Abstract
The zoonotic outbreak of H7N9 subtype avian influenza virus that occurred in eastern China in the spring of 2013 resulted in 135 confirmed human cases, 44 of which were lethal. Sequencing of the viral genome revealed a number of molecular signatures associated with virulence or transmission in mammals. We report here that, in the guinea pig model, a human isolate of novel H7N9 influenza virus, A/Anhui/1/2013 (An/13), is highly dissimilar to an H7N1 avian isolate and instead behaves similarly to a human seasonal strain in several respects. An/13 was found to have a low 50% infectious dose, grow to high titers in the upper respiratory tract, and transmit efficiently among cocaged guinea pigs. The pH of fusion of the hemagglutinin (HA) and the binding of virus to fixed guinea pig tissues were also examined. The An/13 HA displayed a relatively elevated pH of fusion characteristic of many avian strains, and An/13 resembled avian viruses in terms of attachment to tissues. One important difference was seen between An/13 and both the H3N2 human and the H7N1 avian viruses: when inoculated intranasally at a high dose, only the An/13 virus led to productive infection of the lower respiratory tract of guinea pigs. In sum, An/13 was found to retain fusion and attachment properties of an avian influenza virus but displayed robust growth and contact transmission in the guinea pig model atypical of avian strains and indicative of mammalian adaptation.
INTRODUCTION
On 31 March 2013, three human cases of H7N9 influenza virus infection were reported to the World Health Organization (1, 2). An outbreak ensued over the subsequent 2 months, with a total of 133 laboratory confirmed cases reported by the end of May. These apparently zoonotic transmission events took place in a broad region of eastern China, comprising eight provinces and the cities of Shanghai and Beijing (3). One case was detected in Taipei but most likely had its source in Jiangsu, China (4, 5). Although the vast majority of cases were associated with exposure to poultry, usually at live bird markets (6), four clusters of two or more close contacts are suggestive of limited human-to-human transmission in these instances (3, 7). Among the confirmed cases, the disease associated with H7N9 influenza virus infection was typically severe, with hospitalization required for all but a few of the laboratory confirmed infections (1, 8).
Following the closure of live bird markets (9) and coinciding with the arrival of summer in the Northern Hemisphere, no new, confirmed, human cases of H7N9 infection occurred over a 6-week period from 22 May to 9 July. Three additional cases were, however, detected in July and August (10), indicating that the H7N9 viruses are still present in avian populations and highlighting the potential for the outbreak to resume in the colder autumn months.
Full genome sequencing of a number of human and avian isolates of the novel H7N9 lineage was reported rapidly during the outbreak. These data revealed several mammalian adaptive polymorphisms in the viral genomes, including PB2 627K, HA 226L, HA 160A, HA 186V (H3 numbering), and a deletion in the NA stalk (1, 11). The presence of genetic changes previously found to support viral growth in mammalian hosts may increase the potential of the H7N9 viruses to transmit from human-to-human and is cause for concern.
In response to the H7N9 outbreak, a number of comprehensive studies have been carried out with the aim of characterizing the virus and assessing the risk of an H7N9 pandemic. The ferret model has been used by several groups to evaluate influenza virus pathogenicity and transmission. In general, mild disease on par with that seen with seasonal influenza viruses in ferrets was reported (12–16). The Fouchier group did, however, note two instances of more severe disease in ferrets infected by a natural, respiratory droplet, route (14), and intratracheal inoculation of ferrets was found to lead to fatal viral pneumonia (17). Growth of H7N9 viruses in the ferret lung was assessed and ranged from inefficient (15) to robust (16). Despite the lack of severe disease, clear pathological changes characteristic of bronchointerstitial pneumonia were reported (12, 16, 18). Transmission among ferrets was found to be efficient by a contact route (12) and also occurred via respiratory droplets, but with more varied efficiency (12–16, 18).
Evaluation of H7N9 virus pathogenicity and/or transmission has also been carried out in mice, pigs, and cynomolgus macaques. In mice, human H7N9 isolates showed robust growth and were lethal at moderate to high doses (13, 15, 16). In pigs or minipigs, disease was mild or absent, viral growth was low to moderate, and transmission was not observed (12, 15). In macaques, disease was typical of that seen with 2009 pandemic H1N1 influenza virus infection; however, the H7N9 viruses tested replicated efficiently in both the upper and the lower respiratory tracts, whereas human seasonal strains are normally confined to the upper respiratory tract in this species (15).
To evaluate the H7N9 pathogenicity and transmissibility, we used the guinea pig system, which we described in 2006 as a highly useful mammalian model for the study of influenza virus infectivity, growth, and transmission (19). A key advantage of the model, compared to mice, is that guinea pigs are highly susceptible to infection with human strains (requiring no prior adaptation) (19–21). In addition, human influenza viruses show tissue tropism and kinetics of viral growth similar to those seen in uncomplicated influenza in humans (19, 22, 23). Importantly, human influenza virus strains transmit among guinea pigs by contact and respiratory droplet routes, whereas swine-adapted and some highly pathogenic avian influenza viruses transmit poorly and low-pathogenicity avian strains do not transmit (19–21, 24–31). A disadvantage is that, to date, overt signs of disease have not been detected in influenza virus-infected guinea pigs. Nevertheless, rhinitis and bronchointerstitial pneumonia have been described (22, 32). The utility of the guinea pig model is underlined by the fact that, since 2006, many additional labs have adopted it for the study of influenza virus transmission (33–38). Since no animal model is perfect, efforts to accurately model human disease are strengthened by the use of multiple animal species, and ideally by the development of practical models in which specific animals are used to model specific aspects of disease. Thus, the data reported herein are a valuable complement to recently published studies in macaque, ferret, swine, and mouse models.
MATERIALS AND METHODS
Ethics statement.
The present study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animal husbandry and experimental procedures were approved by the University of Georgia Institutional Animal Care and Use Committee (IACUC protocol A2013 05-030).
Biocontainment.
All experiments with infectious A/Anhui/1/2013 (H7N9) virus were conducted under BSL3/ABSL3 enhanced containment at the Animal Health Research Center according to protocols approved by the institutional biosafety committee at the University of Georgia.
Cells.
Madin-Darby canine kidney (MDCK) cells were used for virus titration by plaque assay and 50% tissue culture infective dose(s) (TCID50). Baby hamster kidney-21 (BHK-21) Cells were used in the syncytium formation assay to determine pH of fusion. BSR-T7/5 cells (originally obtained from Karl-Klaus Conzelmann), and Vero cells were used for the luciferase reporter-based fusion assay.
Guinea pigs.
Female, Hartley strain, guinea pigs weighing 250 to 300 g were obtained from Charles River Laboratories. Prior to intranasal inoculation, nasal lavage, or euthanasia, guinea pigs were sedated with a mixture of ketamine and xylazine (30 and 4 mg/kg, respectively). Inoculation and nasal lavage were performed as described previously (19), with phosphate-buffered saline (PBS) as the diluent or collection fluid in each case.
Viruses.
All viruses were amplified in 9- to 11-day-old embryonated hens' eggs, and titers were determined by plaque assay on MDCK cells. A/Anhui/1/2013 (H7N9) virus was kindly provided by Richard Webby (Saint Jude Children's Research Hospital). A/Panama/2007/1999 (H3N2) virus was generated by reverse genetics with wild-type plasmids, as previously described (30). A/rhea/North Carolina/39482/1993 (H7N1), A/duck/Ukraine/1963 (H3N8), A/duck/Alberta/35/1976, and A/turkey/Oregon/1971 (H7N3) viruses were kindly provided from the collection of Peter Palese at the Icahn School of Medicine at Mount Sinai.
Guinea pig transmission experiments.
To evaluate transmission, four guinea pigs were inoculated intranasally with 103 PFU of virus in 300 μl of PBS. At 24 h postinoculation, each infected animal was placed in the same cage with one naive guinea pig. Standard rat cages with wire tops and filter lids were used. In accordance with containment protocols required for the H7N9 virus, each rat cage was housed in a ventilated, negative-pressure ferret containment cage (Allentown). Nasal washes were collected from all eight guinea pigs on days 2, 4, 6, and 8 postinfection, as previously described (19).
Determination of virus titers in guinea pig tissues.
Nasal turbinate and lung tissue were collected from two guinea pigs per group at each time point. For guinea pigs infected with 103 PFU, turbinates were removed, and the mucosa was scraped from the right side of the nasal septum and placed in 1 ml of PBS. For lung tissue collection, ca. 25% of each right and left cranial, medial, and caudal lung lobe was removed and placed in 1 ml of PBS. For guinea pigs infected with 106 PFU, all turbinate and mucosal tissue was removed and placed in 1 ml of PBS. The entirety of the lung was placed in 3 ml of PBS. Tissues were then homogenized using disposable handheld tissue grinders (Fisher), and supernatants clarified by low-speed centrifugation. Virus titers were determined through 50% tissue culture infectious dose assay as previously described (39), and titers were calculated using the Reed and Muench method (40).
Determination of 50% infectious dose.
Groups of four guinea pigs were infected intranasally with 1,000, 100, 10, or 1 PFU in 300 μl of PBS. Nasal washings were collected on days 2 and 4 postinoculation, and virus therein was titrated by TCID50 on MDCK cells. Productive infection was defined as detection of >10 TCID50/ml in nasal washings. The 50% infectious dose was calculated by the method of Reed and Muench (40).
Histopathology.
At necropsy the proximal trachea was clamped prior to opening the chest to keep the lungs inflated. The chest was opened, and the lungs were viewed in situ for lesions. The trachea was then severed above the clamp, and the trachea, lungs, and heart were removed. Approximately 25% of each right and left cranial, medial, and caudal lung lobe was removed and placed in medium for virus isolation. The lungs were then inflated with 10% buffered formalin via the trachea and immersed in 10% buffered formalin. The nasal cavity was opened by removing the bones dorsally. Once nasal tissue to be used for virus detection was removed, the head was then immersed in 10% buffered formalin. After fixation, the head was placed in Kristensen's decalcification solution for 24 h, rinsed, and returned to 10% buffered formalin. Sections of all six lung lobes, proximal and distal trachea, and six transverse sections through the nasal cavity were routinely processed and embedded in paraffin within 12 days of necropsy. All tissues were stained with hematoxylin and eosin and for influenza virus by immunohistochemistry (IHC). For immunohistochemistry, tissues were incubated with two anti-influenza A virus antibodies (mouse monoclonal antibody to NP protein, catalog no. C65331M [Meridian Life Sciences, Inc., Memphis, TN]; goat polyclonal serum, catalog no. B65141G [Meridian Life Sciences]) after antigen retrieval with proteinase K (Dako, Carpinteria, CA). Detection was with biotinylated secondary antibodies and horseradish labeled-streptavidin (4 Plus; Biocare, Concord, CA) with diaminobenzidine as the chromogen, and sections were counterstained with Mayer's hematoxylin.
Nasal cavity changes were scored by the degree of intraepithelial and lamina proprial inflammation (0 = none, 1 = mild, 2 = moderate, and 3 = severe) and mucosal epithelial cell necrosis (0 = none, 1 = <10%, 2 = <50%, and 3 = >50%) in most severely affected areas at ×400 for a total score of 6. Immunohistochemistry for influenza virus was scored 0 to 3 in areas of greatest immunopositivity based on the number of epithelial cells staining in positive ×400 fields and the intensity of that staining compared to the positive control (pig infected with H1N1): 0 = no staining, 1 = rare cells, faint staining; 2 = <50%, moderate to intense staining; and 3 = >50%, moderate to intense staining.
Histological evaluation of virus attachment.
Virus binding to guinea pig tissues was carried out essentially as described before for other species (41). In brief, following concentration through a 30% sucrose cushion and inactivation with 0.05% formaldehyde, viruses were labeled with an equal volume of 0.1 mg of fluorescein isothiocyanate (FITC; Sigma)/ml in bicarbonate buffer. Uninfected guinea pig tissues (n = 2 guinea pigs) were extracted, formalin fixed, paraffin embedded, and sectioned. Paraffin was removed from the tissues with xylene and hydrated using an alcohol gradient. Tissues were incubated with appropriate FITC-labeled viruses overnight at 4°C at a titer of 5 hemagglutinin (HA) units/100 μl. The FITC label was detected with horseradish peroxidase-labeled rabbit anti-FITC antibody (Dako). The signal was amplified with a tyramide signal amplification system (Perkin-Elmer). The peroxidase was revealed with 3-amino-9-ethyl-carbazole (Sigma), producing a red precipitate. Tissues were counterstained with Mayer's hematoxylin and embedded in Kaiser's glycerol gelatin (Merck). Attachment of influenza virus was visualized by light microscopy as a red precipitate on the surface of epithelial cells.
Determination of fusion pH.
HA-mediated membrane fusion was examined using two independent assays, a qualitative syncytia assay and a quantitative cell-cell fusion luciferase reporter assay, both of which were performed as described previously (42). Briefly, for the syncytia assay, BHK-21 cells were transfected with 1.0 μg of HA plasmid using Lipofectamine (Invitrogen), according to the manufacturer's recommendation. At 16 to 18 h posttransfection, cells were washed with PBS and treated with 5 μg of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Sigma)/ml at 37°C for 15 min, followed by treatment with 20 μg of soybean trypsin inhibitor/ml at 37°C for 30 min. Trypsin-treated HA-expressing cells were subsequently exposed to 1.0 ml of PBS that had been pH adjusted using 100 mM citric acid to the indicated pH and incubated at 37°C for 5 min. The pH-adjusted PBS was removed, and the cells were neutralized by the addition of growth medium and incubated at 37°C for 2 h to allow syncytium formation. Cells were then stained with a Hema3 Stat Pak (Fisher, USA) according to the manufacturer's recommendation. Syncytia were visualized and photographed using a Zeiss Axio Observer inverted microscope with attached digital camera.
For the luciferase cell-cell fusion assay, Vero cells were cotransfected with 1.0 μg of HA plasmid and 1.0 μg of a plasmid expressing firefly luciferase under the control of the T7 bacteriophage promoter using Lipofectamine (Invitrogen), according to the manufacturer's recommendation. At 16 h posttransfection, cells were washed once with PBS and treated with 5 μg of TPCK-trypsin/ml at 37°C for 30 min. Trypsin-treated HA-expressing cells were subsequently overlaid with BSR-T7/5 target cells, which constitutively express the bacteriophage T7 RNA polymerase. After 1 h at 37°C, the cells were washed with PBS, exposed to 1.0 ml of PBS that had been pH adjusted using 100 mM citric acid to the indicated pH, and incubated at 37°C for 5 min. After the removal of pH-adjusted PBS, HA-expressing cells were neutralized with growth medium and incubated at 37°C for 6 h to allow for HA-expressing cells to fuse with BSR-T7/5 cells, mediating transfer and expression of the T7-luciferase plasmid in BSR-T7/5 target cells. At 6 h postneutralization, the cells were washed with PBS, lysed with 0.5 ml of reporter lysis buffer (Promega), and clarified of cell debris by centrifugation at 15,000 × g at 4°C. From each lysate, 150 μl was transferred to a 96-well plate (white polystyrene; Corning), and the luciferase activity was quantified by using a BioTek Synergy 2 Luminometer, with 50 μl of luciferase assay substrate (Promega) injected into each sample.
RESULTS
Transmission among cocaged guinea pigs.
Toward evaluating the risk of human-to-human spread of the novel H7N9 influenza viruses, we tested the transmissibility of An/13 by a contact route in the guinea pig model. The human seasonal strain A/Panama/2007/1999 (H3N2) [Pan/99], which we have previously shown to transmit efficiently among guinea pigs (19, 21, 27–31, 43), was used as a positive control and a low-pathogenicity avian influenza virus, A/rhea/North Carolina/39482/1993 (H7N1) [rhea/NC], was used in parallel as an H7 subtype avian virus control. Since there are few published data on low-pathogenicity avian influenza viruses in the guinea pig model, we furthermore included the results of previously performed contact transmission experiments with A/duck/Alberta/35/1976 (H1N1; dk/AB) and A/duck/Ukraine/1963 (H3N8; dk/Ukr) viruses. In each case, four guinea pigs were inoculated intranasally with 103 PFU (or 104 PFU in the case of dk/AB virus). One day after inoculation, a naive guinea pig was placed in the same cage with each infected guinea pig. On days 2, 4, 6, and 8 postinoculation, nasal washings were collected from all eight guinea pigs in order to monitor for transmission and track viral growth in the nasal passages.
As shown in Fig. 1, the human seasonal Pan/99 virus was shed into nasal washings at high titers (∼106 TCID50/ml at the peak of shedding, day 2 postinfection) and transmitted to all four contact guinea pigs, as expected. The H7N9 subtype An/13 strain exhibited higher nasal wash titers than Pan/99 virus and was also transmitted to all four exposed cagemates. In contrast, the rhea/NC, dk/AB, and dk/Ukr avian influenza virus controls were shed at markedly lower titers than Pan/99 virus and did not transmit.
FIG 1.
A/Anhui/1/2013 (H7N9) and A/Panama/2007/1999 (H3N2) influenza viruses transmit efficiently among cocaged guinea pigs, while avian influenza viruses do not. The virus titers in nasal washes are plotted as a function of day postinoculation, with inoculated guinea pigs represented by dashed lines and contact guinea pigs represented by solid lines. An/13, Pan/99, and rhea/NC experiments were performed in parallel. dk/Ukr/63 and dk/AB/76 experiments were performed previously.
Susceptibility of guinea pigs to infection.
The minimal dose of virus required to initiate a productive infection is likely a key determinant of transmission efficiency. We therefore determined the 50% infectious dose (ID50) of An/13 and rhea/NC viruses in guinea pigs. Groups of four guinea pigs were infected intranasally with 10-fold serial dilutions of virus and nasal washes were collected on days 2 and 4 postinoculation to identify animals infected productively. The ID50 of An/13 was found to be 3 PFU, comparable to that previously reported for Pan/99 virus in guinea pigs (19). In contrast, the ID50 of rhea/NC virus was ∼6-fold higher, at 17 PFU.
Viral tropism within the guinea pig respiratory tract.
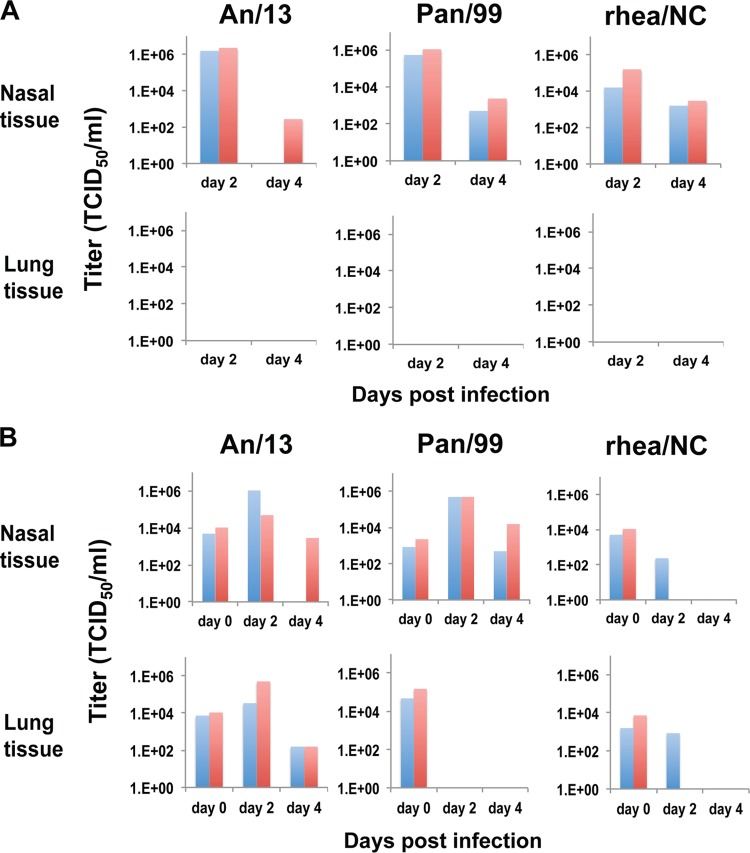
Since the novel H7N9 viruses have led to severe lower respiratory tract disease in the majority of confirmed human cases, we wanted to evaluate the relative potential of Pan/99, An/13, and rhea/NC viruses to replicate in the tissues of the upper versus lower respiratory tracts. To this end, guinea pigs were first infected intranasally with 103 PFU of Pan/99, An/13, or rhea/NC viruses. Nasal turbinate and lung tissues were collected from two guinea pigs on day 2 and an additional two guinea pigs on day 4 after infection. Viral growth was evaluated by preparing homogenates of each tissue and titrating the supernatant by TCID50 in MDCK cells. As summarized in Fig. 2, all three viruses showed productive infection of the nasal turbinates, with titers comparable to those obtained by nasal lavage. After infection with 103 PFU, however, none of the three viruses established infection in the lung. Staining for viral antigen within fixed sections of nasal turbinate and lung confirmed these findings: staining was seen in the nasal turbinates of both guinea pigs infected with An/13 and Pan/99, and one of two guinea pigs infected with rhea/NC (Fig. 3) on day 2 postinfection, whereas a lack of staining was observed in the lungs of each animal tested. The most robust staining was observed in guinea pigs infected with An/13.
FIG 2.
A/Anhui/1/2013 (H7N9) virus shows robust growth in guinea pig nasal turbinates and in lungs when administered at a high dose. The virus titers in tissues collected on day 2 or 4 postinfection are plotted. Each bar represents an individual guinea pig (n = 2 guinea pigs at each time point, for each virus and dose). (A) Guinea pigs were infected intranasally with 103 PFU of the indicated viruses. (B) Guinea pigs were infected intranasally with 106 PFU of the indicated viruses. Tissues collected on day 0 were removed promptly after infection; thus, virus present at this time point is derived from the inoculum.
FIG 3.
The greatest severity of histopathology and antigen staining was seen in A/Anhui/1/2013-infected guinea pigs compared to A/Panama/2007/1999- and A/rhea/39482/NC/1993-infected guinea pigs. Images of nasal tissue stained with hematoxylin and eosin (A, C, and E) and antibody to influenza A virus (B, D, and F) are shown. (A and B) Guinea pigs infected with A/Anhui/1/2013 day 2 postinfection. The lamina propria (LP) is greatly expanded by heterophils admixed with fewer lymphocytes. The respiratory epithelium (E) is extensively infiltrated by moderate numbers of similar cells and appears to be extensively vacuolated. Histopathology score 6. Immunopositive staining is widespread and intense, confined to the epithelium, and mostly nuclear (arrow). IHC score 3. (C and D) Guinea pigs infected with A/Panama/2007/1999 (H3N2) at day 2 postinfection. Inflammation in the lamina propria (LP) is minimal and moderate in the epithelium (E) composed of granulocytes; the epithelium appears vacuolated, likely due to necrosis. Histopathology score 3. Immunohistochemistry staining in the epithelium (arrow) is widespread and moderately intense. IHC score 3. (E and F) Guinea pigs infected with A/rhea/NC/1993 (H7N1) at day 2 postinfection. There is moderate infiltration of granulocytes in the respiratory epithelium (E) with mild necrosis. The lamina propria (LP) is infiltrated with small numbers of mostly heterophils. Histopathology score 2. Only one cell has an immunopositive nucleus (arrow). IHC score 1. Scale bar, 100 μm.
Growth in the lung was then evaluated after inoculation with a high dose of 106 PFU of Pan/99, An/13, or rhea/NC viruses. To confirm that the intranasally applied inocula were reaching the lung, in this experiment we furthermore collected tissues on day 0, as well as days 2 and 4. As shown in Fig. 2, all three viruses were detected in the lungs directly after inoculation. Nevertheless, neither Pan/99 nor rhea/NC viruses established infection in the lung even from the 106 dose. The day 2 and 4 lung titers obtained with the An/13 strain did, however, indicate growth in the lower respiratory tract.
Pathology and immunohistochemistry within the guinea pig respiratory tract.
Histopathology and influenza virus antigen staining in nasal, tracheal and lung tissues were evaluated for guinea pigs infected with 103 PFU of An/13, Pan/99 or rhea/NC viruses. The resultant lesion and immunohistochemistry scores are reported in Table 1.
TABLE 1.
Histopathology and immunohistochemistry of respiratory tissues collected from guinea pigs infected with 103 PFU of influenza virus
| Guinea pig no. | Day postinfection | IHC scorea |
Nasal histopathology scoreb | |
|---|---|---|---|---|
| Lung and trachea | Nasal | |||
| An/13 | ||||
| 1 | 2 | 0 | 3 (R) | 6 |
| 2 | 2 | 0 | 3 (R) | 6 |
| 3 | 4 | 0 | 0 | 2 |
| 4 | 4 | 0 | 0 | 2 |
| Pan/99 | ||||
| 1 | 2 | 0 | 2 (R) | 4 |
| 2 | 2 | 0 | 3 (R) | 3 |
| 3 | 4 | 0 | 1 (R) | 3 |
| 4 | 4 | 0 | 1 (O,R) | 3 |
| Rhea/NC | ||||
| 1 | 2 | 0 | 1 (R) | 2 |
| 2 | 2 | 0 | 0 | 3 |
| 3 | 4 | 0 | 0 | 3 |
| 4 | 4 | 0 | 0 | 2 |
Scoring: 0, no staining; 1, rare cells, faint staining; 2, <50%, moderate to intense staining; 3, >50%, moderate to intense staining. Sites are indicated parenthetically: R, respiratory epithelium; O, olfactory epithelium.
Nasal cavity changes were scored by the degree of intraepithelial and lamina proprial inflammation (0, none; 1, mild; 2, moderate; 3, severe) and mucosal epithelial cell necrosis (0, none; 1, <10%; 2, <50%; 3, >50%) for a total score of 6.
In the nasal cavity, microscopic changes were almost entirely confined to respiratory epithelial regions and rarely adjacent squamous epithelium of the vestibule or olfactory epithelium (Fig. 3). The severity of changes was most severe in guinea pigs infected with An/13 and least severe in animals infected with rhea/NC. Changes included mild to severe intraepithelial and lamina proprial inflammation and mild to severe epithelial cell layer vacuolation on day 2 postinfection, along with regeneration on day 4 postinfection. Lesions were multifocal and varied in severity within an individual animal. Inflammation on day 2 postinfection was heterophilic and admixed with mild to moderate numbers of eosinophils and lymphocytes. By day 4, inflammation was more lymphocytic and plasmacytic, necrosis was subsiding, and there were multifocal regions of epithelial regeneration where previously denuded areas of necrosis are replaced by one to three layers of nonciliated cells with mitoses.
Immunopositivity for influenza virus was only detected in the nasal cavity and was primarily detected in respiratory epithelium and was seen predominantly in areas of inflammation and necrosis (Fig. 3). Limited staining was observed in adjacent squamous epithelium and was rare and faint in the olfactory region of only one animal. Immunopositivity was most pronounced on day 2 postinfection and in animals infected with An/13. Staining was negligible in those animals infected with rhea/NC. Very little staining was present on day 4 and was present only in the animals infected with Pan/99.
Virus binding to guinea pig respiratory and olfactory tissues.
Viral attachment is an important determinant of the cell and tissue tropism of influenza viruses in animals. Although the guinea pig model is increasingly used to study the growth and transmission of influenza viruses in a mammalian host, the tissues to which these viruses bind in guinea pigs are not fully understood. To gain a better understanding of influenza virus binding in guinea pig tissues and to compare the attachment patterns of the An/13 virus to our human and avian control viruses, we used a virus histochemistry-based technique (26, 41, 44, 45). Thus, the binding of Pan/99, An/13, rhea/NC, and dk/Ukr viruses to fixed sections of nasal turbinate, trachea, bronchioles, and alveoli was assessed. Since the airways in the guinea pig lungs are not lined by cartilage, bronchus was not included. The results are summarized in Table 2 and illustrated in Fig. 4 with representative images of selected tissues.
TABLE 2.
Attachment of Pan/99, An/13, rhea/NC, and dk/Ukr influenza viruses to tissues of the guinea pig respiratory tracta
| Virus | GP | Extent of virus attachmentb |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Nasal turbinates |
Trachea | Bronchioli |
Alveoli |
||||||
| Stratified epithelium | Respiratory epithelium | Olfactory epithelium | Ciliated cells | Ciliated cells | Clara cells | Epithelial cells | Alveolar macrophages | ||
| Pan/99 | GP1 | +/– | ++ | ++ | +/– | – | – | +/– | + |
| GP2 | ++ | ++ | +/– | – | – | – | + | ||
| An/13 | GP1 | + | + | ++ | ++ | +/– | +/– | ++ | + |
| GP2 | +/– | + | ++ | ++ | NP | +/– | ++ | + | |
| Rhea/NC | GP1 | + | ++ | ++ | ++ | + | +/– | ++ | + |
| GP2 | +/– | ++ | ++ | ++ | NP | + | ++ | + | |
| dk/Ukr | GP1 | + | ++ | ++ | ++ | ++ | +/– | ++ | + |
| GP2 | ++ | ++ | ++ | ++ | +/– | ++ | + | ||
All viruses included in this study attached also to endothelial cells of larger blood vessel and blood capillaries in the alveolar walls.
Symbols indicate the following prevalence of positive cells: –, <1% cells positive for virus attachment; –/+, <10% cells positive for virus attachment; +, <50% cells positive for virus attachment; ++, >50% cells positive for virus attachment. NP, none present.
FIG 4.
Attachment of A/Anhui/1/2013 virus to tissues of the guinea pig respiratory tract is similar to that of avian influenza viruses. The viruses indicated at the left were incubated with fixed sections of guinea pig respiratory tissues. Red staining is indicative of virus binding.
The attachment pattern of the An/13 virus was comparable to that of the avian strains, rhea/NC and dk/Ukr. The only consistent differences among these three viruses were (i) less attachment of An/13 to the respiratory epithelium of the nasal cavity compared to rhea/NC and dk/Ukr and (ii) lower levels of attachment of both H7 subtype viruses (An/13 and rhea/NC) compared to dk/Ukr at the level of the bronchioles. The Pan/99 (H3N2) human strain, in contrast, showed a number of differences in binding relative to the avian and H7N9 isolates. These differences included markedly decreased binding to tracheal cells and alveolar epithelial cells, including both type I and II pneumocytes, and a lack of binding to the cells of the bronchioles. Thus, overall, the human influenza virus studied attached mainly to the upper respiratory tract, while the avian strains, including An/13, bound throughout the respiratory tract of the guinea pig.
In general, all viruses also attached to endothelial cells of the larger blood vessels and the capillaries in the alveolar wall. Although not likely to be important during infection (since endothelial cells are not exposed to influenza viruses), this characteristic is unique to guinea pigs among the mammalian species examined to date by this method (41, 44).
pH of fusion of viral HA proteins.
The pH at which the HA protein is triggered to undergo conformational changes leading to fusion has been found to differ between avian and mammalian adapted influenza viruses (42), and this viral property has been found to be important for transmission (46, 47). We therefore tested the pH of fusion of the An/13 HA, compared to those of the Pan/99, rhea/NC, and tk/OR HA proteins. Two independent assays were used. In the first assay, the formation of syncytia in BHK cells transiently expressing each of the HA proteins was evaluated microscopically after incubation in buffers of decreasing pH. In the second assay, Vero cells expressing HA and carrying a T7-driven luciferase plasmid were overlaid with BSR-T7/5 cells prior to incubation in buffers of decreasing pH; luminescence in cell lysates was measured as an indicator of fusion. The highest pH values at which syncytia were observed and at which luciferase was detected were taken as the pH of fusion in each assay, respectively. In general, the results of both assays were in close agreement for all four HAs tested (Fig. 5). The fusion pH for the An/13 HA was 5.8, higher than that of the Pan/99 HA (5.5) and HA proteins characterized previously from human influenza viruses (42). This relatively high pH of fusion for An/13 was similar to those observed for avian influenza viruses: at pH 6.0 and 5.6, respectively, the tk/OR and rhea/NC HA proteins tested here bracket the range observed previously for avian virus HAs of various subtypes (42).
FIG 5.
A/Anhui/1/2013 hemagglutinin displays a high pH of fusion. (A) Photomicrographs of syncytium formation assay results. BHK cells were transfected with 1.0 μg of each HA plasmid as indicated, and HA protein was activated by the addition of TPCK-trypsin. Photomicrographs corresponding to the pH unit at which syncytia were observed and 0.1 pH unit higher are shown. (B) Luciferase reporter gene assay for fusion. Vero cells were cotransfected with 1.0 μg of the relevant HA plasmid and 1.0 μg of T7-Luciferase plasmid. HA-expressing Vero cells were overlaid with BSR-T7/5 target cells that constitutively express T7 RNA polymerase. Cell monolayers were pulsed with pH-adjusted PBS in 0.2 pH unit increments at the indicated pH and then were neutralized. Luminescence was measured as an indicator of membrane fusion. The graphs show the mean background-adjusted relative luminescences (± the standard deviations) as a function of pH obtained from three independent experiments.
DISCUSSION
Influenza viral growth and transmission in the guinea pig model is generally representative of the available epidemiological data in humans. Characterization of the novel H7N9 isolate, An/13, in this animal model has demonstrated that this avian-like virus is already partially adapted to a mammalian host: titers of An/13 in the URT exceeded those of the human isolate Pan/99 and transmission proceeded with high efficiency among cocaged guinea pigs (Fig. 1 and Table 3). These results reinforce data generated by others in the ferret model: in ferrets, An/13 was shed at high titers in nasal washes and transmitted efficiently by contact (12). A number of groups have also tested respiratory droplet transmission in ferrets, with various results: with the An/13 strain transmission to as many as 6/6 and to as few as 1/3 exposed ferrets was reported (12–16, 18). We attempted respiratory droplet transmission experiments in guinea pigs with An/13 and Pan/99 viruses. However, due to the need for BSL3-enhanced containment for this work and the lack of an environmental chamber in our BSL3 facility, we were not able to perform these experiments under tightly controlled environmental conditions and transmission was not observed with either virus (data not shown). Temperature and humidity levels present during the respiratory droplet transmission experiment spanned 20 to 23°C and 44 to 62% relative humidity and were similar to conditions that we have previously shown to prevent respiratory droplet transmission of Pan/99 virus (43). This lack of transmission, even for Pan/99 virus, highlights the importance of humidity and temperature to respiratory droplet transmission and underscores the value of running positive-control samples in parallel when studying transmission. We have also previously shown that contact transmission of Pan/99 virus is relatively insensitive to the effects of temperature and humidity (28).
TABLE 3.
Titers of H7N1 and H7N9 influenza viruses collected from nasal washes of guinea pigs infected at various inoculation doses
| Virus and animal no. | Viral titer (log10TCID50/ml)a after inoculation with: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1,000 PFU |
100 PFU |
10 PFU |
1 PFU |
|||||
| Day 2 | Day 4 | Day 2 | Day 4 | Day 2 | Day 4 | Day 2 | Day 4 | |
| An/13 | ||||||||
| 1 | 6.7 | 3.7 | 6.2 | 4.2 | 2.4 | 4.0 | 6.0 | |
| 2 | 6.4 | 4.9 | 5.4 | 5.4 | 5.4 | 4.2 | ||
| 3 | 6.2 | 4.0 | 6.5 | 5.2 | 6.7 | 4.7 | ||
| 4 | 6.4 | 3.2 | 6.4 | 4.7 | ||||
| Rhea/NC | ||||||||
| 1 | 4.2 | 3.0 | 4.4 | 4.2 | 4.2 | 4.4 | ||
| 2 | 4.0 | 2.4 | 4.7 | 3.4 | 3.7 | 4.2 | ||
| 3 | 5.0 | 3.1 | 3.9 | 3.2 | ||||
| 4 | 3.7 | 3.5 | ||||||
Viral titers reported for guinea pigs infected with rhea/NC at 1,000 PFU are taken from the inoculated animals used in the contact transmission experiment (Fig. 1).
Growth in the lung of guinea pigs was unique to An/13 among the strains tested: neither Pan/99 nor the avian H7 rhea/NC established infection in the LRT. This finding is reflective of the lower respiratory disease seen frequently among individuals with confirmed H7N9 virus infection. It is interesting that a high dose was required to initiate viral growth in the lungs of guinea pigs. LRT infection of ferrets and macaques has also been reported, and in both species high doses (105 to 107 PFU or TCID50) were used for inoculation (12–17). It is possible that, also in humans, high doses of H7N9 virus underlie the severe disease seen in hospitalized patients.
Viral growth in the LRT of mammals has previously been attributed to changes in receptor binding preferences of the HA protein (26). Our data suggest that this mechanism does not, however, account for the differing phenotypes of An/13 and rhea/NC viruses in the guinea pig LRT. Attachment to the cells and tissues of the guinea pig respiratory tract was found to be similar among An/13, rhea/NC, and dk/Ukr viruses. A similar discrepancy between attachment and productive infection has been previously observed with H5 subtype influenza virus strains, such that low-pathogenicity avian influenza viruses of H5N9 and H6N1 subtype displayed a similar pattern of viral attachment to human respiratory tract to that of H5N1 virus, even though low-pathogenicity avian influenza viruses rarely infect or cause disease in humans (41). Thus, the ability of the H5N1 virus or the H7N9 virus to attach to human respiratory tract cells is not sufficient to explain the ability of this virus to infect and induce disease in humans.
The binding of avian and human influenza viruses to sections of guinea pig respiratory tissues herein represents an important in-depth characterization of influenza virus receptors in the guinea pig. Both human and avian viruses were found to bind to respiratory and olfactory epithelia of nasal passages, whereas attachment to the tracheal, bronchiolar, and alveolar surfaces was largely limited to the avian strains. Compared to the results of staining with Maackia amurensis agglutinin II (MAA II) and Sambucus nigra agglutinin (SNA) lectins for the detection of α2,3- and α2,6-linked sialic acids, respectively, similarities and differences are apparent. Lectin staining of guinea pig tissues revealed mainly α2,3-linked receptors in the lung and a mixture of α2,3- and α2,6-linked receptors in nasal respiratory epithelia, a finding consistent with the virus binding data (35, 48, 49). Differences were seen in the nasal olfactory epithelia (bound only by MAA II [35], but both human and avian viruses) and trachea (bound by both MAA II and SNA [48], but only avian viruses). These differences could, of course, be due to differences between the precise glycans targeted by the avian viruses used versus MAA II and the human virus used versus SNA. It is interesting to note the similarities between the attachment patterns of An/13 virus in guinea pig and human tissues: in both species, binding was abundant in tissues of the upper and the lower respiratory tracts (49).
When interpreting HA attachment data, it is important to note that, if a given cell type is positive for binding, this does not mean that it is permissive for productive infection. Attachment is critical to the viral life cycle, but the target cell must also support a number of subsequent viral processes (e.g., replication of the genome) to be permissive for infection. This distinction most likely underlies differences between the pattern of H7N9 virus attachment to fixed human respiratory tissues (13, 15) and tropism seen after infection of viable human lung cells (16, 50–54).
Using glycan arrays (13, 15) or alternative in vitro binding assays (16, 50–52), others have observed binding of An/13 to both α2,3- and α2,6-linked sialic acids, which is atypical of avian and human influenza viruses.
We also assessed the binding of An/13 to arrays from the Center for Functional Glycomics and obtained similar results to those published (data not shown). The HA of An/13 and similar H7N9 viruses appears to be intermediate between human and avian influenza virus HA proteins in terms of the receptor-binding properties (50, 55). Importantly, the H7N9 virus HA proteins do not currently show the extent of human adaptation characteristic of historical pandemic strains (56–60).
In addition to receptor binding specificity, the pH at which the influenza virus HA protein mediates membrane fusion has been found to be an important determinant of host adaptation and transmission (42, 46, 47, 61, 62). In general, influenza viruses adapted to avian hosts have a higher pH of fusion than those adapted for circulation in humans (42, 47). The lower fusion pH of human strains equates to greater stability of the HA protein in its prefusion conformation, a property that is thought to be important for the transmission of influenza viruses via respiratory droplets (61, 62), particularly given the relatively acidic nature of nasal passageways in response to inflammation or infection (63, 64). In line with our results, Xiong et al. found that the An/13 HA possessed a relatively high pH of fusion using an assay based on the susceptibility of HA to proteolysis (50). The high fusion pH of An/13 virus is characteristic of avian influenza virus strains and may play a role in limiting the transmission efficiency of this virus by respiratory droplet (12–15).
In sum, An/13 virus displays a number of mammalian adaptive traits, including efficient replication and transmission in the guinea pig model; nevertheless, this virus retains certain characteristics of avian influenza viruses which may, barring further adaptation, limit its potential for sustained human-to-human transmission.
ACKNOWLEDGMENTS
The Protein-Glycan Interaction Resource of the Consortium for Functional Glycomics, supported by grant R24 GM098791, participated in this study. We thank Global Initiative on Sharing All Influenza Data (GISAID) for making sequences of H7N9 virus genomes available.
This study was funded by the National Institutes of Health/National Institute of Allergy and Infectious Disease Centers of Excellence for Influenza Research and Surveillance contract HHSN266200700006C (to J.S., R.T., S.M.T., and D.S.) and by the European Union FP7 ANTIGONE (contract 278976) (to T.K. and D.V.R.).
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368:1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 2.CDC 2013. Emergence of avian influenza A (H7N9) virus causing severe human illness–China, February-April 2013. MMWR Morb. Mortal. Wkly. Rep. 62:366–371 [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, Meng L, Hong Z, Tu W, Cao Y, Li L, Ding F, Liu B, Wang M, Xie R, Gao R, Li X, Bai T, Zou S, He J, Hu J, Xu Y, Chai C, Wang S, Gao Y, Jin L, Zhang Y, Luo H, Yu H, Gao L, Pang X, Liu G, Shu Y, Yang W, Uyeki TM, Wang Y, Wu F, Feng Z. 2013. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N. Engl. J. Med. [Epub ahead of print.] 10.1056/NEJMoa1304617 [DOI] [Google Scholar]
- 4.Lo YC, Chen WC, Huang WT, Lin YC, Liu MC, Kuo HW, Chuang JH, Yang JR, Liu MT, Wu HS, Yang CH, Chou JH, Chang FY. 2013. Surveillance of avian influenza A (H7N9) virus infection in humans and detection of the first imported human case in Taiwan, 3 April to 10 May 2013. Euro Surveill. 18:20479. [PubMed] [Google Scholar]
- 5.Chang SY, Lin PH, Tsai JC, Hung CC, Chang SC. 2013. The first case of H7N9 influenza in Taiwan. Lancet 381:1621. 10.1016/S0140-6736(13)60943-5 [DOI] [PubMed] [Google Scholar]
- 6.Han J, Jin M, Zhang P, Liu J, Wang L, Wen D, Wu X, Liu G, Zou Y, Lv X, Dong X, Shao B, Gu S, Zhou D, Leng Q, Zhang C, Lan K. 2013. Epidemiological link between exposure to poultry and all influenza A (H7N9) confirmed cases in Huzhou City, China, March to May 2013. Euro Surveill. 18:20481. [PubMed] [Google Scholar]
- 7.Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. 2013. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ 347:f4752. 10.1136/bmj.f4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N. Engl. J. Med. 368:2277–2285. 10.1056/NEJMoa1305584 [DOI] [PubMed] [Google Scholar]
- 9.Murhekar M, Arima Y, Horby P, Vandemaele KA, Vong S, Zijian F, Lee CK, Li A. 2013. Avian influenza A (H7N9) and the closure of live bird markets. Western Pac. Surveill. Response J. 4:4–7. 10.5365/wpsar.2013.4.2.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang P, Wang Q, Pang X, Chen L, Tian L, Deng Y. 2013. A case of avian influenza A (H7N9) virus occurring in the summer season, China. J. Infect. 67:624–625. 10.1016/j.jinf.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 11.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. 2013. Genetic analysis of novel avian A (H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 18:20453. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. 2013. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]
- 13.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2013. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501:560–563. 10.1038/nature12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501:551–555. 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, Wang G, Chen Y, Liu L, Liang L, Li Y, Fan J, Wang J, Li W, Guan L, Li Q, Yang H, Chen P, Jiang L, Guan Y, Xin X, Jiang Y, Tian G, Wang X, Qiao C, Li C, Bu Z, Chen H. 2013. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science 341:410–414. 10.1126/science.1240532 [DOI] [PubMed] [Google Scholar]
- 17.Kreijtz JH, Kroeze EJ, Stittelaar KJ, de Waal L, van Amerongen G, van Trierum S, van Run P, Bestebroer T, Kuiken T, Fouchier RA, Rimmelzwaan GF, Osterhaus AD. 2013. Low pathogenic avian influenza A (H7N9) virus causes high mortality in ferrets upon intratracheal challenge: a model to study intervention strategies. Vaccine 31:4995–4999. 10.1016/j.vaccine.2013.06.071 [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Bao L, Deng W, Dong L, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Li X, Huang W, Zhao X, Lan Y, Guo J, Yong W, Wei Q, Chen H, Zhang L, Qin C. 29 August 2013. Novel avian-origin human influenza A(H7N9) can be transmitted between ferrets via respiratory droplets. J. Infect. Dis. 10.1093/infdis/jit474 [DOI] [PubMed] [Google Scholar]
- 19.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 103:9988–9992. 10.1073/pnas.0604157103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pica N, Chou YY, Bouvier NM, Palese P. 2012. Transmission of influenza B viruses in the guinea pig. J. Virol. 86:4279–4287. 10.1128/JVI.06645-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steel J, Palese P, Lowen AC. 2011. Transmission of a 2009 pandemic influenza virus shows a sensitivity to temperature and humidity similar to that of an H3N2 seasonal strain. J. Virol. 85:1400–1402. 10.1128/JVI.02186-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, Chong KT. 2009. Histopathology and growth kinetics of influenza viruses (H1N1 and H3N2) in the upper and lower airways of guinea pigs. J. Gen. Virol. 90:386–391. 10.1099/vir.0.007054-0 [DOI] [PubMed] [Google Scholar]
- 23.Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ. 2008. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 167:775–785. 10.1093/aje/kwm375 [DOI] [PubMed] [Google Scholar]
- 24.Bouvier NM, Lowen AC, Palese P. 2008. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J. Virol. 82:10052–10058. 10.1128/JVI.01226-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J. Virol. 85:11235–11241. 10.1128/JVI.05794-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJ, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Garcia-Sastre A, Perez DR, Fouchier RA. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J. Virol. 84:11802–11813. 10.1128/JVI.01136-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowen AC, Steel J, Mubareka S, Carnero E, Garcia-Sastre A, Palese P. 2009. Blocking inter-host transmission of influenza virus by vaccination in the guinea pig model. J. Virol. 83:2803–2818. 10.1128/JVI.02424-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowen AC, Steel J, Mubareka S, Palese P. 2008. High temperature (30°C) blocks aerosol but not contact transmission of influenza virus. J. Virol. 82:5650–5652. 10.1128/JVI.00325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mubareka S, Lowen AC, Steel J, Coates AL, Garcia-Sastre A, Palese P. 2009. Transmission of influenza virus via aerosols and fomites in the guinea pig model. J. Infect. Dis. 199:858–865. 10.1086/597073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252. 10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. 2010. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J. Virol. 84:21–26. 10.1128/JVI.01732-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon YK, Lipatov AS, Swayne DE. 2009. Bronchointerstitial pneumonia in guinea pigs following inoculation with H5N1 high pathogenicity avian influenza virus. Vet. Pathol. 46:138–141. 10.1354/vp.46-1-138 [DOI] [PubMed] [Google Scholar]
- 33.Van Hoeven N, Belser JA, Szretter KJ, Zeng H, Staeheli P, Swayne DE, Katz JM, Tumpey TM. 2009. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J. Virol. 83:2851–2861. 10.1128/JVI.02174-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seibert CW, Kaminski M, Philipp J, Rubbenstroth D, Albrecht RA, Schwalm F, Stertz S, Medina RA, Kochs G, Garcia-Sastre A, Staeheli P, Palese P. 2010. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 84:11219–11226. 10.1128/JVI.01424-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Zhang Y, Shinya K, Deng G, Jiang Y, Li Z, Guan Y, Tian G, Li Y, Shi J, Liu L, Zeng X, Bu Z, Xia X, Kawaoka Y, Chen H. 2009. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5:e1000709. 10.1371/journal.ppat.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaminski MM, Ohnemus A, Staeheli P, Rubbenstroth D. 2013. Pandemic 2009 H1N1 influenza A virus carrying a Q136K mutation in the neuraminidase gene is resistant to zanamivir but exhibits reduced fitness in the guinea pig transmission model. J. Virol. 87:1912–1915. 10.1128/JVI.02507-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. 2013. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J. Virol. 87:4330–4338. 10.1128/JVI.02749-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H. 2013. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 340:1459–1463. 10.1126/science.1229455 [DOI] [PubMed] [Google Scholar]
- 39.Soboleski MR, Gabbard JD, Price GE, Misplon JA, Lo CY, Perez DR, Ye J, Tompkins SM, Epstein SL. 2011. Cold-adapted influenza and recombinant adenovirus vaccines induce cross-protective immunity against pH1N1 challenge in mice. PLoS One 6:e21937. 10.1371/journal.pone.0021937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed LJ, Muench H. 1938. A simple method for estimating fifty percent end points. Am. J. Hyg. 27:493–497 [Google Scholar]
- 41.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215–1223. 10.2353/ajpath.2007.070248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 9:e1003151. 10.1371/journal.ppat.1003151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowen AC, Mubareka S, Steel J, Palese P. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 3:1470–1476. 10.1371/journal.ppat.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Riel D, den Bakker MA, Leijten LM, Chutinimitkul S, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2010. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 176:1614–1618. 10.2353/ajpath.2010.090949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. 2006. H5N1 virus attachment to lower respiratory tract. Science 312:399. 10.1126/science.1125548 [DOI] [PubMed] [Google Scholar]
- 46.Zaraket H, Bridges OA, Duan S, Baranovich T, Yoon SW, Reed ML, Salomon R, Webby RJ, Webster RG, Russell CJ. 2013. Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J. Virol. 87:9911–9922. 10.1128/JVI.01175-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed ML, Bridges OA, Seiler P, Kim JK, Yen HL, Salomon R, Govorkova EA, Webster RG, Russell CJ. 2010. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J. Virol. 84:1527–1535. 10.1128/JVI.02069-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y, Bi Y, Pu J, Hu Y, Wang J, Gao H, Liu L, Xu Q, Tan Y, Liu M, Guo X, Yang H, Liu J. 2010. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One 5:e15537. 10.1371/journal.pone.0015537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Riel D, Leijten LM, de Graaf M, Siegers JY, Short KR, Spronken MI, Schrauwen EJ, Fouchier RA, Osterhaus AD, Kuiken T. 2013. Novel avian-origin influenza A (H7N9) virus attaches to epithelium in both upper and lower respiratory tract of humans. Am. J. Pathol. 183:1137–1143. 10.1016/j.ajpath.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. 10.1038/nature12372 [DOI] [PubMed] [Google Scholar]
- 51.Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, Garcia-Sastre A, Fernandez-Sesma A. 2013. H7N9 influenza viruses interact preferentially with α2,3-linked sialic acids and bind weakly to α2,6-linked sialic acids. J. Gen. Virol. 94:2417–2423. 10.1099/vir.0.056184-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. 10.1126/science.1242917 [DOI] [PubMed] [Google Scholar]
- 53.Weinheimer VK, Becher A, Tonnies M, Holland G, Knepper J, Bauer TT, Schneider P, Neudecker J, Ruckert JC, Szymanski K, Temmesfeld-Wollbrueck B, Gruber AD, Bannert N, Suttorp N, Hippenstiel S, Wolff T, Hocke AC. 2012. Influenza A viruses target type II pneumocytes in the human lung. J. Infect. Dis. 206:1685–1694. 10.1093/infdis/jis455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knepper J, Schierhorn KL, Becher A, Budt M, Tonnies M, Bauer TT, Schneider P, Neudecker J, Ruckert JC, Gruber AD, Suttorp N, Schweiger B, Hippenstiel S, Hocke AC, Wolff T. 2013. The novel human influenza A (H7N9) virus is naturally adapted to efficient growth in human lung tissue. mBio 4(5):e00601-13. 10.1128/mBio.00601-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R. 2013. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 153:1486–1493. 10.1016/j.cell.2013.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 79:11533–11536. 10.1128/JVI.79.17.11533-11536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 355:1143–1155. 10.1016/j.jmb.2005.11.002 [DOI] [PubMed] [Google Scholar]
- 58.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360. 10.1126/science.1186430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. 2010. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol. 84:1715–1721. 10.1128/JVI.02162-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc. Natl. Acad. Sci. U. S. A. 106:17175–17180. 10.1073/pnas.0906849106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer H, Widdicombe JH. 2006. Mechanisms of acid and base secretion by the airway epithelium. J. Membr. Biol. 211:139–150. 10.1007/s00232-006-0861-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer H, Widdicombe JH, Illek B. 2002. Acid secretion and proton conductance in human airway epithelium. Am. J. Physiol. Cell Physiol. 282:C736–C743. 10.1152/ajpcell.00369.2001 [DOI] [PubMed] [Google Scholar]