Abstract
The contributions of the five mv4Int- and two mv4Xis arm-binding sites to the spatial intasome organization of bacteriophage mv4 were found not to be equivalent. The 8-bp overlap region was mapped to the left extremity of the core region and is directly flanked by the P2 Int arm-binding site. These results and the absence of characteristic Int core-binding sites suggest that the P2 site is the determinant for integrase positioning and recognition of the core region.
TEXT
During the establishment of the lysogeny, many temperate bacteriophages integrate their genome into the host bacterial chromosome through a site-specific recombination mechanism. This integration is catalyzed by a phage-encoded recombinase (integrase) and occurs between the attB and attP attachment sites located on the bacterial and phage genomes, respectively. Prophage excision during the induction of lytic growth is also catalyzed by the recombinase: it involves recombination between the attR and attL attachment junctions and requires at least one phage-encoded recombination directionality factor (RDF) in addition to the integrase.
In well-characterized site-specific recombination systems, the protein binding sites at the att recombination sites are not all used in the same way for integrative or excisive recombination (1–3). The integrase of phage λ is a heterobivalent recombinase that binds two different families of DNA sequences: the “arm-type” and the “core-type” binding sites (4). The set of proteins and the number and combination of the binding sites used are different in integrative and excisive recombination reactions. During integrative recombination, in the intasome, Int binds to four arm-type binding sites (P1, P′1, P′2, and P′3) on attP whereas the accessory factor IHF binds to all three of its cognate sites (2). During excisive recombination, Xis binds to the X1, X1.5, and X2 sites on attR and Int to three of the arm-type sites (P2 on attR and P′1 and P′2 on attL) (2). In contrast, in phage KplE1, the same combination of IntS arm-type binding sites is used for both excisive and integrative recombination reactions (P1, P2, P′1, P′2, and P′4) and thus, the RDF alone plays a key role in directing the reaction toward excision (3).
The site-specific integration system of the Lactobacillus delbrueckii mv4 bacteriophage has been characterized (5, 6). The mv4 integrase has no absolute requirement for accessory factors (7). The minimal attP site of mv4 is 234 bp long, contains five putative integrase arm-binding sites, and has a 17-bp core sequence in common with the attB site which is located in the 3′ end of the tRNAser gene (5, 7). The RDF, mv4Xis, has been identified and characterized: it acts by binding to two sites within the attR or attP sequences, thereby bending the DNA (8).
Here, we report the use of in vitro recombination assays to analyze the involvement of the various binding sites for proteins (factors) in integrative and excisive recombination reactions. We also identified the location of strand exchange by in vitro recombination with suicide substrates.
To analyze the involvement of each Int- or Xis-binding site (Fig. 1A) in the site-specific recombination of phage mv4, we generated mutated sites each bearing three- to six-base changes (Table 1). Each mutated site was tested individually in in vitro recombination tests as supercoiled DNA (plasmid pRC [see Table S1 in the supplemental material]; 200 ng) or as radiolabeled linear DNA (104cpm) with the appropriate partner (7, 8). Each mutated site was also tested with various mutated partners to analyze the cumulative effect of mutations on the recombination efficiency (RE).
FIG 1.
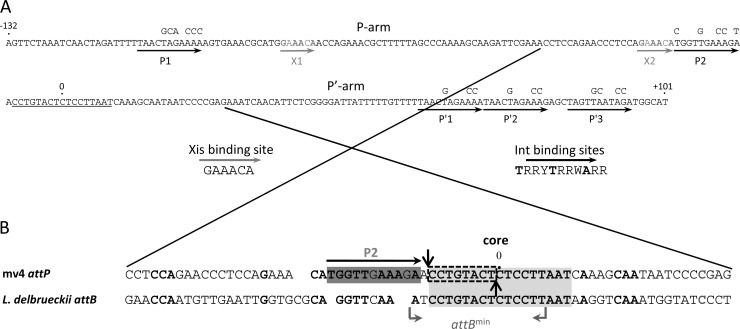
(A) Structure and nucleotide sequence of the mv4 attP region. The core sequence is underlined. 0 corresponds to the central base of the core region. The black arrows indicate the 11-bp consensus sequence 5′-TRRYTRRWARR-3′, which is present in five copies in mv4 attP and corresponds to the Int arm-type binding sites. The gray arrows indicate the 6-bp sequence of the Xis-binding site. The mutations in the Int arm-type binding sites are indicated above the nucleotide sequence. (B) Structure and nucleotide sequence of the P2-core region. The 17-bp core sequence and the P2 Int arm-type binding site are indicated by a light gray box and a dark gray box, respectively. The gray bent arrows indicate the 16-bp minimal attB site (attBmin). The dark vertical arrows indicate the strand exchange loci on the top and bottom strands. The dashed box encircles the overlap region.
TABLE 1.
List of primers used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| DR9 | CGGGATCCAGTTCTAAATCAACTAGATTTTTAACT |
| L1 | CACCATCTTAAAAATAACTT |
| P1mut | ATTTTTAACGCAACCCGGTGAA |
| P1mutC | TTCACCGGGTTGCGTTAAAAAT |
| X1mut | AAACGCAGCAACCCGACCAGA |
| X1mutC | TCTGGTCGGGTTGCTGCGTTT |
| X2mut | AACCCTCGCAACCCGTGGTTG |
| X2mutC | CAACCACGGGTTGCGAGGGTT |
| P2mut | GAAACACGGTGGACCGTACCTG |
| P2mutC | CAGGTACGGTCCACCGTGTTTC |
| P′1mut | TGTTTTTAACGAGACCATAACT |
| P′1mutC | AGTTATGGTCTCGTTAAAAACA |
| P′2mut | AGAAAATAACGAGACCGAGCTA |
| P′2mutC | TAGCTCGGTCTCGTTATTTTCT |
| P′3mutC | ATGCCATGGATGCACTAGCTCT |
| DR3 | ATGCCATCTATTAACTAGCT |
| attB | CATTTGATTTAGATGTCCTT |
| P2core | CATGGTTGAAAGAAcctgtactctccttaatCAAAGCAA |
| coreP2 | TTGCTTTGATTAAGGAGAGTACAGGTTCTTTCAACCATG |
| P2core′ | CATGGTTGAAAGAACCTGTAC |
| ′P2core | TCTCCTTAATCAAAGCAA |
| coreP2′ | TTGCTTTGATTAAGGAGAGTACAG |
| ′coreP2 | GTTCTTTCAACCATG |
The mutations are in bold, the binding sites are underlined, and the core region is in lowercase characters.
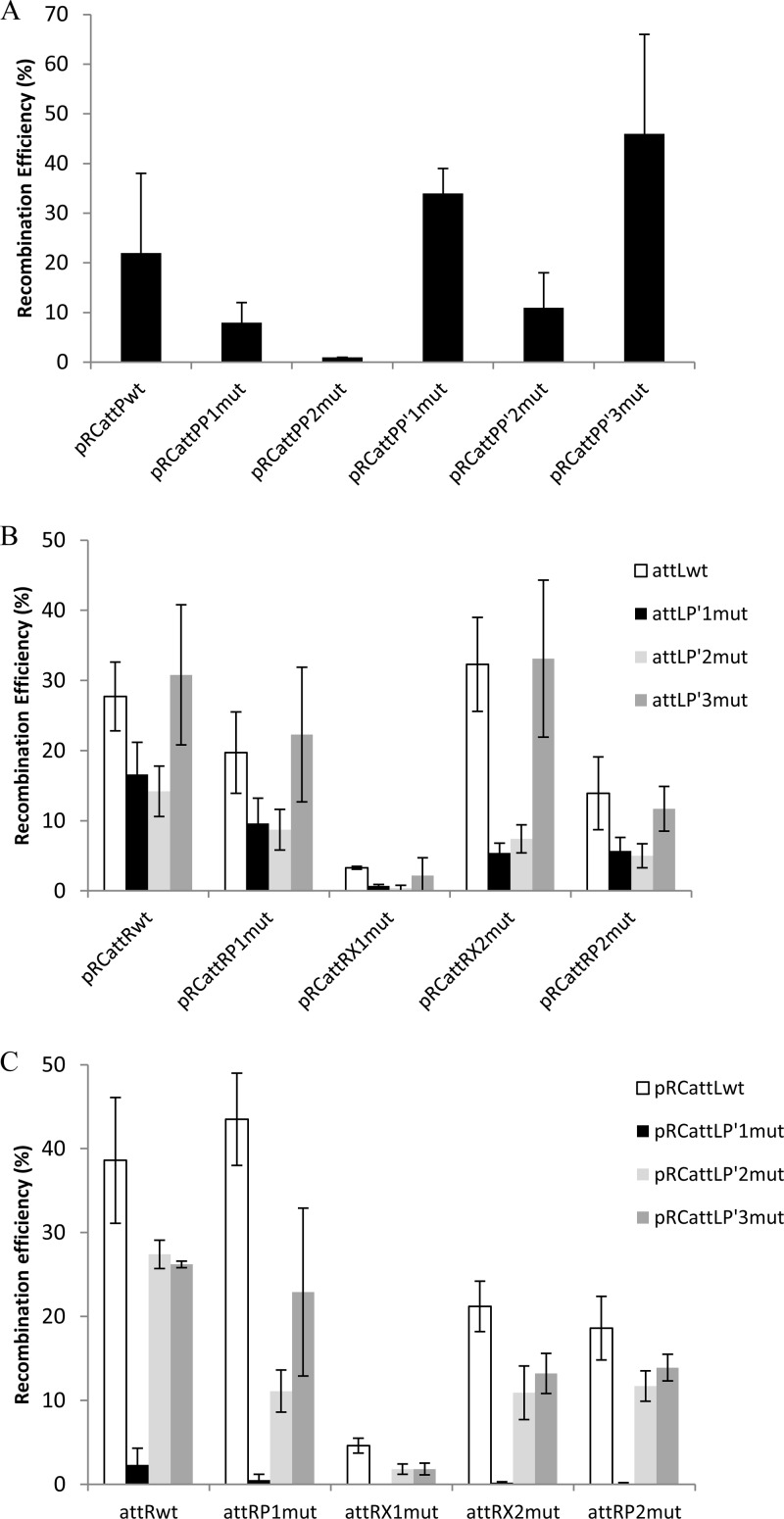
For integrative recombination, the RE value in the presence of linear DNA carrying attBwt was substantially affected for pRCattPP2mut (−95%) compared to pRCattPwt. The RE was also lower than control values for the P1 and P′2 mutations but to a lesser extent (−65% and −50%, respectively). The P′1 and P′3 mutations showed increased efficiency (+50% and +100%, respectively) (Fig. 2A).
FIG 2.
In vitro site-specific recombination. Effects of mutations of the binding sites on integrative (A) and excisive (B and C) recombinations are depicted. The efficiency of recombination between supercoiled plasmid bearing attP or attR or attL (200 ng [0.1 pmol]) and linearly radiolabeled attB or attL or attR (104 cpm) in the presence of mv4Int-enriched cell extract (3 μg [∼20 pmol]), with or without Xis cell extracts (3 μg [∼10 pmol]), was quantified as previously described (7). The values reported in the histograms are means and standard deviations of the results of 4 independent experiments.
In excisive recombination, the values of RE between pRCattRwt and the linear attLP′1mut or attLP′2mut sequences were lower by 40% and 50%, respectively, than that for attLwt (Fig. 2B). In contrast, the RE was not affected by mutant attLP′3mut. The RE between the linear attLwt and pRCattRX1mut sequences was dramatically reduced (−90%) compared to that between the wild-type sequences. Similarly, the values of RE between the attLwt sequence and each pRCattRP2mut and pRCattRP1mut sequence were lower than the control values (−50% and −30%, respectively). No effect was observed when attLwt was recombined with pRCattRX2mut. Thus, the various attR mutated sites behaved in a way similar to the attRwt sequence in assays with the various attL mutated sites. The only exceptions were for attLP′1mut and attLP′2mut sequences tested with pRCattRX2mut: in these cases, the RE was substantially more affected (−85% and −80%) than was the case with the other attR mutated sites.
We then reversed the topological context of the partners: assays were performed with attL on a plasmid (supercoiled) and attR on linear DNA. The RE between pRCattLwt and the linear attRP1mut fragment was the same as that between pRCattLwt and linear attRwt (Fig. 2C), and the RE was 50% lower with attR X2mut fragment (Fig. 2C). The RE with pRCattLP′1mut was much lower than that observed with attLP′1mut (−95% with respect to control values) whatever the attR mutant partner (Fig. 2B and C). The relative RE values for the various attR mutations were similar to those seen with pRCattLP′3mut and pRCattLwt, but the absolute values were about 40% lower with the mutant (Fig. 2C). These results are very different from those observed when attL was in linear form (Fig. 2B). This indicates that the topology of the attL DNA has a noteworthy effect on recombination.
We have shown that the X1 Xis-binding site on the P arm is absolutely required for excisive site-specific recombination and that the P′1 Int-binding site on the P′ arm plays a significant role. The P2 Int-binding site on the P arm is secondarily important for excisive recombination but is absolutely required for integrative recombination.
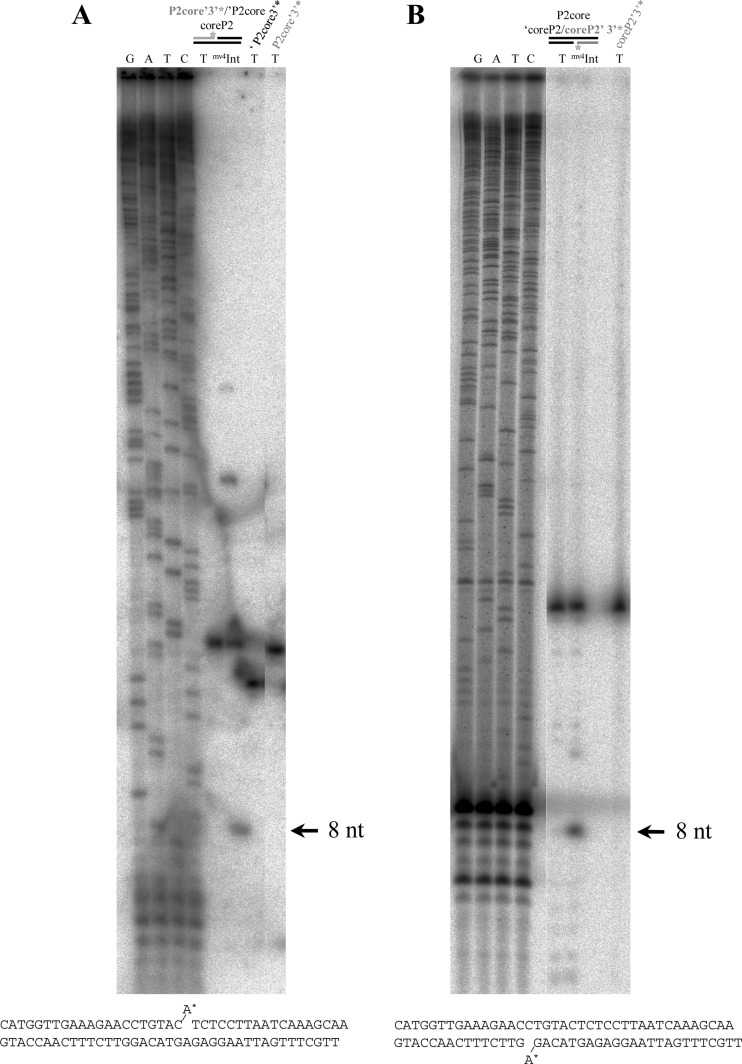
To localize the site of strand exchange, we used suicide substrates (9) corresponding to the core region of phage mv4 recombination sites. We investigated the sites of mv4Int cleavage on both strands: the top strand was cleaved immediately upstream of the C at position −8, which is the first base of the core region (Fig. 1B and 3A); the bottom-strand cleavage was immediately downstream of position −1 (Fig. 1B and 3B). The overlap region was thus 8 bases long, characteristic of the overlap region associated with tyrosine recombinases. The mv4 overlap region is located at the left extremity of the core region and is directly flanked by the P2 Int arm-binding site. These findings will help identify the Int core-binding site necessary for intasome assembly and the strand exchange.
FIG 3.
Identification of the positions of strand exchange. (A) Localization of the position of the cleavage on the top strand. The result of the in vitro recombination assay on the suicide substrate (9) was analyzed on a 12% acrylamide–7 M urea gel in parallel with a sequence scale. At the top of the gel, the suicide substrate is schematized. In lanes T, the reaction mixture contained only the radiolabeled substrate without cell extract. In lane mv4Int, the reaction mixture contained the radiolabeled substrate in the presence of an mv4Int-enriched cell extract (3 μg [∼20 pmol]). The arrow indicates the length of the cleaved fragment. The sequence of the suicide substrate is indicated below the autoradiogram. (B) Localization of the position of the cleavage on the bottom strand. The reactions were analyzed as described for panel A.
In this report, we show that the different binding sites for proteins involved in the site-specific recombination of phage mv4 do not play the same roles in integrative and excisive recombination; this is consistent with findings for other well-characterized recombination systems (1–3). Three binding sites have a key role in the recombination reactions and are absolutely required, two, the X1 Xis-binding site and the P′1 Int-binding site, for excisive recombination and one, the P2 Int arm-type binding site, for integrative recombination. The other protein-binding sites have contributory roles in integrative or excisive recombination and, in some cases, their influence is dependent of the topology of the DNA at the att site.
The overall organization of the mv4 attP site highlights the absence of binding sites for bending proteins between the Int arm-type binding sites and the core region (8); this is unlike what has been found for the attP site in well-characterized phages (3, 10–13). Analysis of the core region sequence does not reveal any Int core-type binding sites (14), and the Int P2 arm-binding site has a noticeable position close to the left border of the core region (Fig. 1B). We studied the site of strand exchange and identified an overlap region of 8 bp which is the sequence that is closest to the P2 arm-binding site (Fig. 1B). In well-characterized integrative intasomes, four monomers of Int recombinase interact with the core binding sites on both sides of the overlap region (15). In the mv4 intasome, Int monomers should similarly be bound on both sides of the mv4 overlap region; this implies that if it exists, one of the Int core-binding sites may overlap the P2 arm-binding site. The attB site shares several nucleotides in common with the P2 arm-binding site (Fig. 1B), and this sequence may be important for Int recognition of the core region. The P2 arm-binding site may therefore be central to the capture of the attB site to form the synapsed intermediate, as has been shown for phage lambda (15).
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Nolivos for advice and helpful discussions.
This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS-UMR5100).
Footnotes
Published ahead of print 20 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02735-13.
REFERENCES
- 1.Moitoso de Vargas L, Landy A. 1991. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc. Natl. Acad. Sci. U. S. A. 88:588–592. 10.1073/pnas.88.2.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbaker D, Azaro MA, Landy A. 2008. A biotin interference assay highlights two different asymmetric interaction profiles for lambda integrase arm-type binding sites in integrative versus excisive recombination. J. Biol. Chem. 283:12402–12414. 10.1074/jbc.M800544200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panis G, Duverger Y, Champ S, Ansaldi M. 2010. Protein binding sites involved in the assembly of the KplE1 prophage intasome. Virology 404:41–50. 10.1016/j.virol.2010.04.027 [DOI] [PubMed] [Google Scholar]
- 4.Moitoso de Vargas L, Pargellis CA, Hasan NM, Bushman EW, Landy A. 1988. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell 54:923–929. 10.1016/0092-8674(88)90107-9 [DOI] [PubMed] [Google Scholar]
- 5.Dupont L, Boizet-Bonhoure B, Coddeville M, Auvray F, Ritzenthaler P. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auvray F, Coddeville M, Ritzenthaler P, Dupont L. 1997. Plasmid integration in a wide range of bacteria mediated by the integrase of Lactobacillus delbrueckii bacteriophage mv4. J. Bacteriol. 179:1837–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auvray F, Coddeville M, Espagno G, Ritzenthaler P. 1999. Integrative recombination of Lactobacillus delbrueckii bacteriophage mv4: functional analysis of the reaction and structure of the attP site. Mol. Gen. Genet. 262:355–366. 10.1007/s004380051094 [DOI] [PubMed] [Google Scholar]
- 8.Coddeville M, Ritzenthaler P. 2010. Control of directionality in bacteriophage mv4 site-specific recombination: functional analysis of the Xis factor. J. Bacteriol. 192:624–635. 10.1128/JB.00986-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blakely GW, Davidson AO, Sherratt DJ. 1997. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J. Mol. Biol. 265:30–39. 10.1006/jmbi.1996.0709 [DOI] [PubMed] [Google Scholar]
- 10.Esposito D, Thrower JS, Scocca JJ. 2001. Protein and DNA requirements of the bacteriophage HP1 recombination system: a model for intasome formation. Nucleic Acids Res. 29:3955–3964. 10.1093/nar/29.19.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis JA, Hatfull GF. 2003. Control of directionality in L5 integrase-mediated site-specific recombination. J. Mol. Biol. 326:805–821. 10.1016/S0022-2836(02)01475-4 [DOI] [PubMed] [Google Scholar]
- 12.Yu A, Haggard-Ljungquist E. 1993. The Cox protein is a modulator of directionality in bacteriophage P2 site-specific recombination. J. Bacteriol. 175:7848–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papagiannis CV, Sam MD, Abbani MA, Yoo D, Cascio D, Clubb RT, Johnson RC. 2007. Fis targets assembly of the Xis nucleoprotein filament to promote excisive recombination by phage lambda. J. Mol. Biol. 367:328–343. 10.1016/j.jmb.2006.12.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auvray F, Coddeville M, Ordonez RC, Ritzenthaler P. 1999. Unusual structure of the attB site of the site-specific recombination system of Lactobacillus delbrueckii bacteriophage mv4. J. Bacteriol. 181:7385–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patsey RL, Bruist MF. 1995. Characterization of the interaction between the lambda intasome and attB. J. Mol. Biol. 252:47–58. 10.1006/jmbi.1995.0474 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.