Abstract
Collagens, the most abundant proteins in animals, also occur in some recently described nucleocytoplasmic large DNA viruses such as Mimiviridae, which replicate in amoebae. To clarify the impact of viral collagens on the immune response of animals exposed to Mimiviridae, we have investigated the localization of collagens in Acanthamoeba polyphaga mimivirus particles and the response of mice to immunization with mimivirus particles. Using protein biotinylation, we have first shown that viral collagen encoded by open reading frame L71 is present at the surface of mimivirus particles. Exposure to mimivirus collagens elicited the production of anti-collagen antibodies in DBA/1 mice immunized intradermally with mimivirus protein extracts. This antibody response also targeted mouse collagen type II and was accompanied by T-cell reactivity to collagen and joint inflammation, as observed in collagen-induced arthritis following immunization of mice with bovine collagen type II. The broad distribution of nucleocytoplasmic large DNA viruses in the environment suggests that humans are constantly exposed to such large virus particles. A survey of blood sera from healthy human subjects and from rheumatoid arthritis patients indeed demonstrated that 30% of healthy-subject and 36% of rheumatoid arthritis sera recognized the major mimivirus capsid protein L425. Moreover, whereas 6% of healthy-subject sera recognized the mimivirus collagen protein L71, 22% of rheumatoid arthritis sera were positive for mimivirus L71. Accordingly, our study shows that environmental exposure to mimivirus represents a risk factor in triggering autoimmunity to collagens.
INTRODUCTION
Nucleocytoplasmic large DNA viruses (NCLDVs) represent a growing group of giant viruses found in various types of aquatic environments (1). NCLDVs include Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycodnaviridae, Mimiviridae, and Marseilleviridae (2). The Paramecium bursaria chlorella virus 1 (PBCV-1) was the first large DNA virus characterized at the molecular level and shown to harbor a complex genome of 330 kbp (3). But the largest NCLDVs described to date belong to the Mimiviridae, which occur in fresh and saline environments and replicate within amoebae (4). Acanthamoeba polyphaga mimivirus was the first member of the Mimiviridae isolated from a cooling water tower and was characterized in 2004 (5). Other members of Mimiviridae include megavirus isolated from a marine environment (6), mamavirus (7), and moumouvirus (8). Mimiviridae feature large capsids exceeding 400 nm in diameter and harbor large genomes of more than 1 Mbp. The genomes of NCLDVs encode structural proteins and enzymes usually not found in viruses, such as aminoacyl-tRNA synthetases, DNA repair enzymes, potassium ion channel, protein kinases, and glycosyltransferases (5, 9, 10).
Interestingly, Mimiviridae also express multiple collagen genes during their infectious life cycle in amoebae. For example, mimivirus expresses seven collagen genes, namely, L71, R196, R239, R240, R241, L668, and L669, already by 6 h postinfection (11). Even the virophage Sputnik includes two collagen genes among its predicted 21 open reading frames (ORFs) (12). The functional relevance of these collagens is, however, presently unknown. First analysis of mimivirus proteins indicated that collagen is hydroxylated in the same way as animal collagen (13). Cryo-electron microscopy and atomic force microscopy studies failed to reveal any collagen-like structures in mimivirus (14, 15) although the dense fibers surrounding mimivirus capsids have been suggested to represent cross-linked glycosylated collagen (14).
The ubiquitous distribution of NCLDVs in aquatic environments (16, 17) suggests that humans are constantly exposed to such viruses. Mimivirus cannot replicate in animal cells but can be internalized by phagocytosis by mouse and human macrophages (18). The uptake of mimivirus particles by human macrophages potentially leads to virus antigen presentation and thereby to the generation of antibodies against virus proteins. Considering the structural similarity between animal and Mimiviridae collagens, we made the hypothesis that antibodies generated against Mimiviridae collagens may cross-react with animal collagens and thereby contribute to an autoimmune response to collagenous structures in animals previously exposed to Mimiviridae. The present study provides evidence supporting this hypothesis by showing that arthritis can be triggered in mice immunized with mimivirus particles and by revealing the increased occurrence of antibodies against mimivirus collagen in rheumatoid arthritis patients.
MATERIALS AND METHODS
Ethics statement.
All mouse experiments were performed in compliance with the Swiss Animal Protection Ordinance and were approved by the local veterinary authority (Kantonales Veterinäramt Zürich, Switzerland). The human sera tested in this study were part of previously existing collection and experimental protocol approved by the Kantonale Ethik-Kommission Zürich (KEK).
Giant virus infection and protein extraction.
Acanthamoeba polyphaga and mimivirus were provided by Didier Raoult (CNRS UMR6020, Université de la Méditerranée, Marseille). Marseillevirus (2) was isolated from a water sample collected from the Lake Zurich. Amoebae were routinely cultured as a monolayer in peptone-yeast-glucose (PYG) medium at 28°C as previously described (5). Mimivirus and marseillevirus were added at a multiplicity of infection (MOI) of 10 to amoebae, and newly formed virus was collected from the culture supernatant at 2 days postinfection. Virus particles were suspended in a mixture of 0.5 M Tris-HCl, pH 8.5, 0.2% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 2 mM Tris-(2-carboxyethyl) phosphine (TCEP), and 6 M guanidine hydrochloride and incubated at 65°C for 10 min. After the mixture was cooled to room temperature, iodoacetamide was added to a final concentration of 3 mM, and the mixture was further incubated at room temperature for 40 min. After dithiothreitol (DTT) was added to a final concentration of 15 mM, protein extracts were centrifuged at room temperature at 17,000 × g, and proteins in the supernatant were precipitated with 12% trichloroacetic acid.
Surface biotinylation of mimivirus proteins.
Purified mimivirus particles were suspended in phosphate-buffered saline (PBS), and sulfo-N-hydroxysuccinimide-biotin (sulfo-NHS-biotin; Thermo Scientific, Waltham, MA, USA) was added to a final concentration of 1 mg/ml. Virions were rotated for 30 min at room temperature, and the reaction was quenched by the addition of an equal volume of 100 mM glycine in PBS. Virions were pelleted and washed twice with 100 mM glycine in PBS, and proteins were extracted with guanidine hydrochloride as described above. Extracts were diluted 10-fold in PBS–0.1% CHAPS containing proteinase inhibitors (Calbiochem Proteinase Inhibitor Cocktail III; Merck Millipore, USA) and subjected to avidin cartridge purification (ABSciex, Framingham, MA, USA). The cartridge was successively washed with 500 μl of PBS–0.1% CHAPS, followed by 1 ml of 650 mM NaCl in 20 mM phosphate buffer (pH 7.2), 0.1% CHAPS, 1 ml of PBS, 0.1% CHAPS, and finally, 1 ml of 0.1% CHAPS in H2O. Biotinylated proteins were then eluted with 800 μl of 0.4% trifluoroacetic acid–0.1% CHAPS. Proteins were precipitated with trichloroacetic acid and subjected to SDS-PAGE. Individual protein bands were excised and subjected to in-gel tryptic digestion, as previously described (19, 20), followed by liquid chromatography-mass spectrometry (LC-MS) protein identification. LC-MS data were analyzed using Mascot (version 2.3.02; Matrix Science, London, United Kingdom). Mascot was set up to search a Swiss-Prot concatenated target-decoy database (accessed 11 January 2011; 1,049,100 entries), assuming that enzyme trypsin is used for protein digestion. Mascot was searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 ppm. An iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Oxidation of methionine and biotinylation of lysine were specified in Mascot as variable modifications. Scaffold (version 3.4.9; Proteome Software Inc., Portland, OR, USA) was used to statistically validate tandem MS (MS/MS)-based peptide and protein identifications.
Collagen-induced arthritis model.
DBA/1 mice were purchased from Charles River (Germany) and bred and maintained in the animal facility of the Institute of Physiology, University of Zurich. All experiments were performed in compliance with the Swiss Animal Protection Ordinance and approved by the local veterinary authority (Kantonales Veterinäramt Zürich, Switzerland). Collagen-induced arthritis was established as described previously (21). Briefly, 6- to 8-week-old mice were immunized intradermally in the tail with either PBS, bovine collagen type II (Chondrex, USA), mimivirus L71 collagen-like protein, mimivirus protein extract, or marseillevirus protein extract emulsified in Complete Freund's Adjuvant (CFA; Chondrex, USA). Each mouse received either 50 μl of PBS, 100 to 120 μg of bovine collagen type II, 150 μg of recombinant L71 collagen-like protein, 120 to 150 μg of mimivirus proteins, or 120 to 150 μg of marseillevirus protein emulsified 1:1 in CFA in a total volume of 50 μl. Thirty days later mice received a booster injection of the same amount of antigen emulsified 1:1 in Incomplete Freund's Adjuvant (IFA; Chondrex, USA). Development of arthritis was monitored daily for 75 days postimmunization. Severity was scored on a level of 0 (no inflammation) to 4 (most severe inflammation) per limb per mouse, thus allowing a maximum score of 16 per mouse (21).
Anticollagen type II antibodies.
Mouse blood sera were collected by heart puncture. Anti-mouse CII antibody titers were measured in blood serum by enzyme-linked immunosorbent assay (ELISA; Chondrex, USA) as per the manufacturer's instructions.
Histology.
Limbs were skinned and fixed overnight in 10% neutral buffered formalin. Tissues were further decalcified using Immunocal solution (Quartett, Germany) for 4 to 5 days, dehydrated, and paraffin embedded. Sections of 5 μm were mounted on glass slides and stained with hematoxylin and eosin (H&E).
Recall assay.
Axillary, lateral axillary, superficial inguinal, and popliteal lymph nodes from mice were collected 8 to 10 days after booster immunization. Aliquots of 100,000 cells in 100 μl of complete RPMI 1640 medium were stimulated with antigens and incubated for 48 h in a CO2 incubator at 37°C with 5% CO2. Cells were stimulated in 100 μl of either medium alone as a negative control or concanavalin-A (Sigma, Switzerland) at 3 μg/ml as a positive control. T-cell proliferation-grade denatured mouse collagen type II at 1 mg/ml (Chondrex, USA), T-cell proliferation-grade denatured bovine collagen type II at 1 mg/ml (Chondrex, USA), and heat-denatured mimivirus collagen L71 at 1.5 mg/ml were used as antigens. After 48 h, 1 μCi of [3H]thymidine (Perkin-Elmer, USA) per well was added and incubated for 16 to 18 h, and cells were harvested on 96-well glass filters (Perkin-Elmer, USA). Radioactivity was counted using a 96-well scintillation beta-counter (Wallac, Perkin-Elmer).
Anti-mimivirus ELISA.
Mimivirus proteins were coated in microtiter plates at 0.1 μg per well in 100 μl of PBS overnight at 4°C. Plates were washed three times with PBS–0.05% Tween and blocked with PBS–0.05% Tween–1% bovine serum albumin at 37°C for 2 h. Plates were washed, 100 μl of diluted human and rabbit sera was added, and the samples were further incubated at room temperature for 1 h. After three wash steps, 100 μl of 1:5,000-diluted biotinylated anti-human or anti-rabbit IgG antibody (BD Biosciences, Switzerland) was added for 2 h. Plates were washed, and 100 μl of 1:1,000-diluted streptavidin-horseradish peroxidase (HRP) conjugate (BD Biosciences) was added for 1 h in dark. Plates were washed, incubated for 2 min with 50 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (BD Biosciences) before the reaction was stopped with 25 μl of 2 N H2SO4. Color development was measured at 440 nm.
Immunoprecipitation of mimivirus proteins.
Aliquots of 25 μl of human serum were incubated with 30 μl of protein G-Sepharose 4 Fast Flow (GE Healthcare, Switzerland) beads along with 80 μl of PBS on a rotating shaker for 1 h at 4°C. After centrifugation at 500 × g for 5 min at 4°C, supernatants were discarded, and beads were incubated with 20 μg of mimivirus protein extract in 80 μl of PBS and further incubated on a rotating shaker for 30 min at 4°C. Beads were washed three times in PBS, and antigen-antibody complexes were eluted from the beads by the addition of 40 μl of 0.1 M glycine, pH 2.7. After neutralization by the addition of 20 μl of 1 M Tris-HCl, pH 9, eluates were separated by SDS-PAGE. Slices of polyacrylamide gel excluding IgG chains were excised and subjected to in-gel tryptic digest as previously described (20), and peptides were identified by tandem mass spectrometry as above.
Cloning, bacterial expression, and purification.
The mimivirus ORFs L71 and L425 were custom synthesized (Genescipt, USA) and subcloned into the expression vector pET16b (Merck Millipore, Switzerland) linearized with XhoI and HindIII (for L71) or with XhoI and BamHI (for L425). His6-tagged recombinant proteins were expressed after transformation into Escherichia coli BL21(DE3) cells (Novagen, Switzerland) under induction of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 32°C for 1.5 h. Recombinant proteins were purified over Ni-Sepharose 6 Fast Flow (GE Healthcare, Switzerland) gravity flow columns.
Western blotting.
Aliquots of 15 μg of recombinant mimivirus L71 and L425 proteins were subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad). A His6-tagged 15-kDa fragment of the human GLT25D2 protein (22) was used as a negative control. A 36-kDa His6-tagged fragment of human collagen type III encompassing 114 G-X-Y repeats and lacking N and C propeptides was provided by Christoph Rutschmann, Institute of Physiology, University of Zurich. Blots were blocked in a solution of 1% polyvinylpyrrolidone (Sigma, Switzerland) and 5% dry milk overnight at 4°C, washed three times for 5 min with TBS–0.1% Tween, and incubated with human serum diluted 1:4,000 for 2 h at room temperature. After four washes of 5 min each, blots were incubated with anti-human IgG-HRP (Promega, Switzerland) at a 1:7,500 dilution at room temperature for 1 h. Blots were developed with SuperSignal chemiluminescent substrate (Thermo Scientific).
Statistical analysis.
One-way analysis of variance (ANOVA) with Dunnett's multiple comparison (GraphPad Prism) was performed to compare experimental groups.
RESULTS
Surface localization of mimivirus collagen.
To assess the possible localization of collagens at the surface of mimivirus, we used a biotinylation approach on mimivirus particles. Biotinylated mimivirus proteins were captured on streptavidin beads, eluted, and identified by tandem mass spectrometry, which revealed 60 surface proteins including the collagen protein L71 (Table 1). The L71 protein has 945 amino acids with four collagen domains encompassing 561 amino acids (Fig. 1). The first collagenous domain of L71 includes a stretch with 73% sequence identity to a major human collagen type II T-cell epitope identified in rheumatoid arthritis (23, 24). Other identified surface proteins comprised the capsid protein L425, the putative glucose-methanol-choline (GMC)-type oxidoreductase R135, and the thioredoxin domain-containing protein R362 among several uncharacterized proteins (Table 1).
TABLE 1.
Mimivirus surface proteins identified by biotinylation
| ORF | Protein annotationa | % Coverage |
|---|---|---|
| R459 | Uncharacterized protein | 92 |
| L724 | Uncharacterized protein | 69 |
| L725 | Uncharacterized protein | 51 |
| R489 | Uncharacterized protein | 49 |
| R714 | Uncharacterized protein | 42 |
| L485 | Uncharacterized protein | 41 |
| L330 | Uncharacterized protein | 36 |
| R305 | Uncharacterized protein | 36 |
| R727 | Uncharacterized protein | 35 |
| R345 | Uncharacterized protein | 35 |
| L53 | Uncharacterized protein | 33 |
| R457 | Uncharacterized protein | 33 |
| R362 | Thioredoxin domain-containing protein | 26 |
| L488 | Uncharacterized protein | 24 |
| R346 | Uncharacterized protein | 24 |
| L586 | Uncharacterized protein | 23 |
| L829 | Uncharacterized protein | 23 |
| R623 | Uncharacterized protein | 22 |
| L550 | Uncharacterized protein | 22 |
| L425 | Capsid protein-1 | 19 |
| L647 | Uncharacterized protein | 19 |
| R306 | Uncharacterized protein | 19 |
| L645 | Uncharacterized protein | 19 |
| L591 | Uncharacterized protein | 19 |
| L719 | Uncharacterized protein | 18 |
| R463 | Uncharacterized protein | 18 |
| R705 | Uncharacterized protein | 17 |
| L585 | Uncharacterized protein | 17 |
| L454 | Uncharacterized protein | 17 |
| R653 | Uncharacterized protein | 16 |
| R610 | Uncharacterized protein | 14 |
| L629 | Uncharacterized protein | 14 |
| R710 | Uncharacterized protein | 13 |
| L399 | Uncharacterized protein | 12 |
| R443 | Thioredoxin domain-containing protein | 12 |
| R253 | Uncharacterized protein | 11 |
| L324 | Uncharacterized protein | 11 |
| R307 | PP2C-like domain-containing protein | 11 |
| L492 | Uncharacterized protein | 11 |
| R596 | Probable FAD-linked sulfhydryl oxidase | 10 |
| R135 | Putative GMC-type oxidoreductase | 9.5 |
| L442 | Uncharacterized protein | 9.5 |
| R347 | Uncharacterized protein | 9.5 |
| R622 | Putative tyrosine-protein phosphatase | 9 |
| L609 | Uncharacterized protein | 8.6 |
| L612 | Uncharacterized protein | 7.5 |
| L309 | Uncharacterized protein | 6.2 |
| R252 | Uncharacterized protein | 6.1 |
| R526 | Putative alpha/beta hydrolase | 6.1 |
| R692 | Uncharacterized protein | 4.8 |
| L236 | Uncharacterized protein | 4.7 |
| L448 | Uncharacterized protein | 4.5 |
| L264 | Uncharacterized WD repeat-containing protein | 4.5 |
| R588 | Uncharacterized protein | 3.3 |
| L605 | Structural PPIase-like protein | 3 |
| R553 | Uncharacterized protein | 2.7 |
| L71 | Collagen-like protein 1 | 2.5 |
| L357 | Uncharacterized protein | 2.4 |
| R643 | Uncharacterized protein | 2.2 |
| L397 | Uncharacterized protein | 2.1 |
Mimivirus surface proteins were identified by mass spectrometric peptide sequencing after biotinylation and analyzed by Mascot software. Results were validated by Scaffold (version 3.4.9; Proteome Software, Inc., Portland, OR), and peptide identifications were accepted if they established >80% probability as specified by the Peptide Prophet algorithm (34). Protein identifications were accepted if they could be established at >99% probability and contained at least two identified peptides as specified by Protein Prophet algorithm (35). GMC, glucose-methanol-choline; PP2C, protein phosphatase 2C; PPIase, peptidyl-prolyl isomerase; FAD, flavin adenine dinucleotide.
FIG 1.
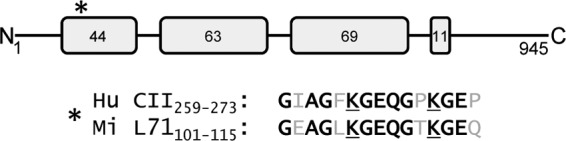
Domain organization of mimivirus L71 protein. The four collagen domains of L71 are shown as gray boxes with the number of G-X-Y repeats given inside. The asterisk shows the position of the sequence motif similar to the epitope human collagen type II recognized as immunodominant in rheumatoid arthritis (24). The sequence of this human collagen type II (Hu CII) T-cell epitope encompassing amino acids 259 to 273 is shown aligned with the corresponding sequence of mimivirus L71 (Mi L71) amino acids 101 to 115.
Mimivirus proteins promote arthritis in mice.
Considering the surface expression of collagen L71, we have addressed the potential of mimivirus and recombinant L71 protein to induce joint inflammation in DBA/1 mice using the standard protocol for collagen-induced arthritis (21), which closely resembles rheumatoid arthritis in humans. Bovine collagen type II was used as a positive control to induce arthritis. A protein extract of the giant virus marseillevirus, which lacks collagen-like proteins, was used as a negative control. Intradermal immunization of bovine collagen type II and mimivirus protein extracts led to joint inflammation, as assessed by visual inspection and histological examination of limb tissues. Mice immunized with mimivirus proteins reached clinical scores of 6, whereas those immunized with bovine collagen type II reached a score of 12 by 75 days (Fig. 2A). In contrast, immunization with recombinant L71 protein alone and immunization with marseillevirus proteins failed to elicit joint inflammation (Fig. 2A). Altered cartilage integrity and synovial hyperplasia were evident in joints of mice immunized with bovine collagen type II and, to a lesser extent, in mice immunized with mimivirus proteins (Fig. 2B).
FIG 2.
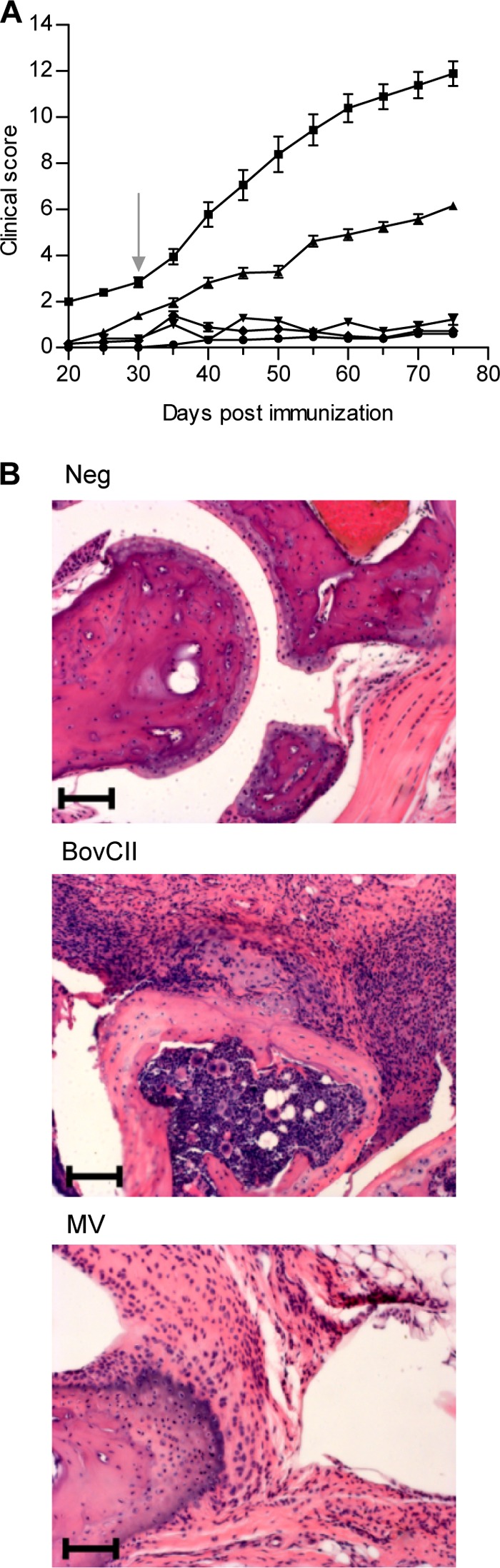
Joint inflammation in DBA/1 mice immunized with mimivirus proteins. (A) Clinical severity of arthritic limbs in the groups of mice immunized with PBS (●), bovine collagen type II (■), recombinant L71 protein (▼), marseillevirus proteins (◆), and mimivirus proteins (▲) are shown as means ± standard errors of the means. The arrow shows the time point of booster immunization. Data represent three independent experiments with 10 to 21 mice per group. (B) Representative H&E-stained sections of hind limbs by day 75 after immunization showing cartilage damage and synovial hyperplasia in mice immunized with bovine collagen type II (BovCII) and mimivirus protein (MV). No signs of pathology were visible in PBS-immunized negative-control mice (Neg). Scale bar, 100 μm.
The breakdown of immune tolerance induced by bovine collagen and mimivirus proteins was confirmed by the detection of elevated serum titers of anti-mouse collagen type II IgG in mice immunized with bovine collagen type II and mimivirus proteins, whereas mice immunized with PBS and marseillevirus did not show elevated anti-mouse collagen type II IgG titers (Fig. 3). Mice immunized with recombinant L71 protein also showed significantly elevated anti-mouse collagen type II IgG titers yet by an order of magnitude lower than the titers observed in mice immunized with mimivirus proteins (Fig. 3). The lower antibody response achieved with L71 immunization may account for the lack of joint inflammation seen in these mice. The cross-reactivity of T cells was investigated in recall assays (25). Cells isolated from draining lymph nodes of the immunized mice were found to proliferate in response to in vitro presentation of denatured fragments of mouse collagen type II (Fig. 4A), bovine collagen type II (Fig. 4B), and mimivirus collagen protein L71 (Fig. 4C), thereby confirming the presence of autoreactive T cells after immunization with mimivirus proteins. The proliferative response to collagen was strongest for cells isolated from mice immunized with mimivirus proteins. This finding was surprising since mice immunized with bovine collagen type II showed the highest score for limb inflammation and for anticollagen IgG titers. The strong proliferative response of T cells from mimivirus protein-immunized mice may reflect the higher antigenicity of mimivirus collagen considering its peptide sequence divergence from mammalian collagen sequences.
FIG 3.
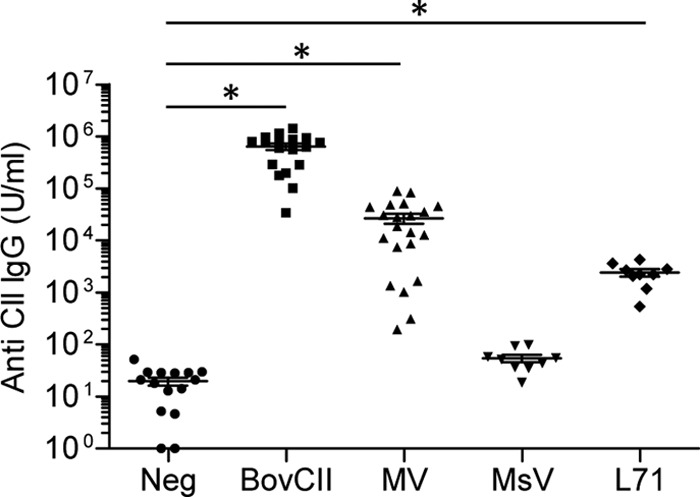
Anti-collagen type II IgG titers in DBA/1 mice immunized with mimivirus proteins. Levels of serum IgG measured by ELISA against endogenous mouse collagen type II (CII) in mice immunized with PBS (Neg), bovine collagen type II (BovCII), mimivirus proteins (MV), marseillevirus proteins (MsV), or recombinant L71 protein (L71). Data represent three independent experiments with 10 to 21 mice per group. Horizontal bars show means ± standard errors of the means (*, P < 0.01).
FIG 4.
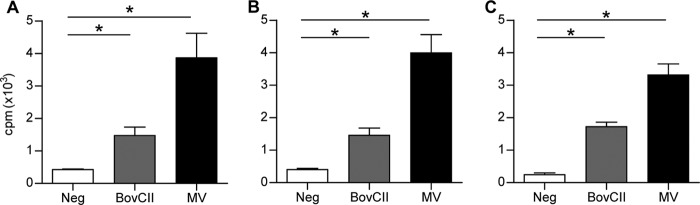
Autoreactive T-cell response in DBA/1 mice immunized with mimivirus proteins. (A) Recall responses in cells isolated from draining lymph nodes of mice immunized with PBS (Neg), bovine collagen type II (BovCII), or mimivirus proteins (MV) after stimulation with denatured mouse collagen type II. (B) Recall responses after stimulation with denatured bovine collagen type II. (C) Recall responses after stimulation with denatured fragmented recombinant mimivirus protein L71. Data represent means ± standard errors of the means of groups of 3 mice (*, P < 0.01).
Immunity to mimivirus in humans.
To determine whether humans are commonly exposed to mimivirus, we first examined the presence of antibodies against mimivirus in 100 healthy subjects by ELISA using whole mimivirus proteins as antigens. Reactivity to mimivirus proteins was variable; 58 human sera showed significant IgG titers in the 5% range of titers observed in the sera of rabbits previously immunized with mimivirus proteins (Fig. 5). To identify the major mimivirus proteins recognized by human sera, we coupled the IgG fraction of sera from healthy subjects (healthy-subject sera) and rheumatoid arthritis patients (rheumatoid arthritis sera) to protein G-Sepharose beads, which were further incubated with preparations of mimivirus proteins. Mimivirus proteins retained on the IgG-protein G beads were identified by mass spectrometric peptide sequencing after trypsin digestion. The major capsid protein L425 was found in all samples, followed by the putative GMC-type oxidoreductase R135 and core protein L410, which were found in 7 of 10 samples (Table 2). Interestingly, the most frequent mimivirus proteins recognized by human sera were surface proteins according to our surface biotinylation study (Table 1). Mimivirus collagens did not appear among the proteins recognized by serum. This absence may be related to the abundance of lysine in mimivirus collagens, thereby yielding very short tryptic peptides that remained below the detection range of mass spectrometric peptide sequencing. The recognition of multiple mimivirus proteins by human serum confirmed the exposure of humans to mimivirus.
FIG 5.
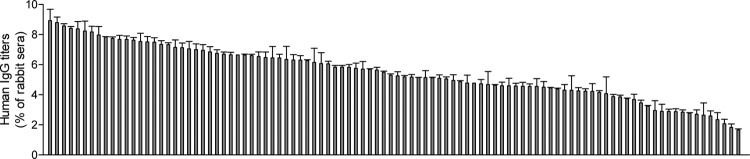
Reactivity of human sera toward mimivirus proteins. Anti-mimivirus IgG titers in sera of 100 healthy subjects were measured by ELISA after dilution to 1:100 and expressed as a ratio to IgG titers measured in 1:1,000-diluted sera from rabbits previously immunized with mimivirus particles. Data represent means ± standard errors of the means from four analyses.
TABLE 2.
Mimivirus proteins recognized by human serum IgG
| ORF | Protein annotation | Mascot score for sera from:a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy subjects |
Rheumatoid arthritis subjects |
||||||||||
| L425 | Capsid protein | 68 | 601 | 463 | 68 | 387 | 983 | 271 | 258 | 70 | 327 |
| R135 | Putative GMC-type oxidoreductase | 370 | 394 | 153 | 774 | 255 | 163 | 161 | |||
| L410 | Core protein | 223 | 284 | 179 | 685 | 123 | 133 | 97 | |||
| R345 | Putative regulator of chromosome condensation | 52 | 123 | 145 | 28 | 58 | 350 | 185 | 77 | 217 | |
| R349 | Uncharacterized protein | 28 | 28 | 30 | 26 | 26 | 35 | 26 | 28 | 32 | |
Scores for viral proteins recognized by five healthy subject sera and five rheumatoid arthritis sera are listed in columns for each serum tested. Values indicate Mascot scores representing the probability of positive matches for the recognized proteins. Scores above 25 were significant at a P value of <0.05.
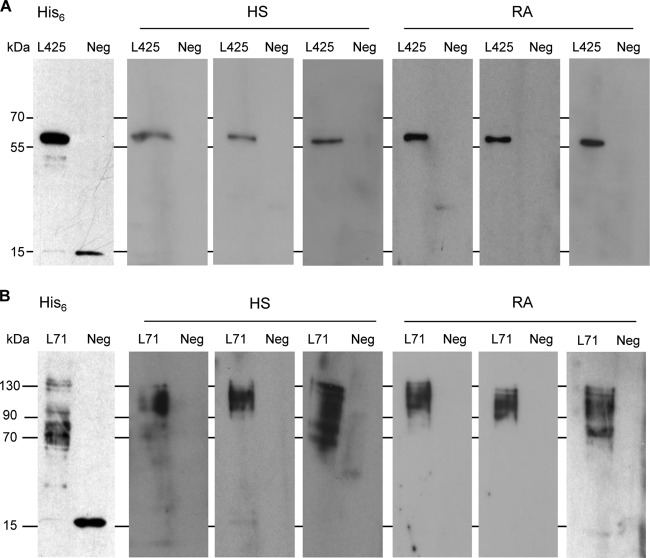
To further validate the occurrence of antibodies against specific mimivirus proteins in human sera, reactivity toward mimivirus L425 and L71 proteins was analyzed by Western blotting. The major capsid L425 and collagen L71 proteins were expressed as His6-tagged recombinant proteins in E. coli and purified on Ni2+-Sepharose columns. Pools of 100 healthy-subject sera and 100 rheumatoid arthritis sera were probed against the recombinant L425 and L71 mimivirus proteins. We examined the reactivity of sera toward surface collagen L71 since this protein was not detected among the mimivirus proteins captured by immobilized serum IgG in our previous experiment. For the 100 healthy-subject and 100 rheumatoid arthritis sera tested, respectively, 30 and 36 sera recognized the capsid L425 protein (Fig. 6A). This result confirmed that exposure to mimivirus is common in the human population. The detection of IgG against the mimivirus capsid protein L425 in 30% of tested sera suggests repeated antigenic challenge, probably caused by repeated contact with mimivirus. Reactivity of human sera toward mimivirus collagen was more discriminatory. Whereas only 6 healthy-subject sera recognized the mimivirus collagen L71, 22 rheumatoid arthritis sera were positive for the mimivirus collagen L71 (Fig. 6B). To exclude nonspecific cross-reactivity of human sera toward polypeptides containing (G-X-Y)n collagen domains, we tested the recognition of L71-positive sera for a fragment of human collagen type III encompassing 114 G-X-Y repeats and lacking N and C propeptides. None of the 28 human sera positive for mimivirus L71 recognized the 36-kDa (G-X-Y)114 construct (Fig. 7), thereby demonstrating the specificity of the antibody response to mimivirus L71 collagen. Accordingly, this work confirmed that the reactivity to mimivirus collagen was 3.5 times more frequent in the pool of rheumatoid arthritis sera than in the sera from healthy subjects, which showed limited reactivity.
FIG 6.
Recognition of mimivirus proteins by human sera. (A) Representative Western blots of sera from healthy subjects (HS) and rheumatoid arthritis (RA) patients recognizing mimivirus capsid protein L425. (B) Representative Western blots of sera from healthy subjects (HS) and rheumatoid arthritis (RA) patients recognizing mimivirus collagen L71. Sera were diluted 1:4,000. Positions of recombinant L425 and L71 proteins in the blots are shown at the left of each panel using an anti-His6 antibody (His6). A 15-kDa fragment of His6-tagged human GLT25D2 protein was used as a negative control (Neg).
FIG 7.
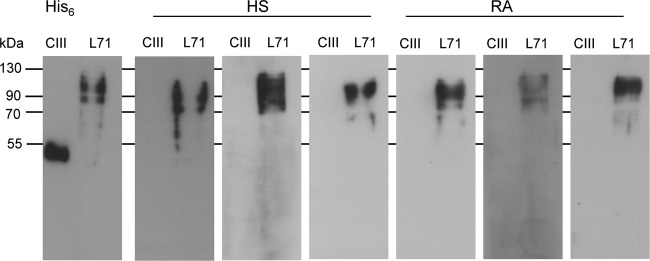
Specific recognition of mimivirus collagen L71 by human sera. Representative Western blots of L71-positive sera from healthy subjects (HS) and rheumatoid arthritis (RA) patients recognizing mimivirus collagen L71 but not a fragment of human collagen type III containing 114 G-X-Y repeats (CIII). Sera were diluted 1:4,000. Positions of recombinant L71 and CIII proteins in the blots are shown at the left of the panel using an anti-His6 antibody (His6).
DISCUSSION
The present study demonstrated that mice generated autoreactive anticollagen antibodies after immunization with mimivirus proteins including viral collagens. A possible relationship between exposure to mimivirus collagen and the development of autoimmunity was corroborated by the occurrence of IgG against mimivirus collagen among rheumatoid arthritis patients. These findings suggested that repeated exposure to mimivirus leads to antibody formation to virus collagen and to a breakdown of immune tolerance for endogenous collagens. Giant viruses like mimivirus are ubiquitous in the environment (16, 17), thereby supporting the frequent contact of humans with such viruses. Mimivirus is most likely ingested by water uptake and captured by dendritic cells and macrophages lining the gastrointestinal mucosa. Alternatively, virus particles may enter the airways as aerosol and be taken up by alveolar macrophages. In fact, mimivirus can be phagocytized by human and mouse macrophages although the virus cannot replicate in these cells (18). In line with this observation, mimivirus infection has been related to pneumonia in isolated cases, although without evidence for virus particles in disease cases (26, 27). This putative pathogenicity, however, does not preclude a more general effect of mimivirus on priming an autoimmune response.
The sequence similarity between a stretch of mimivirus L71 and human collagen type II (Fig. 1) supports a possible cross-reactivity of antibodies due to antigenic mimicry. This was indeed confirmed in our study by the detection of anti-mouse collagen type II IgG in mice immunized with recombinant L71 protein. A similar case of antigenic mimicry occurs in Campylobacter jejuni infection, which causes gastroenteritis but can lead to Guillain-Barré syndrome when antibodies against Campylobacter lipooligosaccharides cross-react with endogenous GM1 gangliosides on nerve cells (28). Likewise, the detection of antibodies toward mimivirus L425 capsid protein in some Francisella tularensis-infected patients suggested cross-reactivity of mimivirus antigens with other microorganisms (29). But, surprisingly, no reactivity to mimivirus antigens was found in sera from healthy subjects in the study of Pelletier et al. (29).
The main factors involved in the pathogenicity of rheumatoid arthritis could be either genetic or environmental. Rheumatoid arthritis is an autoimmune disease with a significant environmental component, as supported by twin studies (30, 31). Repeated contact to collagen antigens found in the environment may promote the development of cross-reactive anticollagen antibodies and to inflammation in collagen-rich tissues. Antibodies against collagens can recognize either the triple helical conformation or the peptide sequences in the triple helical domain or in telopeptides. The specificity of antibodies against collagen depends on the activation of a humoral response alone or on a combination of cell-mediated and humoral responses (32). The reactivity of human serum to mimivirus collagen L71 shown by Western blotting indicates that epitopes based on amino acid sequence and not three-dimensional (3D) conformations are being recognized. This notion was supported by finding no reactivity of the L71-positive human sera for the human collagen type III (G-X-Y)114 polypeptide used as a negative control. The lack of recognition for the (G-X-Y)114 polypeptide also indicated that the reactivity toward L71 was specific to mimivirus exposure and not the result of cross-reactivity to collagen domain-containing proteins, such as those found in some Gram-positive bacteria. We did find that mimivirus L71 protein was immunogenic and led to the production of anti-mouse collagen type II IgG, but immunization of DBA/1 mice with recombinant L71 protein failed to induce arthritis. This recombinant protein likely did not mimic the native conformation of collagen-like proteins, which is essential for arthritogenicity. In fact, denaturation of collagen prior to immunization abrogates the arthritic response (33).
Based on these results and in view of the structural similarity between mimivirus collagen and human collagen sequences, we propose that giant viruses expressing collagen represent a potential environmental risk factor contributing to the development of rheumatoid arthritis. A systematic survey of mimivirus distribution in the environment will contribute to a better appreciation of the environmental risk associated with such giant viruses in relation to the geographical incidence of rheumatoid arthritis.
ACKNOWLEDGMENTS
We are grateful to Lubor Borsig, Daniel Rhyner, Carole Oertli, Jürg Cabalzar, Christoph Rutschmann, and Charlotte Burger for technical assistance. We are grateful to the Functional Genomics Center Zurich for its support with mass spectrometric analysis.
This work was supported by a Swiss National Science Foundation Grant 310030-129633 to T.H.
Footnotes
Published ahead of print 30 October 2013
REFERENCES
- 1.Wilson WH, Van Etten JL, Allen MJ. 2009. The Phycodnaviridae: the story of how tiny giants rule the world. Curr. Top. Microbiol. Immunol. 328:1–42. 10.1007/978-3-540-68618-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colson P, Pagnier I, Yoosuf N, Fournous G, La Scola B, Raoult D. 2013. “Marseilleviridae,” a new family of giant viruses infecting amoebae. Arch. Virol. 158:915–920. 10.1007/s00705-012-1537-y [DOI] [PubMed] [Google Scholar]
- 3.Van Etten JL, Burbank DE, Kuczmarski D, Meints RH. 1983. Virus infection of culturable chlorella-like algae and development of a plaque assay. Science 219:994–996. 10.1126/science.219.4587.994 [DOI] [PubMed] [Google Scholar]
- 4.Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34:281–294. 10.1111/j.1574-6976.2009.00209.x [DOI] [PubMed] [Google Scholar]
- 5.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. 10.1126/science.1101485 [DOI] [PubMed] [Google Scholar]
- 6.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. U. S. A. 108:17486–17491. 10.1073/pnas.1110889108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. 2011. Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes. Genome Biol. Evol. 3:737–742. 10.1093/gbe/evr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol. Evol. 4:1324–1330. 10.1093/gbe/evs109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Etten JL, Gurnon JR, Yanai-Balser GM, Dunigan DD, Graves MV. 2010. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta 1800:152–159. 10.1016/j.bbagen.2009.07.024 [DOI] [PubMed] [Google Scholar]
- 10.Wang IN, Li Y, Que Q, Bhattacharya M, Lane LC, Chaney WG, Van Etten JL. 1993. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. U. S. A. 90:3840–3844. 10.1073/pnas.90.9.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legendre M, Santini S, Rico A, Abergel C, Claverie JM. 2011. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 8:99. 10.1186/1743-422X-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100–104. 10.1038/nature07218 [DOI] [PubMed] [Google Scholar]
- 13.Luther KB, Hulsmeier AJ, Schegg B, Deuber SA, Raoult D, Hennet T. 2011. Mimivirus collagen is modified by bifunctional lysyl hydroxylase and glycosyltransferase enzyme. J. Biol. Chem. 286:43701–43709. 10.1074/jbc.M111.309096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. 2005. Cryo-electron microscopy of the giant Mimivirus. J. Mol. Biol. 353:493–496. 10.1016/j.jmb.2005.08.060 [DOI] [PubMed] [Google Scholar]
- 15.Kuznetsov YG, Xiao C, Sun S, Raoult D, Rossmann M, McPherson A. 2010. Atomic force microscopy investigation of the giant mimivirus. Virology 404:127–137. 10.1016/j.virol.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 16.Boughalmi M, Saadi H, Pagnier I, Colson P, Fournous G, Raoult D, La Scola B. 2013. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 15:2000–2007. 10.1111/1462-2920.12068 [DOI] [PubMed] [Google Scholar]
- 17.La Scola B, Campocasso A, N′Dong R, Fournous G, Barrassi L, Flaudrops C, Raoult D. 2010. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53:344–353. 10.1159/000312919 [DOI] [PubMed] [Google Scholar]
- 18.Ghigo E, Kartenbeck J, Lien P, Pelkmans L, Capo C, Mege JL, Raoult D. 2008. Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 4:e1000087. 10.1371/journal.ppat.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, Zhao R, Puravs E, Tra J, Michael CW, Misek DE, Hanash SM. 2003. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J. Biol. Chem. 278:7607–7616. 10.1074/jbc.M210455200 [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 21.Brand DD, Latham KA, Rosloniec EF. 2007. Collagen-induced arthritis. Nat. Protoc. 2:1269–1275. 10.1038/nprot.2007.173 [DOI] [PubMed] [Google Scholar]
- 22.Schegg B, Hülsmeier AJ, Rutschmann C, Maag C, Hennet T. 2009. Core glycosylation of collagen is initiated by two β(1-O)galactosyltransferases. Mol. Cell. Biol. 29:943–952. 10.1128/MCB.02085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snir O, Backlund J, Bostrom J, Andersson I, Kihlberg J, Buckner JH, Klareskog L, Holmdahl R, Malmstrom V. 2012. Multifunctional T cell reactivity with native and glycosylated type II collagen in rheumatoid arthritis. Arthritis Rheum. 64:2482–2488. 10.1002/art.34459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backlund J, Carlsen S, Hoger T, Holm B, Fugger L, Kihlberg J, Burkhardt H, Holmdahl R. 2002. Predominant selection of T cells specific for the glycosylated collagen type II epitope (263–270) in humanized transgenic mice and in rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 99:9960–9965. 10.1073/pnas.132254199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becher B, Durell BG, Miga AV, Hickey WF, Noelle RJ. 2001. The clinical course of experimental autoimmune encephalomyelitis and inflammation is controlled by the expression of CD40 within the central nervous system. J. Exp. Med. 193:967–974. 10.1084/jem.193.8.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Scola B, Marrie TJ, Auffray JP, Raoult D. 2005. Mimivirus in pneumonia patients. Emerg. Infect. Dis. 11:449–452. 10.3201/eid1103.040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanspauwen MJ, Franssen FM, Raoult D, Wouters EF, Bruggeman CA, Linssen CF. 2012. Infections with mimivirus in patients with chronic obstructive pulmonary disease. Respir. Med. 106:1690–1694. 10.1016/j.rmed.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 28.Yuki N, Susuki K, Koga M, Nishimoto Y, Odaka M, Hirata K, Taguchi K, Miyatake T, Furukawa K, Kobata T, Yamada M. 2004. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barre syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:11404–11409. 10.1073/pnas.0402391101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier N, Raoult D, La Scola B. 2009. Specific recognition of the major capsid protein of Acanthamoeba polyphaga mimivirus by sera of patients infected by Francisella tularensis. FEMS Microbiol. Lett. 297:117–123. 10.1111/j.1574-6968.2009.01675.x [DOI] [PubMed] [Google Scholar]
- 30.Jarvinen P, Aho K. 1994. Twin studies in rheumatic diseases. Semin. Arthritis Rheum. 24:19–28. 10.1016/0049-0172(94)90096-5 [DOI] [PubMed] [Google Scholar]
- 31.Svendsen AJ, Holm NV, Kyvik K, Petersen PH, Junker P. 2002. Relative importance of genetic effects in rheumatoid arthritis: historical cohort study of Danish nationwide twin population. BMJ 324:264–266. 10.1136/bmj.324.7332.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynn AK, Yannas IV, Bonfield W. 2004. Antigenicity and immunogenicity of collagen. J. Biomed. Mater. Res. B Appl. Biomater. 71:343–354. 10.1002/jbm.b.30096 [DOI] [PubMed] [Google Scholar]
- 33.Stuart JM, Townes AS, Kang AH. 1982. Nature and specificity of the immune response to collagen in type II collagen-induced arthritis in mice. J. Clin. Invest. 69:673–683. 10.1172/JCI110495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 35.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]