Abstract
In February 2013, zoonotic transmission of a novel influenza A virus of the H7N9 subtype was reported in China. Although at present no sustained human-to-human transmission has been reported, a pandemic outbreak of this H7N9 virus is feared. Since neutralizing antibodies to the hemagglutinin (HA) globular head domain of the virus are virtually absent in the human population, there is interest in identifying other correlates of protection, such as cross-reactive CD8+ T cells (cytotoxic T lymphocytes [CTLs]) elicited during seasonal influenza A virus infections. These virus-specific CD8+ T cells are known to recognize conserved internal proteins of influenza A viruses predominantly, but it is unknown to what extent they cross-react with the newly emerging H7N9 virus. Here, we assessed the cross-reactivity of seasonal H3N2 and H1N1 and pandemic H1N1 influenza A virus-specific polyclonal CD8+ T cells, obtained from HLA-typed study subjects, with the novel H7N9 virus. The cross-reactivity of CD8+ T cells to H7N9 variants of known influenza A virus epitopes and H7N9 virus-infected cells was determined by their gamma interferon (IFN-γ) response and lytic activity. It was concluded that, apart from recognition of individual H7N9 variant epitopes, CD8+ T cells to seasonal influenza viruses display considerable cross-reactivity with the novel H7N9 virus. The presence of these cross-reactive CD8+ T cells may afford some protection against infection with the new virus.
INTRODUCTION
Influenza viruses are an important cause of respiratory tract infections. Occasionally, animal influenza viruses cross the species barrier and infect humans after zoonotic transmission. In the past 2 decades, several avian influenza A viruses, like those of the H9N2 subtype (1), the H7N7 subtype (2, 3), and the H5N1 subtype (4–9), have infected humans. In 2009, H1N1 influenza A viruses of swine origin (H1N1pdm09) caused a pandemic outbreak, and these viruses continue to circulate in the human population (10).
In February 2013, the first human cases of infection with a novel avian influenza A virus of the H7N9 subtype were reported in China. As of September 2013, 135 laboratory-confirmed cases had been reported, 44 of which had a fatal outcome (11). Older male individuals especially seem to be at risk for developing severe disease upon infection (12–15). Most hospitalized patients developed severe viral pneumonia and acute respiratory distress syndrome (ARDS) (16–19).
Influenza A viruses with hemagglutinin (HA) and neuraminidase (NA) of subtypes H7 and N9, respectively, circulate in wild bird species (20, 21). The newly emerged H7N9 virus is most likely the result of multiple reassortment events of at least three avian viruses (17, 22, 23). Although the H7N9 virus has been classified as a low-pathogenic virus based on the intravenous pathogenicity index (IVPI) in chickens and the absence of a multibasic cleavage site in the HA, it is quite pathogenic in humans (17). The virus also replicates efficiently in the airways of other mammalian species, including mice, ferrets, and cynomolgus macaques (24, 25). It is more pathogenic than seasonal influenza A H3N2 (sH3N2) viruses or pandemic 2009 H1N1 (pH1N1) viruses and after intratracheal inoculation causes fatal disease in ferrets (26). The high pathogenicity in mammals correlates with the presence of known pathogenicity markers. Several human isolates of the H7N9 virus contain the E627K substitution in PB2, which allows avian influenza viruses to replicate at lower temperatures (27). A deletion of 5 amino acids in the NA of H7N9 virus is associated with enhanced virus replication (17). The presence of the Q226L substitution in the HA (17, 28) is associated with binding to alpha(2,6)-linked sialic acids found in the human upper respiratory tract (24) and has been associated with airborne transmission of avian H5N1 virus in ferrets (29). In the case of the novel H7N9 virus, only limited transmission between ferrets was observed (24, 25, 30, 31). Acquisition of gene segments from human influenza A viruses by the avian influenza H7N9 virus through genetic reassortment may lead to further adaptation to humans (10, 32–37). The detection of an H7N9 patient who was coinfected with an sH3N2 virus underscores this possible scenario (38). Although at present no sustained human-to-human transmission of the H7N9 virus has been reported (39), the pandemic potential of H7N9 virus should be considered seriously, especially since virus-neutralizing antibodies directed to the HA globular head domain of the virus are virtually absent in the human population (18), though low concentrations of stalk region-specific antibodies might be present (40, 41).
On the other hand, virus-specific CD8+ T cells (cytotoxic T lymphocytes [CTLs]), induced after infection with seasonal influenza A viruses, are mainly directed to the conserved internal proteins of influenza A viruses (33, 42–51). The presence of these cross-reactive CD8+ T cells may afford a certain degree of heterosubtypic immunity against infection with novel H7N9 viruses. Using various combinations of influenza A virus subtypes for primary and secondary infection, this type of immunity and the contribution of virus-specific CD8+ T cells were demonstrated in various animal models (52–57). Evidence for heterosubtypic immunity and the role of CD8+ T cells in humans is limited (58–61), though the presence of CD8+ T cells cross-reactive with avian H5N1 and swine origin triple-reassortant A H3N2 (vH3N2) viruses has been demonstrated (49–51, 62). It is unknown to what extent CD8+ T cells elicited by a seasonal or 2009 pH1N1 influenza A virus infection cross-react with the novel H7N9 virus. Here, we show that polyclonal CD8+ T cell populations specific for seasonal H1N1 (sH1N1), sH3N2, or pH1N1 virus cross-react with the H7N9 virus by determining their gamma interferon (IFN-γ) response upon in vitro stimulation with the novel H7N9 virus and their lytic activity toward H7N9 virus-infected human leukocyte antigen (HLA)-matched target cells. The preexisting cross-reactive CD8+ T cells may afford some level of protection and may reduce morbidity and mortality caused by infections with the novel H7N9 virus.
MATERIALS AND METHODS
Cells.
Peripheral blood mononuclear cells (PBMCs) were obtained from 6 HLA-typed healthy blood donors (35 to 50 years of age) between 2008 and 2013 (Sanquin Bloodbank, Rotterdam, The Netherlands). Lymphoprep (Axis-Shield PoC, Oslo, Norway) gradient centrifugation was used to isolate PBMCs, which were subsequently cryopreserved at −135°C. Donors were selected based on their HLA class I alleles for which functionally confirmed influenza A virus HLA class I epitopes have been identified and had the following HLA haplotypes: subjects 1 and 2, HLA-A*0101, -A*0201, -B*0801, and -B*3501; subjects 3 and 4, HLA-A*0101, -A*0201, -B*0801, -B*2705; and subjects 5 and 6, HLA-A*0101, -A*0301, -B*0801, and -B*3501. The use of PBMCs for scientific research was approved by the Sanquin Bloodbank after informed consent was obtained from the blood donors.
Peptides.
The amino acid sequences of confirmed influenza A virus HLA class I epitopes were aligned with their H7N9 analogues from human isolates between February 2013 and 22 April 2013 (Table 1). Sequences were obtained from the influenza virus resource database (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database). In addition, conservation of these epitope sequences in the prototype viruses used in the present study, sH3N2 (A/Netherlands/348/07), sH1N1 (A/Netherlands/26/07), and pH1N1 (A/Netherlands/602/09), was determined (Table 1). The H7N9 variant epitopes for which the HLA restriction was compatible with the HLA type of the study subjects were ordered as synthetic immunograde peptides (>85% purity) (Eurogentec, Seraing, Belgium).
TABLE 1.
Variant amino acid sequences of known CD8+ T cell epitopes in the influenza A H7N9 virusa
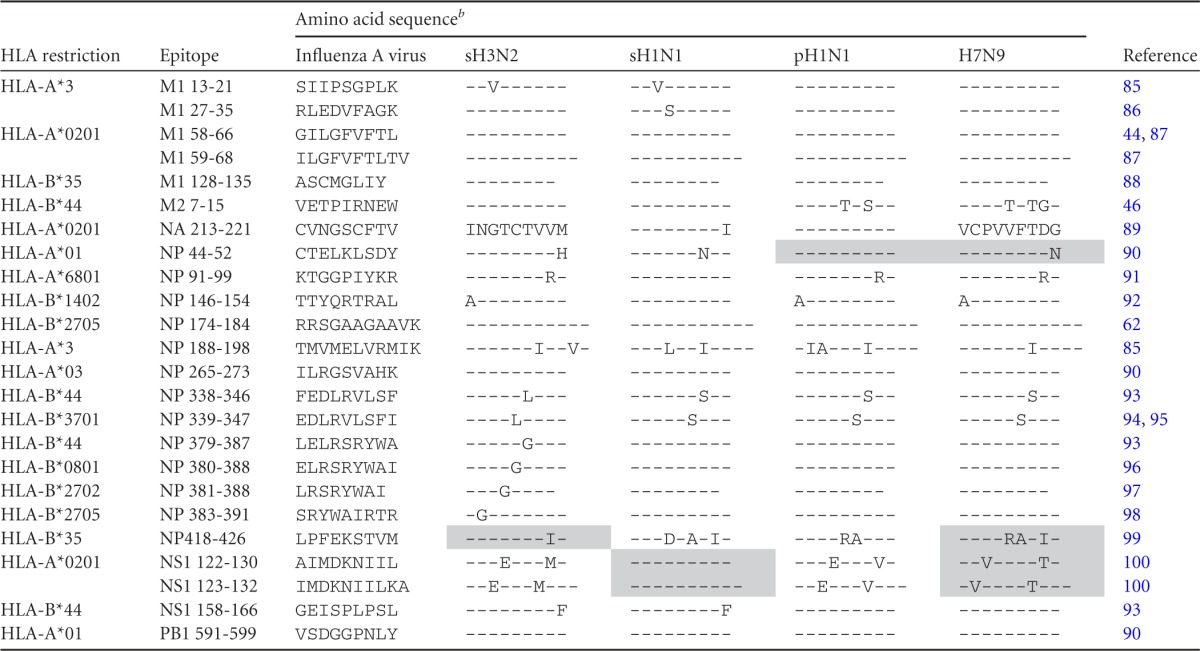
The A/Anhui/1/13 (H7N9) sequence was unavailable at the time of ordering the peptides. All epitopes, except LPFEKSTVM (H7N9 LPFERARIM), were conserved between the H7N9 viruses present in the database at April 22th and the A/Anhui/1/13 virus used in this study.
Peptides used in the present study are shaded and were selected based on variation in the H7N9 sequence and correspondence to the HLA alleles of the study subjects. Synthetic immunograde peptides were ordered with >85% purity. The dashes indicate identity with the amino acids in the influenza A virus sequence.
Viruses.
Influenza virus A/Anhui/1/2013 (H7N9) was isolated from a fatal human case (Anhui Province, People's Republic of China) and was kindly provided through the WHO Pandemic Influenza Preparedness (PIP) framework and subsequently passaged once in Madin Darby Canine Kidney (MDCK) cells. Prototypic seasonal influenza A viruses A/Netherlands/348/07 (sH3N2), A/Netherlands/26/07 (sH1N1), and A/Netherlands/602/09 (pH1N1) were propagated in MDCK cells. Culture supernatants were clarified by low-speed centrifugation and subsequently purified by ultracentrifugation through a sucrose gradient. Their infectious-virus titers were determined as described previously (63).
Amino acid sequence identity.
The amino acid sequence identity of the viral proteins of influenza viruses A/Anhui/1/2013 (H7N9) and the prototype sH3N2, sH1N1, and pH1N1 was determined using BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Table 2). The consensus sequence of A/Anhui/1/2013 was obtained from the GISAID database (http://platform.gisaid.org), and the consensus sequence of the influenza virus A/Anhui/1/2013 preparation used in the present study was confirmed by sequence analysis (30).
TABLE 2.
Percent amino acid sequence identity with A/Anhui/1/2013 (H7N9)
| Gene segment | % Identity |
||
|---|---|---|---|
| sH3N2 | sH1N1 | pH1N1 | |
| PB2 | 94 | 94 | 97 |
| PB1 | 97 | 95 | 96 |
| PA | 94 | 95 | 96 |
| HA | 47 | 41 | 41 |
| NP | 91 | 92 | 93 |
| NA | 45 | 43 | 45 |
| M1 | 91 | 91 | 92 |
| M2 | 82 | 78 | 89 |
| NS1 | 76 | 80 | 78 |
| NS2 | 93 | 90 | 88 |
In vitro expansion of influenza A virus-specific CD8+ T cells.
PBMCs obtained from HLA-typed study subjects were stimulated with sH3N2, sH1N1, and pH1N1 viruses at a multiplicity of infection (MOI) of 3, as described previously (64). Eight days after stimulation, polyclonal CD8+ T cells were isolated from the expanded PBMC cultures by means of CD8+ magnetically activated cell sorting (MACS) bead sorting according to the manufacturer's recommendations (Miltenyi Biotec, Bergisch Gladbach, Germany) and subsequently used as effector cells in IFN-γ enzyme-linked immunosorbent spot (ELISpot) and lytic assays (see below).
Target cells.
HLA-matched B lymphoblastoid cell lines (BLCLs) were prepared as described previously (65). The cells (106) were incubated with or without 100 μM peptide for 16 h at 37°C and subsequently washed and resuspended in RPMI 1640 medium (Lonza, Basel, Switzerland) containing antibiotics and 10% fetal bovine serum (Sigma-Aldrich, Zwijndrecht, The Netherlands) (R10F medium). Virus-infected target cells were prepared by inoculating BLCLs at an MOI of 3 with sH3N2, sH1N1, pH1N1, or H7N9 virus. After 1 h, the cells were washed and resuspended in R10F medium and cultured for 16 to 18 h at 37°C before being used for the stimulation of T cells or as target cells.
IFN-γ ELISpot assay.
The IFN-γ responses of in vitro-expanded polyclonal CD8+ T cells were determined by ELISpot assays, which were performed according to the manufacturer's instructions (Mabtech, Nacka Strand, Sweden). In brief, 10,000 or 5,000 in vitro-expanded polyclonal CD8+ T cells were used as effector cells and incubated for 16 to 18 h with 30,000 peptide-loaded, virus-infected, or untreated HLA class I-matched target cells, in triplicate. The average number of spots was determined using an ELISpot reader and image analysis software (Aelvis, Sanquin Reagents, Amsterdam, The Netherlands).
CTL assay.
The lytic capacity of the in vitro-expanded polyclonal CD8+ T cells was determined using a CTL assay with carboxyfluorescein succinimidyl ester (CFSE)-labeled target cells. In brief, 5 × 106 cells of HLA class I-matched BLCLs were incubated with 50 μM CFSE (Sigma-Aldrich, Zwijndrecht, The Netherlands) for 5 min at 37°C. Subsequently, these cells were inoculated with sH3N2, sH1N1, pH1N1, or H7N9 virus at an MOI of 3 for 16 to 18 h. The infected and CFSE-labeled BLCL target cells were cocultured with the in vitro-expanded polyclonal CD8+ effector T cells in effector-to-target cell (E:T) ratios of 5, 2.5, and 1.25. After a 3-h incubation period, the cells were fixed using Cytofix/Cytoperm (BD Biosciences, Breda, The Netherlands), and lysis in the target cell population was determined by flow cytometry using BD FACSDiva software (Becton, Dickinson B.V., Breda, The Netherlands). Experiments were performed in triplicate.
Statistical analysis.
The data were analyzed using an independent t test, and differences were considered significant at a P value of <0.05.
RESULTS
Comparison of amino acid sequences of CD8+ T cell epitopes.
The amino acid sequences of 24 confirmed influenza A virus HLA class I epitopes were compared with their influenza A H7N9 virus analogues. As shown in Table 1, most epitopes (>50%) were fully conserved in H7N9 viruses. Based on these results, four variant H7N9 epitopes that were conserved in our prototypic sH3N2, sH1N1, and/or pH1N1 viruses (Table 1) and were compatible with the HLA type of the study subjects under investigation were further tested for cross-recognition in the ELISpot assay. All epitopes except NP418-426 were conserved among H7N9 viruses available in the influenza virus resource database (22 April 2013) and the A/Anhui/1/13 (H7N9) virus used in this study.
Cross-recognition of influenza A (H7N9) analogues of known influenza A HLA class I epitopes.
In vitro-expanded CD8+ T cell preparations specific for sH3N2, sH1N1, and pH1N1 influenza viruses were tested for their cross-reactivity with the selected H7N9 variant epitopes listed in Table 1 using peptide-loaded HLA-matched BLCLs.
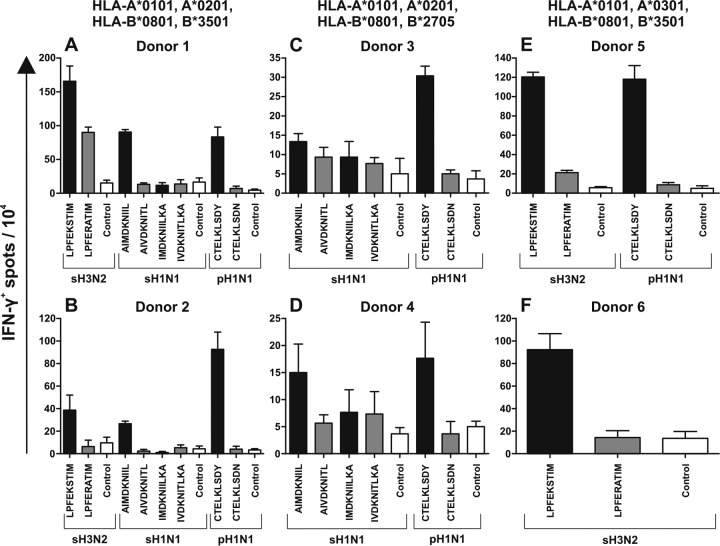
Virus-specific CD8+ T cells obtained from study subjects 1 and 2 (HLA-A*0101, -A*0201, -B*0801, and -B*3501) displayed strong reactivity with the homologous epitopes, except for epitope NS123-132 (IMDKNIILKA) (Fig. 1A and B). The H7N9 variant of the NP418-426 (LPFERATIM) epitope was recognized by sH3N2-specific CD8+ T cells derived from subject 1, although the IFN-γ response was lower than the response to the homologous epitope (LPFEKSTIM) (Fig. 1A). None of the other H7N9 variant epitopes were recognized by virus-specific CD8+ T cells of these HLA-A*0101, -A*0201, -B*0801, and -B*3501 study subjects.
FIG 1.
Epitope-specific IFN-γ production by seasonal influenza virus-specific CD8+ T cells after stimulation with peptide-loaded BLCLs. Polyclonal CD8+ T cells were isolated from PBMCs in vitro stimulated with sH3N2, sH1N1, or pH1N1, as indicated. No pH1N1 in vitro stimulation was performed for subject 6, since those PBMCs were isolated in 2008, prior to the 2009 pandemic outbreak. The polyclonal CD8+ T cells were subsequently stimulated with peptide-loaded and untreated HLA class I-matched BLCLs. Stimulation with homologous peptides is indicated by black bars, stimulation with H7N9 variant peptides is indicated by gray bars, and control cells without peptide are indicated by white bars. The number of IFN-γ-producing cells per 10,000 polyclonal CD8+ T cells was determined by ELISpot assay. The results represent the averages of triplicate wells. Peptides were selected based on the variation in the H7N9 sequence and their compatibility with the HLA haplotypes of our study subjects. The error bars indicate standard deviations of results from the triplicate wells.
Virus-specific CD8+ T cells obtained from study subjects 3 and 4 (HLA-A*0101, -A*0201, -B*0801, and -B*2705) displayed a minor response to homologous epitopes NS122-130 and NS123-132 (Fig. 1C and D). This is in agreement with the subdominant nature of the responses to these epitopes in these subjects (data not shown). CD8+ T cells from both subjects did not display any response to the H7N9 variant of the NS122-130 and NS123-132 epitopes (Fig. 1C and D). Although CD8+ T cells of the two subjects displayed reactivity with the homologous NP44-52 epitope, they did not respond to the H7N9 variant of the epitope (CTELKLSDN).
Virus-specific CD8+ T cells from study subjects 5 and 6 (HLA-A*0101, -A*0301, -B*0801, and -B*3501) displayed a strong response to the homologous sH3N2 variant of the NP418-426 epitope (Fig. 1E and F). Some minor cross-reactivity with the H7N9 variant (LPFERATIM) was observed with CD8+ T cells derived from subject 5 (Fig. 1E). As for the other subjects, no cross-reactivity was observed with the H7N9 variant of the NP44-52 (CTELKLSDN) epitope with CD8+ T cells obtained from subject 5.
Overall, the extent of cross-reactivity of influenza virus-specific CD8+ T cells against individual H7N9 variant epitopes was low and dependent on the study subjects and peptides tested.
CD8+ T cells cross-react with influenza A H7N9 virus-infected cells.
Since more than 50% of previously identified influenza virus HLA class I epitopes were present in the H7N9 virus, we wished to compare the overall amino acid sequence identities between the H7N9 virus and the prototypic sH3N2, sH1N1, and pH1N1 viruses used in the present study. BLAST analysis revealed that the sequence identity of most viral proteins was high (>76%), except for hemagglutinin and neuraminidase (Table 2).
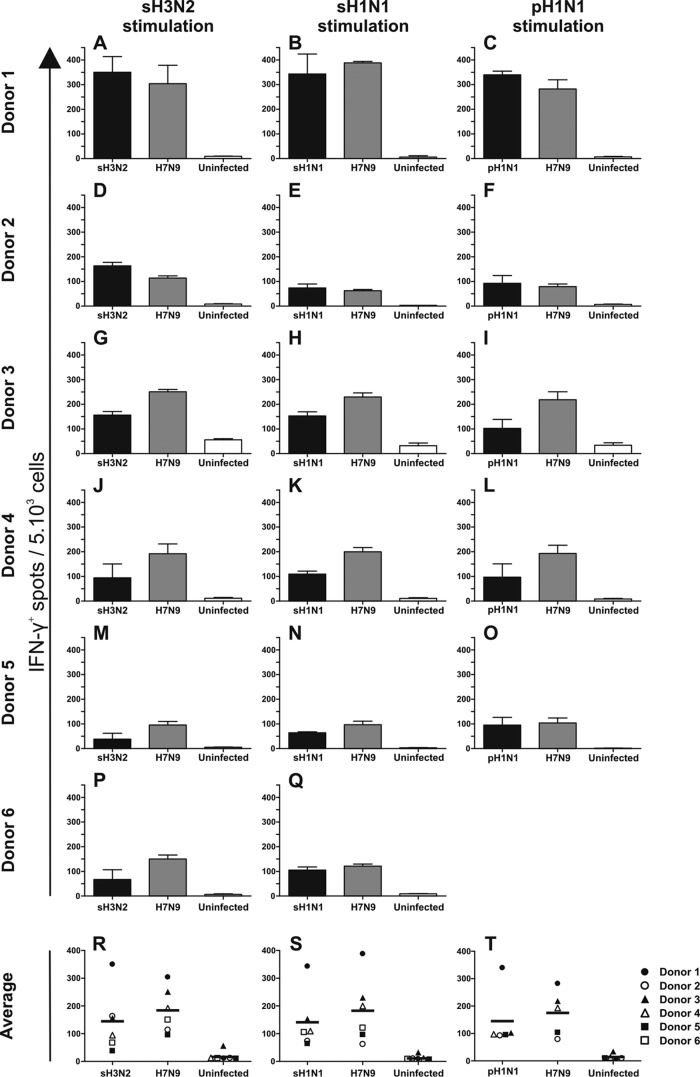
Since the sequence identity between seasonal influenza viruses used in this study and H7N9 virus is high, we wished to determine the cross-reactivity of polyclonal CD8+ T cells specific for sH3N2, sH1N1, or pH1N1 viruses with H7N9 virus. To this end, in vitro-expanded seasonal influenza virus-specific polyclonal CD8+ T cells were stimulated with HLA class I-matched BLCLs infected with the homologous seasonal influenza A virus or H7N9 virus (A/Anhui/1/2013). The number of IFN-γ-producing cells per 5,000 CD8+ T cells was determined in an IFN-γ ELISpot assay (Fig. 2).
FIG 2.
Virus-specific IFN-γ production by polyclonal CD8+ T cells after stimulation with BLCLs infected with homologous or H7N9 virus. (A to Q) Seasonal influenza virus-specific polyclonal CD8+ T cells were isolated from PBMCs stimulated with sH3N2 (A, D, G, J, M, and P), sH1N1 (B, E, H, K, N, and Q), or pH1N1 (C, F, I, L, and O). PBMCs of subject 6 were not stimulated in vitro with pH1N1, since they were isolated prior to the pH1N1 outbreak. The CD8+ T cells were subsequently cocultured with BLCLs infected with homologous virus (sH3N2, sH1N1, or pH1N1) (black bars) or the heterologous novel H7N9 virus (gray bars). The number of IFN-γ-producing cells per 5,000 polyclonal CD8+ T cells was determined by ELISpot assay. Uninfected BLCLs were used as negative controls (white bars). Experiments were performed in triplicate. The error bars indicate standard deviations for the triplicates. (R, S, and T) The symbols represent the averages of triplicate experiments for each individual subject, and the horizontal bars represent the average responses of all study subjects combined.
Study subject 1 showed a high response to homologous seasonal influenza viruses (sH3N2, sH1N1, and pH1N1), but also after stimulation with H7N9 virus-infected cells (Fig. 2A, B, and C). Although the frequency of seasonal influenza virus-specific CD8+ T cells derived from subject 2 was lower than that of cells derived from subject 1, these T cells also cross-reacted with H7N9 virus-infected cells (Fig. 2D, E, and F). Subjects 3 and 4 responded to both the homologous viruses and H7N9 virus (Fig. 2G, H, I, J, K, and L). Subjects 5 and 6 (who lack the HLA-A*0201 allele) showed the lowest response to stimulation with homologous viruses. However, the virus-specific CD8+ T cells of these two subjects also displayed cross-reactivity with H7N9 virus (Fig. 2M, N, O, P, and Q).
Thus, although the frequency of virus-specific IFN-γ-producing T cells varied between the study subjects, the cells cross-reacted with the H7N9 virus. This was independent of the sH3N2, sH1N1, or pH1N1 virus used for the in vitro expansion of the polyclonal CD8+ T cells (Fig. 2). The average number of spots tended to be higher after restimulation with H7N9 virus than after restimulation with the homologous viruses, although the difference was not statistically significant (Fig. 2R, S, and T).
Cross-recognition of CD8+ T cells with influenza A H7N9 virus assessed by lytic activity.
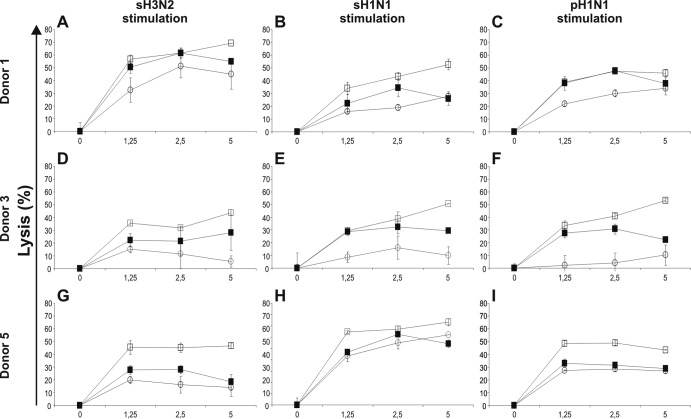
Based on the IFN-γ ELISpot results, we selected high-responding study subjects from each HLA group to test the lytic capacity of the CD8+ T cells against HLA class I-matched BLCLs infected with the homologous or H7N9 virus. To this end, polyclonal CD8+ T cells derived from sH3N2, sH1N1, or pH1N1 virus-stimulated PBMC cultures from study subjects 1, 3, and 5 were incubated with CFSE-labeled BLCLs infected with the sH3N2, sH1N1, pH1N1, or H7N9 virus.
CD8+ T cells from subject 1 obtained after sH3N2, sH1N1, and pH1N1 virus stimulation not only displayed lytic activity to the respective homologous viruses, but also displayed similar or even stronger lytic activity to H7N9 virus-infected cells, as was observed for sH1N1 virus-specific CD8+ T cells (Fig. 3A, B, and C). A similar trend was observed for virus-specific CD8+ T cells obtained from subject 3. Again, the lytic activity to H7N9 virus-infected cells exceeded that to cells infected with the homologous viruses to various extents (Fig. 3D, E, and F). Virus-specific CD8+ T cells of subject 5 displayed minor lytic activity to target cells infected with the respective homologous viruses. Again, the lytic activity to target cells infected with H7N9 virus exceeded that to cells infected with the homologous viruses and uninfected control cells (Fig. 3G, H, and I). The background lytic activity of T cells derived from subjects 1 and 5 was high, which may be related to bystander proliferation of Epstein-Barr virus (EBV)-specific T cells.
FIG 3.
Lytic activity of virus-specific polyclonal CD8+ T cells against BLCLs infected with the homologous or H7N9 virus. Seasonal influenza virus-specific polyclonal CD8+ T cells from study subjects 1, 3, and 5 were isolated after stimulation with sH3N2 (A, D, and G), sH1N1 (B, E, and H), or pH1N1 (C, F, and I) virus, as indicated. Lytic activity against CFSE-labled BLCLs infected with the homologous virus (sH3N2, sH1N1, or pH1N1) (solid squares) or the heterologous novel H7N9 virus (open squares) was assessed as lytic background activity against uninfected cells (open circles). Experiments were performed in triplicate. The error bars indicate standard deviations for the triplicates.
DISCUSSION
Here, we assessed the cross-reactivity of seasonal influenza A virus-specific CD8+ T cells with the newly emerging H7N9 virus. This study showed that a significant proportion of the polyclonal CD8+ T cells specific for sH3N2 (A/Netherlands/384/07), sH1N1 (A/Netherlands/26/07), and pH1N1 (A/Netherlands/602/09) cross-react with the novel H7N9 virus (A/Anhui/1/2013).
Comparison of epitope sequences revealed that the majority of the currently known HLA class I epitopes are conserved in the novel H7N9 viruses. Several studies have shown that the conservation of these HLA class I epitopes is responsible for cross-reactivity of influenza A virus-specific CD8+ T cells with influenza A viruses of another subtype (43, 45, 49–51). However, variation in some of the known epitopes was observed. We demonstrated that there is very little cross-reactivity of seasonal influenza A virus-specific CD8+ T cells with four individual H7N9 variant epitopes, although CD8+ T cells of subject 1 displayed some cross-reactivity with the H7N9 NP418-426 (HLA-B*35-restricted) epitope (Fig. 1A). The magnitudes of the responses to individual peptides varied between study subjects (Fig. 1). These differences may reflect differences in HLA class I makeup (64) and/or differences in the history of influenza A virus infections.
Although it has been suggested that the novel H7N9 virus is poorly immunogenic based on the in silico predictions of T cell epitopes in HA (66), we clearly demonstrate that the presence of most conserved HLA class I epitopes in the novel H7N9 virus contributes to the high cross-reactivity of the polyclonal CD8+ T cell populations with the H7N9 virus (Fig. 2 and 3). The low IFN-γ responses of study subjects 5 and 6 (A*0101, -A*0301, -B*0801, and -B*3501) to stimulation with the homologous seasonal influenza viruses and the H7N9 virus (Fig. 2) might be attributed to the absence of the HLA-A*0201 allele, which is required for a dominant CD8+ T cell response to the conserved and M158-66 epitopes (64). All study subjects displayed cross-reactive responses to H7N9 virus equal to or greater than those against the homologous viruses (Fig. 2 and 3), which could not be attributed to differences in infection rates (data not shown). These results correspond to previous assessments of cross-reactive CD8+ T cells with avian influenza A viruses of the H5N1 subtype (49). The strong reactivity to avian influenza A viruses might be the result of differences in antigen processing in infected cells, allowing more peptides to be liberated and presented from viral proteins of avian viruses than from those of human influenza viruses. It can be hypothesized that since these avian viruses have not circulated in the human population extensively, they have not yet had a chance to acquire mechanisms to escape from human epitope processing (33, 67–72).
Although we have studied the cross-reactivity of CD8+ T cells of study subjects with selected HLA types, it is likely that individuals with other HLA types also possess cross-reactive CD8+ T cells. The conservation of HLA class I epitopes restricted by other HLA alleles (Table 1) and the high amino acid sequence identity between the seasonal influenza viruses and the H7N9 virus underscores this (Table 2).
Cross-reactive influenza A virus-specific CD8+ T cells are found in individuals who have experienced an influenza A virus infection at least once. In contrast, a seroprevalence study indicated that a large proportion of children under the age of 4 years had not experienced an influenza A virus infection and therefore may not have developed virus-specific T cell responses (73). This age group may therefore be at higher risk of developing severe disease during a pandemic outbreak than adults. This was indeed the case during the 2009 H1N1 pandemic (74) and the localized outbreaks of the H5N1 subtype (75) and the vH3N2 subtype (76, 77). However, in the case of the novel H7N9 virus, mainly older (male) individuals were at risk for developing severe disease (12–15). The reason for this discrepancy is unknown at present. It has been suggested that differences in cell-mediated immunity between different age groups are the basis for this predilection (78). Elderly people who had experienced an H1N1 infection before 1957 were serologically protected during the 2009 pandemic outbreak and in the following years, whereas many unprotected individuals, including children, suffered from a pH1N1 infection in recent years (74, 79). Recent influenza A virus infections in children and young adults most likely boosted their cellular immune responses, which may afford some protection from infection with viruses of novel subtypes, including those of the H7N9 subtype (78). Others have suggested that preexisting immunity consisting of low levels of weakly heterosubtypic antibodies may result in antibody-dependent enhancement (ADE) of the infection (14). Instead of neutralizing the virus, these antibodies would enhance uptake of the virus and thus promote its replication. The possibility that other confounding factors may have contributed to the predilection of H7N9 disease for older individuals cannot be excluded. Elderly people are more likely to suffer from underlying diseases (80) and are known to have altered T cell immunity, which is likely to influence the outcome of an influenza A virus infection (81, 82).
It is difficult to predict to what extent preexisting influenza A virus-specific CD8+ T cells will afford protection against novel pandemic influenza viruses. Several animal studies have shown that virus-specific CD8+ T cells contribute to heterosubtypic immunity (52–57). However, evidence for heterosubtypic protection by CD8+ T cells in humans is sparse (59, 83). Epidemiologic studies showed that individuals who had experienced a seasonal H1N1 infection prior to the 1957 H2N2 pandemic were partially protected (60, 61), which could be attributed to cross-reactive T cells and/or antibodies to, e.g., the stalk region of HA. A similar trend was observed in isolated H5N1 infections (75). However, recent studies performed during the 2009 H1N1 pandemic provide better insight into the protective role of CD8+ T cells during an infection with an antigenically distinct influenza virus in serologically naive humans. It was shown that patients developed less severe illness when they had a high frequency of preexisting virus-specific CD8+ T cells before the onset of the pandemic (58). Another study showed that infected patients developed strong and rapid cross-reactive recall T cell responses, which in most cases coincided with the disappearance of clinical symptoms (84).
In conclusion, we have demonstrated that CD8+ T cells that can cross-react with the newly emerging H7N9 influenza virus and that may afford some protection in the absence of virus-neutralizing antibodies are present in the human population. Cross-reactive CD8+ T cells do not establish sterile immunity; they do, however, contribute to more rapid clearance of the H7N9 virus infection. Immunity afforded by the presence of cross-reactive CD8+ T cells may not only reduce the severity of disease caused by H7N9 virus infection, it may also contribute to reduction of virus spread in the population, since infected individuals may be infectious for a shorter time. Induction of cross-reactive virus-specific T cell responses may be a promising approach for the development of universal influenza vaccines that can elicit broadly protective immunity against influenza A viruses of various subtypes.
Footnotes
Published ahead of print 20 November 2013
REFERENCES
- 1.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U. S. A. 97:9654–9658. 10.1073/pnas.160270697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361. 10.1073/pnas.0308352100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat Vennema HH, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587–593. 10.1016/S0140-6736(04)15589-X [DOI] [PubMed] [Google Scholar]
- 4.Gambotto A, Barratt-Boyes SM, de Jong MD, Neumann G, Kawaoka Y. 2008. Human infection with highly pathogenic H5N1 influenza virus. Lancet 371:1464–1475. 10.1016/S0140-6736(08)60627-3 [DOI] [PubMed] [Google Scholar]
- 5.de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. 1997. A pandemic warning? Nature 389:554. 10.1038/39218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, Uyeki TM. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261–273. 10.1056/NEJMra0707279 [DOI] [PubMed] [Google Scholar]
- 7.WHO 2013. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003-2013. http://www.who.int/influenza/human_animal_interface/EN_GIP_20130705CumulativeNumberH5N1cases_2.pdf Accessed 14 August 2013 [Google Scholar]
- 8.de Jong MD, Bach VC, Phan TQ, Vo MH, Tran TT, Nguyen BH, Beld M, Le TP, Truong HK, Nguyen VV, Tran TH, Do QH, Farrar J. 2005. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N. Engl. J. Med. 352:686–691. 10.1056/NEJMoa044307 [DOI] [PubMed] [Google Scholar]
- 9.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353:1374–1385. 10.1056/NEJMra052211 [DOI] [PubMed] [Google Scholar]
- 10.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO 2013. Number of confirmed human cases of avian influenza A(H7N9) reported to WHO. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/09_ReportWebH7N9Number.pdf Accessed 27 September 2013. 10.1016/S0140-6736(13)61904-2 [DOI] [Google Scholar]
- 12.WHO 2013. Overview of the emergence and characteristics of the avian influenza A(H7N9) virus, 31 May 2013. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/WHO_H7N9_review_31May13.pdf Accessed 14 August 2013 [Google Scholar]
- 13.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, Gao L, Pang X, Liu G, Shu Y, Yang W, Uyeki TM, Wang Y, Wu F, Feng Z. 24 April 2013. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N. Engl. J. Med. [Epub ahead of print.] 10.1056/NEJMoa1304617 [DOI] [Google Scholar]
- 14.Skowronski D, Janjua N, Kwindt T, De Serres G. 2013. Virus-host interactions and the unusual age and sex distribution of human cases of influenza A(H7N9) in China, April 2013. Euro Surveill. 18:20465. [PubMed] [Google Scholar]
- 15.Arima Y, Zu R, Murhekar M, Vong S, Shimada T, World Health Organization Outbreak Response Team 2013. Human infections with avian influenza A(H7N9) virus in China: preliminary assessment of the age and sex distribution. Western Pac. Surveill. Response J. 4:1–3. 10.5365/wpsar.2013.4.2.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Sun J, Cai J, Miao Z, Lu M, Qin S, Wang X, Lv H, Yu Z, Amer S, Chai C. 2013. Epidemiological, clinical and viral characteristics of fatal cases of human avian influenza A (H7N9) virus in Zhejiang Province, China. J. Infect. 67:595–605. 10.1016/j.jinf.2013.08.007 [DOI] [PubMed] [Google Scholar]
- 17.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 18.CDC 2013. Emergence of avian influenza A(H7N9) virus causing severe human illness---China, February-April 2013. MMWR Morb. Mortal. Wkly. Rep. 62:366–371 [PMC free article] [PubMed] [Google Scholar]
- 19.Yu L, Wang Z, Chen Y, Ding W, Jia H, Chan JF, To KK, Chen H, Yang Y, Liang W, Zheng S, Yao H, Yang S, Cao H, Dai X, Zhao H, Li J, Bao Q, Chen P, Hou X, Li L, Yuen KY. 2013. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin. Infect. Dis. 57:1449–1457. 10.1093/cid/cit541 [DOI] [PubMed] [Google Scholar]
- 20.Belser JA, Bridges CB, Katz JM, Tumpey TM. 2009. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg. Infect. Dis. 15:859–865. 10.3201/eid1506.090072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown IH. 2010. Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis. 54:187–193. 10.1637/8949-053109-Reg.1 [DOI] [PubMed] [Google Scholar]
- 22.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. 10.1038/nature12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. 10.1016/S0140-6736(13)60938-1 [DOI] [PubMed] [Google Scholar]
- 24.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreijtz JH, Kroeze EV, Stittelaar KJ, de Waal L, van Amerongen G, van Trierum S, van Run P, Bestebroer T, Kuiken T, Fouchier RA, Rimmelzwaan GF, Osterhaus AD. 2013. Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: a model to study intervention strategies. Vaccine 31:4995–4999. 10.1016/j.vaccine.2013.06.071 [DOI] [PubMed] [Google Scholar]
- 27.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonges M, Meijer A, Fouchier RA, Koch G, Li J, Pan JC, Chen H, Shu YL, Koopmans MP. 2013. Guiding outbreak management by the use of influenza A(H7Nx) virus sequence analysis. Euro Surveill. 18:20460. [PubMed] [Google Scholar]
- 29.Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2013. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501:560–563. 10.1038/nature12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]
- 32.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13–20. 10.1016/0042-6822(78)90153-8 [DOI] [PubMed] [Google Scholar]
- 33.van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. 2012. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 4:1438–1476. 10.3390/v4091438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorrell EM, Schrauwen EJ, Linster M, De Graaf M, Herfst S, Fouchier RA. 2011. Predicting ‘airborne' influenza viruses: (trans-) mission impossible? Curr. Opin. Virol. 1:635–642. 10.1016/j.coviro.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS, Guan Y. 2009. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 106:11709–11712. 10.1073/pnas.0904991106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- 37.Bodewes R, Nieuwkoop NJ, Verburgh RJ, Fouchier RA, Osterhaus A, Rimmelzwaan GF. 2012. The use of influenza A viruses expressing reporter genes to assess the frequency of double infections in vitro. J. Gen. Virol. 93:1645–1648. 10.1099/vir.0.042671-0 [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Qi X, Cui L, Zhou M, Wang H. 2013. Human co-infection with novel avian influenza A H7N9 and influenza A H3N2 viruses in Jiangsu province, China. Lancet 381:2134. 10.1016/S0140-6736(13)61135-6 [DOI] [PubMed] [Google Scholar]
- 39.Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. 2013. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ 347:f4752. 10.1136/bmj.f4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, Garcia-Sastre A, Palese P, Treanor JJ, Krammer F. 2013. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J. Virol. 87:4728–4737. 10.1128/JVI.03509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yewdell JW, Bennink JR, Smith GL, Moss B. 1985. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 82:1785–1789. 10.1073/pnas.82.6.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotch F, McMichael A, Smith G, Moss B. 1987. Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J. Exp. Med. 165:408–416. 10.1084/jem.165.2.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bednarek MA, Sauma SY, Gammon MC, Porter G, Tamhankar S, Williamson AR, Zweerink HJ. 1991. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J. Immunol. 147:4047–4053 [PubMed] [Google Scholar]
- 45.Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. 2008. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82:12241–12251. 10.1128/JVI.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jameson J, Cruz J, Ennis FA. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J. Virol. 72:8682–8689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berkhoff EG, de Wit E, Geelhoed-Mieras MM, Boon AC, Symons J, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2005. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J. Virol. 79:11239–11246. 10.1128/JVI.79.17.11239-11246.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berkhoff EG, de Wit E, Geelhoed-Mieras MM, Boon AC, Symons J, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2006. Fitness costs limit escape from cytotoxic T lymphocytes by influenza A viruses. Vaccine 24:6594–6596. 10.1016/j.vaccine.2006.05.051 [DOI] [PubMed] [Google Scholar]
- 49.Kreijtz JH, de Mutsert G, van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2008. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J. Virol. 82:5161–5166. 10.1128/JVI.02694-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee LY, Ha DO, Simmons LAC, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, Dong T. 2008. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118:3478–3490. 10.1172/JCI32460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hillaire ML, Vogelzang-van Trierum SE, Kreijtz JH, de Mutsert G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2013. Human T-cells directed to seasonal influenza A virus cross-react with 2009 pandemic influenza A (H1N1) and swine-origin triple-reassortant H3N2 influenza viruses. J. Gen. Virol. 94:583–592. 10.1099/vir.0.048652-0 [DOI] [PubMed] [Google Scholar]
- 52.Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. 1998. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 8:683–691. 10.1016/S1074-7613(00)80573-7 [DOI] [PubMed] [Google Scholar]
- 53.O'Neill E, Krauss SL, Riberdy JM, Webster RG, Woodland DL. 2000. Heterologous protection against lethal A/HongKong/156/97 (H5N1) influenza virus infection in C57BL/6 mice. J. Gen. Virol. 81:2689–2696 [DOI] [PubMed] [Google Scholar]
- 54.Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2007. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25:612–620. 10.1016/j.vaccine.2006.08.036 [DOI] [PubMed] [Google Scholar]
- 55.Kreijtz JH, Bodewes R, van den Brand JM, de Mutsert G, Baas C, van Amerongen G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2009. Infection of mice with a human influenza A/H3N2 virus induces protective immunity against lethal infection with influenza A/H5N1 virus. Vaccine 27:4983–4989. 10.1016/j.vaccine.2009.05.079 [DOI] [PubMed] [Google Scholar]
- 56.Weinfurter JT, Brunner K, Capuano SV, III, Li C, Broman KW, Kawaoka Y, Friedrich TC. 2011. Cross-reactive T cells are involved in rapid clearance of 2009 pandemic H1N1 influenza virus in nonhuman primates. PLoS Pathog. 7:e1002381. 10.1371/journal.ppat.1002381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hillaire ML, van Trierum SE, Kreijtz JH, Bodewes R, Geelhoed-Mieras MM, Nieuwkoop NJ, Fouchier RA, Kuiken T, Osterhaus AD, Rimmelzwaan GF. 2011. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol. 92:2339–2349. 10.1099/vir.0.033076-0 [DOI] [PubMed] [Google Scholar]
- 58.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A. 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 19:1305–1312. 10.1038/nm.3350 [DOI] [PubMed] [Google Scholar]
- 59.McMichael AJ, Gotch FM, Noble GR, Beare PA. 1983. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 309:13–17. 10.1056/NEJM198307073090103 [DOI] [PubMed] [Google Scholar]
- 60.Epstein SL. 2006. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 193:49–53. 10.1086/498980 [DOI] [PubMed] [Google Scholar]
- 61.Slepushkin AN. 1959. The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bull. World Health Organ. 20:297–301 [PMC free article] [PubMed] [Google Scholar]
- 62.Jameson J, Cruz J, Terajima M, Ennis FA. 1999. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J. Immunol. 162:7578–7583 [PubMed] [Google Scholar]
- 63.Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J. Virol. Methods 74:57–66. 10.1016/S0166-0934(98)00071-8 [DOI] [PubMed] [Google Scholar]
- 64.Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582–590. 10.1128/JVI.76.2.582-590.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rimmelzwaan GF, Nieuwkoop N, Brandenburg A, Sutter G, Beyer WE, Maher D, Bates J, Osterhaus AD. 2000. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine 19:1180–1187. 10.1016/S0264-410X(00)00310-8 [DOI] [PubMed] [Google Scholar]
- 66.De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, Martin W. 2013. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum. Vaccin. Immunother. 9:950–956. 10.4161/hv.24939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eisenlohr LC, Yewdell JW, Bennink JR. 1992. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J. Exp. Med. 175:481–487. 10.1084/jem.175.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gould KG, Bangham CR. 1998. Virus variation, escape from cytotoxic T lymphocytes and human retroviral persistence. Semin. Cell Dev. Biol. 9:321–328. 10.1006/scdb.1998.0241 [DOI] [PubMed] [Google Scholar]
- 69.Iwasaki A, Stiernholm BJ, Chan AK, Berinstein NL, Barber BH. 1997. Enhanced CTL responses mediated by plasmid DNA immunogens encoding costimulatory molecules and cytokines. J. Immunol. 158:4591–4601 [PubMed] [Google Scholar]
- 70.Milicic A, Price DA, Zimbwa P, Booth BL, Brown HL, Easterbrook PJ, Olsen K, Robinson N, Gileadi U, Sewell AK, Cerundolo V, Phillips RE. 2005. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J. Immunol. 175:4618–4626 [DOI] [PubMed] [Google Scholar]
- 71.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH. 1991. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 66:1145–1153. 10.1016/0092-8674(91)90037-Y [DOI] [PubMed] [Google Scholar]
- 72.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. 1997. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J. Immunol. 158:3227–3234 [PubMed] [Google Scholar]
- 73.Bodewes R, de Mutsert G, van der Klis FR, Ventresca M, Wilks S, Smith DJ, Koopmans M, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2011. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 18:469–476. 10.1128/CVI.00396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945–1952. 10.1056/NEJMoa0906453 [DOI] [PubMed] [Google Scholar]
- 75.Smallman-Raynor M, Cliff AD. 2007. Avian influenza A (H5N1) age distribution in humans. Emerg. Infect. Dis. 13:510–512. 10.3201/eid1303.060849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skowronski DM, De Serres G, Janjua NZ, Gardy JL, Gilca V, Dionne M, Hamelin ME, Rheaume C, Boivin G. 2012. Cross-reactive antibody to swine influenza A(H3N2) subtype virus in children and adults before and after immunisation with 2010/11 trivalent inactivated influenza vaccine in Canada, August to November 2010. Euro Surveill. 17:20066. [DOI] [PubMed] [Google Scholar]
- 77.CDC 2012. Update: influenza A (H3N2)v transmission and guidelines—five states, 2011. MMWR Morb. Mortal. Wkly. Rep. 60:1741–1744 [PubMed] [Google Scholar]
- 78.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2013. Age distribution of cases caused by different influenza viruses. Lancet Infect. Dis. 13:646–647. 10.1016/S1473-3099(13)70181-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skountzou I, Koutsonanos DG, Kim JH, Powers R, Satyabhama L, Masseoud F, Weldon WC, Martin MP, Mittler RS, Compans R, Jacob J. 2010. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J. Immunol. 185:1642–1649. 10.4049/jimmunol.1000091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu X, Zhang X, He Y, Wu H, Gao X, Pan Q, Shen J, Zhu J, Chen H, Zhu Y, Wu F, Wang J, Yuan Z. 2013. Mild infection of a novel H7N9 avian influenza virus in children in Shanghai. Emerg. Microbes Infect. 2:e41. 10.1038/emi.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee N, Shin MS, Kang I. 2012. T-cell biology in aging, with a focus on lung disease. J. Gerontol. 67:254–263. 10.1093/gerona/glr237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang J, Fisher EM, Murasko DM. 2013. Intrinsic defects in CD8 T cells with aging contribute to impaired primary antiviral responses. Exp. Gerontol. 48:579–586. 10.1016/j.exger.2013.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sridhar S, Begom S, Bermingham A, Ziegler T, Roberts KL, Barclay WS, Openshaw P, Lalvani A. 2012. Predominance of heterosubtypic IFN-gamma-only-secreting effector memory T cells in pandemic H1N1 naive adults. Eur. J. Immunol. 42:2913–2924. 10.1002/eji.201242504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hillaire ML, van Trierum SE, Bodewes R, van Baalen CA, van Binnendijk RS, Koopmans MP, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2011. Characterization of the human CD8+ T cell response following infection with 2009 pandemic influenza H1N1 virus. J. Virol. 85:12057–12061. 10.1128/JVI.05204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. 2000. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum. Immunol. 61:438–452. 10.1016/S0198-8859(00)00105-1 [DOI] [PubMed] [Google Scholar]
- 86.Trojan A, Urosevic M, Hummerjohann J, Giger R, Schanz U, Stahel RA. 2003. Immune reactivity against a novel HLA-A3-restricted influenza virus peptide identified by predictive algorithms and interferon-gamma quantitative PCR. J. Immunother. 26:41–46. 10.1097/00002371-200301000-00005 [DOI] [PubMed] [Google Scholar]
- 87.Gotch F, McMichael A, Rothbard J. 1988. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. Use of analogues to orientate the matrix peptide in the HLA-A2 binding site. J. Exp. Med. 168:2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dong T, Boyd D, Rosenberg W, Alp N, Takiguchi M, McMichael A, Rowland-Jones S. 1996. An HLA-B35-restricted epitope modified at an anchor residue results in an antagonist peptide. Eur. J. Immunol. 26:335–339. 10.1002/eji.1830260210 [DOI] [PubMed] [Google Scholar]
- 89.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. 2001. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J. Virol. 75:11392–11400. 10.1128/JVI.75.23.11392-11400.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DiBrino M, Tsuchida T, Turner RV, Parker KC, Coligan JE, Biddison WE. 1993. HLA-A1 and HLA-A3 T cell epitopes derived from influenza virus proteins predicted from peptide binding motifs. J. Immunol. 151:5930–5935 [PubMed] [Google Scholar]
- 91.Guo HC, Jardetzky TS, Garrett TP, Lane WS, Strominger JL, Wiley DC. 1992. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360:364–366. 10.1038/360364a0 [DOI] [PubMed] [Google Scholar]
- 92.DiBrino M, Parker KC, Margulies DH, Shiloach J, Turner RV, Biddison WE, Coligan JE. 1994. The HLA-B14 peptide binding site can accommodate peptides with different combinations of anchor residues. J. Biol. Chem. 269:32426–32434 [PubMed] [Google Scholar]
- 93.DiBrino M, Parker KC, Margulies DH, Shiloach J, Turner RV, Biddison WE, Coligan JE. 1995. Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry 34:10130–10138. 10.1021/bi00032a005 [DOI] [PubMed] [Google Scholar]
- 94.Gatfield J, Lammert E, Nickolaus P, Munz C, Rothenfusser S, Fisch P, Stevanovic S, Schild H, Rammensee HG, Arnold D. 1998. Cell lines transfected with the TAP inhibitor ICP47 allow testing peptide binding to a variety of HLA class I molecules. Int. Immunol. 10:1665–1672. 10.1093/intimm/10.11.1665 [DOI] [PubMed] [Google Scholar]
- 95.Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. 1986. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell 44:959–968. 10.1016/0092-8674(86)90019-X [DOI] [PubMed] [Google Scholar]
- 96.Sutton J, Rowland-Jones S, Rosenberg W, Nixon D, Gotch F, Gao XM, Murray N, Spoonas A, Driscoll P, Smith M, Willis A, McMichael A. 1993. A sequence pattern for peptides presented to cytotoxic T lymphocytes by HLA B8 revealed by analysis of epitopes and eluted peptides. Eur. J. Immunol. 23:447–453. 10.1002/eji.1830230222 [DOI] [PubMed] [Google Scholar]
- 97.Tussey LG, Rowland-Jones S, Zheng TS, Androlewicz MJ, Cresswell P, Frelinger JA, McMichael AJ. 1995. Different MHC class I alleles compete for presentation of overlapping viral epitopes. Immunity 3:65–77. 10.1016/1074-7613(95)90159-0 [DOI] [PubMed] [Google Scholar]
- 98.Bowness P, Moss PA, Rowland-Jones S, Bell JI, McMichael AJ. 1993. Conservation of T cell receptor usage by HLA B27-restricted influenza-specific cytotoxic T lymphocytes suggests a general pattern for antigen-specific major histocompatibility complex class I-restricted responses. Eur. J. Immunol. 23:1417–1421. 10.1002/eji.1830230702 [DOI] [PubMed] [Google Scholar]
- 99.Boon AC, de Mutsert G, van Baarle D, Smith DJ, Lapedes AS, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. 2004. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J. Immunol. 172:2453–2460 [DOI] [PubMed] [Google Scholar]
- 100.Man S, Newberg MH, Crotzer VL, Luckey CJ, Williams NS, Chen Y, Huczko EL, Ridge JP, Engelhard VH. 1995. Definition of a human T cell epitope from influenza A non-structural protein 1 using HLA-A2.1 transgenic mice. Int. Immunol. 7:597–605. 10.1093/intimm/7.4.597 [DOI] [PubMed] [Google Scholar]