular differentiation leads to the concomitant loss of ZFP809-TRIM28 recruitment to the provirus, thereby mitigating PBS-dependent retroviral silencing (13). Therefore, to rule out the possibility that depletion of endogenous EBP1 in embryonic cells may lead to either cellular differentiation or changes in the levels of ZFP809, we prepared total RNA from NIH 3T3, F9, F9-control KD, and F9-EBP1-KD cells and quantified the mRNA levels of Nanog, a marker for embryonic cells, as well as ZFP809 using quantitative reverse transcription-PCR (qRT-PCR) (Fig. 3F). Consistently, EBP1-depleted cells did not show reduced levels of Nanog or ZFP809, while NIH 3T3 cells show reduced levels of both mRNAs. Collectively, these observations suggest a role for EBP1 in mediating ZFP809-TRIM28-dependent retroviral restriction in embryonic cells.
FIG 3.
EBP1 depletion impairs PBS-dependent retroviral silencing. (A) Total cell lysates prepared from HEK-293, 293-ZFP809, 293-ZFP809-control KD, and 293-ZFP809-EBP1 KD cells were immunoblotted with antisera as indicated. Note the sequence of the human EBP1 KD shRNA is TATTTACCGAAATGCTGGTGG (as per Thermo Scientific). (B) HEK-293, 293-ZFP809, 293-ZFP809-control KD, and 293-ZFP809-EBP1 KD cells were infected with vesicular stomatitis virus glycoprotein (VSV G)-pseudotyped MLV particles containing either MLV-PBSpro-Zeo or MLV-PBSB2-Zeo constructs. Reporter gene expression in each cell line was assessed by colony counting after Zeocin selection. Graph shows the ratio of B2/Pro infection efficiency, termed fold repression. Results shown are means ± standard errors of the means (SEMs) from three independent experiments performed in triplicate. Student's t test was used for statistical analysis. * denotes P < 0.05. (C) Total cell lysates prepared from WT F9 or F9 cells stably transduced with either control shRNA or EBP1-specific shRNA were immunoblotted with antisera as indicated. Note the sequence of the mouse EBP1 KD shRNA is TTATACTTGGTCACGACCAGG (as per Thermo Scientific). (D) Rat2, F9, F9-control KD, F9-EBP1 KD, and F9-TRIM28 KD cells were infected with VSVG-pseudotyped MLV particles containing either MLV-PBSpro-Neo or MLV-PBSB2-Neo constructs. The reporter gene expression in each cell line was assessed by colony counting after G418 selection. Graph shows the ratio of B2/Pro infection efficiency, termed fold repression. Results shown are means ± SEMs from seven independent experiments performed in triplicate. Student's t test was used for statistical analysis. ** denotes P < 0.01. (E) Chromatin immunoprecipitations (ChIP) assays using anti-Flag (positive control) or anti-EBP1 antibodies in Flag-ZFP809-expressing HEK-293 cells infected with either MLV-PBSpro or MLV-PBSgln reporter viruses. Results shown are mean enrichments at the PBS locus ± SEMs from 3 independent experiments relative to input chromatin and normalized to the signal of the negative-control gene (GAPDH). Student's t test was used for statistical analysis. * denotes P < 0.05. (F) Total RNA prepared from NIH 3T3, WT F9, or F9 cells stably transduced with either control shRNA or EBP1-specific shRNA were subjected to quantitative RT-PCR using the indicated primers. The RNA levels were normalized to WT F9 cells (set to 1). Results shown are means ± SEMs from four independent experiments performed in duplicate.
The ability of mouse embryonic cells to silence MLV DNA via the PBS lies in its expression of the sequence-specific KRAB-ZFP ZFP809. ZFP809, through its interaction with the PBSpro of MLV, serves to coordinate the assembly of macromolecular silencing complexes containing TRIM28, ESET, HP1, and others, thereby establishing a transcriptionally inactive provirus. In this report, we expand our current understanding of retroviral silencing by identifying a potential link between EBP1, a known transcriptional repressor of E2F1 and androgen receptor-regulated promoters (20–24), to the ZFP809-TRIM28 silencing complex in embryonic cells. Previously, in breast and prostate cancer cell lines, EBP1, in association with RB (25), HDAC2 (23), and Sin3A (20), has been shown to prevent cellular proliferation via the transcriptional repression of E2F1- and AR-regulated genes (20). Interestingly, our observation of the interaction between EBP1 and TRIM28 coincides with previous findings showing the involvement of both proteins at the E2F target promoter (26). Therefore, it remains plausible that on other promoters, specific DNA binding proteins, acting in a manner similar to that of ZFP809, may recruit both EBP1 and TRIM28 to the target promoter and modulate gene expression epigenetically. We note that both EBP1 and TRIM28 are ubiquitously expressed and could regulate target genes in many cell types, but the ES cell-specific regulation will be determined by ES cell-specific zinc finger proteins, such as ZFP809, that tether them to the DNA.
EBP1 has recently been implicated in the biology of RNA viruses in other settings. Honda et al. identified EBP1 as a novel binding partner for the PB1 subunit of influenza virus RNA polymerase and showed that overexpression of EBP1 blocked the transcription of influenza virus and led to reduced viral titers (27). In another study, it was reported that infection of Vero cells by rinderpest virus led to the downregulation of EBP1 expression and that overexpression of EBP1 led to the inhibition of rinderpest virus transcription (28). EBP1's role in antagonizing retroviral infection in embryonic cells (Fig. 3B and D) expands the repertoire of RNA viruses sensitive to EBP1 regulation. Preliminary tests suggest that cellular EBP1 expression is not altered by retroviral infection (data not shown) and that the course of viral DNA synthesis by reverse transcriptase was not affected.
Mouse embryonic cells suppress the expression of both incoming exogenous retroviruses and endogenous retroelements. The loss of either TRIM28 (19) or SETDB1 (29) in mouse ES cells led to robust activation of endogenous retroviruses, as well as failure to block new retroviral infections. In F9 EC cells, depletion of EBP1 did not result in the upregulation of various endogenous retroelements (data not shown). Therefore, we suggest that although EBP1 is required for the silencing of newly integrated provirus, it may be dispensable for the silencing of endogenous retroviral elements. Such division of labor has also been observed in the case of the histone methyltransferase G9a (30).
In summary, the present study revealed EBP1 to be a novel component of the PBS-dependent silencing machinery in mouse embryonic cells. It should be noted that retroviral silencing is mediated through both PBS-dependent and PBS-independent pathways involving the host transcription factor Yin Yang 1 (31), and further studies will be necessary to address the potential cross talk between these two arms. It will also be of interest to determine whether TRIM28 in concert with other KRAB-ZFPs also requires EBP1 to mediate silencing in other settings.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NCI grant R37 CA 30488 from the National Cancer Institute of Health. S.P.G. is an investigator of the Howard Hughes Medical Institute.
We thank Anne Hamburger (University of Maryland School of Medicine) for her generosity with reagents.
Footnotes
Published ahead of print 13 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02578-13.
REFERENCES
- 1.Hilberg F, Stocking C, Ostertag W, Grez M. 1987. Functional analysis of a retroviral host-range mutant: altered long terminal repeat sequences allow expression in embryonal carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 84:5232–5236. 10.1073/pnas.84.15.5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linney E, Davis B, Overhauser J, Chao E, Fan H. 1984. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. Nature 308:470–472. 10.1038/308470a0 [DOI] [PubMed] [Google Scholar]
- 3.Akgun E, Ziegler M, Grez M. 1991. Determinants of retrovirus gene expression in embryonal carcinoma cells. J. Virol. 65:382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flanagan JR, Krieg AM, Max EE, Khan AS. 1989. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol. Cell. Biol. 9:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukiyama T, Niwa O, Yokoro K. 1989. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol. Cell. Biol. 9:4670–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa O, Yokota Y, Ishida H, Sugahara T. 1983. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell 32:1105–1113. 10.1016/0092-8674(83)90294-5 [DOI] [PubMed] [Google Scholar]
- 7.Barklis E, Mulligan RC, Jaenisch R. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47:391–399. 10.1016/0092-8674(86)90596-9 [DOI] [PubMed] [Google Scholar]
- 8.Feuer G, Taketo M, Hanecak RC, Fan H. 1989. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 63:2317–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loh TP, Sievert LL, Scott RW. 1987. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol. Cell. Biol. 7:3775–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh TP, Sievert LL, Scott RW. 1990. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonal carcinoma cells. Mol. Cell. Biol. 10:4045–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen R, Kempler G, Barklis E. 1991. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell. Biol. 11:1214–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J. Virol. 82:4675–4679. 10.1128/JVI.02445-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46–57. 10.1016/j.cell.2007.07.026 [DOI] [PubMed] [Google Scholar]
- 14.Wolf D, Hug K, Goff SP. 2008. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc. Natl. Acad. Sci. U. S. A. 105:12521–12526. 10.1073/pnas.0805540105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204. 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz DC, Friedman JR, Rauscher FJ., III 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 15:428–443. 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamauchi M, Freitag B, Khan C, Berwin B, Barklis E. 1995. Stem cell factor binding to retrovirus primer binding site silencers. J. Virol. 69:1142–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Akinmade D, Hamburger AW. 2005. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 33:6024–6033. 10.1093/nar/gki903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Fondell JD, Wang Q, Xia X, Cheng A, Lu ML, Hamburger AW. 2002. Repression of androgen receptor mediated transcription by the ErbB-3 binding protein, Ebp1. Oncogene 21:5609–5618. 10.1038/sj.onc.1205638 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, Goloubeva O, Ross DD, Brodie A, Hamburger AW. 2005. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl. Acad. Sci. U. S. A. 102:9890–9895. 10.1073/pnas.0503829102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Woodford N, Xia X, Hamburger AW. 2003. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res. 31:2168–2177. 10.1093/nar/gkg318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A, Awasthi S, Hamburger AW. 2013. ErbB3-binding protein EBP1 decreases ErbB2 levels via a transcriptional mechanism. Oncol. Rep. 29:1161–1166. 10.3892/or.2012.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia X, Cheng A, Lessor T, Zhang Y, Hamburger AW. 2001. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell. Physiol. 187:209–217. 10.1002/jcp.1075 [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Rauscher FJ, III, Cress WD, Chen J. 2007. Regulation of E2F1 function by the nuclear corepressor KAP1. J. Biol. Chem. 282:29902–29909. 10.1074/jbc.M704757200 [DOI] [PubMed] [Google Scholar]
- 27.Honda A, Okamoto T, Ishihama A. 2007. Host factor Ebp1: selective inhibitor of influenza virus transcriptase. Genes Cells 12:133–142. 10.1111/j.1365-2443.2007.01047.x [DOI] [PubMed] [Google Scholar]
- 28.Gopinath M, Raju S, Honda A, Shaila MS. 2010. Host factor Ebp1 inhibits rinderpest virus transcription in vivo. Arch. Virol. 155:455–462. 10.1007/s00705-010-0599-y [DOI] [PubMed] [Google Scholar]
- 29.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- 30.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, Rossi F, Lorincz MC. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl. Acad. Sci. U. S. A. 108:5718–5723. 10.1073/pnas.1014660108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. 2013. Proviral silencing in embryonic cells is regulated by yin yang 1. Cell Rep. 4:50–58. 10.1016/j.celrep.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.