Abstract
Epstein-Barr virus (EBV) and rhesus lymphocryptovirus (rLCV) are closely related gammaherpesviruses in the lymphocryptovirus subgroup that express viral microRNAs (miRNAs) during latent infection. In addition to many host mRNAs, EBV miRNAs are known to target latent viral transcripts, specifically those encoding LMP1, BHRF1, and EBNA2. The mRNA targets of rLCV miRNAs have not been investigated. Using luciferase reporter assays, photoactivatable cross-linking and immunoprecipitation (PAR-CLIP), and deep sequencing, we demonstrate that posttranscriptional regulation of LMP1 expression is a conserved function of lymphocryptovirus miRNAs. Furthermore, the mRNAs encoding the rLCV EBNA2 and BHRF1 homologs are regulated by miRNAs in rLCV-infected B cells. Homologous to sites in the EBV LMP1 and BHRF1 3′-untranslated regions (UTRs), we also identified evolutionarily conserved binding sites for the cellular miR-17/20/106 family in the LMP1 and BHRF1 3′UTRs of several primate LCVs. Finally, we investigated the functional consequences of LMP1 targeting by individual EBV BART miRNAs and show that select viral miRNAs play a role in the previously observed modulation of NF-κB activation.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) noncoding RNAs, expressed by all metazoans, that posttranscriptionally inhibit gene expression. Most miRNAs originate from stem-loop RNA structures that are cleaved by the RNase III enzyme Drosha in the nucleus to liberate ∼60-nt RNA hairpins, termed precursor miRNAs (pre-miRNAs) (reviewed in reference 1). Pre-miRNAs are exported to the cytoplasm by Exportin 5 (2), where they are cleaved by a second RNase III enzyme, Dicer, thereby liberating ∼22-bp imperfect miRNA-miRNA* duplexes (reviewed in reference 3). The miRNA strand of this duplex is incorporated into the RNA-induced silencing complex (RISC) to guide RISC to partially complementary target sites located predominantly in mRNA 3′-untranslated regions (UTRs) while the second, passenger miRNA* strand is degraded. The seed sequence of the mature miRNA (nt 2 to 8) typically has full complementarity to the target mRNA and plays a key role in target site recognition (4). RISC binding to a target mRNA can inhibit its translation and/or lead to mRNA degradation (reviewed in reference 5). miRNAs have been shown to play important roles in a number of diverse biological processes, and at least one-third of all human genes are predicted to be under miRNA regulation (6, 7).
A number of viruses, particularly DNA tumor viruses such as the gammaherpesviruses, encode miRNAs (reviewed in reference 8). Epstein-Barr virus (EBV), a ubiquitous human gammaherpesvirus commonly associated with infectious mononucleosis, exploits the cellular miRNA biogenesis machinery to process 25 viral pre-miRNAs expressed during latent infection (9–12). EBV miRNAs are transcribed from two discrete genomic loci: three pre-miRNAs are encoded within the BHRF1 locus, while the BART region encompasses 22 BART pre-miRNAs. The closely related rhesus lymphocryptovirus (rLCV; also called macacine herpesvirus 4 or cercopithecine herpesvirus 15), which naturally infects rhesus macaques (Macaca mulatta), expresses 36 pre-miRNAs from the homologous BHRF1 and BART regions (9, 13, 14).
EBV and rLCV are both members of the lymphocryptovirus subgroup and are evolutionarily separated by at least 13 million years (15, 16). Nevertheless, the EBV and rLCV genomes exhibit colinear organization and, overall, ∼65% nucleotide conservation. Regions encoding structural proteins or homologs of cellular proteins, such as the Bcl-2 homolog BHRF1 or the CD40-mimic LMP1, are more highly conserved (16, 17). Remarkably, sequence analysis has revealed that 22 orthologous pre-miRNA hairpins exist between EBV and rLCV (9, 14). This is in contrast to miRNAs encoded by other gammaherpesviruses. In particular, members of the rhadinovirus subgroup of gammaherpesviruses, such as Kaposi's sarcoma-associated herpesvirus (KSHV) and rhesus rhadinovirus (RRV), each have multiple miRNAs within similar latency-associated regions, yet only one KSHV and one RRV miRNA exhibit any sequence conservation (18, 19). Computational analysis of 14 different gammaherpesvirus genomes from both human and nonhuman primates has suggested that gammaherpesvirus miRNA sequence conservation is rare, although the organization of miRNA loci appears to be conserved (14).
With few exceptions (20–23), gammaherpesvirus miRNAs also lack overall sequence homology to cellular miRNAs. There are, however, several examples of viral miRNAs mimicking cellular miRNA seed sequences (nt 2 to 7, i.e., the minimal functional unit of an miRNA); consequently, these viral and cellular miRNAs have common target mRNAs (21, 23, 24). Several EBV BART miRNAs share common seeds with cellular miRNAs (miR-BART5-5p, miR-18a/b, miR-BART1-3p, miR-29a/b/c, miR-BART9-3p, and miR-141/200a) (20, 23); however, the extent to which these miRNA seed mimics target and/or potentially compete for common target sites on mRNAs has not yet been determined.
Approximately one-third of the mature EBV and rLCV miRNAs share conserved seed sequences (14), implying that these lymphocryptovirus miRNAs share conserved mRNA targets. Recently, we analyzed the viral and cellular miRNA targetome in several latency III EBV B95-8 transformed lymphoblastoid cell lines (LCLs) using photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) and bioinformatic tools (25–27). The B95-8 laboratory strain of EBV, in particular, bears an ∼12-kb deletion which removes 17 of the 22 BART miRNAs, leaving only five BART miRNAs and the BHRF1 miRNAs intact. In addition to viral miRNAs, latency III cells express all latent viral gene products, including six EBV nuclear antigens (EBNAs), three latent membrane proteins (LMPs), and other noncoding RNAs, such as EBERs. Our analysis revealed binding sites for several cellular miRNAs, particularly the myc-regulated miR-17/20/106 family, in the LMP1 and BHRF1 3′UTRs (27). Furthermore, in silico analysis showed that the miR-17/20/106 binding sites are conserved in the rLCV LMP1 and BHRF1 3′UTRs (27). Intriguingly, EBNA2, BHRF1, and LMP1 mRNAs were also found to be RISC associated by high-throughput sequencing and cross-linking immunoprecipitation (HITS-CLIP) analysis of latency III EBV+ Burkitt's lymphoma (BL) cells (28). In addition to the miR-17/20/106 binding sites, HITS-CLIP revealed binding sites for several BART miRNAs not present in EBV B95-8 LCLs, namely, binding sites for miR-BART5-5p and 19-5p in the LMP1 3′UTR and miR-BART10-3p in the BHRF1 3′UTR (28). As EBV miR-BART5-5p and miR-BART10-3p are both conserved in rLCV (9, 14), we asked whether any rLCV miRNAs and/or the rhesus macaque miR-17/20/106 family targets the rLCV LMP1 and/or BHRF1 homologs.
Here, we investigated miRNA targeting of lymphocryptovirus mRNAs in depth by performing PAR-CLIP analysis on human and rhesus macaque B cells infected with either wild-type EBV or rLCV. Reporter assays were used to further investigate viral miRNA targeting of the EBV and rLCV LMP1 and BHRF1 3′UTRs in greater detail. These experiments define the individual miRNA target sites on the LMP1 and BHRF1 mRNA homologs; furthermore, they demonstrate conserved miRNA targeting of viral transcripts during lymphocryptovirus infection. Lastly, we explored the downstream consequences of LMP1 targeting by viral miRNAs, which uncovered a role for select EBV BART miRNAs in modulating NF-κB signaling pathways.
MATERIALS AND METHODS
Cell culture and plasmids.
Akata-LCLd3 and IBL1-LCLd3 (LCLs) were generated by infecting human peripheral blood mononuclear cells (PBMCs) with wild-type EBV derived from IgG-stimulated Akata cells or IBL-1 diffuse large B cell lymphoma (DLBCL) cells (29). EF3D-MigW LCLs were generated in parallel with EF3D-Ago2 LCLs as previously described (27) and were infected with EBV B95-8. rLCV-infected rhesus macaque rLCLs (211-98 and 309-98) and baboon S594 LCLs (kind gifts of F. Wang) have been described (30, 31). Established LCLs were maintained in log phase in RPMI 1640 supplemented with heat-inactivated 12% fetal bovine serum (FBS) and antibiotics. 293T and 293T-IκBα cells were grown in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. All cells were cultured at 37°C in 5% CO2.
EBV miRNA expression plasmids were generated in the pLCE lentiviral vector backbone as previously described (27). Briefly, an ∼200-nt region surrounding the pre-miRNA was PCR amplified from Akata genomic DNA and cloned into the enhanced green fluorescent protein 3′UTR at XhoI and XbaI. miRNA expression was confirmed by indicator assays as previously described (27), as well as by deep sequencing of small RNAs from 293T cells ectopically expressing EBV miRNAs (not shown). rLCV miRNA expression plasmids were generated in pcDNA3. An ∼200-nt region surrounding each pre-miRNA was PCR amplified from 309-98 rLCL genomic DNA and cloned downstream of the cytomegalovirus (CMV) promoter at XhoI and XbaI.
To generate luciferase 3′UTR reporters, the LMP1 and BHRF1 3′UTRs from EBV B95-8 and rLCV 309-98 LCL genomic DNA were PCR amplified and cloned into pLSG at XhoI and XbaI. To generate LMP1 expression plasmids, the full-length LMP1 gene and the LMP1 coding region were PCR amplified from EF3D LCL genomic DNA and cloned into pcDNA3; LMP1 transcripts are expressed from the CMV promoter. Oligonucleotides used for cloning can be provided upon request.
Western blot analysis.
293T cells transfected with BART miRNAs and pcDNA3-LMP1-FL were lysed in NP-40 lysis buffer. Cell lysates were separated by SDS-PAGE and blotted onto nitrocellulose membranes. Western blots were probed using monoclonal antibodies to LMP1 (S11) or beta-actin (C4; Santa Cruz Biotechnology) and developed with peroxidase-conjugated anti-mouse IgG (Sigma) and WesternBright Sirius chemiluminescent substrate (Advansta).
Generation of deep sequencing libraries for smRNA-seq, RIP-seq, and PAR-CLIP.
To generate small RNA sequence (smRNA-seq) libraries, total RNA was isolated from EBV and rLCV LCLs using TRIzol, and smRNAs (<200 nt) were enriched from the total RNA using the mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. To generate RISC-bound miRNA sequence (RIP-seq) libraries, ∼100 million cells were lysed on ice in NP-40 lysis buffer, and RISC-bound RNAs were immunoprecipitated using pan-Argonaute (Ago) monoclonal antibody (ab57113; Abcam) and protein G Dynabeads (Invitrogen). Beads were washed 10 times in NT2 buffer (32) and then treated with proteinase K (Roche). RISC-bound RNAs were recovered by acid phenol-chloroform extraction. PAR-CLIP libraries for deep sequencing were prepared as previously described (27). Briefly, LCLs were grown overnight in the presence of 4-thiouridine and cross-linked at a UV wavelength of 365 nm. RISC-bound RNAs were immunoprecipitated using pan-Ago monoclonal antibodies (ab57113) and recovered by proteinase K treatment and acid phenol-chloroform extraction prior to adapter ligation.
Small RNAs (smRNA-seq), RISC-bound miRNAs (RNA immunoprecipitation [RIP-seq]), and RISC-bound, cross-linked RNAs (PAR-CLIP) were sequentially ligated to Illumina adapter sequences using the TruSeq small RNA sample prep kit (Illumina) according to the manufacturer's instructions. Following ligation to adapters, RNAs were reverse transcribed by SsIII (Invitrogen) using a primer complementary to the 3′ adapter and cDNAs were PCR amplified. A pilot PCR was performed for each library to ensure that amplification occurred in the linear range. High-throughput sequencing (50-bp, single end) was performed on an Illumina HiSeq 2000 at the Duke Genome Sequencing & Analysis Core.
Bioinformatics.
Sequencing reads were preprocessed using the FAST-X toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to remove 3′ adapter sequences. Reads ≥15 nt in length were aligned concurrently to the human (HG19) and EBV1 (accession no. AJ507799) genomes (human LCLs) or Macaca mulatta (Mmu1_051212) and macacine herpesvirus 4 (accession no. NC_006146) genomes (rhesus macaque LCLs) using Bowtie (33). Up to two mismatches were allowed for smRNA-SEQ and RIP-SEQ libraries; three mismatches (including thymidine-to-cytidine [T→C] conversions) were allowed for PAR-CLIP libraries. Genomic locations with reads falling in the best stratum were kept for further analysis. To annotate miRNAs, reads were next aligned to human, macaque, or viral pre-miRNA stem-loops plus flanking sequences present in miRBASE v19.0.
Analysis of PAR-CLIP reads was performed using PARalyzer as described previously (25, 27). Briefly, aligned reads were grouped and analyzed for T→C conversions to generate clusters. Clusters, which represent RISC interaction sites, were then interrogated for canonical miRNA seed matches (minimally 7mer1A) using the top ∼200 unique miRNA sequences present in matched smRNA-SEQ and/or RIP-SEQ libraries.
qRT-PCR for RISC-associated BHRF1 mRNAs.
EF3D-MigW and EF3D-Ago2 LCLs were lysed on ice in NP-40 lysis buffer, and RISC-associated RNAs were immunoprecipitated using monoclonal antibodies to FLAG (clone M2; Sigma) or green fluorescent protein (GFP; clone B-2; Santa Cruz Biotechnology) and protein G Dynabeads (Invitrogen). Beads were washed 10 times in NT2 buffer, and RNAs were purified and recovered following proteinase K treatment and phenol-chloroform extraction. RNA was reverse transcribed using SsIII reverse transcriptase (Invitrogen) and oligo(dT) or random primers. Previously described primers F2, F3, and F6, which detect the BHRF1 intron, open reading frame (ORF), or 3′UTR (34, 35), were used to detect BHRF1 cDNAs by SYBR green real-time quantitative PCR (qRT-PCR; Applied Biosystems). The EBV BHRF1 gene was cloned into pcDNA3 using previously described primers (34, 35) and serially diluted to generate a standard curve for real-time PCR.
Luciferase 3′UTR reporter assays.
Dual-luciferase assays were performed as previously described (27). Briefly, 293T cells were seeded in 24-well plates and cotransfected with a firefly luciferase reporter (pLSG-based), a Renilla internal control plasmid (pLSR), and an miRNA expression vector using Fugene 6 according to the manufacturer's protocol (Roche). Forty-eight to 72 h posttransfection, cells were lysed in 1× passive lysis buffer and luciferase activity was measured using the dual reporter luciferase assay system (Promega). All assays are reported as the averages from at least two experiments. We considered >20% knockdown (P < 0.05 by Student's t test) as significant for miRNA-mediated inhibition of a 3′UTR luciferase reporter.
NF-κB-reporter and IκBα-luciferase assays.
293T-IκBα cells were generated by transducing 293T cells with vIRES-puro/IκBα-photinus (kind gift of B. Gewurz and E. Davis) (36) and selecting with 1 μg/ml puromycin. 293T and 293T-IκBα cells were transfected using Fugene 6 with the indicated amounts of pcDNA3-LMP1 or pcDNA3-LMP1d3UTR and miRNA expression vectors. For NF-κB assays, 48 h later, 293T cells were transfected with an NF-κB-regulated firefly luciferase vector (Stratagene), a Renilla internal control plasmid, and 400 ng filler DNA and incubated overnight prior to harvesting lysates. For tumor necrosis factor alpha (TNF-α) experiments, transfected cells were incubated with human recombinant TNF-α (10 or 20 ng/ml) for 3 h prior to analysis.
Deep sequence accession number.
Sequencing data have been deposited in the NCBI BioProject database under accession number 218007.
RESULTS
Small RNAs expressed in lymphocryptovirus-infected LCLs.
We have previously analyzed the small RNAs (18 to 24 nt) in EBV B95-8 LCLs by deep sequencing to obtain the exact sequences of all mature miRNAs and determine the relative levels of EBV miRNAs present in infected cells (27). The EBV B95-8 laboratory strain bears an ∼12-kb deletion that removes a significant portion of the BART region; thus, only five of the 22 BART pre-miRNAs are expressed. Therefore, we were interested in the miRNAs present in LCLs infected with wild-type, full-length EBV, particularly the expression levels of the BART miRNAs that have been previously reported to target latent viral gene products (27, 28, 37).
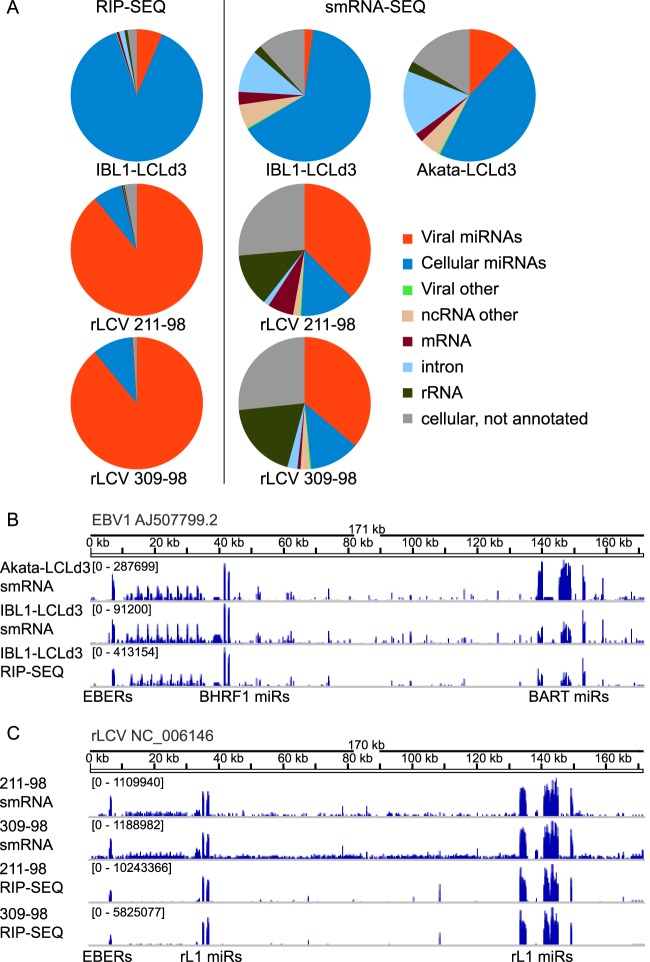
We established two LCLs infected with Akata-derived or IBL1-derived viruses (Akata-LCLd3 and IBL1-LCLd3, respectively). Small RNAs (<200 nt) and RISC-associated RNAs from these LCLs were isolated and analyzed by deep sequencing. Sequencing reads (∼20 to 30 million per sample) were processed as previously described (27) and aligned concurrently to the human and EBV1 genomes, allowing up to two mismatches. Forty-eight to 66% of the aligned small RNA reads (smRNA-seq) mapped to viral and cellular miRNA hairpins present in miRBase v19, while >95% of the aligned RISC-associated (RIP-seq) reads mapped to miRNAs (Fig. 1A). The most highly expressed, RISC-associated cellular miRNAs included hsa-miR-155, miR-181a, miR-21, miR-92a, and miR-191 (not shown).
FIG 1.
Deep sequencing analysis of lymphocryptovirus miRNAs. (A) Distribution of aligned, deep sequencing reads from RISC-immunoprecipitations (RIP-SEQ) or size-selected total RNA (smRNA). Reads from IBL-LCLd3 and Akata-LCLd3 (human LCLs infected with wild-type EBV) were aligned to the human genome (HG19) and EBV1, while reads from 211-98 and 309-98 rLCLs were aligned to the rhesus macaque genome (Macaca mulatta; Mmu1_051212) and rLCV (NC_006146.1). (B) Whole-genome view of deep sequencing reads from EBV LCLs aligned to EBV1. Seg files (containing start position, end position, and read count for each unique read) were generated for each deep sequencing library and visualized using the Integrative Genomics Viewer (IGV). Read counts are normalized for each library and shown on a log scale. (C) Same as panel B, except reads from the rLCL libraries are shown aligned to the rLCV genome.
We identified mature viral miRNAs expressed from all 25 EBV pre-miRNAs, including four different BHRF1 miRNAs and >44 different BART miRNAs (Fig. 1B). Similar to the variable viral miRNA levels observed in EBV B95-8 LCLs (27), EBV miRNAs accounted for ∼4 to 21% of the miRNAs in the small RNA libraries (3.8% for IBL1-LCLd3 and 20.8% for Akata-LCLd3). In IBL1-LCLd3, 6.6% of the RISC-associated miRNAs were viral (Fig. 1A, shown in orange). The most highly expressed viral miRNAs in IBL1-LCLd3 cells were the BHRF1 miRNAs and miR-BART2, which account for >75% of the viral miRNAs in these cells. The BHRF1 miRNAs were moderately expressed in Akata-LCLd3 (accounting for 11% of EBV miRNAs); however, we observed high levels of miR-BART6, miR-BART7, miR-BART8, miR-BART11, miR-BART17, miR-BART19, and miR-BART22. Together, these seven BART miRNAs accounted for 64% of the EBV miRNAs and 12% of all miRNAs detected in Akata-LCLd3. miR-BART3, miR-BART5, and miR-BART16 were present at moderate levels (∼3.3% of the EBV miRNAs).
Multiple EBV miRNAs are highly conserved in rLCV (9, 13, 14), including miR-BART3 and miR-BART5, which are reported to target the EBV LMP1 3′UTR (27, 28), as well as miR-BART10-3p, which is reported to target the EBV BHRF1 3′UTR (28). To investigate rLCV miRNAs in depth, we also deep sequenced the small RNAs and RISC-associated RNAs present in two rhesus macaque LCLs (rLCLs), 211-98 and 309-98, which are derived from rLCV infection of two individual rhesus macaques (31). Small RNAs in rLCV-infected 309-98 rLCLs have previously been deep sequenced (13), while miRNAs in 211-98 rLCLs have been investigated by TA cloning and shallow sequencing (9, 14). As described above, we obtained ∼20 to 30 million reads per sample which were aligned concurrently to the rhesus macaque and rLCV genomes. Congruent with previous studies, we identified rLCV miRNAs expressed from a total of 36 distinct pre-miRNAs (Fig. 1C and Table 1). No RRV miRNAs or lymphocryptovirus snoRNAs (38) were detected.
TABLE 1.
rLCV miRNAs expressed in LCLs as determined by deep sequencing
| rLCV pre-miR | % of rLCV miRs (present in 211-98) | miRNA sequencea |
EBV homolog/orthologb | % of EBV miRs (present in Akata-LCLd3) | |
|---|---|---|---|---|---|
| 5′ | 3′ | ||||
| rL1-1 | 0.6 | TAACCTGATCAGCCCCGGGGTT | CTCCGGGCCTGAAGAGGTTGAC | BHRF1-1 | 8.9 |
| rL1-2 | 0.04 | AAATTCTGCCACAGAAGATAGC | TATCTTTTGCGGGGGAATTTC | BHRF1-2 | 0.4 |
| rL1-17 | 0.2 | TGATGGACAGCGGGGAAGTGCACT | TGCTTCGCCCTCTCCATCAT | BHRF1-3** | 0.7 |
| rL1-18 | 0.2 | TAGATGATTGGGGGCGGCGTATC | TTAGCCCCTCCCCATCATCTTG | None | |
| rL1-3 | 27.9 | CGGTGGAGACCGCGGGTGCGGTG* | TCGCACCTCGCCGTCTCTACTGC | None | |
| rL1-4 | 0.6 | ACCTAGTAATTGTGCGGTGTT | CACCACACGATCCACTAGGTC | None | |
| rL1-5 | 0.01 | AACCTAGTGCCGGTGATGTGCT | CGCACCACTTTTCACTAGGTG | BART3 | 1.2 |
| rL1-6 | 0.05 | TTTTAGTGGAAGTGACGTGCTGT | TAGCACCGCTATCCACTATGTCT | BART1 | 1.4 |
| rL1-7 | 0.5 | CGAGGTAAACATCGGCTTACTG | AGTGGGCCTGTTTCCCTCACA | BART15** | 0.5 |
| rL1-8 | 0.006 | TAAGGTGAATATAGCTGCCCATT | TGTCTGTTTAGAACCCGGTGGGCCGCAGTTCACCTACA | BART5 | 0.3 |
| rL1-19 | 0.3 | (T)ATAGATAGCGTGGGTGTGTGACC* | ATCACCACCCCCTGTCTGCT* | BART16 | 1.8 |
| rL1-9 | 0.01 | TAAGAGGGGGGCCTGCACGCCGAG* | TCGATGCATGGTCCCCCCTTAGT | BART17 | 10.9 |
| rL1-20 | 0.1 | TAAAGGGTAGTGTGGTTCACAGC | TGTGGATCTAACTTCCCTTAGT | BART6** | 11.6 |
| rL1-34 | 0.03 | TAGATGGCGTAACAGGCAACT | TTAGCCCGAACCGCTATCCATA | None | |
| rL1-21 | 0.01 | TTGATTGTGGGGGGAGGGCTCT | AGGCCTCTTCTCACAATTCTAAT | None | |
| rL1-22 | 0.01 | TCGTGGGGGGAAACGGTGGAT* | TCACCGTTTCATCCCCACGATT | None | |
| rL1-23 | 10.4 | TCACTAGTGCTGGCACCTAAGA | TTAGTTGTCTGCACTGGAGAGT | BART21 | 0.1 |
| rL1-10 | 9.8 | ATCGGGTTTCCGCTGTACTTGCA | TAGTGCGCCGGTGACCTGATAG | None | |
| rL1-24 | 1.3 | CTCAAGTTCTCATTTCCAATAC | TATCGGAGATAGGACTTGATACC | BART18 | 1.6 |
| rL1-11 | 0.1 | (T)CCGTATCCTATCAAGTGTGACG* | TGACACTCGATAGGATACGGGG | None | |
| rL1-12 | 0.3 | AGACCAGACCATGCACAGTGGG | AACGGTGCATGGACTGGCTAGA | None | |
| rL1-13 | 0.1 | CCTGGGCATGGGCTATGAAACA | GATCATAGCCAGTGTCCAGGGA | BART7 | 5.1 |
| rL1-25 | 0.002 | TGCTAGACCCTAAATTGGAAAC | TGACAATTTAATGGGTCTAGTAGT | BART22 | 3.9 |
| rL1-26 | 0.3 | AGTCATTGGGCCTCTGC* | TACGATTCTCCGGGTTTAGC | None | |
| rL1-27 | 0.005 | GCCGCCTCCTTGGTTCTGTC | CACATAACCATGGAGGTGGTTG | BART10 | 1.1 |
| rL1-14 | 44.9 | TCGGACGGTCTGGTGCGCTTGAT | TCGCACATCAGGCTGAACGACT | BART11 | 20.2 |
| rL1-15 | 0.3 | ACCCACCCTTCTCGACGGGGC | TCCTGTAGAGTATGGGTGTGGTTT | BART12** | 0.3 |
| rL1-28 | 0.003 | TGGAAGGCGTTGCTTTCAGCTG | GAGGAAAGTATCGCCTTCTAG | None | |
| rL1-29 | 0.1 | CCAGTCCCAGCAGACAAAAC | TTTTGTTTGCTTGGGACTGCAGA | BART19 | 5.6 |
| rL1-16 | 0.1 | AAGCAGGCATGTCTTCATTC | CATGAAACACATGGCCTGTTCCT | BART20 | 0.03 |
| rL1-30 | 0.1 | ACCGGTCCTTTGAGGTACGAAAC | TTTCGTTTTCGAAGGTCGGCTGT | None | |
| rL1-35 | 0.001 | CACCGGCTCACAGCTAGTACGGACGATTATTGGCTC | TGTAACTTGTCGTGGGACGGGA | BART13 | 2.4 |
| rL1-31 | 1.6 | TCGCTTACGCCGACCGCGTAAC | TAGGCGTGTCGTCGTAAGCAGC | None | |
| rL1-32 | 0.1 | TAACTTACTACTGCCGCTTTACA | TAAATGCGAGCAGTAGTAGGCGTC | BART14 | 2.4 |
| rL1-33 | 0.2 | CATTGTCTTCATTTGCCCTTGC | AAAGAGCAAATGGGGGGAATAGC | BART2** | 3.2 |
| None | BART4 | 0.8 | |||
| None | BART8 | 14.3 | |||
| None | BART9 | 1.3 | |||
Novel sequences or nucleotide changes that affect the previously annotated 5′ seed sequences are denoted with an asterisk.
Double asterisks indicate orthologs.
Similar to EBV-infected LCLs, the most highly expressed, RISC-associated cellular miRNAs in rLCLs included mml-miR-155, miR-181a, miR-21, miR-92a, and miR-191 (not shown). EBV infection of B lymphocytes is known to induce the expression of several cellular miRNAs, particularly miR-155 and miR-21, which are thought to be growth promoting (39–42). Indeed, sustained miR-155 expression is critical to the survival and proliferation of EBV-infected LCLs (43). The high levels of mml-miR-155 and mml-miR-21 observed here in rLCLs suggest that lymphocryptoviruses employ similar, conserved mechanisms to influence host miRNA expression patterns during latent infection.
In contrast to the levels of EBV miRNAs present in LCLs, rLCV miRNAs accounted for ∼74% of the identified miRNAs in the smRNA libraries and ∼90% of the RISC-associated miRNAs in rLCLs (Fig. 1A, shown in orange). miR-rL1-3-3p, miR-rL1-23-3p, miR-rL1-10-3p, and miR-rL1-14-3p were the most abundant, accounting for >80% of the RISC-associated miRNA population in rLCLs. Surprisingly, miR-rL1-5, miR-rL1-8, and miR-rL1-19 (homologs of BART miRNAs targeting LMP1; see below) were weakly expressed, accounting for only ∼0.4% of RISC-associated viral miRNAs (Table 1).
We detected both the mature miRNAs and passenger strands from all known and predicted rLCV pre-miRNAs. Table 1 lists the identified rLCV miRNA sequences and their EBV homologs/orthologs; novel sequences (i.e., miR-rL1-9-5p and miR-rL1-3-5p) or nucleotide changes that affect the previously annotated 5′ seed sequences are denoted with an asterisk. Sequences for the major miRNAs are generally consistent with what has previously been reported (9, 13, 14); however, we did find differences in the major sequences that are annotated for miR-rL1-19, miR-rL1-26-5p, and miR-rL1-22-5p. Furthermore, miR-rL1-19-5p and miR-rL1-11-5p each have two mature sequences offset by single 5′ nucleotides; thus, there are two seed sequences, which expands the number of potential mRNA targets for these miRNAs. In addition to viral miRNAs, we also detected several reads mapping to the EBERs in both EBV-infected and rLCV-infected LCLs (Fig. 1B and C). While EBER-derived small RNAs were found to be associated with RISC, presumably these small RNAs arise from breakdown products of the highly expressed, structured EBERs, as they lack a concise size or 5′ end.
BHRF1 pre-miRNAs are conserved in primate lymphocryptoviruses.
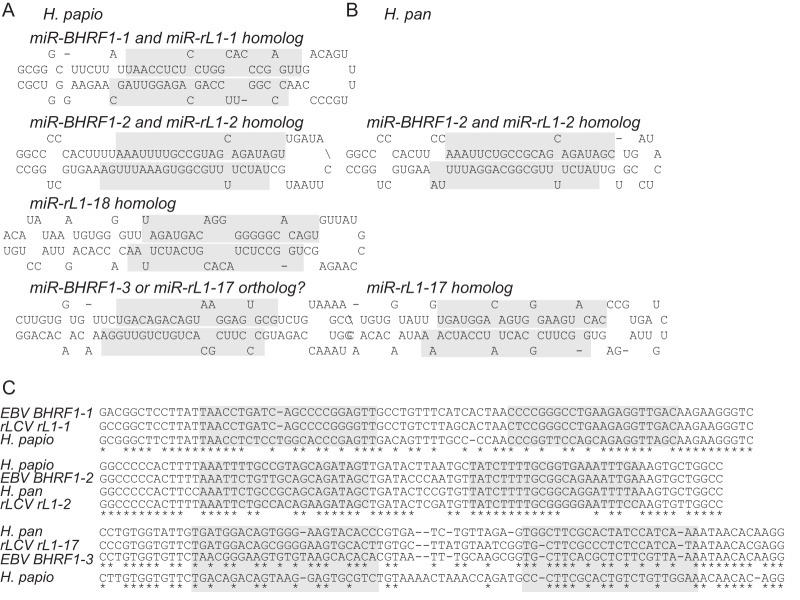
Previous work by Walz et al. has suggested that evolutionary conservation of viral pre-miRNAs is rare among gammaherpesviruses, and genomes with <60% sequence identity lack evidence of miRNA conservation (14). Their study was limited to the 14 gammaherpesvirus genomes that are fully sequenced, however, and gammaherpesvirus subgroups were not investigated in depth. There are at least eight reported viruses in the genus Lymphocryptoviridae, including EBV, rLCV, herpesvirus papio (cercopithecine herpesvirus 12), and herpesvirus pan (panine herpesvirus 1; also called pongine herpesvirus 1) (44). Herpesvirus papio and herpesvirus pan infect Old World baboons and chimpanzees, respectively. Since the sequence of the BHRF1 region is known for herpesvirus papio (AF231983.1 and AF200364) and a partial sequence is known for herpesvirus pan (AF231984.1) (45), we asked whether these LCVs encode BHRF1 miRNA homologs. Based on sequence homology to the rLCV and EBV BHRF1 pre-miRNAs and mature miRNAs (Table 1) and the known requirements for Drosha substrates (46–48), we predicted four BHRF1 pre-miRNA candidates for herpesvirus papio and two BHRF1 pre-miRNA candidates for herpesvirus pan. Figure 2 shows the predicted structures of the pre-miRNA stem loops with additional flanking regions; the putative mature miRNA sequences are shaded.
FIG 2.
BHRF1 miRNAs are encoded by primate lymphocryptoviruses. (A and B) Predicted hairpin structures for novel herpesvirus papio and herpesvirus pan BHRF1 precursor miRNAs generated using RNAfold (52). The predicted mature miRNA sequences, based on homology to known EBV and rLCV BHRF1 miRNAs, are shaded. (C) Clustal2.1 multiple sequence alignment of predicted and known BHRF1 pre-miRNA hairpin and flanking sequences from EBV, rLCV, herpesvirus papio, and herpesvirus pan.
We used ClustalW 2.0 to align the pre-miRNAs plus flanking sequences from the four LCVs (Fig. 2C). Unlike the other BHRF1 miRNAs, the seed sequence of miR-BHRF1-2-3p is highly conserved, suggesting that the miR-BHRF1-2 miRNA in particular is important for the LCV life cycle. With the exception of a single, nonconserved site for miR-rL1-2-3p in rLCV BLRF2 (see Table 3; also described further below), no viral targets for miR-rL1-2-3p or miR-BHRF1-2-3p have been identified to date (27, 28). These data suggest that miR-BHRF1-2-3p binds to cellular mRNA targets that drive the conservation of its seed sequence.
TABLE 3.
RISC-associated rLCV transcripts identified by PAR-CLIP
| Gene | No. of clustersa | miRNAs with seed match(es) |
|
|---|---|---|---|
| Viral | Cellular | ||
| BNRF1 | 13 (5) | None | miR-25/92a/92b/363, miR-25/92a/92b/155, miR-25/92a/92b/142-3p |
| EBNA2 | 15 (4) | miR-rL1-3-5p, miR-rL1-33-3p, miR-rL1-2-3p, miR-rL1-7-5p | miR-221, miR-361-5p |
| BHRF1 | 13 (8) | miR-rL1-5-3p, miR-rL1-34-5p, miR-rL1-7-3p | miR-106a/17-5p, miR-106a/423-5p |
| BMRF1 | 6 (2) | miR-rL1-18-5p | None |
| BMRF2 | 7 (2) | miR-rL1-34-5p, miR-rL1-4-3p | None |
| BLRF2 | 3 (1) | miR-rL1-2-3p | None |
| BKRF3 | 4 (1) | None | miR-15a/15b/16, miR-15a/15b/128a |
| BBRF3 | 9 (2) | miR-rL1-18-5p, miR-rL1-17-5p | miR-106a/17-5p |
| BVRF2 | 5 (2) | miR-rL1-34-5p, miR-rL1-17-5p | None |
| gp350 | 11 (4) | miR-rL1-27-3p | None |
| BZLF1 | 5 (2) | miR-rL1-33-5p, miR-rL1-8-5p | None |
| BBLF4 | 3 (1) | miR-rL1-10-3p | None |
| BDLF1 | 5 (1) | miR-rL1-28-3p, miR-rL1-23-3p | None |
| BcLF1 | 16 (4) | miR-rL1-28-5p, miR-rL1-3-5p, miR-rL1-26-5p, miR-rL1-32-3p | miR-449a |
| BXLF2 | 7 (1) | miR-rL1-9-5p | miR-484 |
| BALF2 | 5 (2) | miR-rL1-9-5p, miR-rL1-31-3p | None |
| LMP1 | 11 (4) | miR-rL1-28-3p, miR-rL1-28-5p, miR-rL1-29-3p, miR-rL1-31-3p, miR-rL1-22-5p, miR-rL1-23-3p, miR-rL1-8-5p, miR-rL1-5-3p, miR-rL1-7-5p | miR-103/107, miR-103/98, miR-103/17-5p |
Numbers in parentheses are the number of overlapping clusters between 211-98 and 309-98 PAR-CLIP libraries.
Intriguingly, we observed conservation of the flanking sequences for the miR-BHRF1-3 hairpin precursor as well as conservation of its position within the BHRF1 3′UTR region of herpesvirus papio and herpesvirus pan, but not of the actual miRNA sequence itself (Fig. 2C). Thus, while the structure and location for a viral miRNA hairpin is conserved among these four primate LCVs, the mature miRNA sequences have diverged within each species. One possibility is that the miR-BHRF1-3 orthologs have species-specific cellular targets driving changes in the miRNA sequence. Nonetheless, these data illustrate that viral pre-miRNA sequences, specifically within the lymphocryptovirus genus, are much more conserved than previously reported (14) and imply that other related primate LCVs, such as those that infect gorillas or orangutans (44), also encode BHRF1 miRNA homologs.
PAR-CLIP analysis of LCV-infected LCLs reveals RISC-associated viral mRNAs.
Having identified all viral miRNA homologs and orthologs expressed in wild-type EBV LCLs and rLCV rLCLs, we were next interested in evolutionarily conserved miRNA targets. EBV LMP1 and BHRF1 mRNAs are cotargeted by both viral and cellular miRNAs, such as EBV BART miRNAs and the miR-17/20/106 family, in EBV B95-8 LCLs and type III BL cells (27, 28). This prompted us to ask whether the rLCV BART miRNA homologs or the rhesus macaque miR-17/20/106 family also targets rLCV gene products. To globally identify miRNA binding sites on viral RNAs in situ, we analyzed one wild-type EBV LCL (Akata-LCLd3) and the two rLCV rLCLs (211-98 and 309-98) using photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) (26) to isolate RISC-associated mRNAs. Cells were cultured in log phase, pulsed overnight with 4-thiouridine (4SU), and UV cross-linked at 365 nm. Cross-linked, RISC-bound RNAs were purified from RISC immunoprecipitations, ligated to adapters, and sequenced on the Illumina HiSeq2000 instrument. The introduction of 4SU into cells and, as a result, nascent RNA causes final sequencing reads to contain a T→C mutation at cross-linked sites (26). PAR-CLIP reads were mapped concurrently to the human or macaque genomes and respective viral genomes and clustered using the T→C signal, by PARalyzer, to define RISC-associated sites (25). The majority of reads aligned to cellular locations, and we identified >32,000 clusters with T→C conversions in each PAR-CLIP library that represent cross-linked, RISC-bound regions. Since we were interested in RISC-associated viral transcripts, we focused on reads mapping exclusively to the viral genomes (∼5% of all aligned reads for rLCL 211-98 and ∼2% of all aligned reads for Akata-LCLd3).
As expected, most RISC-associated viral regions with high read counts in the three PAR-CLIP libraries represented noncoding RNAs such as viral miRNAs or fragments of the EBERs. Excluding the reads aligning to noncoding RNAs, clusters with the highest read counts in the EBV PAR-CLIP library mapped to regions encoding BHRF1, EBNA2, and LMP1 (Fig. 3A). In addition to these three RISC-associated hot spots, which have been observed in EBV B95-8 LCLs and latency III BL cells (27, 28), we identified clusters mapping to lytic mRNAs encoding BNRF1 and BALF2 (Table 2). In the rLCV PAR-CLIP libraries, we identified clusters mapping to the rLCV BHRF1, EBNA2, and LMP homologs, demonstrating conserved miRNA regulation of these three viral gene products (Fig. 3B and C). We also identified clusters mapping to several lytic transcripts, such as BZLF1 (immediate early transactivator), BcLF1 (capsid protein), BNRF1 (tegument protein), gp350 (envelope glycoprotein), and BALF2 (single-stranded DNA binding protein), among others (Table 3), indicating a low level of lytic replication occurring in the rLCLs. This level of lytic replication likely also explains the high levels of rLCV miRNAs observed in rLCLs (Fig. 1). Despite capturing RISC-associated lytic transcripts in both the EBV-infected and rLCV-infected LCLs, we did not detect any reads mapping to BALF5 or the rLCV BALF5 homolog, which have previously been shown or predicted to be targeted by miR-BART2 (11, 49) or miR-rL1-33 (14), respectively.
FIG 3.
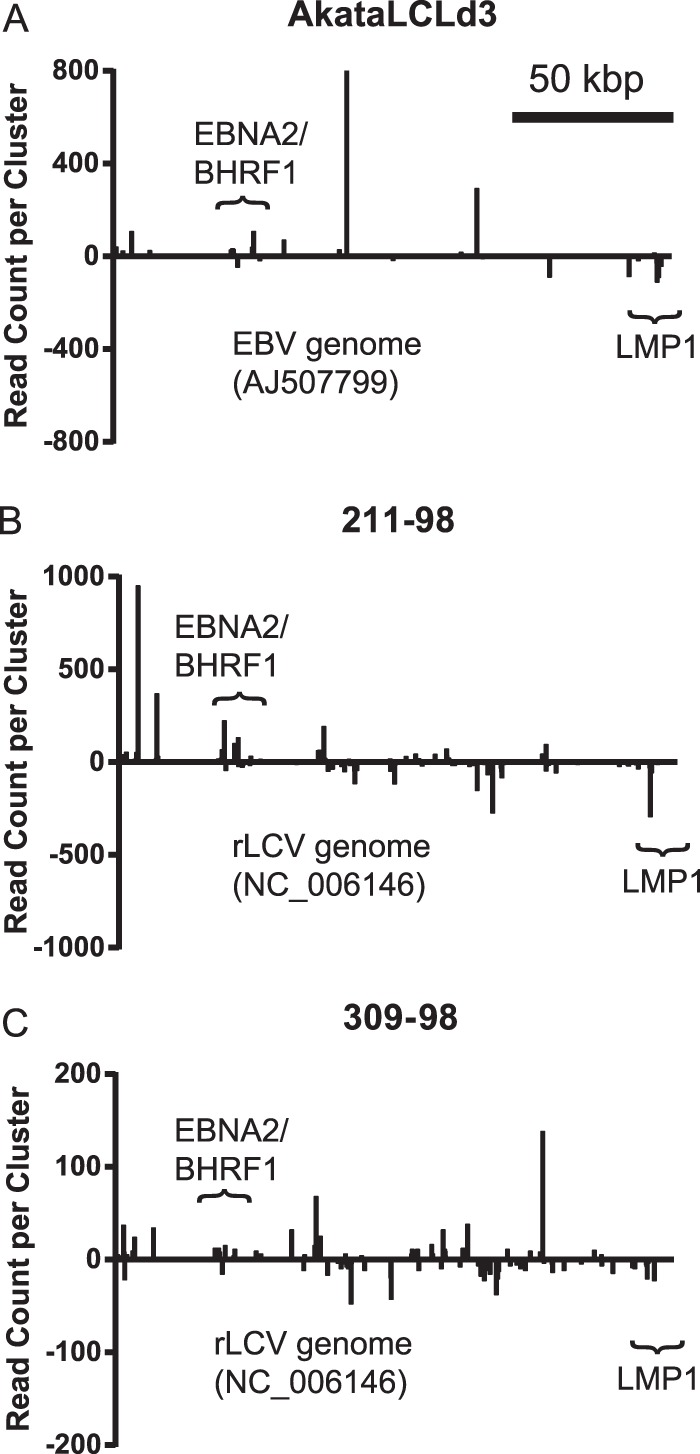
PAR-CLIP analysis of LCLs. Whole-genome view of PAR-CLIP/PARalyzer clusters aligning to EBV1 for AkataLCLd3 (A) or rLCV for 211-98 and 309-98 rLCLs (B and C). The y axis shows the read count per cluster. Read counts below the x axis (negative y axis values) indicate reads aligned to the complementary strand. Nucleotide positions in the viral genome are shown on the x axis. Clusters mapping to noncoding RNAs (i.e., miRNAs and EBERs) are not shown. Scale, 50 kbp.
TABLE 2.
RISC-associated EBV transcripts identified by PAR-CLIP
| Gene | No. of clusters | miRNAs with seed match(es) |
|
|---|---|---|---|
| Viral | Cellular | ||
| BNRF1 | 3 | miR-BART5-5p, miR-BART17-3p | None |
| EBNA2 | 4 | None | miR-423-5p |
| BHRF1 | 11 | miR-BART10-3p | miR-142-3p |
| BALF2 | 3 | miR-BART1-5p | None |
| LMP1 | 5 | miR-BART5-5p, miR-BART16-5p, miR-BART19-3p | miR-17-5p/93-5p, miR-17-5p/186-5p |
To determine which miRNAs are targeting the RISC-associated sites on viral transcripts, we scanned viral clusters for seed matches to all EBV or rLCV miRNAs as well as the top ∼200 mature cellular miRNAs (cutoff of >0.01% of reads aligned to cellular miRNAs) identified by RIP-SEQ and/or smRNA-SEQ (Fig. 1). Tables 2 and 3 list the miRNAs with seed matches to viral clusters (in coding regions and 3′UTRs). Latent viral transcripts, in particular those encoding LMP1, BHRF1, and rLCV EBNA2, are cotargeted by both viral and cellular miRNAs in LCLs (Tables 2 and 3), albeit at discrete sites (Fig. 3 to 5). The rLCV EBNA2 mRNA, for example, contains predicted binding sites for four different viral miRNAs and two cellular miRNAs (Table 3). Intriguingly, these data demonstrate that, in addition to latent viral transcripts, lytic transcripts are also subject to miRNA-mediated regulation. Unlike sites on latent transcripts, the majority of RISC-associated sites on lytic transcripts contained seed matches to viral, not cellular, miRNAs (Table 3). These data suggest that lytic viral mRNAs have not evolved to be targeted by host cellular miRNAs, and that one function of lymphocryptovirus miRNAs is to attenuate the expression of specific lytic viral gene products.
FIG 5.
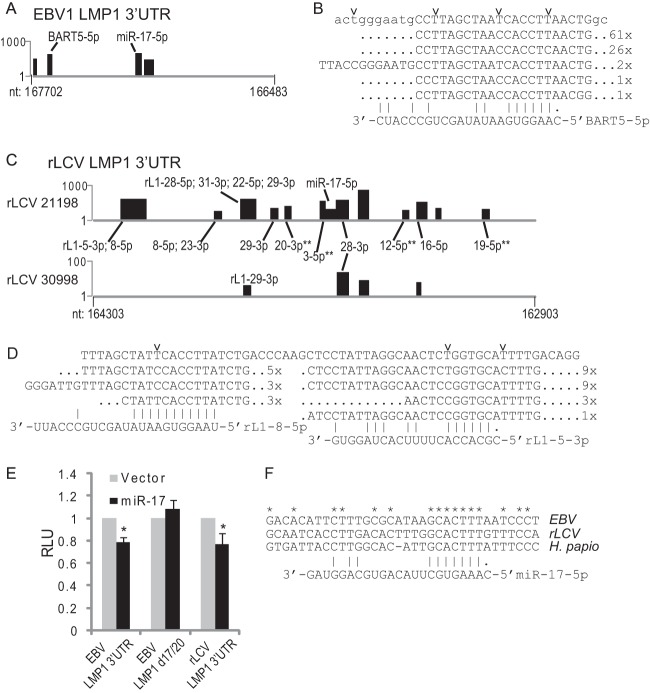
LMP1 transcript is a conserved target of viral and cellular miRNAs. (A) The EBV LMP1 3′UTR (nt 166483 to 167702; EBV1) contains multiple RISC-associated sites. Highlighted are clusters (black bars) with seed matches to miR-BART5-5p and the cellular miR-17/20/106 family. (B) miR-BART5-5p targets EBV LMP1 3′UTR. Shown is the cluster containing the miR-BART5-5p seed match site and associated reads with read counts. Individual T→C conversions are marked. (C) The rLCV LMP1 3′UTR (nt 162903 to 164303; rLCV) contains multiple RISC-associated sites (black bars). Highlighted are rLCV miRNAs with seed matches (minimally 7mer1A) to individual clusters. Two asterisks indicate a miRNA with a 6mer (nt 2 to 7) match. For panels A and C, the y axis indicates read count per cluster on a log scale. (D) miR-rL1-8-5p (miR-BART5 homolog) and miR-rL1-5-3p (miR-BART3 homolog) target the rLCV LMP1 3′UTR. Shown is a cluster containing rLCV miRNA seed matches and associated reads; individual T→C conversions are marked. (E) 293T cells were cotransfected with 1 μg hsa-miR-17 expression vector, 10 ng pLSR, and 10 ng pLSG-LMP1, pLSG-LMP1d17/20 (contains a mutation in the miR-17/20/106 binding site), or pLSG-rLMP1 3′UTR reporter. Luciferase activity was measured 48 to 72 h posttransfection. Shown are the averages from three independent experiments performed in triplicate; error bars indicate standard deviations. All firefly luciferase units are normalized to Renilla activity (RLU, relative light units) and are reported relative to the empty vector. *, P < 0.05 relative to empty vector (Student's t test). (F) The miR-17 binding site in the LMP1 3′UTR is conserved in primate lymphocryptoviruses. Clustal2.0 multiple sequence alignment of the miR-17 binding region in EBV, rLCV, and herpesvirus papio LMP1 mRNAs.
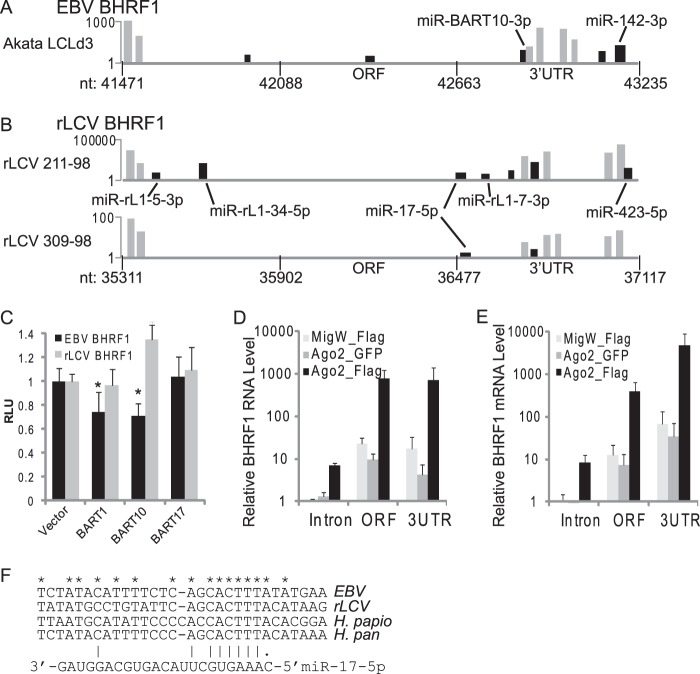
BHRF1 mRNAs are RISC associated in LCLs.
Multiple BHRF1 RNAs are transcribed from lytic or latent promoters during EBV infection and either spliced to generate BHRF1 mRNAs or cleaved by Drosha to generate BHRF1 miRNAs (34, 35). PAR-CLIP analysis of Akata-LCLd3 detected binding sites for miR-BART10-3p and cellular miR-142-3p within the BHRF1 3′UTR (Fig. 4A) as well as several other sites which could not be assigned to miRNAs with complementary seed matches. Although we and others have previously identified a binding site for the miR-17/20/106 family in the BHRF1 3′UTR in EBV-infected B cells (27, 28), we did not detect any reads encompassing this site in the Akata-LCLd3 PAR-CLIP library. The most likely explanation for this absence is that PAR-CLIP libraries are not saturating; therefore, we do not capture every miRNA-mRNA interaction. We examined the miR-17/20/106, miR-142-3p, and miR-BART10-3p binding sites in the BHRF1 3′UTR of six different EBV isolates, including those in Akata, MutuI, and Raji BL cells, and found that the sites are fully conserved in EBV isolates (not shown).
FIG 4.
BHRF1 mRNA is targeted by miRNAs. (A) PAR-CLIP/PARalyzer clusters (black bars) from Akata-LCLd3 aligned with the EBV BHRF1 region (nt 41471 to 43235; EBV1). Light gray bars indicate clusters mapping to the EBV BHRF1 miRNAs. Highlighted are clusters in the BHRF1 3′UTR containing seed matches to miR-BART10-3p and miR-142-3p. (B) Clusters from rLCV 211-98 and 309-98 PAR-CLIP/PARalyzer analysis (black bars) that align to the rLCV BHRF1 region. Light gray bars indicate clusters mapping to the rLCV BHRF1 miRNA homologs. Highlighted are clusters with seed matches to viral and cellular miRNAs. Of note, the miR-rL1-5-3p and miR-rL1-34-5p binding sites occur in clusters that presumably map to a lytic BHRF1 mRNA. For panels A and B, the y axis indicates the read count per cluster on a log scale. (C) 293T cells were cotransfected with 1 μg miRNA expression vector, 10 ng pLSR, and 10 ng pLSG-EBV-BHRF1 3′UTR reporter or pLSG-rLCV-BHRF1 3′UTR reporter. Luciferase activity was measured 48 to 72 h posttransfection. Shown are the averages from three independent experiments performed in triplicate; error bars indicate standard deviations. All firefly luciferase units are normalized to Renilla activity (RLU, relative light units) and are reported relative to the empty vector. *, P < 0.0 relative to empty vector (Student's t test). (D and E) EBV BHRF1 mRNA is RISC associated. RISC-associated RNAs were immunoprecipitated from EF3D-Ago2 LCLs using anti-FLAG or anti-GFP (negative-control) antibody. EF3D-MigW (expressing only GFP) cells were used as an additional negative control. RISC-associated RNAs were purified and reverse transcribed using random hexamers (D) or oligo(dT) (E) as the primer. BHRF1 cDNA was detected by quantitative PCR using primers against the BHRF1 5′ intron, ORF, or 3′UTR. Shown are the levels of BHRF1 RNA or mRNA relative to EF3D-MigW (control) and pcDNA3-BHRF1 standards (see Materials and Methods) (n = 2). Error bars indicate standard deviations.
Ectopic expression of miR-BART10 in 293T cells inhibited luciferase activity from an EBV BHRF1 3′UTR reporter (Fig. 4C), confirming miR-BART10 targeting of the BHRF1 mRNA. Interestingly, the miR-BART10-3p homolog, miR-rL1-27-3p, lacks a seed match to the rLCV BHRF1 3′UTR. We did not observe knockdown of an rLCV BHRF1 3′UTR reporter with miR-BART10 expression (Fig. 4C), indicating that miR-BART10 targeting of the BHRF1 3′UTR is specific to EBV. In addition to miR-BART10, we observed significant inhibition of the EBV BHRF1 3′UTR reporter in the presence of miR-BART1. miR-BART1-3p and its rLCV homolog, miR-rL1-6-3p, have seed matches (nt 2 to 8) to the EBV and rLCV BHRF1 3′UTRs, although the positions of the seed match sites and the miRNA inhibition (Fig. 4C and not shown) are not conserved. We did not observe clusters in the EBV or rLCV BHRF1 3′UTRs with seed matches to miR-BART1-3p or miR-rL1-6-3p (Fig. 4A); however, the knockdown of the luciferase reporter raises the possibility that miR-BART1-3p also targets the EBV BHRF1 mRNA.
Notably, we captured miR-17/20/106 binding sites in both of the rLCL PAR-CLIP samples (Fig. 4B), demonstrating that targeting of the BHRF1 3′UTR by the miR-17 family is evolutionarily conserved. Homologous to EBV BHRF1, the miR-17/20/106 binding site in the rLCV BHRF1 RNA sits adjacent to the stop codon at the 5′ start of the 3′UTR. This location encompasses several BHRF1 transcripts, including the lytic and latent BHRF1 mRNAs and the RNAs arising from cleavage by Drosha to generate BHRF1 miRNAs (34, 35). To confirm that it is a BHRF1 mRNA that is associated with RISC, we performed RIP-quantitative PCR analysis on EBV B95-8 LCLs expressing Ago2-Flag. Briefly, RISC-bound RNAs were immunopurified from LCLs and reverse transcribed using either random primers (Fig. 4D) or oligo(dT) (Fig. 4E). The cDNAs from Ago2-bound BHRF1 RNAs and mRNAs were analyzed by quantitative PCR using primers against the intron, ORF, or 3′UTR of EBV BHRF1. We observed an ∼100-fold enrichment (over controls) for BHRF1 mRNAs in the Ago2-Flag immunoprecipitations (Fig. 4E).
The BHRF1 3′UTR is highly conserved between EBV and rLCV (16). This prompted us to ask if the miR-17 binding site is conserved in additional primate lymphocryptoviruses. Homologous to EBV and rLCV, an miR-17 binding site is present in the BHFR1 3′UTRs of herpesvirus papio and herpesvirus pan (45) and sits adjacent to the BHRF1 stop codon (Fig. 4F), providing further evidence of a connection between the miR-17 family and the BHRF1 mRNA(s) that has been conserved during lymphocryptovirus evolution.
LMP1 is regulated by conserved and nonconserved lymphocryptovirus miRNAs.
Examination of the EBV LMP1 3′UTR by PAR-CLIP revealed four RISC-associated regions in Akata-LCLd3, two of which contained binding sites for miR-BART5-5p (7mer1A seed match) and the cellular miR-17/20/106 family (Fig. 5A and B). The other two sites could not be assigned to any viral or cellular miRNAs with complementary seed matches; presumably, these sites arise from noncanonical miRNA interactions which are difficult to decipher.
EBV miR-BART5-5p shares a common seed sequence (nt 2 to 7) with cellular miR-18a-5p (20, 23). While Riley et al. did not observe repression of an LMP1 3′UTR reporter in the presence of an miR-18a-5p mimic (28), our prior PAR-CLIP analysis of EBV B95-8 LCLs (27) (which do not express miR-BART5) uncovered a RISC-associated region overlapping the miR-BART5-5p/miR-18a-5p seed match site identified here in Akata-LCLd3. These data suggest that this site in the LMP1 3′UTR also is occupied by cellular miR-18a/b.
Multiple RISC-associated sites were identified in the rLCV LMP1 3′UTR containing seed matches to several different rLCV and host miRNAs (Fig. 5C and Table 3). We identified two seed match sites for miR-rL1-8-5p (miR-BART5 homolog), one of which is located in a conserved position. Similar to the RISC-associated LMP1 region captured for miR-BART5 (Fig. 5B), the maximum T→C signal in the most 5′ RISC-associated rLMP1 site for miR-rL1-8-5p occurs directly adjacent to the miRNA seed match site (Fig. 5D), which is characteristic of authentic seed match miRNA target sites (25–27). We also observed clusters with seed matches to miR-rL1-23-3p (miR-BART21-3p homolog) and miR-rL1-29-3p (miR-BART19-3p homolog), among others (Fig. 5C). These data indicate that LMP1 mRNA homologs are targeted by conserved and nonconserved viral miRNAs.
LMP1 is a conserved target of the miR-17/20/106 family.
Notably, we observed a cluster with nine reads that overlaps the miR-17-5p seed match site in 211-98 rLCLs (Fig. 5C). Due to the overwhelming presence of rLCV miRNAs in rLCLs (Fig. 2), the levels of cellular miR-17-5p are very low in these cells, which may explain why we do not capture more reads at this site. To confirm miR-17 targeting of LMP1 mRNA, we cotransfected 293T cells with a miR-17 expression vector and a luciferase reporter containing either the full EBV LMP1 3′UTR cloned from EBV B95-8 LCLs (pLSG-LMP1) (27), the full EBV LMP1 3′UTR with a mutated miR-17 binding site (pLSG-LMP1d17/20), or the full (1347 nt) rLCV LMP1 3′UTR cloned from rLCV rLCLs. Overexpression of miR-17 (Fig. 5E) resulted in >20% knockdown of luciferase activity from the EBV and rLCV LMP1 reporters containing intact miR-17 binding sites, demonstrating that miR-17 targeting of lymphocryptovirus LMP1 mRNAs is conserved. Importantly, inhibition of luciferase activity by miR-17 was alleviated upon mutation of the miR-17/20/106 binding site in the EBV LMP1 3′UTR (Fig. 5E).
To ask whether the miR-17 binding site is conserved in other lymphocryptoviruses, we sequenced the LMP1 3′UTR from baboon LCLs (S594 cells) infected with herpesvirus papio (50). The predicted miR-17 site was indeed present in the herpesvirus papio LMP1 3′UTR, further supporting the hypothesis that these viral LMP1 mRNA homologs have coevolved with host miR-17 and that miR-17-dependent regulation of LMP1 is important to the lymphocryptovirus life cycle (Fig. 5G). We also queried the herpesvirus papio LMP1 3′UTR for putative viral miRNA seed match sites using the conserved rLCV and EBV miRNA seed sequences as well as the rLCV miRNAs that have PAR-CLIP-identified sites in the rLCV LMP1 3′UTR. Intriguingly, seed matches for miR-rL1-8-5p, rL1-23, rL1-16, and rL1-28 were present in the herpesvirus papio LMP1 3′UTR sequence (not shown). The presence of these sites implies that herpesvirus papio encodes BART miRNA homologs that presumably target the herpesvirus papio LMP1 mRNA in a manner homologous to EBV and rLCV miRNAs.
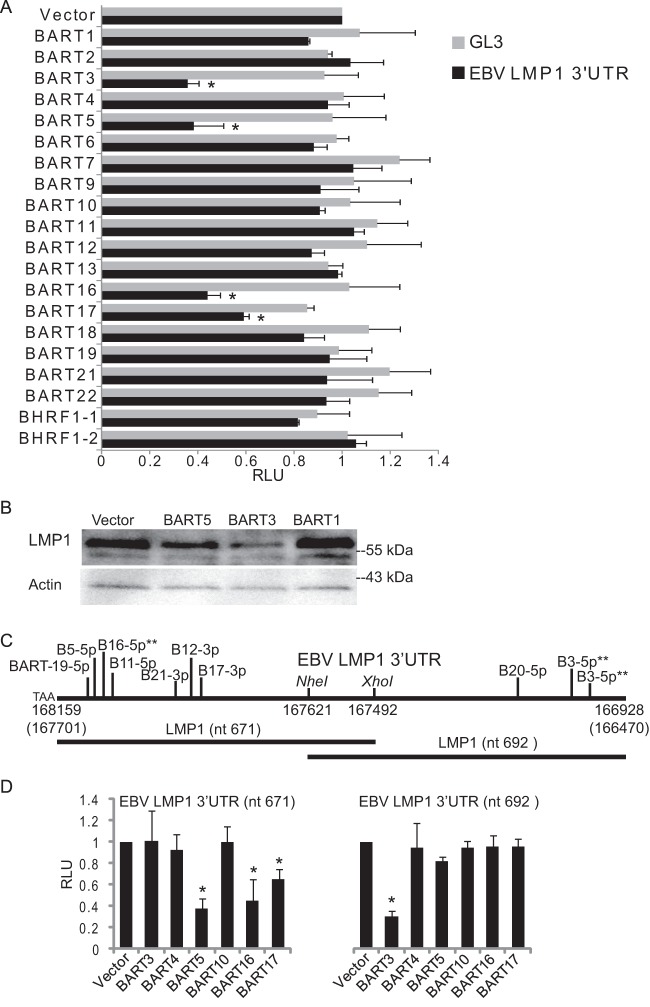
Multiple BART miRNAs target the LMP1 3′UTR.
Lo et al. initially described targeting of the LMP1 3′UTR by EBV BART cluster I miRNAs, specifically miR-BART1-5p, miR-BART16, and miR-BART17-5p (37). At that time, however, the EBV miRNA sequences were not fully characterized, and several of the miRNA mimics used in their study differ from the authentic EBV BART miRNAs by one or more nucleotides in the miRNA seed region. We have been unable to confirm miR-BART1-5p targeting of the LMP1 3′UTR by PAR-CLIP analysis of EBV B95-8 LCLs (27) and wild-type LCLs (Fig. 5) or using luciferase reporter assays (27) (Fig. 6); however, we have previously observed knockdown of an LMP1 3′UTR reporter by miR-BART3 (27). Recently, HITS-CLIP analysis of latency III EBV+ BL cells revealed binding sites for other BART miRNAs in the LMP1 3′UTR, specifically, miR-BART5-5p (confirmed above) (Fig. 5) and miR-BART19-5p (not confirmed) (28). As none of these studies have been in agreement, we decided to investigate viral miRNA targeting of the LMP1 3′UTR in greater detail.
FIG 6.
Multiple EBV BART miRNAs target the LMP1 3′UTR. (A) 293T cells were cotransfected with 1 μg of the indicated miRNA expression vector (BART or BHRF1 miRNA), 10 ng pLSR (Renilla internal control plasmid), and 10 ng either pLSG control luciferase (GL3) or pLSG-LMP1 luciferase reporter containing the full EBV LMP1 3′UTR cloned from EBV B95-8 LCLs. For all luciferase assays (A and D), luciferase activity was measured 48 to 72 h posttransfection. Shown are the averages from 2 to 4 independent experiments performed in triplicate; error bars indicate standard deviations. All firefly luciferase units are normalized to Renilla activity (RLU, relative light units) and are reported relative to empty vector. *, P < 0.05 relative to empty vector (Student's t test). (B) Western blot analysis of LMP1 levels in the presence of BART miRNAs. 293T cells were transfected with 1 μg individual BART miRNA expression vectors. Twenty-four h later, cells were transfected again with 5 ng pLMP1 and 1 μg filler DNA (pK-GST). Lysates were harvested for Western blotting after 20 h. LMP1 and beta-actin were detected using monoclonal antibodies. (C) The EBV LMP1 3′UTR contains seed match sites (minimally, 7mer1A) to seven BART miRNAs. The double asterisks indicate positions of predicted, noncanonical sites for miR-BART16-5p and miR-BART3-5p. Numbers below indicate genomic coordinates relative to EBV B95-8 or, in parentheses, wild-type EBV1. Two truncated LMP1 3′UTR luciferase reporters were generated in pLSG and contain either the first 5′ 671 nt (nt 1 to 671) or the last 3′ 694 nt (nt 563 to the end) of the EBV LMP1 3′UTR. (D) Same as panel A, except 293T cells were cotransfected with 1 μg miRNA expression vector, 10 ng pLSR (Renilla internal control plasmid), and 10 ng truncated LMP1 3′UTR luciferase reporter. Lysates were analyzed after 72 h.
To definitively identify EBV miRNAs that can target the LMP1 3′UTR, we generated expression vectors for 20 of the 25 EBV pre-miRNAs and cotransfected these vectors into 293T cells with either a control luciferase vector (pLSG) or the EBV LMP1 3′UTR luciferase reporter (pLSG-LMP1) (Fig. 6A). Ectopic expression of miR-BART5 significantly inhibited the LMP1 3′UTR reporter, confirming results from our PAR-CLIP analysis of Akata-LCLd3 (Fig. 5). Additionally, expression of three other BART miRNAs, miR-BART3, miR-BART16, and miR-BART17, significantly inhibited the reporter (Fig. 6A). Contrary to previous reports, which used artificial miRNA mimics that bypass the miRNA biogenesis machinery, we did not observe inhibition of the LMP1 3′UTR reporter by miR-BART1 (37) or miR-BART19 (28). We next cloned full-length LMP1 into the expression plasmid pcDNA3 and performed Western blot analysis on 293T cells cotransfected with pLMP1 and BART miRNAs. LMP1 protein expression was reduced in the presence of miR-BART3 or miR-BART5, but not miR-BART1, compared to the vector control (Fig. 6B), confirming targeting of LMP1 by miR-BART3 and miR-BART5.
To identify binding sites for the four miRNAs inhibiting the LMP1 3′UTR reporter, we analyzed the LMP1 3′UTR sequence for seed matches to EBV miRNAs. There are seven predicted seed match sites (minimally 7mer, nt 2 to 8, or minimally 7mer1A, nt 2 to 7, with an A across from position 1 of the miRNA) for BART miRNAs (Fig. 6C). Comparison of the EBV B95-8 LMP1 3′UTR sequence to sequences from other EBV isolates, including those present in Akata (KC207813), Raji (NC_007605.1), and MutuI (KC207814) BL cells (51), C666 nasopharyngeal epithelial carcinoma (NPC) cells (EF103558), and other NPC isolates (37), showed that six of the seven seed match sites are 100% conserved (data not shown). A predicted miR-BART17-3p seed match site is potentially affected by a single point mutation in MutuI cells (not shown). For the four EBV miRNAs inhibiting the LMP1 3′UTR reporter (Fig. 6A), we identified putative seed match sites for miR-BART5-5p and miR-BART17-3p but not miR-BART3 or miR-BART16 (Fig. 6C). We next used RNAhybrid (52) to scan the LMP1 3′UTR for miR-BART3 and miR-BART16 binding sites. A potential noncanonical site (minimal free energy [MFE], −33.2 kcal/mol), located within the first 115 nt of the 3′UTR, was predicted for miR-BART16-5p, while two noncanonical sites (MFE, <−20.2) for miR-BART3-5p were predicted in the 3′ 220 nt of the 3′UTR (Fig. 6C). All of these sites contained G:U base pairs in the 5′ seed regions (nt 2 to 8) with extensive 3′ compensatory pairing (not shown).
To map BART miRNA binding sites within the LMP1 3′UTR, we cloned the 5′ 671 nt or 3′ 692 nt of the LMP1 3′UTR into pLSG and tested these truncated 3′UTR reporters against BART miRNA expression vectors. Consistent with the location of their seed match sites, miR-BART5 and miR-BART17 inhibited pLSG-LMP1nt671 but not pLSG-LMP1nt692 (Fig. 6D). miR-BART16 also inhibited pLSG-LMP1nt671 but not pLSG-LMP1nt692, indicating that miR-BART16 targets the 5′ region of the LMP1 3′UTR. Congruent with the predicted, noncanonical binding sites for miR-BART3-5p, miR-BART3 inhibited pLSG-LMP1nt692 but not pLSG-LMP1nt671 (Fig. 6D). These data indicate that miR-BART5 and miR-BART17 target the LMP1 3′UTR via canonical seed match sites while miR-BART3 and miR-BART16 utilize noncanonical sites. Interestingly, we did not capture sites for miR-BART3, miR-BART16, or miR-BART17 in our PAR-CLIP analysis of Akata-LCLd3 (Fig. 5). While the most likely explanation is that we do not capture the sites due to nonsaturating conditions and incomplete coverage, it is also possible that these viral miRNAs do not directly interact with the LMP1 3′UTR in LCLs.
Viral miRNA targeting of the LMP1 3′UTR is conserved in rLCV-infected rLCLs.
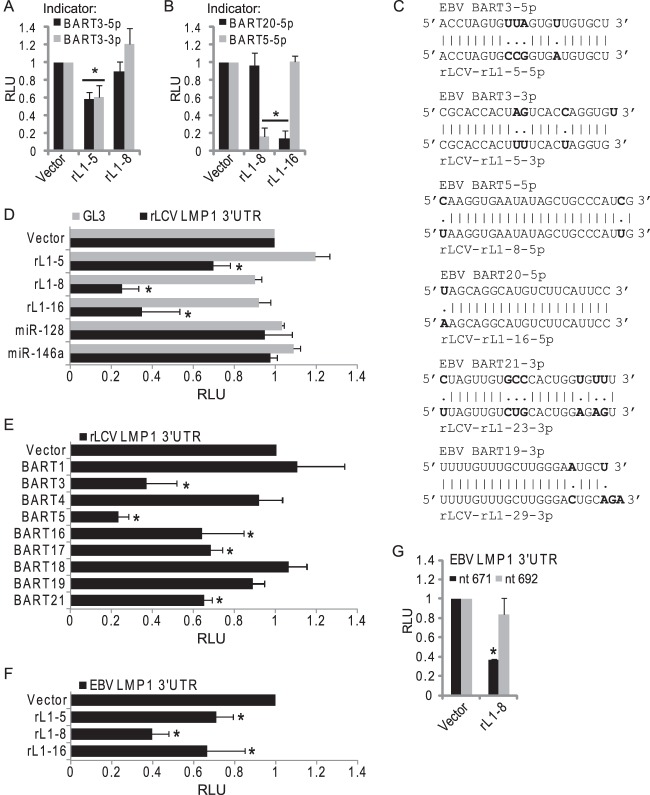
Having identified several EBV miRNAs that can target the EBV LMP1 3′UTR, we next analyzed the rLCV LMP1 3′UTR sequence for seed matches (minimally 7mer1A) to rLCV miRNAs. At least 26 potential binding sites exist for rLCV miRNAs, including sites for the miR-BART5-5p homolog (miR-rL1-8-5p) and the miR-BART3-5p homolog (miR-rL1-5-5p), as well as miR-rL1-16-5p (miR-BART20-5p homolog), miR-rL1-23-3p (miR-BART21-3p homolog), and miR-rL1-29-3p (mir-BART19-3p homolog) (data not shown). Notably, sites for the rLCV miRNAs listed above were identified in our PAR-CLIP analysis (Fig. 3). For the remaining two EBV BART miRNAs that target the LMP1 3′UTR (Fig. 6), miR-rL1-19-5p is a homolog of miR-BART16-5p while miR-rL1-9 is an ortholog of miR-BART17-3p (Table 1). Neither of these two rLCV miRNAs has a predicted binding site in the rLMP1 3′UTR, indicating that either the miRNA binding site or the miRNA itself has diverged. Indeed, miR-rL1-9 and miR-BART17-3p are conserved in their 3′ regions but have entirely different 5′ seed sequences (14) (Table 1).
We generated miRNA expression vectors for miR-rL1-5, miR-rL1-8, and miR-rL1-16. To test miRNA activity, we cotransfected the rLCV miRNA expression vectors with EBV BART miRNA indicator vectors with perfect matches to a given miRNA (termed perfect indicators) (Fig. 7A). The seed sequences for miR-BART3-3p and miR-BART3-5p are identical to rLCV miR-rL1-5-3p and miR-rL1-5-5p (Fig. 7C); however, nucleotide differences occur in the middle and 3′ ends of the mature miRNAs, which cause the rLCV miRNAs to have partial complementarity to the EBV miRNA perfect indicators. As expected, expression of miR-rL1-5 resulted in partial knockdown of both the miR-BART3-3p and miR-BART3-5p perfect indicators, demonstrating that miR-rL1-5-3p and miR-rL1-5-5p are both expressed (Fig. 7A). miR-rL1-8-5p and miR-rL1-16-5p exhibit near-perfect sequence identity to miR-BART5-5p and miR-BART20-5p, respectively (Fig. 7C); expression of these two rLCV miRNAs resulted in >85% knockdown of their respective EBV miRNA homolog indicators (Fig. 7B).
FIG 7.
Conserved targeting of LCV LMP1 by viral miRNAs. (A and B) 293T cells were cotransfected with 1 μg rLCV miRNA expression vector, 10 ng pLCR (Renilla internal control plasmid), and 10 ng EBV BART miRNA indicator. EBV BART miRNA indicators contain two perfect, complementary binding sites for a BART miRNA in the firefly luciferase 3′UTR of pLCG. (C) Sequence comparison of the mature miRNA sequences for select EBV and rLCV miRNAs that bear seed matches to lymphocryptovirus LMP1 3′UTRs. Nucleotide differences are noted in boldface. (D to G) 293T cells were cotransfected with 1 μg of the indicated miRNA expression vector, 10 ng pLSR (Renilla internal control plasmid), and 10 ng either pLSG control luciferase (GL3) (D), pLSG-rLMP1 3′UTR reporter containing 1,347 nt of the rLCV LMP1 3′UTR (D and E), pLSG-LMP1 3′UTR reporter containing the full EBV LMP1 3′UTR (F), or pLSG-LMP1nt671 or pLSG-LMP1nt692 containing truncated EBV LMP1 3′UTRs (G). hsa-miR-146a and hsa-miR-128 are shown as negative controls for panels D and E. For all luciferase assays, luciferase activity was measured 48 to 72 h posttransfection. Shown are the averages from 2 to 4 independent experiments performed in triplicate; error bars indicate standard deviations. All firefly luciferase units are normalized to Renilla activity (RLU, relative light units) and are reported relative to empty vector. *, P < 0.05 relative to empty vector (Student's t test).
We next assayed luciferase activity from the rLMP1 3′UTR reporter (pLSG-rLMP1) in the presence of the three rLCV miRNAs. Expression of each rLCV miRNA significantly inhibited the rLMP1 3′UTR reporter (Fig. 7D), confirming results from our PAR-CLIP analysis described above (Fig. 5C). We next asked if the EBV and rLCV miRNAs could functionally substitute for each other. rLCV miRNAs were tested against the EBV LMP1 3′UTR reporter(s) and vice versa. As predicted, expression of individual, homologous viral miRNAs resulted in significant inhibition of the lymphocryptovirus LMP1 3′UTR reporters to various degrees (Fig. 7E to G). miR-BART16 and miR-BART17 lack seed match sites in the rLCV LMP1 3′UTR; however, these two miRNAs also moderately inhibited the rLCV LMP1 3′UTR reporter (Fig. 7E), presumably via noncanonical interactions. Intriguingly, ectopic expression of miR-BART21 also inhibited luciferase expression from the rLCV LMP1 3′UTR reporter (Fig. 7E), consistent with our PAR-CLIP results demonstrating that the miR-BART21 homolog (miR-rL1-23) binds the rLMP1 mRNA (Fig. 5C). We also observed significant repression of the EBV LMP1 reporter by miR-rL1-16, indicating that the miR-rL1-16 homolog (miR-BART20) can target EBV LMP1 (Fig. 7F). Together, these data demonstrate that LMP1 mRNA homologs are targeted by multiple, conserved viral miRNAs.
Functional consequences of LMP1 targeting by viral miRNAs: EBV miRNAs attenuate LMP1-mediated NF-κB activation.
LMP1 is a transmembrane protein that contains two C-terminal activation domains (CTAR1 and CTAR2) which recruit TRAFs (TNF receptor-associated factors) and TRADD (TNF receptor 1-associated death domain protein) to activate cell signaling pathways, such as the induction of nuclear factor kappa B (NF-κB) activity. CTAR2 predominantly activates canonical, IKK-dependent NF-κB signaling, while CTAR1 activates both canonical and noncanonical, NIK-dependent NF-κB signaling pathways (53, 54). Activation of NF-κB prosurvival signals via both LMP1 CTAR1 and CTAR2 is required for B cell transformation, cell survival, and proliferation (54, 55). Overexpression of LMP1 and/or dysregulation of NF-κB signaling pathways during EBV infection, however, can induce apoptosis and inhibit cell proliferation (56, 57). Notably, the ability of LMP1 to induce NF-κB activity via C-terminal cytoplasmic domains is conserved in the rhesus and baboon LCV LMP1 protein homologs (50).
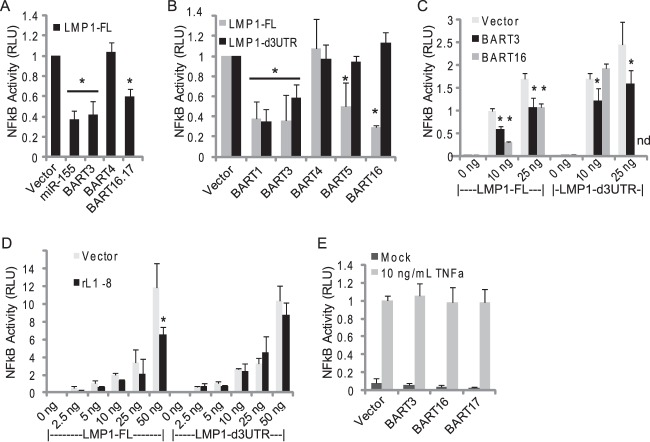
To explore the downstream consequences of LMP1 targeting by viral miRNAs, we assayed LMP1-induced NF-κB activation in the presence of EBV BART miRNAs. Full-length EBV LMP1, including the 3′UTR, was cotransfected into 293T cells with EBV miRNA expression vectors and an NF-κB luciferase reporter containing multiple NF-κB-responsive sites (Fig. 8). miR-155, which has previously been shown to disrupt LMP1-mediated NF-κB activation (41), was tested in parallel. Congruent with prior studies, ectopic expression of miR-155 or coexpression of miR-BART16 and miR-BART17 together (37) inhibited LMP1-mediated NF-κB activation (Fig. 8A). We also asked whether the other BART miRNAs that target the LMP1 mRNA (Fig. 5 and 6) could confer a similar phenotype. Indeed, miR-BART3, miR-BART5, and miR-BART16 each inhibited NF-κB activation when cotransfected with full-length LMP1 (Fig. 8A to C). Deletion of the LMP1 3′UTR rescued the inhibitory effects of miR-BART5 and miR-BART16 (Fig. 8B and C), consistent with targeting of the LMP1 3′UTR. We also tested the miR-BART5 homolog, rLCV miR-rL1-8. Ectopic expression of miR-rL1-8 inhibited NF-κB activation in the presence of full-length EBV LMP1, while deletion of the LMP1 3′UTR rescued the inhibition (Fig. 8D). These data demonstrate that miR-rL1-8 can functionally substitute for miR-BART5 and that miR-BART5 and miR-BART16 negatively regulate NF-κB signaling by directly targeting the LMP1 3′UTR.
FIG 8.
EBV BART miRNAs attenuate LMP1-mediated NF-κB activation. (A) 293T cells were cotransfected with 25 ng NF-κB luciferase reporter, 10 ng Renilla internal control, 5 ng pcDNA3-LMP1-FL, and 1 μg the indicated miRNA expression vector. Lysates were harvested 48 h posttransfection and assayed for luciferase activity. (B) 293T cells were cotransfected with 10 ng either pcDNA3-LMP1-FL or pcDNA3-LMP1d3UTR and 1 μg the indicated miRNA expression vector. After 48 h, cells were cotransfected with 25 ng NF-κB luciferase reporter and 10 ng Renilla internal control. Lysates were harvested 18 h later and assayed for luciferase activity. (C and D) Same as panel B, except the indicated amounts of EBV LMP1 expression vectors were used. For panels A to D, shown are the averages from 2 to 4 independent experiments performed in triplicate; error bars indicate standard deviations. All firefly luciferase units are normalized to Renilla activity (RLU, relative light units) and are reported relative to empty vector. *, P < 0.05 relative to empty vector (Student's t test). (E) EBV BART miRNAs do not attenuate TNF-α-induced NF-κB activation. 293T cells were cotransfected with 1 μg of the indicated miRNA expression vector, 15 ng NF-κB luciferase reporter, and Renilla internal control. Forty-eight h posttransfection, 10 ng/ml TNF-α was added for 3 h and lysates were assayed for luciferase activity.
Interestingly, deletion of the LMP1 3′UTR partially, but not significantly, rescued the inhibitory effect of miR-BART3 (Fig. 8B and C). Similarly, miR-BART1, which does not target the LMP1 3′UTR or impact LMP1 protein expression (Fig. 6), inhibited NF-κB-activation irrespective of the presence of LMP1 3′UTR (Fig. 8B). While we have established that miR-BART3 can target the LMP1 3′UTR (Fig. 6), we cannot rule out the possibility that miR-BART3 also targets the LMP1 coding region, which could impact LMP1 expression and downstream signaling. However, the most likely explanation for the observed inhibitory effect is that these two BART miRNAs negatively regulate cellular genes involved in NF-κB signaling pathways.
LMP1 can induce both canonical and noncanonical NF-κB signaling. To investigate whether BART miRNAs modulate canonical NF-κB signaling independently of LMP1, we treated BART-miRNA-expressing 293T cells with TNF-α for 3 h. In contrast to LMP1-induced NF-κB activation, the presence of miR-BART3, miR-BART16, or miR-BART17 had no significant effect on TNF-α-induced NF-κB activation (Fig. 8E). These data indicate that the inhibitory effects of these BART miRNAs on NF-κB activation are specific to the activities of LMP1.
EBV BART miRNAs stabilize IκBα in the presence of LMP1.
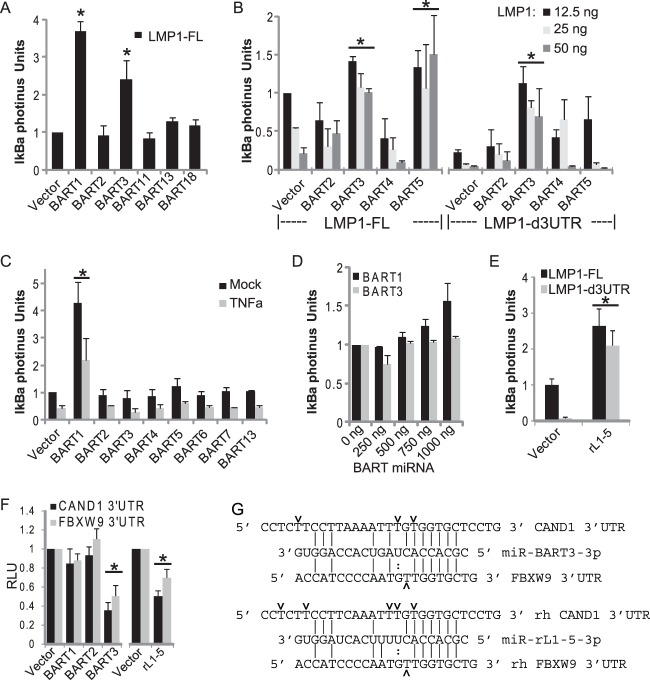
To further explore the role of BART miRNAs in NF-κB signaling, we generated 293T cells stably expressing an IκBα-photinus luciferase fusion protein (293T-IκBα). In canonical NF-κB signaling, NF-κB transcription factors (p50 and p65) are sequestered by IκBα in the cytoplasm. Following a stimulatory signal, IKKs phosphorylate IκBα and induce IκBα degradation, thereby allowing nuclear accumulation of NF-κB transcription factors (reviewed in reference 58). To assay IκBα stability in the presence of viral miRNAs, we monitored the luciferase activity in 293T-IκBα cells cotransfected with pLMP1 and individual BART miRNA expression vectors. Congruent with earlier results (Fig. 8), expression of miR-BART1, miR-BART3, or miR-BART5 stabilized IκBα in the presence of full-length LMP1, while expression of miR-BART2, miR-BART4, miR-BART11, miR-BART13, and miR-BART18 had no effect (Fig. 9A and B). Deletion of the LMP1 3′UTR alleviated the effect of miR-BART5 on IκBα stability, while miR-BART3 continued to stabilize IκBα (Fig. 9B).
FIG 9.
LCV miRNAs regulate IκBα stability. (A) 293T-IκBα cells (stably expressing IκBα-photinus luciferase fusion protein) were cotransfected with 12.5 ng pcDNA3-LMP1-FL and 1 μg BART miRNA expression vector. Luciferase activity was measured 48 h posttransfection. (B) 293T-IκBα cells were cotransfected with 1 μg miRNA expression vector and the indicated amounts of pcDNA3-LMP1-FL or pcDNA3-LMP1-d3UTR (without the 3′UTR). Luciferase activity was measured 48 h posttransfection. (C and D) 293T-IκBα cells were transfected with 1 μg the indicated miRNA expression vector (C) or increasing amounts of miRNA expression vector (D), using empty vector as a filler. In both panels C and D, at 48 h posttransfection cells were treated with 20 ng/ml TNF-α for 3 h, and lysates were assayed for IκBα-photinus luciferase activity. (E) 293T-IκBα cells were cotransfected with 1 μg rLCV miR-rL1-5 expression vector and 12.5 ng pcDNA3-LMP1-FL or pcDNA3-LMP1-d3UTR. Luciferase activity was measured 48 h posttransfection. For panels A to E, shown are the averages from two or more experiments; error bars indicate standard deviations. (F) EBV miR-BART3 and rLCV rL1-5 target the 3′UTRs of CAND1 and FBXW9. 293T cells were cotransfected with 1 μg the indicated miRNA expression vector (BART or rLCV miRNA), 10 ng pLSR (Renilla internal control plasmid), and 10 ng either pLSG-CAND1 or pLSG-FBXW9 3′UTR luciferase reporter. Luciferase activity was measured 48 to 72 h posttransfection, and all firefly luciferase units are normalized to Renilla activity (RLU, relative light units). Shown are averages from 2 to 4 independent experiments performed in triplicate; error bars indicate standard deviations. *, P < 0.05 relative to empty vector (Student's t test). (G) PAR-CLIP/PARalyzer-identified clusters mapping to the CAND1 and FBXW9 3′UTRs with seed matches to miR-BART3-3p (top) or miR-rL1-5-3p (bottom); individual T→C conversions are marked. Rh, rhesus macaque.
To ask whether miR-BART1 and miR-BART3 altered IκBα stability independently of LMP1, 293T-IκBα cells were transfected with BART miRNAs and treated with TNF-α. The presence of miR-BART1 stabilized basal IκBα levels ∼4-fold over vector control and modestly inhibited TNF-α-mediated IκBα degradation in a dose-dependent manner (Fig. 9C and D). In contrast, miR-BART3 expression had no effect on basal IκBα levels or IκBα levels following TNF-α treatment (Fig. 9C and D), suggesting that miR-BART3 function is coupled to the activity of LMP1. Together, these data indicate a role for BART miRNAs in attenuating NF-κB induction and suggest that miR-BART1 and miR-BART3, in particular, target cellular genes involved in IκBα stability and canonical NF-κB signaling. To determine whether the miR-BART3 homolog, rLCV miR-rL1-5, functions similarly in stabilizing IκBα, we transfected 293T-IκBα cells with the rLCV miRNA and EBV LMP1. Homologous to miR-BART3, expression of miR-rL1-5 enhanced IκBα stability in the presence of LMP1 irrespective of the LMP1 3′UTR (Fig. 9E).
IκBα stability is regulated, in part, by β-TRCP, a Skp1/Cullin1/F-box protein complex (SCF) that possesses E3-ubiquitin ligase activity. Following IKK-dependent Ser32 and Ser36 phosphorylation, IκBα is polyubiquitinated by SCF/β-TRCP, which marks it for proteasomal degradation (reviewed in reference 58). One attractive hypothesis is that miR-BART1 and miR-BART3 target components or regulators of the SCF/β-TRCP complex, thereby stabilizing IκBα. A query of our Akata-LCLd3 and prior EBV B95-8 PAR-CLIP libraries (27) for candidate targets revealed RISC-associated sites in the UBXN7/UBXD7, USP47, FBXW9, and CAND1 3′UTRs with seed matches to miR-BART1 and miR-BART3 miRNAs, respectively (Fig. 9G and data not shown). Notably, our PAR-CLIP analysis of rLCV rLCLs revealed RISC-associated sites in the rhesus macaque homologs of these genes with seed matches to rLCV miRNAs (Fig. 9G and data not shown). UBXD7 interacts with neddylated cullins and negatively regulates cullin-RING ubiquitin ligase activity (59, 60) while USP47, a deubiquitylating cysteine protease, interacts directly with the SCF/β-TRCP complex (61). The evolutionarily conserved FBXW9 (called MEC-15 in Caenorhabditis elegans) is one of >100 F-box proteins in mammals that mediates the specificity of the SCF complex (reviewed in references 58 and 62). Finally, CAND1 is a critical exchange factor that regulates the assembled components of the SCF complex (reviewed in reference 62).
We cloned the human CAND1 and FBXW9 3′UTRs downstream of luciferase and tested reporter activity in the presence of LCV miRNAs. Ectopic expression of miR-BART3 or its homolog, miR-rL1-5, resulted in ∼2- to 3-fold luciferase knockdown for each 3′UTR (Fig. 9F). Thus, these data demonstrate an evolutionarily conserved function for LCV miRNAs in the regulation of SCF complex components which consequently may influence IκBα stability and NF-κB activity during viral infection.
DISCUSSION
In this report, we demonstrate that LMP1 and BHRF1 homologs are evolutionarily conserved targets of viral and cellular miRNAs during primate LCV infection. Our previous studies uncovered binding sites for the miR-17/20/106 family and EBV BART miRNAs in latent EBV transcripts. Furthermore, our in silico predictions following analysis of the rLCV genomic sequence (16) suggested that these miRNA binding sites were not limited to EBV; rather, they are conserved in the closely related rLCV (27). Using luciferase reporter assays and by performing PAR-CLIP analysis of LCLs latently infected with EBV or rLCV, we show here that five EBV BART miRNAs (miR-BART3, miR-BART5, miR-BART16, miR-BART17, and miR-BART20) and at least four rLCV BART miRNA homologs (miR-rL1-5, miR-rL1-8, miR-rL1-16, and miR-rL1-23) target LMP1 3′UTRs. Thus, our detailed study establishes a functionally conserved role for LCV miRNAs in the targeting of LMP1 transcripts. In addition to this conserved function for LCV miRNAs, we show that the macaque miR-17 miRNA family targets the rLCV LMP1 and BHRF1 mRNAs homologous to the human miR-17 miRNA family in EBV+ LCLs.
Is miRNA regulation of the LMP1 and BHRF1 3′UTRs conserved by other LCVs?
In addition to rLCV, a number of closely related, Old World LCVs (i.e., herpesvirus papio, herpesvirus pan, herpesvirus gorilla, herpesvirus pongo, and cynomolgus EBV) have been isolated in the past 30 years (30, 44, 63–65). LCV infection in nonhuman primates is similar to EBV infection in humans, and in vitro, simian LCVs exhibit transforming activity comparable to that of EBV; indeed, latently infected LCLs can be established from simian peripheral blood lymphocytes in a manner homologous to the establishment of EBV+ LCLs (30, 31, 44). Given these similarities, it is not surprising that rLCV+ rLCLs also exhibit cellular miRNA expression patterns comparable to those of EBV+ LCLs shown here (i.e., high levels of miR-155 and miR-21 and moderate levels of miR-17 family members), which suggests that LCVs employ similar mechanisms to modulate the cellular miRNA environment and have encountered a similar set of conserved host miRNAs throughout evolution. Most Old World LCVs are known to encode LMP1 and BHRF1 homologs (44, 45, 50, 66), which we predict to be subject to the same viral and cellular miRNA regulation as that observed for EBV and rLCV homologs. In an attempt to determine the extent to which the miR-17 family binding sites are conserved in other LCVs, we sequenced the LMP1 3′UTR region for herpesvirus papio and analyzed published BHRF1 sequences for herpesvirus papio and herpesvirus pan. Encouragingly, both of these simian LCVs contained the miR-17 family binding sites in the BHRF1 and LMP1 3′UTRs, highlighting the potential significance of these sites.
Our analysis of the herpesvirus papio LMP1 3′UTR sequence further revealed the presence of conserved, putative seed match binding sites for viral miRNAs with conserved seeds (carried by EBV and rLCV), leading us to hypothesize that herpesvirus papio and related LCVs also carry their own viral miRNAs. The genomic sequences for most simian LCVs are not yet known; however, using partial genomic and cDNA sequences, we were able to predict several BHRF1 pre-miRNA homologs for herpesvirus papio and herpesvirus pan (Fig. 2). Hence, it is reasonable to presume that BART miRNA homologs are also carried by these viruses and that targeting of LMP1 transcripts is an overall evolutionarily conserved function of LCV miRNAs.
Intriguingly, viral and cellular miRNA regulation of the LMP1 and BHRF1 transcripts appears to be restricted to Old World LCVs. Callitrichine herpesvirus 3 (CalHV-3), a much more distant New World LCV first isolated from common marmoset B cell lymphomas, encodes functional, but not sequence, homologs of LMP1 and BHRF1 (C1 and ORF64, respectively) (67). Similar to EBV LMP1, C1 exhibits transforming activity in vitro and induces NF-κB activation via interactions with TRAFs, although it does so via different regions than EBV, rLCV, and herpesvirus papio LMP1 proteins (44, 50, 67). CalHV-3 is also predicted to encode 37 viral pre-miRNAs which lack sequence identity to the EBV and rLCV pre-miRNAs (14). We have examined the sequences of CalHV-3 C1 and ORF64 3′UTRs for potential viral miRNA and/or cellular miR-17/20/106 binding sites; remarkably, we found no candidates. Interestingly, the predicted length of the CalHV-3 C1 3′UTR is only 133 nt (following the stop codon to a canonical polyadenylation signal), whereas the 3′UTRs of the EBV, rLCV, and herpesvirus papio LMP1 mRNAs are >1,200 nt, suggesting that the CalHV-3 C1 mRNA actually avoids miRNA regulation, as has been observed for cellular mRNAs with short 3′UTRs (68). It remains to be determined whether the viral miRNAs predicted to be encoded by CalHV-3 or other cellular miRNAs counteract or modulate the levels and/or activities of C1 and ORF64 proteins indirectly.
At this point, we can only speculate as to the functional significance of the miR-17 family binding sites conserved in LCV LMP1 and BHRF1 mRNAs and, furthermore, why these transcripts are targeted at multiple, discrete locations by viral and cellular miRNAs in latently infected cells. Clearly, the targeting of LMP1 mRNA by these miRNAs alters LMP1 protein production (Fig. 6) (27, 28), which can translate into changes in the overall, pleiotropic effects of LMP1 in cells (Fig. 8 and 9) (37). miRNA-dependent changes in BHRF1 protein levels have also been observed (28), consistent with our findings that it is a BHRF1 mRNA that is associated with RISC (Fig. 5). BHRF1 is a Bcl-2 homolog with antiapoptotic activity that is required for LCL outgrowth early after infection (69), while LMP1 is a CD40 mimic that critically activates NF-κB signaling pathways (described above). Presumably, miRNA regulation of these viral mRNAs directs the timing, amplitude, and/or duration of the host cell signaling events initiated and/or sustained through the expression of these proteins. Indeed, recent studies analyzing EBV-induced gene expression changes following primary B cell infection showed a delay in the expression of LMP1 protein and NF-κB target genes; arguably, this delay is due, in part, to the higher levels of the myc-regulated miR-17 family observed very early after infection (i.e., from EBV EBNA2-driven myc induction), which would suppress LMP1 protein expression (57, 70).
The high conservation of the miR-17 family binding sites among LCVs suggests that the activities of LMP1 and BHRF1 are further linked to the cellular pathways intrinsically regulated by this miRNA family. Intriguingly, PAR-CLIP and HITS-CLIP analysis of miR-17 targets in EBV-infected B cells has shown that the miR-17 family as well as other miRNAs expressed from the polycistronic, myc-regulated miR-17/92 cluster inhibit several proapoptotic genes, with the most prominent being BCL2L11/Bim, an antagonist of Bcl2 (27, 28, 71). These miRNAs were also found to target negative regulators of canonical NF-κB signaling (i.e., CYLD and components of the A20 ubiquitin-editing complex), and recent findings in transgenic mouse models demonstrate this leads to enhancement of canonical NF-κB signaling upon miR-17/92 overexpression (72). Notably, these miR-17/92 targets are also conserved in rLCV+ LCLs, as determined from our PAR-CLIP studies (data not shown). Thus, in LCV-infected B cells, miR-17, BHRF1, and LMP1 are targeting similar apoptotic and NF-κB signaling pathways. As the combination of these activities may prove detrimental to an infected cell (i.e., elicit inflammatory responses [73]), it is possible that LCVs have resolved the issue(s) and counterbalance the activities by placing LMP1 and BHRF1 under miR-17 regulation.
What mechanisms govern IκBα stabilization in the presence of LCV miRNAs?
In exploring the functional consequences of LMP1 targeting by viral miRNAs, we identified two EBV miRNAs, miR-BART1 and miR-BART3, that stabilized IκBα and, consequently, inhibited NF-κB activation in the presence of LMP1. Since the attenuation of NF-κB signaling by these BART miRNAs occurred independently of LMP1 targeting, we postulated that cellular targets likely are responsible for the phenotype, with the most likely targets being components of the SCF/β-TRCP E3-ubiquitin ligase complex, which mediates IκBα degradation (58). Our PAR-CLIP experiments revealed evolutionarily conserved LCV miRNA binding sites in SCF-associated genes (CAND1 and FBXW9, among others), which were confirmed by reporter assays. While more experiments are required to decipher the direct consequences of CAND1 and FBXW9 targeting, our experiments clearly demonstrate an evolutionarily conserved function for LCV miRNAs in the regulation of SCF components.
Interestingly, a number of viral proteins, including a lytic EBV protein, have been shown to bind or target the SCF complex, thereby indirectly suppressing NF-κB activation pathways that elicit innate immune responses (74–76). The question remains as to whether the LCV miRNA involvement in NF-κB signaling observed here translates into cell growth and prosurvival signals by potentially directing the specificity of LMP1-induced signal transduction; alternatively, it may be a mechanism for evading innate immune responses (or both). Targeting of NF-κB signaling components has been observed for other gammaherpesvirus miRNAs. For example, the KSHV miRNA miR-K1 targets the IκBα 3′UTR, thereby reducing IκBα protein levels and activating canonical NF-κB pathways; presumably, this enhances cell growth and survival (77). Another KSHV miRNA, miR-K11, targets IκB kinase epsilon (IKKe), thereby attenuating type I interferon signaling induced via innate antiviral responses (21, 78). Additionally, components of the Toll-like receptor and interleukin-1 signaling pathways, IRAK1 and MYD88, are targeted by miR-K9 and miR-K5, respectively, inhibiting IL-6 and IL-8 secretion (79). Two EBV miRNAs (miR-BHRF1-3 and miR-BART15), neither of which are conserved in rLCV, also have demonstrated activity in modulating innate immune responses (80, 81). Further studies are required to dissect the roles of LCV miRNAs in NF-κB signaling pathways.
In summary, our data indicate that primate lymphocryptoviruses have coevolved with miRNAs expressed in host cells and adapted to utilize both viral and cellular miRNAs to regulate latent viral gene products. Presumably, the presence of these cellular miRNA target sites in viral mRNAs provides a biological advantage to the virus, and it remains to be determined how such miRNA-mRNA interactions modulate LCV infection and viral persistence. rLCV infection of rhesus macaques is an important animal model for studying EBV infection in humans (44); thus, our study provides an important stepping stone toward the goal of understanding viral and cellular miRNA functions in vivo.
ACKNOWLEDGMENTS
We thank Fred Wang (Harvard University) for providing the 211-98, 309-98, and S594 cell lines, members of the Duke Genome Sequencing & Analysis Core for assistance with high-throughput sequencing, and Micah Luftig (Duke University) for critical reading of the manuscript.
This work was supported by an award from the National Institutes of Health (R01-AI067968) to B.R.C. R.L.S. was additionally supported by a Duke Center for AIDS Research (CFAR) small grant award (P30-AI064518).
Footnotes
Published ahead of print 20 November 2013
REFERENCES
- 1.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2.Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011–3016. 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, Doudna JA. 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457:405–412. 10.1038/nature07755 [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105. 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500. 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 8.Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141. 10.1146/annurev.micro.112408.134243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. 10.1371/journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundhoff A, Sullivan CS, Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750. 10.1261/rna.2326106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. 10.1126/science.1096781 [DOI] [PubMed] [Google Scholar]
- 12.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333–3341. 10.1128/JVI.01689-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley KJ, Rabinowitz GS, Steitz JA. 2010. Comprehensive analysis of rhesus lymphocryptovirus microRNA expression. J. Virol. 84:5148–5157. 10.1128/JVI.00110-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walz N, Christalla T, Tessmer U, Grundhoff A. 2010. A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J. Virol. 84:716–728. 10.1128/JVI.01302-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerner CS, Dolan A, McGeoch DJ. 2004. Phylogenetic relationships in the Lymphocryptovirus genus of the Gammaherpesvirinae. Virus Res. 99:187–192. 10.1016/j.virusres.2003.10.011 [DOI] [PubMed] [Google Scholar]
- 16.Rivailler P, Jiang H, Cho YG, Quink C, Wang F. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421–426. 10.1128/JVI.76.1.421-426.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogg MH, Garry D, Awad A, Wang F, Kaur A. 2006. The BZLF1 homolog of an Epstein-Barr-related gammaherpesvirus is a frequent target of the CTL response in persistently infected rhesus macaques. J. Immunol. 176:3391–3401 [DOI] [PubMed] [Google Scholar]
- 18.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. 2007. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology 364:21–27. 10.1016/j.virol.2007.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umbach JL, Strelow LI, Wong SW, Cullen BR. 2010. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology 405:592–599. 10.1016/j.virol.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. 10.1371/journal.pone.0012745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099. 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343. 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845. 10.1128/JVI.01804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12:R79. 10.1186/gb-2011-12-8-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. 2012. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 8:e1002484. 10.1371/journal.ppat.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. 2012. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 31:2207–2221. 10.1038/emboj.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein U, Gloghini A, Gaidano G, Chadburn A, Cesarman E, Dalla-Favera R, Carbone A. 2003. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood 101:4115–4121. 10.1182/blood-2002-10-3090 [DOI] [PubMed] [Google Scholar]
- 30.Falk L, Deinhardt F, Nonoyama M, Wolfe LG, Bergholz C. 1976. Properties of a baboon lymphotropic herpesvirus related to Epstein-Barr virus. Int. J. Cancer 18:798–807. 10.1002/ijc.2910180611 [DOI] [PubMed] [Google Scholar]
- 31.Rivailler P, Carville A, Kaur A, Rao P, Quink C, Kutok JL, Westmoreland S, Klumpp S, Simon M, Aster JC, Wang F. 2004. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood 104:1482–1489. 10.1182/blood-2004-01-0342 [DOI] [PubMed] [Google Scholar]
- 32.Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1:302–307. 10.1038/nprot.2006.47 [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing L, Kieff E. 2011. cis-Acting effects on RNA processing and Drosha cleavage prevent Epstein-Barr virus latency III BHRF1 expression. J. Virol. 85:8929–8939. 10.1128/JVI.00336-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing L, Kieff E. 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J. Virol. 81:9967–9975. 10.1128/JVI.02244-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gewurz BE, Towfic F, Mar JC, Shinners NP, Takasaki K, Zhao B, Cahir-McFarland ED, Quackenbush J, Xavier RJ, Kieff E. 2012. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc. Natl. Acad. Sci. U. S. A. 109:2467–2472. 10.1073/pnas.1120542109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. U. S. A. 104:16164–16169. 10.1073/pnas.0702896104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Huttenhofer A. 2009. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 5:e1000547. 10.1371/journal.ppat.1000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. 2008. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382:257–266. 10.1016/j.virol.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forte E, Salinas RE, Chang C, Zhou T, Linnstaedt SD, Gottwein E, Jacobs C, Jima D, Li QJ, Dave SS, Luftig MA. 2012. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growth promoting in EBV-infected B cells. J. Virol. 86:6889–6898. 10.1128/JVI.07056-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. 2008. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J. Virol. 82:10436–10443. 10.1128/JVI.00752-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. 2007. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 35:e73. 10.1093/nar/gkm244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. 2010. Virally induced cellular miR-155 plays a key role in B-cell immortalization by EBV. J. Virol. 84:11670–11678. 10.1128/JVI.01248-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Rivailler P, Rao P, Cho Y. 2001. Simian homologues of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:489–497. 10.1098/rstb.2000.0776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams T, Sale D, Hazlewood SA. 2001. BHRF1 is highly conserved in primate virus analogues of Epstein-Barr virus. Intervirology 44:55–58. 10.1159/000050031 [DOI] [PubMed] [Google Scholar]
- 46.Zeng Y, Cullen BR. 2005. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 280:27595–27603. 10.1074/jbc.M504714200 [DOI] [PubMed] [Google Scholar]
- 47.Zeng Y, Cullen BR. 2006. Recognition and cleavage of primary microRNA transcripts. Methods Mol. Biol. 342:49–56. 10.1385/1-59745-123-1:49 [DOI] [PubMed] [Google Scholar]
- 48.Zeng Y, Cullen BR. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9:112–123. 10.1261/rna.2780503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36:666–675. 10.1093/nar/gkm1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z, Wang X, Strong MJ, Concha M, Baddoo M, Xu G, Baribault C, Fewell C, Hulme W, Hedges D, Taylor CM, Flemington EK. 2013. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J. Virol. 87:1172–1182. 10.1128/JVI.02517-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517. 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc. Natl. Acad. Sci. U. S. A. 101:141–146. 10.1073/pnas.2237183100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soni V, Cahir-McFarland E, Kieff E. 2007. LMP1 TRAFficking activates growth and survival pathways. Adv. Exp. Med. Biol. 597:173–187. 10.1007/978-0-387-70630-6_14 [DOI] [PubMed] [Google Scholar]
- 55.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. 2004. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108–4119. 10.1128/JVI.78.8.4108-4119.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratt ZL, Zhang J, Sugden B. 2012. The latent membrane protein 1 (LMP1) oncogene of Epstein-Barr virus can simultaneously induce and inhibit apoptosis in B cells. J. Virol. 86:4380–4393. 10.1128/JVI.06966-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price AM, Tourigny JP, Forte E, Salinas RE, Dave SS, Luftig MA. 2012. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-kappaB activation. J. Virol. 86:11096–11106. 10.1128/JVI.01069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh G, Wang VY, Huang DB, Fusco A. 2012. NF-kappaB regulation: lessons from structures. Immunol. Rev. 246:36–58. 10.1111/j.1600-065X.2012.01097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G. 2012. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1alpha accumulation. BMC Biol. 10:36. 10.1186/1741-7007-10-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. 2012. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat. Struct. Mol. Biol. 19:511–516. 10.1038/nsmb.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peschiaroli A, Skaar JR, Pagano M, Melino G. 2010. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene 29:1384–1393. 10.1038/onc.2009.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olma MH, Dikic I. 2013. Cullins getting undressed by the protein exchange factor Cand1. Cell 153:14–16. 10.1016/j.cell.2013.03.014 [DOI] [PubMed] [Google Scholar]
- 63.Gerber P, Pritchett RF, Kieff ED. 1976. Antigens and DNA of a chimpanzee agent related to Epstein-Barr virus. J. Virol. 19:1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neubauer RH, Rabin H, Strnad BC, Nonoyama M, Nelson-Rees WA. 1979. Establishment of a lymphoblastoid cell line and isolation of an Epstein-Barr-related virus of gorilla origin. J. Virol. 31:845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rasheed S, Rongey RW, Bruszweski J, Nelson-Rees WA, Rabin H, Neubauer RH, Esra G, Gardner MB. 1977. Establishment of a cell line with associated Epstein-Barr-like virus from a leukemic orangutan. Science 198:407–409. 10.1126/science.198878 [DOI] [PubMed] [Google Scholar]
- 66.Faucher S, Dimock K, Wright KE. 2002. Characterization of the Cyno-EBV LMP1 homologue and comparison with LMP1s of EBV and other EBV-like viruses. Virus Res. 90:63–75. 10.1016/S0168-1702(02)00144-2 [DOI] [PubMed] [Google Scholar]
- 67.Cho Y, Ramer J, Rivailler P, Quink C, Garber RL, Beier DR, Wang F. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc. Natl. Acad. Sci. U. S. A. 98:1224–1229. 10.1073/pnas.98.3.1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. 10.1371/journal.pbio.0030404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9:405–414. 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin HY, Oda H, Lai M, Skalsky RL, Bethel K, Shepherd J, Kang SG, Liu WH, Sabouri-Ghomi M, Cullen BR, Rajewsky K, Xiao C. 2013. MicroRNA-17∼92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J. 32:2377–2391. 10.1038/emboj.2013.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Oliveira DE, Ballon G, Cesarman E. 2010. NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol. 18:248–257. 10.1016/j.tim.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 74.Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 83:4140–4152. 10.1128/JVI.01835-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gastaldello S, Callegari S, Coppotelli G, Hildebrand S, Song M, Masucci MG. 2012. Herpes virus deneddylases interrupt the cullin-RING ligase neddylation cycle by inhibiting the binding of CAND1. J. Mol. Cell Biol. 4:242–251. 10.1093/jmcb/mjs012 [DOI] [PubMed] [Google Scholar]
- 76.Hayden MS, West AP, Ghosh S. 2006. NF-kappaB and the immune response. Oncogene 25:6758–6780. 10.1038/sj.onc.1209943 [DOI] [PubMed] [Google Scholar]
- 77.Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. 2010. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell. Biol. 12:193–199. 10.1038/ncb2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang D, Gao Y, Lin X, He Z, Zhao Q, Deng Q, Lan K. 2011. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKepsilon. Cell Res. 21:793–806. 10.1038/cr.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. 2012. KSHV microRNAs target two components of the TLR/IL-1R signaling cascade, IRAK1 and MYD88, to reduce inflammatory cytokine expression. J. Virol. 86:11663–11674. 10.1128/JVI.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O'Neill LA, Masters SL. 2012. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J. Immunol. 189:3795–3799. 10.4049/jimmunol.1200312 [DOI] [PubMed] [Google Scholar]
- 81.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442. 10.1158/0008-5472.CAN-07-5126 [DOI] [PMC free article] [PubMed] [Google Scholar]