Abstract
Foamy viruses (FV) are complex retroviruses that naturally infect all nonhuman primates (NHP) studied to date. Zoonotic transmission of Old World NHP simian foamy viruses (SFV) has been documented, leading to nonpathogenic persistent infections. To date, there have been no reports concerning zoonotic transmission of New World monkey (NWM) SFV to humans and resulting infection. In this study, we developed a Western blot assay to detect antibodies to NWM SFV, a nested PCR assay to detect NWM SFV DNA, and a β-galactosidase-containing indicator cell line to assay replication of NWM SFV. Using these tools, we analyzed the plasma and blood of 116 primatologists, of whom 69 had reported exposures to NWM. While 8 of the primatologists tested were seropositive for SFV from a NWM, the spider monkey, none had detectable levels of viral DNA in their blood. We found that SFV isolated from three different species of NWM replicated in some, but not all, human cell lines. From our data, we conclude that while humans exposed to NWM SFV produce antibodies, there is no evidence for long-term viral persistence.
INTRODUCTION
Foamy viruses (FV) are unusual complex retroviruses that infect cattle, horses, cats, and all species of nonhuman primates (NHP) examined to date (reviewed in reference 1). Simian foamy viruses (SFV) can cause life-long infections in natural hosts without any apparent pathogenicity (2). In cell culture models, SFV can establish latent infections in some cell types and lytic infections in others, resulting in cytopathic effects (CPE) that include syncytium formation (reviewed in reference 3). In infected macaques, FV DNA is found in almost all tissues, while FV RNA and replicating virus are limited to the superficial epithelial cells of the oral mucosa in immunocompetent animals (4, 5). Consistent with the site of viral replication detected in vivo, it is thought that FV are transmitted via saliva, through grooming, biting, and other behaviors. Studies of natural transmission suggest that infant and young NHP are resistant to infection, presumably because of passive immunity from maternal antibodies, but typically (for macaques) become infected by 3 years of age (6). Epidemiological studies indicate that SFV seroprevalence can reach up to 100% in adults (reviewed in reference 7).
Although FV do not naturally infect humans, SFV have been found to be zoonotically transmitted, most likely through contact with saliva from an infected NHP. Several studies have documented the transmission of SFV to humans who interact directly with Old World (OW) NHP, including Cercopithicus species, baboons, macaques, mandrills, gorillas, and chimpanzees (reviewed in reference 7). SFV antibody-positive humans have been found in a variety of natural settings, including people in Asia who live in areas with free-ranging macaques, villagers in Gabon with known exposure to NHP, and a population of hunters in Cameroon with bites from Old World NHP (6, 8–11). SFV antibody-positive humans have also been documented in various laboratory, veterinary, and zoo settings (12–17). While it is clear that SFV from a wide range of Old World NHP species have the ability to infect humans, little is currently known about zoonotic transmission of SFV from New World Monkey (NWM) species.
NWM are comprised of approximately 60 NHP species that live in the forests of Central and South America (18). A recent study reported phylogenetic analysis of SFV from 14 genera of NWM and found that, similar to Old World SFV, the NWM SFV coevolved with their hosts for at least 15 million years (19). Thus, NWM SFV infect their hosts and establish nonpathogenic persistent infections, similar to that seen in OW NHP. Presumably, zoonotic transmission of NWM SFV can occur through direct exposure to NWM saliva, as seen with OW SFV. Throughout North, Central, and South America, several species of NWM are kept as pets, including tamarins, marmosets, spider monkeys, and capuchins. In these domestic contexts, NWM and humans live in close contact with one another, providing opportunity for SFV zoonotic transmission. Other groups of people at higher risk for zoonotic infection with New World SFV are primatologists, laboratory researchers, and veterinarians who work directly with various species of NWM in natural, laboratory, or clinical settings (20). A previous study analyzed 187 individuals who reported occupational exposure to both Old and New World NHP species for zoonotic infection with OWM and ape SFV (12). However, the transmission of NWM SFV to humans was not examined in this group.
In this report, we describe use of Western blotting, nested PCR, and NWM SFV indicator cell assays to specifically investigate New World SFV infection in monkeys and humans. Blood and plasma from NWM and from humans with reported contact with NWM were analyzed for SFV seroreactivity and viral DNA in the blood. We confirmed that SFV infect New World monkeys and establish persistent infection, similar to Old World SFV. However, while some humans have detectable antibody to NWM SFV in their blood, we found no evidence of viral DNA or persistent infection.
MATERIALS AND METHODS
Human subject sampling.
Biological samples were collected from subjects recruited for participation in this study at the 2009 conference of the American Society of Primatologists (ASP), held in San Diego, CA, 18 to 21 September 2009. A total of 380 individuals attended the conference, and 116 volunteered to participate in the study. ASP officers made conference attendees aware of the research, and an area adjacent to meeting auditoriums was designated for research activities. Authors L. Jones-Engel and G. A. Engel were available in this area several hours a day during the conference. Participants could approach the authors and ask questions about the study. Potential subjects were informed of the possible risks and benefits of participation. Those agreeing to participate signed an informed consent form and filled out a questionnaire. Upon completion of the questionnaire, 10 ml of whole blood was collected from the subject into an EDTA tube as described previously (8) and refrigerated for no more than 6 h prior to being aliquoted and frozen on dry ice. Sociodemographic data, including occupational information describing the location, type, and duration of all employment relating to NHP, were collected and have been reported elsewhere (20). Additionally, subjects were asked to list and describe all occupational exposures to NHP and/or NHP tissues and body fluids. The human subjects' research protocols were reviewed and approved by the University of Washington Institutional Review Board (23055). Additionally, whole blood from a subject in Bangladesh (BGH 31), previously collected and reported (8), was also used in this study.
Primate biological samples.
Whole blood and plasma samples from captive squirrel monkeys with no reported contact with other NWM species were purchased from Southwest National Primate Research Center (SQM 28, 2120, 2809, and 4028). Whole blood and plasma samples from free-ranging rhesus macaques in Bangladesh were also used in this study. Animal handling, sampling, and shipping protocols for the rhesus macaques have been published elsewhere (8). Genomic DNA isolated from the blood of free-ranging squirrel monkey, spider monkey, howler monkey, and capuchin were provided by one of the authors (G.A.G.). These research protocols have been reviewed and approved by the University of Washington Institutional Animal Care and Research Committee (4233-01).
Construction of NFAB indicator cells.
To clone the squirrel monkey SFV (SFVsqm) long terminal repeat (LTR) into a β-galactosidase (β-Gal) reporter plasmid, 831 bp of the 3′-LTR of squirrel monkey SFV (nucleotides 10621 to 11151; GenBank accession number GU356394.1) was PCR amplified from pCR4-SFVsqu3′ (obtained from J. Sodroski). This fragment was digested with SmaI and PstI and inserted into pHSRV5LG (21), after removing the lab-adapted prototype foamy virus (PFV) LTR by cutting with SmaI and PstI. The resultant plasmid, p5LGSqm, contained the SFVsqm LTR, the β-Gal gene with a 5′ nuclear localization signal, and a simian virus 40 poly(A) signal. To generate the NFAB indicator cell line, BHK-21 baby hamster kidney cells were cotransfected with p5LGSqm and pCMVhgh, which contains a hygromycin resistance gene under the control of the cytomegalovirus promoter (22), at a 10:1 molar ratio. Hygromycin (Corning, NY) at 200 μg/ml was added after 24 h, and 9 resistant clones were isolated after approximately 3 weeks. These clones were tested for β-Gal expression 48 h postinfection with SFVsqm. Eight of the clones showed cells that stained blue in our assay only after infection with virus. We chose the clone that had the lowest background staining in the absence of virus to use in subsequent experiments.
Cell culture and infections.
HT1080 (human fibrosarcoma) (23), BHK-21 (baby hamster kidney fibroblast) (24), Telo-RF (telomerase-immortalized rhesus fibroblast) (25), Cf2Th (fetal canine thymus) (26), LNCaP (human prostate adenocarcinoma) (27), AGS (human stomach adenocarcinoma) (28), C33A (human cervical epithelial) (29), LoVo (human colon adenocarcinoma) (30), and MDA-MB-231 (human breast adenocarcinoma) (31) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin (10,000 units/ml) and streptomycin (10,000 μg/ml). FAB (21), SFAB (6), and NFAB cells were cultured in DMEM supplemented with 5% bovine growth serum, penicillin, streptomycin, and hygromycin (200 μg/ml). Cells were infected with FV stocks, including the lab-adapted prototype foamy virus (PFV), which was isolated from a human zoonotically infected with SFV from a chimpanzee (32), the lab-adapted SFV isolated from Macaca mulatta (SFVmac) (33), an SFV isolated from an infected squirrel monkey (SFVsqm), an SFV isolated from an infected spider monkey (SFVspm), or an SFV isolated from a marmoset (SFVmar). SFVspm was obtained from W. Switzer (Centers for Disease Control and Prevention), SFVmar was obtained from J. Sodroski (Dana Farber Cancer Institute), and SFVsqm was obtained from the ATCC (VR-843). Cultures were monitored for cytopathic effects, such as formation of vacuoles and syncytium formation, and harvested at approximately 90% infection, as previously described (34). Cell lysates for Western blot analysis were generated by removing the medium, rinsing cells in cold phosphate-buffered saline (PBS), scraping cells from the plates in 1× SDS sample buffer (12.5% 50 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.01% bromophenol blue), and processing lysates with Qiashredder (Qiagen) in accordance with the manufacturer's protocol. To generate virus stocks, cells were scraped from plates, and the cells and supernatants were collected and subjected to three rounds of freezing in liquid nitrogen and thawing at 37°C, followed by low-speed centrifugation.
Western blot analysis.
Cell lysates from uninfected and SFV-infected BHK-21, HT1080, and Telo-RF cells were prepared, and the proteins were separated by using 10% SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride membranes (Millipore) and incubated with blocking buffer (5% dry milk powder and 0.05% Tween 20 in PBS) and antibodies as previously described (35). Human plasma samples were diluted 1:10, squirrel monkey and macaque plasma samples were diluted 1:200, and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Sigma-Aldrich, St. Louis, MO) was diluted 1:3,000 in blocking buffer. These diluted plasma and antibodies served as primary antibodies in Western blot analyses. A goat monoclonal anti-human IgG (Rockland Immunochemicals, Gilbertsville, PA), goat anti-monkey IgG (Rockland Immunochemicals), and goat monoclonal anti-mouse IgG (Rockland Immunochemicals) were used at a 1:3,000 dilution as secondary antibodies. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Thermo Fisher, Waltham, MA) was used to visualize proteins in accordance with the manufacturer's protocol.
Nested PCR and real-time PCR.
Genomic DNA (gDNA) was extracted from whole blood by using the QIAamp DNA blood minikit (Qiagen, Valencia, CA). Nested PCR for the detection of OWM SFV DNA was performed using pol primers as previously described (36). A nested NWM PCR protocol was used to amplify a first-round 978-bp region and a second-round 403-bp region of New World SFV pol. The first round of PCR used the outer forward Primer 1 (SFVsqm GenBank GU356394 positions 5077 to 5101; 5′-TAAAAATCCAAGCCTGGATGCAGAG-3′) and outer reverse Primer 2 (SFVsqm positions 6035 to 6054; 5′-ACCCTCTCCTGGACAAGAAG-3′). A 200-ng aliquot of template gDNA was amplified for each sample at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 52°C for 1 min, and 72°C for 30 s, with extension at 72°C for 7 min. The second round of the nested PCR used inner forward Primer 3 (SFVsqm positions 5307 to 5325; 5′-ACTGGTGGCCAAACATGAG-3′) and inner reverse Primer 4 (SFVsqm positions 5683 to 5709; 5′-GGGTGGTAAGGAGTACTGAATTCCAAT-3′). A 2-μl aliquot of the first-round product was amplified at 95°C for 3 min, followed by 40 cycles at 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s, and then extension at 72°C for 7 min. Various quantities of pSFVsqu3′, an SFVsqm plasmid containing bp 5083 to 11484 (obtained from J. Sodroski), were used as the positive control. Second-round PCR products were separated on 1% agarose gels and visualized by ethidium bromide staining. PCR of gDNA was also performed to detect the gapdh gene as an internal control for DNA quality, as described above (GAPDH forward primer, 5′-CTACTGGCGCTGCCAAGGCTGT-3′; GAPDH reverse primer, 5′-GCCATGAGGTCCACCACCCTGT-3′).
First-round template DNA was also used for second-round real-time PCR with the 7900HT real-time PCR system (Applied Biosystems). Second-round real-time PCR was performed using the NWM SFV pol inner forward Primer 3 and inner reverse Primer 4 at 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s. Amplification was detected by using a TaqMan 6-carboxyltetramethylrhodamine (TAMRA) probe (SFVsqm positions 5613 to 5637; 5′-TGCATTCCGATCAAGGATCAGCATT-3′).
Quantitative PCR was also performed on gDNA from whole blood of squirrel monkeys, macaques, and humans to measure levels of viral and cellular genes, by using the iTaq universal probes supermix (Bio-Rad). Samples were analyzed using the 7900 real-time PCR system (Applied Biosystems). The SFV gag primers used for Old World monkeys and human samples were the following: gag1759F, 5′ACGAACATCTGGTGCGGG; gag1835R, 5′CTGCGTTTCCACCAGCTGA. The probe was gag1789, 6-carboxyfluorescein (FAM)–AGGAAGAGGGAACCAAAACCGAAACCA–TAMRA. The primers used for squirrel monkey samples were the NWM SFV pol inner forward and inner reverse primers described above for the second-round nested PCR. A c-myc cellular DNA quantitative PCR (qPCR) was performed for all samples in each assay to normalize for cell equivalents. The primers for c-myc were as follows: c-mycF1, 5′ GCCCCTCAACGTTAGCTTCA; c-mycR1, CGCATGAGAAATACGGCTGCA; probe, FAM-CAACAGGAACTATGACCTCGACTACGACTCG-TAMRA. For all qPCRs, the conditions were as follows: 95°C for 3 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. The plasmid pSFV-1 containing the gag gene from M. mulatta, the pSFVsqu3′ plasmid described above, and Telo-RF DNA was used to generate standard curves in assays for macaque, squirrel monkey, and human samples. Each assay also included a nontemplate control and an SFV-negative control (Telo-RF genomic DNA).
RESULTS
Identification of SFV-seropositive New World monkeys.
Monkey plasma samples were analyzed for antibodies that reacted with the major SFV structural protein, Gag. In all SFV, the Gag protein migrates as a doublet at approximately 70 kDa. SFV isolated from a spider monkey (SFVspm) was used to infect HT1080 (human fibrosarcoma) cells to generate Gag antigen for use in Western blot analyses. The SFVspm Gag protein in our studies migrated as a doublet at 68/70 kDa. Plasma samples from four captive squirrel monkeys with no known exposure to spider monkeys were tested against uninfected and SFV-infected cell lysates. On Western blots, plasma diluted 1:200 from each monkey detected the Gag doublet in SFVspm-infected cells but not in cells infected with SFVmac from M. mulatta or uninfected cell lysates (Fig. 1A). Thus, SFVsqm antibodies cross-react with SFVspm Gag but not Old World SFVmac Gag.
FIG 1.
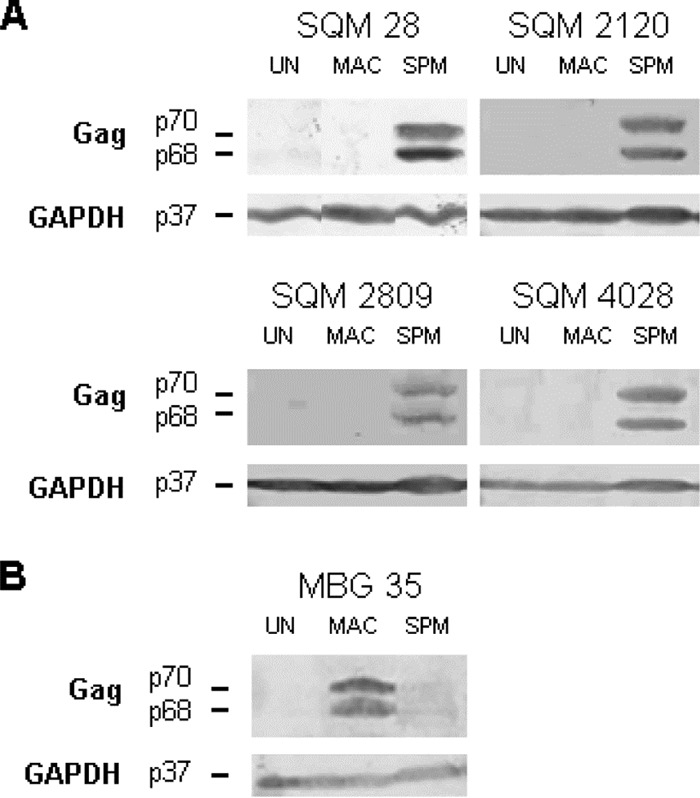
Reactivity of Old and New World monkey plasma to SFVspm and SFVmac. Cell lysates were generated from uninfected HT1080 cells (UN), cells infected with SFVmac (MAC), or cells infected with SFVspm (SPM). (A) Western blot assays were performed using plasma (1:200) from four different squirrel monkeys (SQM 28, 2120, 2809, and 4028) and an anti-GAPDH antibody (1:3,000). (B) Western blot assays were performed using plasma (1:200) from a macaque known to be infected with SFVmac (MBG 35); an anti-GAPDH antibody was also used.
To determine whether the reverse is also true, plasma diluted 1:200 from an SFV-infected Bangladeshi macaque (MBG 35) was tested against SFVspm Gag in infected cell lysates. The antibodies from the infected macaque showed some cross-reactivity to SFVspm Gag; however, it appeared to be weaker than the level of reactivity seen with the SFVmac Gag (Fig. 1B) (8). Although the level of cross-reactivity has not been formally quantitated, analyses of the identical SFVspm-infected lysates with MBG 35 plasma (Fig. 1B) showed less reactivity than was seen with the NWM plasma at the same dilution (Fig. 1A).
Detection of SFV DNA in blood cells from New World monkeys.
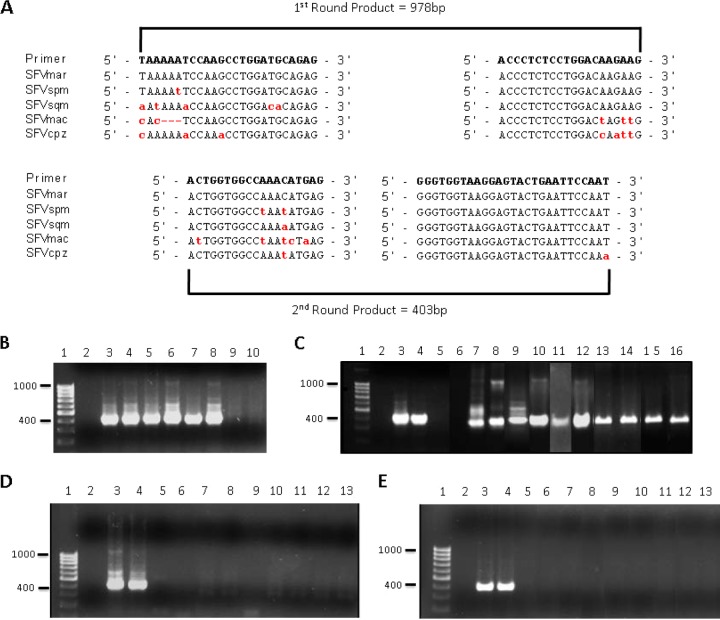
The sequences of SFVspm (GenBank EU010385), SFVsqm (GenBank GU356394), and SFV from a marmoset (SFVmar; GenBank GU356395) have been reported (37, 38). In order to design PCR primers that had the potential to detect NWM SFV from as many different species of monkeys as possible, a nucleotide alignment of the three available NWM SFV was performed. This alignment was used to design a set of nested PCR primers specific to highly conserved sequences within the integrase region of the pol gene (Fig. 2A). A plasmid containing the SFVsqm pol gene, pCR4-SFVsqu3′, was used to test the pol primer pairs and optimize the conditions of our nested PCR assay. When plasmid DNA was added to genomic DNA isolated from uninfected cells, our assay was able to detect as little as one copy of the SFVsqm pol gene (Fig. 2B, lanes 7 and 8). In the absence of genomic DNA, our assay could reliably detect 5 copies of the SFVsqm pol gene per PCR run (Fig. 2C, lane 4, and data not shown). We also found that the NWM SFV primers were able to detect SFVsqm, SFVspm, and SFVmar pol in genomic DNA from infected Cf2Th (fetal canine thymus) cells (Fig. 2B, lanes 7 to 9).
FIG 2.
Nested PCR detection of SFV DNA in New World monkey and human blood. (A) A set of nested PCR primers were designed to specifically amplify a region of the SFV pol gene from NWM. Alignment of available New World SFV sequences from marmoset, spider, and squirrel monkeys allowed the design of primers in highly conserved regions. The red, lowercase letters indicate nonconsensus nucleotides. The nested PCR protocol amplified a 978-bp DNA fragment in the first round of PCR and a 403-bp DNA fragment in the second round. (B to E) Nested PCRs were performed, and second-round PCR products were visualized using gel electrophoresis and ethidium bromide staining. (B) Sensitivity of the NWM Pol primers was assessed by using known amounts of the SFVspm pol gene in the plasmid pSFVsqu3′ added to 200 ng of genomic DNA isolated from uninfected HT1080 cells. Samples included a DNA molecular size marker (lane 1), a no-DNA control (lane 2), and 10 (lanes 3 and 4), 5 (lanes 5 and 6), 1 (lanes 7 and 8), or no (lanes 9 and 10) copy equivalents of SFVsqu3′ mixed with 200 ng of HT1080 genomic DNA. (C) The ability of the NWM pol primers to detect SFV DNA extracted from infected cells in culture and from blood of New World monkeys was examined. Samples included a DNA molecular size marker (lane 1); no-template control (lane 2); 10 (lane 3), 5 (lane 4), or 1 (lane 5) copy equivalents of the plasmid pSFVsqu3′; 200 ng of genomic DNA extracted from uninfected (lane 6); SFVsqu-infected (lane 7); SFVspm-infected (lane 8) or SFVmar-infected (lane 9) Cf2Th cells; 200 ng of genomic DNA extracted from the whole blood of captive squirrel monkeys 2120 (lane 10), 2809 (lane 11), and 4028 (lane 12) and wild monkeys from Costa Rica: Howler monkey AP170 (lane 13) and AP171 (lane 14), capuchin CC74 (lane 15), and squirrel monkey SO39 (lane 16). (D and E) The presence of New and Old World SFV DNA in the blood of antibody-positive APH individuals was determined. (D) Samples included a DNA molecular size marker (lane 1), a no-template control (lane 2), or 10 (lane 3), 5 (lane 4), or 1 (lane 5) copy equivalents of the plasmid pSFVsqu3′. (E) Additional samples for pSFV-1 included 200 ng of genomic DNA extracted from whole blood of APH 3 (lane 6), APH 10 (lane 7), APH 25 (lane 8), APH 48 (lane 9), APH 71 (lane 10), APH 75 (lane 11), APH 84 (lane 12), and APH 111 (lane 13). NWM SFV pol primers were used in panel D; previously described primers specific to OW SFV pol (55) were used in panel E.
A panel of samples collected from various NWM from both captive and wild environments was used to determine whether our primers were able to detect New World SFV viral DNA in blood cells from diverse host species. Genomic DNA was extracted from these whole-blood samples and analyzed using our nested PCR protocol. SFV sequences were successfully amplified from all NWM species tested (squirrel monkey, spider monkey, howler monkey, and capuchin) (Fig. 2B, lanes 10 to 16). The PCR amplicons from each reaction mixture were isolated, cloned, and sequenced to confirm the specificity of our primers. In each case, sequences showed between 76 and 85% identity with previously published NWM SFV sequences (data not shown). Additionally, the New World SFV pol primers were tested by using a control plasmid containing the pol gene from SFV-1, a molecular clone derived from M. mulatta (33). While Old World SFV pol primers amplified this sequence readily, the New World SFV pol primers failed to amplify it (data not shown). Taken together, these results show that the New World SFV pol primers are sensitive, specific to NWM, and can amplify sequences from a broad range of NWM species.
Identification of primatologists seropositive for New World SFV.
In September 2009, blood samples were collected from 116 attendees at the meeting of the American Society of Primatologists (20). Study participants reported contacts with multiple NHP species, including Old and New World NHP, in the context of careers that spanned decades and work that took place on multiple continents and in multiple contexts (field research, laboratory work, zoos). Of the 116 primatologists studied, 12 interacted exclusively with NWM, 47 interacted exclusively with OW NHP, and 57 interacted with both Old and New World NHP. This unique cohort had a particularly high level of reported exposure to NWM, which allowed us to investigate transmission of NWM SFV to humans.
The plasma samples from the 116 primatologists and control plasmas from a nonexposed human and a Bangladeshi human infected with SFVmac (NHS and BGH63, respectively) were first screened by Western blotting for seroreactivity to New World SFV by using lysates from either Telo-RF (rhesus monkey fibroblast) or HT1080 cells that were either uninfected or infected with SFVspm. After repeated Western blot analyses, eight individuals were confirmed seroreactive to SFVspm Gag, showing the characteristic doublet seen with the macaque and squirrel monkey plasma (Fig. 3). Although the human plasma samples showed a higher overall background signal than seen with the monkey plasma, the Gag doublet was still visible. The eight NWM SFV-seropositive samples were also analyzed for seroreactivity to SFV from M. mulatta, and none of them was reactive (Fig. 3). This represents the first report of any humans seropositive for NWM SFV, with prevalence rates of 6.9% in this cohort and 11.6% for individuals with exposure to NWM.
FIG 3.
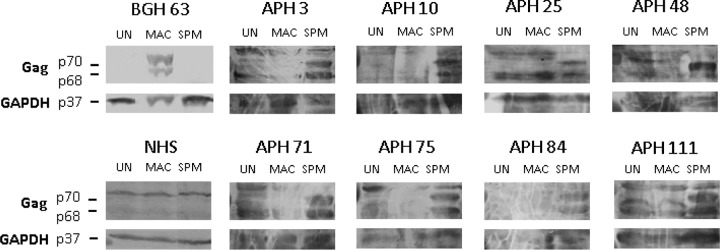
Reactivity of sera from humans occupationally exposed to SFVspm and SFVmac. Cell lysates were generated from uninfected HT1080 cells (UN), cells infected with SFVmac (MAC), or cells infected with SFVspm (SPM). Western blot assays were performed using plasma (1:20) collected from primatologists (APH) with documented exposures to NWM; an anti-GAPDH antibody (1:3,000) was also used. Representative Western blots are shown for the 8 of the 116 APH samples that were seropositive (APH 3, 10, 25, 48, 71, 75, 84, and 111) in addition to a Western blot showing results with plasma (1:20) from an individual with no known exposures to nonhuman primates (NHS) and an individual from Bangladesh known to be infected with SFVmac only (BGH 63).
In order to further characterize the types of exposures that the seropositive primatologists had had to NWM, the questionnaire data are reviewed and summarized in Table 1. All 8 of the NWM SFV-seropositive individuals reported some contact with NWM. However, only 4 of the 8 reported direct bites, scratches, or needle sticks involving NWM species. The remaining 4 individuals reported contact with NHP body fluids but not parenteral exposure, suggesting parenteral contact may not be necessary for zoonotic transmission. While all 8 seropositive primatologists had antibodies that reacted with SFVspm, only 2 of the 8 reported contact with spider monkeys. Seropositive individuals reported contact with 7 additional NWM genera. This result suggests that antibodies to different NWM SFV may be cross-reactive and is consistent with our observation that plasma from infected squirrel monkeys reacted with antigen from a spider monkey (Fig. 1). Despite 4 of the 8 seropositive primatologists reporting contact with 6 different OW genera, including Macaca, none of them was seroreactive to SFVmac Gag.
TABLE 1.
Summary of seropositive primatologist nonhuman primate exposures
| Subject ID | NWM bite/scratch/needle stick | Other NWM contacta | OWM contactb |
|---|---|---|---|
| APH 3 | + | − | + |
| APH 10 | + | − | − |
| APH 25 | − | + | + |
| APH 48 | − | + | + |
| APH 71 | − | + | + |
| APH 75 | + | − | − |
| APH 84 | + | − | − |
| APH 111 | − | + | − |
Includes handling NW monkeys or contact with their urine/feces.
For the purposes of this table, “OWM” includes Old World monkeys and apes.
New World SFV DNA is undetectable in seropositive primatologists.
Genomic DNA extracted from whole-blood samples of the 8 seropositive humans was analyzed by nested PCR with our NWM-specific pol primers and OWM-specific pol primers in order to confirm SFV infection and determine viral sequences. All PCR experiments included a negative control (no genomic DNA) and a positive control (SFVsqu or SFVmac plasmid at 10, 5, or 1 copies per reaction mixture). None of the samples was positive for OWM SFV DNA (Fig. 2E), consistent with the observed lack of seroreactivity of these individuals to SFVmac (Fig. 3). Surprisingly, after screening between 5.5 × 105 and 5.1 × 106 nucleated blood cells/person, none of the samples was found to be positive for NWM SFV by PCR (Fig. 2D). Given that our PCR assay is sensitive, our controls performed as expected, and the quality of the extracted DNA was verified by PCR amplification of the cellular gapdh gene (data not shown), these results suggest that, unlike most Old World SFV-seropositive individuals (39), New World SFV-seropositive individuals have undetectable levels of SFV DNA in their blood.
Old World and New World monkeys have similar amounts of SFV DNA in blood cells.
Given the absence of detectable persistent SFV infection in primatologists exposed to NWM, we wanted to compare the levels of viral DNA present in monkeys infected with New World SFV to the levels in monkeys and humans infected with SFVmac. We did not have the necessary buccal swab samples to compare levels of replicating SFV found in the oral mucosa. We did have access to whole-blood samples for both the NWM SFV- and OWM SFV-seropositive humans and monkeys, which allowed us to measure and compare the levels of viral DNA in blood cells. Real-time PCR was performed with the genomic DNA from blood cells of either infected humans or monkeys and with primers specific to either NWM SFV or SFVmac. Four macaques from Bangladesh (MBG) were analyzed using primers specific to the Old World SFV gag gene and their data were normalized to results for the cellular c-myc gene. We found a range of 7.6 × 10−5 to 9.0 × 10−4 FV DNA copies/cell equivalent (Table 2). Similar analyses of two captive squirrel monkeys, based on primers specific to the New World SFV pol gene and c-myc, revealed 2.2 × 10−3 and 1.6 × 10−4 FV DNA copies/cell equivalent. This indicated that the level of SFV DNA in the blood of infected New World monkeys is similar to or higher than levels seen in Old World monkeys, based on this limited sampling.
TABLE 2.
Quantitation of FV DNA in SFV-infected monkeys and humans
| DNA source | FV DNA (no. of copies/cell equivalent) |
|---|---|
| Old World monkeys | |
| MBG 205 | 6.0 × 10−4 |
| MBG 228 | 9.0 × 10−4 |
| MBG 232 | 7.6 × 10−5 |
| MBG 241 | 9.0 × 10−4 |
| New World monkeys | |
| SQM 2120 | 2.2 × 10−3 |
| SQM 4028 | 1.6 × 10−4 |
| Human infected with Old World SFV | |
| BGH 31 | 9.1 × 10−5 |
| Humans seropositive for New World SFV | |
| APH 3 | <1 × 10−6 |
| APH 10 | <1 × 10−6 |
| APH 25 | <1 × 10−6 |
| APH 48 | <1 × 10−6 |
| APH 71 | <1 × 10−6 |
| APH 75 | <1 × 10−6 |
| APH 84 | <1 × 10−6 |
| APH 111 | <1 × 10−6 |
SFV infections have been frequently detected by nested PCR in a group of humans exposed to macaques in Bangladesh (39). For example, our previous work monitoring zoonotic transmission of SFV from macaques to humans in Bangladesh found that approximately 61% of seropositive humans are also PCR positive (8). Quantitative real-time PCR analysis of genomic DNA extracted from a human known to be infected with SFVmac (BGH 31) found that the FV DNA copy number was 9.1 × 10−5 per cell equivalent, approximately 5 times lower than that seen in the infected macaques (Table 2). In contrast, the seropositive primatologists (APH) did not have detectable levels of NWM SFV DNA when analyzed by quantitative PCR, consistent with our earlier nested PCR findings.
Generation of a New World SFV-specific indicator cell line.
Information about NWM SFV replication is limited to a single published study, which describes replication of these viruses in Cf2Th cells (38). Given the lack of detectable viral DNA in the seropositive primatologists, we wanted to determine if New World SFV has the ability to replicate efficiently in human cells. In order to quantitate NWM SFV, we created an indicator cell line called NFAB (New World monkey FAB), based on the previously described FAB and SFAB cells used for assays of PFV and SFVmac, respectively (6, 21). NFAB cells contain a copy of the β-Gal gene containing a nuclear localization signal, driven by the SFVsqm LTR. Foamy viruses encode the LTR transcriptional activator protein Tas, which is expressed from an internal promoter and is absolutely required for viral transcription (40). Infection of NFAB cells results in expression of Tas and activation of the LTR promoter which drives the expression of both viral genes and the β-Gal reporter gene. Individually infected cells can be visualized and counted by staining the cells with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Creation of the NFAB cells is described in detail in Materials and Methods.
Infection of the NFAB cells with SFVsqm transactivated β-Gal expression and produced an intense blue staining of the infected cells in our assay, while no β-Gal activity was observed in uninfected NFAB cells (Fig. 4). We also tested the ability of SFVspm and SFVmar to transactivate the β-Gal gene in NFAB cells. SFVspm-infected cells showed blue staining that was slightly less intense than the color seen in the SFVsqm-infected NFAB cells (Fig. 4). However, SFVmar-infected NFAB cells showed very weak blue staining that was variable in intensity. These observations suggest that the SFVsqm LTR is compatible with its own Tas protein as well as the SFVspm Tas, but it is less compatible with the SFVmar Tas. Therefore, NFAB cells can be used to detect these three NWM SFV, but they can only be used to reliably determine titers for SFVsqm and SFVspm virus stocks. We have not tested SFV from any other NWM species.
FIG 4.
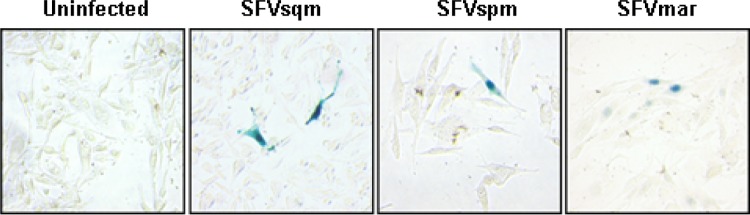
Visualization of New World SFV-infected NFAB cells. The newly described NFAB indicator cells were infected with SFVsqm, SFVspm, SFVmar, or medium containing no virus. At 48 h postinfection, cells were fixed, incubated in X-Gal staining solution, and visualized at 200× magnification with an inverted microscope.
Specificity of FAB, SFAB, and NFAB indicator cells.
The specificities of NFAB, FAB, and SFAB indicator cell lines for both New World (SFVsqm, SFVspm, and SFVmar) and Old World SFV (PFV, a lab-adapted human isolate from a chimpanzee, and SFVmac, a lab-adapted virus from M. mulatta) were examined. PFV and SFVmac were only able to transactivate the reporter gene being driven by their homologous LTRs in FAB and SFAB cells, respectively (Table 3). Similarly, the three NWM SFV exclusively transactivated the reporter gene driven by the SFVsqm LTR in NFAB cells but did not activate β-Gal expression in FAB or SFAB cells (Table 3). Thus, Old World monkey, Old World ape, and New World monkey Tas proteins only activate their respective LTRs.
TABLE 3.
Selective activation of β-Gal in FAB indicator cell lines by Old and New World SFV
| Virus | β-Gal activitya in indicator cell line |
||
|---|---|---|---|
| FAB | SFAB | NFAB | |
| None | − | − | − |
| PFV-1 | +++* | − | − |
| SFVmac | − | +++ | − |
| SFVsqm | − | − | +++ |
| SFVspm | − | − | ++ |
| SFVmar | − | − | +/− |
*, the number of + signs indicates the intensity of the staining visualized.
New World SFV replicate in human cell lines.
Given the lack of detectable provirus in the blood of the NWM-seropositive primatologists, we next asked whether NWM SFV can replicate in human cell lines. In addition to the HT1080 and BHK cell lines, the growth of NWM SFV, SFVmac, and PFV was compared in five different human cell lines, at a multiplicity of infection of 0.02, based on titers obtained from our BHK-21-derived indicator cells The cell lines tested were LNCaP (human prostate adenocarcinoma), AGS (human stomach adenocarcinoma), C33A (human cervical epithelial), LoVo (human colon adenocarcinoma), and MDA-MB-231 (human breast adenocarcinoma). Supernatants were collected 5 days postinfection, and titers were determined by using the appropriate FAB cells. Unexpectedly, we found that while PFV and SFVmac were able to replicate in HT1080 cells, they were each only able to replicate to detectable levels in one of the additional human cell lines; PFV replicated in MDA cells and SFVmac replicated in AGS cells (Table 4). SFVspm showed the broadest tropism, with replication in 3 of the 6 human cell lines tested, while SFVsqm showed replication in 1 of the 6. For each virus, the highest titers were observed in the BHK and HT1080 cell lines. These results demonstrated that SFV from different NWM species do have the ability to replicate in some, but not all, human cell lines and suggest that the absence of detectable persistent infection after zoonotic transmission of NWM SFV, as measured by antibody production, is not caused by a general block to replication in human cells.
TABLE 4.
NWM SFV replication in human cell lines
| Virus | Viral titer in indicated cell line |
||||||
|---|---|---|---|---|---|---|---|
| BHK | HT1080 | MDA | AGS | C33A | LN | LoVo | |
| PFV | 2.5 × 106 | 3.7 × 105 | 6.6 × 102 | <1 | <1 | <1 | <1 |
| MAC | 1.4 × 106 | 2.6 × 104 | <1 | 2.0 × 102 | <1 | <1 | <1 |
| SPM | 1.6 × 106 | 2.2 × 104 | 1.7 × 103 | <1 | 8.0 × 101 | <1 | <1 |
| SQM | 2.6 × 106 | 5.2 × 102 | <1 | <1 | <1 | <1 | <1 |
DISCUSSION
SFV can be zoonotically transmitted to humans (reviewed in reference 7), but prior to this study, all reported human infections were from Old World monkey and ape SFV. The presumed route of transmission is from infectious virions or infected cells in NHP saliva to human blood (10, 41). It is not well understood how SFV establishes infection after transmission, but a current model involves SFV entering blood cells where a latent infection is established. These infected blood cells can then transit throughout the body, ultimately reaching the oral mucosa, where viral replication is known to occur in natural hosts. During this process, it is likely that antibodies are generated against SFV, but it is not clear if viral replication is required for immune recognition and antibody production. Here we have described the first humans with seroreactivity to New World SFV. One-half of the NWM SFV-seropositive individuals (4/8) did not report exposures to NWM involving a break in their skin. Previous studies monitoring exposures and transmission of Old World SFV also described infected individuals with no reported direct parenteral exposure to Old World species (6, 12). Reliable recollection of historical parenteral exposures can be a complicating factor, particularly for the APH cohort, which had had decades of contact with NHP (20). However, all of our NWM SFV-infected humans reported at least some contact with NWM and/or their feces or urine. As in previous studies with Old World SFV, the portal of entry is not clear for these cases (2).
Despite the detection of New World SFV-seropositive individuals in our study, we could find no evidence of viral DNA in human blood. While it is thought that latent infections are established in the blood, it remains possible that NWM SFV in humans may persist in cells other than blood cells. This is in contrast to most previously characterized OW SFV infections, which did show established latent infections in the blood, and our findings suggest that zoonotic transmission of New World SFV may be restricted or limited by some mechanism yet to be defined.
One possible mechanism of New World SFV restriction in humans could involve the host retroviral restriction factor TRIM5-α. A previous study looking at the ability of TRIM5-α to block the replication of NWM SFV found that the TRIM5-α protein from the same host species as the virus did not block replication of that virus (38). However, the TRIM5-α proteins from other primate species were able to block the replication of NWM SFV to varying extents. In this study, human TRIM5-α was found to significantly reduce SFVsqm replication in Cf2Th canine cells but had little effect on SFVmar or SFVspm replication. Another study found that TRIM5-α proteins from a variety of NWM species inhibit replication of SFV from Old World NHP (42). Clearly, there is precedence for the idea of TRIM5 host restriction factors preventing replication, and possibly transmission, of retroviruses between different NWM species and between Old and New World NHP species. Although we don't know the levels of viral replication in NWM or the human host, it is possible that a host restriction factor, such as human TRIM5-α, blocks NWM SFV replication in the context of zoonotic transmission. It will be interesting to determine whether TRIM5-α is expressed in the human cell lines that did not support NWM SFV replication.
A role for host factors of the APOBEC3 family of cytidine deaminases in the restriction of New World SFV in humans is another intriguing possibility. Much work has shown that APOBEC3 activities lead to mutations in the genomes of orthoretroviruses (reviewed in reference 43). While an initial study found no effect of the nonstructural SFV protein Bet on APOBEC3 (44), subsequent studies demonstrated that Bet can counteract APOBEC3 activity, similar to lentiviral Vif (45, 46, 47, 48). Recent work analyzing the SFV sequences from macaques and infected humans found evidence of hypermutation in both, although much stronger in humans, and suggested that APOBEC3 may be important within the human host to limit Old World SFV replication (F. A. Matsen IV, C. T. Small, K. Soliven, G. A. Engel, M. M. Feeroz, X. Wang, K. L. Craig, M. Kamrul Hasan, M. Emerman, M. L. Linial, and L. Jones-Engel, submitted for publication). However, another recent study found that viral genetic changes did not necessarily account for FV restriction in humans (49). It remains possible that human APOBEC3 activity against New World SFV is particularly potent, or that New World SFV are unable to counteract human APOBEC3 activity, creating an interspecies barrier to establishment of persistent latent infections in humans.
Virus-host cospeciation appears to be a common characteristic of SFV evolution (50). An endogenous SFV has been reported in the prosimian aye-aye genome (51), suggesting an ancient evolution of FV in simian species for at least 85 million years. Recent work comparing sequences of NWM SFV from 14 different genera supported the coevolution of NWM SFV and their hosts and estimated that NWM SFV are at least 15 million years old (19). SFV from OW monkeys and apes are known to infect humans, but these species are more closely related to humans than NWM species. Given the strong coevolutionary history of SFV with their hosts, it is possible that humans are too divergent from NWM species to support the replication of SFV from these animals. A study examining zoonotic transmission of feline foamy virus (FFV) in a cohort of exposed veterinarians found no evidence of zoonotic infection (52), despite the ability of FFV to replicate in human cell lines. However, unlike FFV, we found serological evidence of NWM SFV infection in humans in the absence of detectable levels of viral DNA in the blood. It remains to be determined if NWM SFV infections in humans are cleared prior to the establishment of latent proviruses or whether these viruses establish latency in cells other than blood cells.
Our initial characterization of NWM SFV replication in an established human fibrosarcoma cell line found the typical foamy virus replication, leading to CPE with formation of syncytia. This is similar to what is seen for Old World SFV in human cells and suggests that the lack of New World SFV latent infection in humans is not caused by an intrinsic inability of these viruses to infect human cells. Interestingly, when we tested several other human cell lines we found that all FV tested showed varying abilities to replicate, and in some instances they were unable to replicate at all. Given that the proteoglycan heparin sulfate is known to be an important factor for FV attachment and entry, it is presumed that almost all human cells are permissive for FV entry (53). Our results provide indirect evidence that there are likely host factors involved in either supporting or inhibiting FV replication in humans.
In conclusion, we have established several assays specific to NWM SFV that identify seroreactivity, detect viral DNA, and quantitate replication of these viruses. By using these tools, a group of primatologists with documented exposure histories were analyzed for infection with NWM SFV. We have described the first evidence for transmission of New World SFV to humans and demonstrated that these individuals were antibody positive for SFVspm and negative for SFVmac. Despite being seropositive for SFVspm, none of the individuals had detectable levels of NWM SFV viral DNA in the blood. This differs from infections with Old World NHP SFV, which have been shown to frequently cause persistent latent infections in humans. Similar levels of viral DNA were present in the blood of both Old World and New World infected monkeys. It would be of interest to measure the level of SFV RNA in buccal swabs of NWM, to see if there is as much presumptive viral replication as in OW (macaque) NHP buccal swabs (54). It remains to be seen if the human immune system is able to specifically restrict NWM SFV in certain cell types.
ACKNOWLEDGMENTS
We thank J. Sodroski for sharing the SFVmar virus stock and DNA fragments of SFVsqu, SFVspm, and SFVmar, W. Switzer for sharing the SFVspm virus stock, N. Salama and D. Galloway for sharing human cell lines, and Eun-Gyung Lee for helpful discussions. We also sincerely thank the 116 primatologists who volunteered to participate in this study.
This work was supported by NIH NIAID grants R21AI097832 and R01 AI078229, NIH NCI grant CA18282, and M.J. Murdock Charitable Trust grant 2010183:JVZ2/24/2011.
We declare no conflicts of interest.
Footnotes
Published ahead of print 6 November 2013
REFERENCES
- 1.Meiering CD, Linial ML. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165–176. 10.1128/CMR.14.1.165-176.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boneva RS, Switzer WM, Spira TJ, Bhullar VB, Shanmugam V, Cong Me Lam L, Heneine W, Folks TM, Chapman LE. 2007. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res. Hum. Retroviruses 23:1330–1337. 10.1089/aid.2007.0104 [DOI] [PubMed] [Google Scholar]
- 3.Linial ML. 2007. Foamy viruses, p 2245–2263 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 4.Murray SM, Picker LJ, Axthelm MK, Linial ML. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 80:663–670. 10.1128/JVI.80.2.663-670.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray SM, Picker LJ, Axthelm MK, Hudkins K, Alpers CE, Linial ML. 2008. Replication in a superficial epithelial cell niche explains the lack of pathogenicity of primate foamy virus infections. J. Virol. 82:5981–5985. 10.1128/JVI.00367-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones-Engel L, Steinkraus KA, Murray SM, Engel GA, Grant R, Aggimarangsee N, Lee BPY-H, May C, Schillaci MA, Somgird C, Sutthipat T, Vojtech L, Zhao J-Y, Linial ML. 2007. Sensitive assays for simian foamy viruses reveal a high prevalence of infection in commensal, free-ranging, Asian monkeys. J. Virol. 81:7330–7337. 10.1128/JVI.00343-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gessain A, Rua R, Betsem E, Turpin J, Mahieux R. 2013. HTLV-3/4 and simian foamy retroviruses in humans: discovery, epidemiology, cross-species transmission and molecular virology. Virology 435:187–199. 10.1016/j.virol.2012.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feeroz M, Soliven K, Small C, Engel G, Pacheco M, Yee J, Wang X, Hasan K, Oh G, Levine K, Alam S, Craig K, Jackson D, Lee E-G, Barry P, Lerche N, Escalante A, Matsen F, Linial M, Jones-Engel L. 2013. Population dynamics of rhesus macaques and associated foamy virus in Bangladesh. Emerg. Microbes Infect. 2:e29. 10.1038/emi.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calattini S, Betsem EB, Froment A, Mauclere P, Tortevoye P, Schmitt C, Njouom R, Saib A, Gessain A. 2007. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg. Infect. Dis. 13:1314–1320. 10.3201/eid1309.061162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouinga-Ondeme A, Caron M, Nkogh D, Telfer P, Marx P, Saib A, Leroy E, Gonzalez JP, Gessain A, Kazanji M. 2012. Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J. Virol. 86:1255–1260. 10.1128/JVI.06016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7(10):e1002306. 10.1037/journal.ppat.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780–2789. 10.1128/JVI.78.6.2780-2789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, Khan AS, Lerche NW, Schweizer M, Neumann-Haefelin D, Chapman LE, Folks TM. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403–407 [DOI] [PubMed] [Google Scholar]
- 14.Schweizer M, Falcone V, Gänge J, Turek R, Neumann-Haefelin D. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandstrom PA, Phan KO, Switzer WM, Fredeking T, Chapman L, Heneine W, Folks TM. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551–552. 10.1016/S0140-6736(99)05292-7 [DOI] [PubMed] [Google Scholar]
- 16.Brooks JI, Rud EW, Pilon RG, Smith JM, Switzer WM, Sandstrom PA. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387–388. 10.1016/S0140-6736(02)09597-1 [DOI] [PubMed] [Google Scholar]
- 17.Huang F, Wang H, Jing S, Zeng W. 2012. Simian foamy virus prevalence in Macaca mulatta and zookeepers. AIDS Res. Hum. Retroviruses 28:591–593. 10.1089/aid.2011.0305 [DOI] [PubMed] [Google Scholar]
- 18.Hershkovitz P. 1977. Living New World monkeys (Platyrrhini). University of Chicago Press, Chicago, IL [Google Scholar]
- 19.Muniz CP, Troncoso LL, Moreira MA, Soares EA, Pissinatti A, Bonvicino CR, Seuanez HN, Sharma B, Jia H, Shankar A, Switzer WM, Santos AF, Soares MA. 2013. Identification and characterization of highly divergent simian foamy viruses in a wide range of New World primates from Brazil. PLoS One 8(7):e67568. 10.1371/journal.pone.0067568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel GA, Jones-Engel L. 2012. Primates and primatologists: social contexts for interspecies pathogen transmission. Am. J. Primatol. 75:543–550. 10.1002/ajp.20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu SF, Linial ML. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronoff R, Linial ML. 1991. Specificity of retroviral RNA packaging. J. Virol. 65:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027–1033 [DOI] [PubMed] [Google Scholar]
- 24.Macpherson I, Stoker M. 1962. Polyoma transformation of hamster cell clones: an investigation of genetic factors affecting cell competence. Virology 16:147–151. 10.1016/0042-6822(62)90290-8 [DOI] [PubMed] [Google Scholar]
- 25.Kirchoff V, Wong S, St Jeor S, Pari GS. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321–333. 10.1007/s705-002-8322-9 [DOI] [PubMed] [Google Scholar]
- 26.Nelson-Rees WA, Owens RB, Arnstein P, Kniazeff AJ. 1976. Source, alterations, characteristics and use of a new dog cell line (Cf2Th). In Vitro 12:665–669. 10.1007/BF02797468 [DOI] [PubMed] [Google Scholar]
- 27.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. 1983. LNCaP model of human prostatic carcinoma. Cancer Res. 43:1809–1818 [PubMed] [Google Scholar]
- 28.Barranco SC, Townsend CM, Jr, Casartelli C, Macik BG, Burger NL, Boerwinkle WR, Gourley WK. 1983. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 43:1703–1709 [PubMed] [Google Scholar]
- 29.Yee C, Krishnan-Hewlett I, Baker CC, Schlegel R, Howley PM. 1985. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am. J. Pathol. 119:361–366 [PMC free article] [PubMed] [Google Scholar]
- 30.Drewinko B, Romsdahl MM, Yang LY, Ahearn MJ, Trujillo JM. 1976. Establishment of a human carcinoembryonic antigen-producing colon adenocarcinoma cell line. Cancer Res. 36:467–475 [PubMed] [Google Scholar]
- 31.Cailleau R, Young R, Olivé M, Reeves WJ., Jr 1974. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 53:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herchenroder O, Renne R, Loncar D, Cobb EK, Murthy KK, Schneider J, Mergia A, Luciw PA. 1994. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology 201:187–199. 10.1006/viro.1994.1285 [DOI] [PubMed] [Google Scholar]
- 33.Mergia A, Wu M. 1998. Characterization of provirus clones of simian foamy virus type 1. J. Virol. 72:817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E-G, Sinicrope A, Jackson DL, Yu SF, Linial ML. 2012. Foamy virus Pol protein expressed as a Gag-Pol fusion retains enzymatic activities, allowing for infectious virus production. J. Virol. 86:5992–6001. 10.1128/JVI.06979-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin DN, Linial ML. 1998. The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72:3658–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweizer M, Neumann-Haefelin D. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577–582. 10.1006/viro.1995.1120 [DOI] [PubMed] [Google Scholar]
- 37.Thümer L, Rethwilm A, Holmes EC, Bodem J. 2007. The complete nucleotide sequence of a New World simian foamy virus. Virology 369:191–197. 10.1016/j.virol.2007.07.018 [DOI] [PubMed] [Google Scholar]
- 38.Pacheco B, Finzi A, McGee-Estrada K, Sodroski J. 2010. Species-specific inhibition of foamy viruses from South American monkeys by New World monkey TRIM5α proteins. J. Virol. 84:4095–4099. 10.1128/JVI.02631-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel GA, Small CT, Soliven K, Feeroz MM, Wang X, Hasan MK, Oh G, Alam SMR, Craig KL, Jackson DL, Matsen FA, IV, Linial ML, Jones-Engel L. 2013. Zoonotic simian foamy virus in Bangladesh reflects diverse patterns of transmission and co-infection. Emerg. Microbes Infect. 2:e58. 10.1038/emi.2013.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodem J. 2011. Regulation of foamy viral transcription and RNA export, p 1–31 In Maramorosch K, Shatkin AJ, Murphy FA. (ed), Advances in virus research, vol 78 Academic Press, London, England: [DOI] [PubMed] [Google Scholar]
- 41.Khan AS, Kumar D. 2006. Simian foamy virus infection by whole-blood transfer in rhesus macaques: potential for transfusion transmission in humans. Transfusion 46:1352–1359. 10.1111/j.1537-2995.2006.00862.x [DOI] [PubMed] [Google Scholar]
- 42.Yap MW, Lindemann D, Stanke N, Reh J, Westphal D, Hanenberg H, Ohkura S, Stoye JP. 2008. Restriction of foamy viruses by primate Trim5α. J. Virol. 82:5429–5439. 10.1128/JVI.02462-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118–128. 10.1016/j.tibs.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 44.Perkovic M, Schmidt S, Marino D, Russell RA, Stauch B, Hofmann H, Kopietz F, Kloke BP, Zielonka J, Strover H, Hermle J, Lindemann D, Pathak VK, Schneider G, Lochelt M, Cichutek K, Munk C. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein Bet. J. Biol. Chem. 284:5819–5826. 10.1074/jbc.M808853200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell RA, Wiegand HL, Moore MD, Schafer A, McClure MO, Cullen BR. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731. 10.1128/JVI.79.14.8724-8731.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lochelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rosler U, Battenberg M, Saib A, Flory E, Cichutek K, Munk C. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987. 10.1073/pnas.0501445102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, Wain-Hobson S, Gessain A, Vartanian JP, Schwartz O. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614. 10.1128/JVI.80.2.605-614.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasudevan A, Perkovi-ç Bulliard MY, Cichutek K, Trono D, Haussinger D, Munk C. 2013. Prototype foamy virus Bet impairs the dimerization and cytosolic solubility of human APOBEC3G. J. Virol. 87:9030–9040. 10.1128/JVI.03385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rua R, Betsem E, Calattini S, Saib A, Gessain A. 2012. Genetic characterization of simian foamy viruses infecting humans. J. Virol. 86:13350–13359. 10.1128/JVI.01715-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Switzer WM, Parekh B, Shanmugam V, Bhullar V, Phillips S, Ely JJ, Heneine W. 2005. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: evidence for a recent introduction following chimpanzee divergence. AIDS Res. Hum. Retroviruses 21:335–342. 10.1089/aid.2005.21.335 [DOI] [PubMed] [Google Scholar]
- 51.Han GZ, Worobey M. 2012. An endogenous foamy virus in the aye-aye (Daubentonia madagascariensis). J. Virol. 86:7696–7698. 10.1128/JVI.00650-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butera ST, Brown J, Callahan ME, Owen SM, Matthews AL, Weigner DD, Chapman LE, Sandstrom PA. 2000. Survey of veterinary conference attendees for evidence of zoonotic infection by feline retroviruses. J. Am. Vet. Med. Assoc. 217:1475–1479 [DOI] [PubMed] [Google Scholar]
- 53.Plochmann K, Horn A, Gschmack E, Armbruster N, Krieg J, Wiktorowicz T, Weber C, Stirnnagel K, Lindemann D, Rethwilm A, Scheller C. 2012. Heparan sulfate is an attachment factor for foamy virus entry. J. Virol. 86:10028–10035. 10.1128/JVI.00051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soliven K, Wang X, Small CT, Feeroz MM, Lee E-G, Craig KL, Hasan K, Engel GA, Jones-Engel L, Matsen FA, IV, Linial ML. 2013. Simian foamy virus infection of rhesus macaques in Bangladesh: relationship of latent proviruses and transcriptionally active viruses. J. Virol. 87:13628–13639. 10.1128/JVI.01989-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweizer M, Schleer H, Pietrek M, Liegibel J, Falcone V, Neumann-Haefelin D. 1994. Genetic stability of foamy viruses: long-term study in an African green monkey population. J. Virol. 73:9256–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]