Abstract
Bacteriophage T5 represents a large family of lytic Siphoviridae infecting Gram-negative bacteria. The low-resolution structure of T5 showed the T=13 geometry of the capsid and the unusual trimeric organization of the tail tube, and the assembly pathway of the capsid was established. Although major structural proteins of T5 have been identified in these studies, most of the genes encoding the morphogenesis proteins remained to be identified. Here, we combine a proteomic analysis of T5 particles with a bioinformatic study and electron microscopic immunolocalization to assign function to the genes encoding the structural proteins, the packaging proteins, and other nonstructural components required for T5 assembly. A head maturation protease that likely accounts for the cleavage of the different capsid proteins is identified. Two other proteins involved in capsid maturation add originality to the T5 capsid assembly mechanism: the single head-to-tail joining protein, which closes the T5 capsid after DNA packaging, and the nicking endonuclease responsible for the single-strand interruptions in the T5 genome. We localize most of the tail proteins that were hitherto uncharacterized and provide a detailed description of the tail tip composition. Our findings highlight novel variations of viral assembly strategies and of virion particle architecture. They further recommend T5 for exploring phage structure and assembly and for deciphering conformational rearrangements that accompany DNA transfer from the capsid to the host cytoplasm.
INTRODUCTION
Bacteriophage T5 is a member of the Siphoviridae family infecting Escherichia coli. It consists of an icosahedral capsid containing a large molecule of double-stranded DNA (dsDNA) (121.75 kbp) attached to a long flexible noncontractile tail. The complete genomes of two wild-type T5 strains (GenBank accession numbers AY587007 [1] and AY543070) and of the heat-stable deletion mutant T5st0 (GenBank accession number AY692264 [this study]) have been sequenced. Moreover, the genomes of T5-related phages H8 (2), EPS7 (3), SPC35 (4), AKVF3 (5), pVp-1 (6), and My1 (7) exhibit high sequence similarity to T5. Of the 159 to 174 genes predicted in each genome, about 70 were assigned functions on the basis of similarity searches and/or previous genetic studies. Most of the identified genes are related to nucleotide metabolism, DNA replication, recombination, and various enzymatic functions. Despite the fact that the major structural proteins of T5 have been identified (8), the functions of 13 of the 23 late genes encoding the structural and morphogenesis proteins remain to be ascertained.
The overall structure of T5 was solved by cryo-electron microscopy (cryo-EM) and image reconstruction (9). The large icosahedral capsid consists of the coat protein pb8, arranged as 11 pentamers at the vertices and 120 hexamers on the faces. The 12th vertex is occupied by a dodecamer of the portal protein pb7. The early events of T5 capsid assembly have been partly deciphered (10). The initial shell (prohead I) is assembled from the precursor form of pb8 (51 kDa), which includes a 159-residue N-terminal scaffolding domain. This extension is cleaved by the T5-encoded head protease pb11, leaving the 32-kDa mature pb8 and yielding prohead II. Packaging of DNA into prohead II is accompanied by expansion of the capsid, which involves large structural rearrangements of the coat protein subunits and allows accommodation of the full-length genome (11). The mature capsid is decorated with the 17.3-kDa protein pb10, which binds as a monomer to the center of the hexamers (9).
The T5 tail consists of a flexible noncontractile tail tube 160 nm in length. Its distal tail tip is formed by a collar structure that serves as a baseplate for three long L-shaped fibers (LTF) (12–14) and a cone ending in a central fiber. The T5 tail tube is composed of a stack of trimeric rings of the tail tube protein (TTP) pb6 and exhibits an unusual 3-fold symmetry while a hexameric organization is shared by most other Siphoviridae (9). The tail tube contains in its central core the multifunctional and oligomeric protein pb2, whose long N-terminal coiled-coil domain plays the role of tape measure protein (TMP). In vitro, the C-terminal end of pb2 has the dual role of pore-forming protein and peptidoglycan hydrolase and most likely participates in the formation of the channel through which the phage genome crosses the host envelope (15, 16). On the basis of biochemical studies, pb2 was designated the major component of the central fiber (17). However, this previous assignment must be revisited as pb2 is not long enough to extend from the bottom of the tail tube to the end of the central fiber (18). pb5, the receptor binding protein (RBP), ensures the binding of T5 to its receptor, the E. coli outer membrane protein FhuA (19, 20). The FhuA-pb5 complex has been characterized in vitro (21–23), but the exact location of pb5 at the tail tip remains uncertain. Thus, in spite of the significant information accumulated on T5, knowledge of the detailed organization of its viral particle and of the factors engaged in its assembly remains fragmented. In this work, we combined a proteomic analysis of T5 particles with bioinformatics and electron microscopic immunolocalization to assign a function to the genes encoding the structural and morphogenesis proteins, providing the first complete description of the effectors of T5 viral particle assembly. We report the topology of the proteins forming the distal tail tip complex and highlight their function in adsorption of T5 onto its host.
MATERIALS AND METHODS
Bacterial and phage strains.
The heat-stable deletion mutant T5st0 (24) was used for sequencing of the complete genome. T5HER28 (Reference Center for Bacterial Viruses, Laval University, Quebec, Canada) was used as a reference wild-type strain to determine the position of the deletion of T5st0. The high-density mutant T5hd1 lacking the L-shaped tail fibers (13) was a gift from K. Heller (Max Rubner-Institut, Kiel, Germany). T5D17am34d and T5D20am30d mutants were used as tail and head donors, respectively; T5amN5 was used as a donor for empty proheads II; and T5amHA911 was used for the production of phage T5 lacking the single-strand interruptions. E. coli F, a fast-adsorbing strain for T5 (25), was used for the propagation of T5st0 and T5hd1 and as a nonsuppressive host for the production of phage subparticles from T5 amber mutants. E. coli CR63 was used as a permissive host for replication of amber mutants.
Production and purification of T5 particles and phage subparticles.
T5st0, T5hd1, and T5 amber mutants as well as DNA-filled capsids, proheads, and tails were produced by infecting exponential-phase cultures of the appropriate E. coli strain grown at 37°C in LB medium. Phages and mature capsids were purified by precipitation with polyethylene glycol (PEG)-NaCl and equilibrium centrifugation in CsCl gradients according to standard protocols (26). Empty proheads and tails were purified by PEG-NaCl precipitation, followed by centrifugation through a 10 to 40% glycerol step gradient and anion-exchange chromatography as described in reference 10. This protocol was modified for tail purification: after PEG-NaCl precipitation, the tail pellet was suspended in the presence of detergent (1% lauryldimethylamine oxide [LDAO]) to solubilize membrane vesicles resulting from bacterial lysis. This treatment is required to prevent copurification of membranes with T5 tails.
T5st0 DNA sequencing.
Genomic DNA was isolated from CsCl-purified phage T5st0 by SDS release followed by phenol-chloroform extraction and purified by centrifugation on a continuous CsCl gradient (Beckmann 70Ti rotor; 450,000 rpm, 10°C, for 48 h) in the presence of ethidium bromide (EtBr) (5 μg/ml). Purified DNA was extracted with isoamyl alcohol and dialyzed against Tris-EDTA (TE) buffer. The genome was sequenced by Integrated Genomics (Jena, Germany). A random T5 plasmid library (1,438 clones) covering >10-fold the size of the genome was generated using the TOPO shotgun kit (Invitrogen) and subjected to sequencing with a multicapillary ABI 3700 sequencer, using fluorescent dye-terminator chemistry. The T5 sequence was assembled using the Pred-Phrap package (University of Washington), and finishing was achieved by primer walking. Genome annotation was performed as follows: all open reading frames (ORFs) larger than 29 amino acids were searched for similarity in GenBank, Swiss-Prot, and COG (NCBI) sequence databases with the BLASTP tool, and motif/domains were searched in the PFAM database using hmmpfam (HMMER 2.3.1; University of Washington). Candidate coding sequences (CDS) were identified with the Glimmer 2 program (27), and graphical maps of the genome were used as a support for gene validation, annotation, and start codon determination (http://www-archbac.u-psud.fr/genomes/BPHAG/T5/T5.html). Promoter element prediction was done by scanning the T5 sequence with hmmsearch (University of Washington), using a profile constructed from an alignment of published T5 promoter sequences (28).
DNA-less or “ghost” particle preparation.
Phage T5st0 and mature heads were emptied of their DNA before their protein content was analyzed. A T5st0 sample (1012 PFU/ml) was mixed with 1 volume of 10 M LiCl, incubated for 10 min at 46°C, and then diluted 10 times in 20 mM Tris buffer, pH 7.5, 10 mM MgCl2, and 1 mM CaCl2. The T5D17am34d head sample was incubated with 50 mM EDTA for 30 min at 37°C, after which 100 mM MgCl2 was added. Both samples were then incubated for 40 min at 37°C with DNase I at 15 U/ml (GE Healthcare), concentrated by ultracentrifugation for 30 min at 100,000 × g, and finally resuspended in 50 mM Tris buffer, pH 7.5, for SDS-PAGE analysis.
Protein identification by N-terminal sequencing.
Phage proteins were resolved on 12% SDS-PAGE gels, transferred onto a polyvinylidene difluoride (PVDF) membrane, briefly stained with Coomassie blue R-250 in 1% acetic acid, and destained with a solution of 1% acetic acid and 50% ethanol. Bands of interest were excised and submitted to N-terminal sequencing by automated Edman degradation using a Procise sequencer (Applied Biosystems) equipped with an online phenylthiohydantoin amino acid analysis system.
NanoLC-MS/MS analyses for protein identification.
Phage proteins were resolved on SDS-PAGE gels (4 to 20% Precise protein gels; Thermo Scientific) and stained with Bio-Safe Coomassie blue G-250 (Bio-Rad). Standard enzymatic digestion of excised bands was performed with trypsin (Gold; Promega; 10 ng/μl) using the Progest robot (Genomic Solutions). Peptide mixtures were SpeedVac treated for 10 min and then solubilized with 0.1% formic acid and injected in a quadrupole time of flight (Q-TOF) Premier mass spectrometer coupled to a nanoAcquity liquid chromatograph equipped with a trapping column (Symmetry C18; 180 μm by 20 mm, 5-μm particle size) and an analytical column (BEH130 C18; 75 μm by 100 mm; 1.7-μm particle size; Waters). The aqueous solvent (buffer A) was 0.1% formic acid in water, the organic phase (buffer B) was 0.1% formic acid in acetonitrile, and a 2 to 40% B gradient was set for 25 min. For exact mass measurements, a glufibrinopeptide reference (m/z = 785.8426) was continuously supplied during nano-liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analyses using the lockspray device. Peptide mass measurements were corrected during data processing, and peak lists were generated by PLGS (ProteinLynx Global Server; Waters). Processed data were submitted to Mascot searching using the following parameters: the homemade phage T5 data bank, including head and tail proteins; peptide tolerance, 20 ppm; fragment tolerance, 0.1 Da; digest reagent trypsin with one missed cleavage allowed; variable modifications oxidation (methionine) and fixed modifications carbamidomethylation (cysteine). As pb11 protein was supposed to be processed by proteolytic cleavage at both ends, chymotrypsin and Asp-N proteases (Roche; 10 ng/μl) were used to unambiguously identify N and C termini of pb11. Chymotrypsin and Asp-N peptide mixtures were analyzed by nanoLC-MS/MS as described above.
Purification of tail proteins and antibody preparation.
pb5 was purified as previously described (21). T5p142, T5p132, pb9, and pb3 coding sequences were cloned into the pML14 vector (29) in frame with a six-His tag by using a common PCR and restriction strategy. Plasmids were transformed into the E. coli BL21(DE3) CodonPlus-RIL strain (Stratagene), and recombinant proteins were overproduced by growing bacteria at 37°C in ZY autoinducing medium (pb9 and pb3), at 20°C in LB medium with overnight induction by 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (T5p132), and at 37°C in LB medium with 4 h of induction by 0.1 mM IPTG (T5p142). Each protein was purified by nickel-affinity chromatography followed by anion-exchange chromatography and concentrated by ultrafiltration. The final protein concentrations were in the 0.65- to 1-mg/ml range, in 20 mM Tris-HCl, pH 8.0. As overexpression of the TMP gene pb2 (124 kDa) yielded insoluble material, pb2 was “purified” by electroelution of the protein band from SDS-PAGE of T5 ghosts, concentrated by precipitation with 80% acetone, and resuspended in 50 mM Tris buffer, pH 7.8. Rabbit polyclonal antisera raised against each protein were produced according to standard protocols (30), and IgG fractions were purified by affinity chromatography using HiTrap protein A columns as recommended by the supplier (GE Healthcare).
Electron microscopy and immunolocalization.
For direct EM analysis of phages or tails, samples were adsorbed onto carbon film and imaged after negative staining with 2% uranyl acetate. For immunolocalization, 1 μl of phage T5st0 or T5hd1 (1013 PFU/ml) was mixed with 1 to 10 μl of purified IgG of interest and complemented to 20 μl with T5 buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 1 mM CaCl2). The mixture was incubated 1 h to overnight at 4°C or room temperature and diluted twice with T5 buffer. Aggregates were discarded after centrifugation (2 min at 18,600 × g), and free IgGs were separated from the cross-linked phages by chromatography on a Sephacryl 500 MicroSpin column equilibrated with T5 buffer (400 μl, 75% slurry, spun for 5 min at 700 × g) (31). The phage-IgG complexes were imaged after negative staining with 2% uranyl acetate or additionally labeled with anti-rabbit goat IgG–5-nm gold complexes (British Biocell). Free goat IgGs and unbound gold were separated from phages by spin chromatography as described above. Electron microscopy was performed using a Tecnai G2 Spirit equipped with an Eagle charge-coupled device (CCD) camera (FEI).
For EM analysis of pb5 protein, the sample was diluted to 10 μg/ml and adsorbed to a glow-discharged carbon film-coated grid, washed with 3 droplets of pure water, and subsequently stained with 2% uranyl acetate. EM was performed using a Philips CM10 microscope equipped with a Veleta CCD camera (Olympus). Reference-free alignment was performed on manually selected particles using the EMAN image processing package (32). After using a reference-free alignment procedure, particle projections were classified by multivariate statistical analysis. The class averages with the best signal-to-noise ratio were selected and gathered in a gallery.
Nucleotide sequence accession numbers.
The sequence data reported in the present study were deposited in the GenBank database under accession numbers AY692264 and AY734508.
RESULTS
Identification of phage T5 head and tail proteins.
We sequenced the 113,720-bp DNA molecule of strain T5st0 (GenBank accession number AY692264), a heat-stable deletion mutant that we widely used for various studies on T5 infection (9, 33) but that lacks a nonessential region of 8,208 bp within the early genes (GenBank accession number AY734508). Only 20 polymorphic sites were found in the chromosome when comparing T5st0 with two already-published T5 “wild-type” strains (GenBank accession numbers AY587007 and AY543070). Among these differences, 10 are allelic variants, and the rest are single nucleotide polymorphism sites with no significant impact on chromosome organization and gene structure of T5st0.
In order to identify the as-yet-uncharacterized structural proteins of the T5 particle, we used T5 amber mutants defective in phage assembly (8, 34). We sequenced their DNA to identify the mutated gene (Table 1) and analyzed the protein content of the particles produced upon infection of a nonsuppressive strain (see Fig. 2). T5D17am34d, affected in gene D17 encoding the tail protein pb4, produces mature DNA-filled heads, without a functional tail. In T5amN5, the mutation was located within the gene of the terminase large subunit (TerL) responsible for DNA packaging, yielding immature proheads and separate functional tails. T5D20am30d, affected in the gene encoding the major head protein pb8, produces only tails. In the high-density mutant T5hd1 lacking the L-shaped fibers, we identified a single insertion in the ltf gene encoding the pb1 protein. Proteins from T5st0 and T5hd1 ghosts, proheads II, and mature heads and tails were resolved by SDS-PAGE (Fig. 1). The protein patterns revealed low-molecular-weight and low-abundance proteins that were not identified previously. LC-MS/MS spectrometry and/or N-terminal sequencing by Edman degradation (see Materials and Methods) allowed their identification and subsequent assignment to specific T5 genes (Table 2 ). We identified six new components of the T5 particle: the head protein T5p144 and the tail proteins T5p132, T5p135, T5p140, T5p142, and T5p143, bringing the number of different proteins present in the T5 virion to 16. By combining published data with our results, we provide a comprehensive genetic map of the 23 putative morphogenesis genes, which are organized into five putative operons embedded in two distinct modules encoding the head and tail genes, respectively (Fig. 2). This shows that T5 gene order in the morphogenetic block resembles the one found in other siphophages, with the exception of the receptor binding protein pb5.
TABLE 1.
Nucleotide sequence of T5 mutations affecting T5 assemblya
| Name | Gene/protein affected | Phenotype | Mutation | Stop codon positionb | Reference |
|---|---|---|---|---|---|
| T5D17am34d | D17 (T5p133), tail protein pb4 | Filled mature head | C268T | 90/689 | 34 |
| T5D20am30d | D20 (T5p145), major head protein pb8 | Functional tail | C1132T | 378/459 | 34 |
| T5amN5 | T5p151, terminase large subunit | Empty prohead II | C1189T | 397/439 | 8 |
| T5amHA911 | T5p149, nicking endonuclease | Nick-less DNA | C337T | 118/151 | 43 |
| T5hd1 | ltf, L-shaped fiber protein | High density | +T873 | 304/1396 | 13 |
Mapping of the mutations was determined by sequencing the late gene region of each T5 mutant genome: 6-kbp fragments amplified by PCR from purified T5 mutant DNA were sequenced using primers designed from the T5st0 genome sequence. In amber mutants, the Stop codon results from a point mutation. In T5hd1, the Stop codon results from an insertion (+) that alters the normal reading frame.
Position of the translation termination relative to the total number of amino acids of the wild-type protein.
FIG 2.
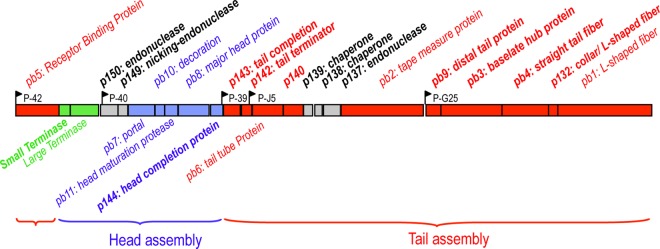
Map of T5 morphogenesis genes. T5 late genes (positions 73825 to 102307 in T5st0 genome, GenBank accession number AY692264) are transcribed from five putative operons encoded by the minus strand of the DNA (1, 76). The annotation of the morphogenesis genes was updated from the combination of genome sequencing data with bioinformatics studies and analysis of the protein composition of T5 entire particles, tails, and heads. Assigned functions issued from this study are noted in bold. The tail proteins, head proteins, and packaging proteins are colored red, blue, and green, respectively. Nonstructural assembly proteins or proteins of unknown functions are colored gray. For clarification, this map is presented in the usual direction from left to right, but it is reversed from the 5′→3′ sequence of the T5 genome. Black arrows indicate the T5 late promoters: P-39, P-40, and P-42 are putative promoters, while P-J5 and P-G25 are identified promoters (28).
FIG 1.
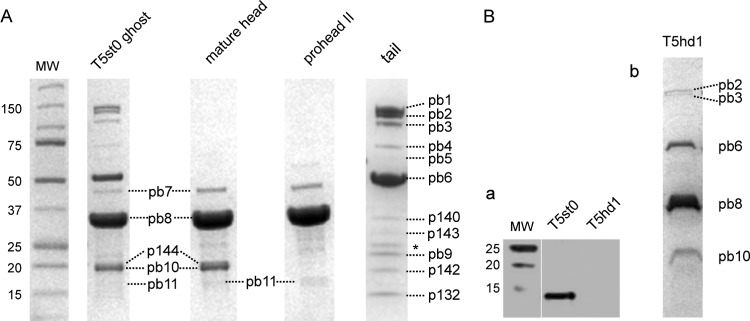
SDS-PAGE of T5 protein components. (A) T5 ghosts were obtained from T5st0. Mature heads were produced from T5D17am34d, empty proheads II were produced from T5amN5, and tails were produced from T5D20am30d. Protein bands are listed by their number according to Table 2. The asterisk indicates the presence of the outer membrane major proteins OmpC and OmpF from contaminating host cell membrane vesicles. (B) (a) Western blot detection of T5p132 inT5st0 and T5hd1. Phage particles (≈50 μl, 1011 PFU/ml, disrupted by repeated cycles of freezing and thawing) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for Western blotting. T5p132 protein was visualized by using rabbit anti-p132 IgGs and goat secondary rabbit antibodies (IRDye 800CW; Odyssey). (b) SDS-PAGE of T5hd1 proteins showing the absence of the L-shaped fiber protein pb1. MW, protein molecular weight markers in kDa.
TABLE 2.
T5 structural proteins identified by LC-MS/MS and Edman N-terminal sequencinga
| Gene | ORF no. a/b/c | Band name | Molecular mass (kDa) | LC-MS/MS % coverage | Processing | Function/location | Reference(s) |
|---|---|---|---|---|---|---|---|
| Capsid | |||||||
| T5p144/T5.148/ORF137 | p144M | 19.3 | 44 | No | Head completion | This study | |
| D20 | T5p145/T5.149/ORF138 | pb8P,M | 32.8 | N: K159 (Ed.) | Major head protein | 8, 9 | |
| T5p146/T5.150/ORF139 | pb11P,M | 15.8 | 64 | Head protease | 10; this study | ||
| N5 | T5p147/T5.151/ORF140 | pb10M | 17.3 | 62 | C: S1264 | Decoration | 8, 9 |
| T5p148/T5.152/ORF141 | pb7P,M | 43.8 | N: K10 (Ed.) | Portal | 8, 9; this study | ||
| Tail | |||||||
| ltf | T5p131/T5.133/ORF124 | pb1 | 148.0 | 33 | C: S1264 | L-shaped fibers | 12–14; this study |
| T5p132/—/ORF125 | p132 | 15.0 | 84 | No | L-fibers/tail tip collar | This study | |
| D17 | T5p133/T5.137/ORF126 | pb4 | 74.9 | Central straight fiber | This study | ||
| D16 | T5p134/T5.138/ORF127 | pb3 | 107.3 | 26 | Baseplate hub protein | This study | |
| T5p135/T5.139/ORF128 | pb9 | 22.7 | 69 | No | Distal tail protein | This study; 56 | |
| D18/19 | T5p136/T5.140/ORF129 | pb2 | 121.9 | 47 | C: V1148 ±2 | Tape measure protein | 15, 16 |
| T5p140/T5.144/ORF133 | p140 | 34.5 | 47 | No | This study | ||
| N4 | T5p141/T5.145/ORF134 | pb6 | 50.3 | Tail tube protein | 8, 9 | ||
| T5p142/T5.146/ORF135 | p142 | 18.4 | 38 | No | Tail terminator | This study | |
| T5p143/T5.147/ORF136 | p143 | 27.8 | 57 | No | Tail completion | This study | |
| oad | T5p153/T5.157/ORF141 | pb5 | 67.8 | No | Receptor binding protein | 17, 19–23 |
Proteins identified in previous studies are indicated with their gene name according to the first mapping of T5 genes (34) and with their band name according to the SDS-PAGE analysis published in reference 8. All proteins are indicated with their ORF numbers in the sequences of the T5 genome deposited in GenBank: a, T5st0 deletion mutant (AY692264); b, T5 wild-type strain from Pushchino collection (AY543070); c, T5 strain ATCC 11303-B5 (AY587007). For the capsid proteins, P or M indicates that they were found in proheads II or in mature capsids, respectively. Processing sites were determined from protein coverage in MS or N-terminal sequencing by Edman degradation (Ed.). When sequence coverage included both N- and C-terminal ends of the protein, no processing was mentioned (No). In the absence of specific indication, we assumed that the protein is not processed due to the agreement between the apparent molecular mass determined on SDS-PAGE and the mass predicted from the corresponding ORF. —, not annotated.
Structure and assembly of T5 capsid. (i) Multiple proteolysis events control the maturation of T5 capsid.
Apart from the major head protein pb8 (32.8 kDa) and the decoration protein pb10 (17.3 kDa) (9, 10), the proteins forming the capsid have not been characterized. The genes encoding the portal protein pb7 and the head maturation protease pb11 were identified by similarity searches through HHpred. We related pb7 to the family of phage HK97 portal-like proteins (Pfam PF04860), with a probability of 100% despite an average sequence identity of 30% over ≈400 residues. We identified the pb7 band (43.8 kDa) on the SDS-PAGE gel of the T5 capsids and ghosts (Fig. 2), and Edman degradation revealed that the pb7 gene product was processed by proteolytic cleavage after lysine K10 (Table 2). HHpred searches related pb11 to the families of serine head proteases of HK97-like phages (U35) and herpesvirus (S21) (35, 36). In SDS-PAGE of T5 ghost, mature heads, and proheads II, we found a minor protein that migrated as a faint band of ca. 16 kDa (Fig. 1). N-terminal sequencing and LC-MS/MS analysis identified this protein as pb11 and revealed that the primary gene product (with a predicted mass of 23.3 kDa) was cleaved at both ends after residues K23 and K166, yielding a mature peptide of 15.8 kDa. These data suggest that pb11 specifically cuts itself, pb7, and pb8 (trimming of the N-terminal scaffolding domain) upon assembly of the T5 procapsid. The cleavage of the three proteins occurs invariably after a lysine; however, the protein sequences surrounding the processing sites do not show a conserved motif. Multiple alignments of pb11 with other phages and herpesvirus head proteases indicate that the catalytic triad consists of residues H76-S122-E148 (data not shown). This H-S-E motif is conserved in the U35 family of phage proteases and remains intact in the processed form of pb11 found in the mature capsid.
(ii) A single head completion protein closes the T5 capsid.
The product of gene T5p144 was identified as a minor protein that comigrates in SDS-PAGE with the decoration protein pb10 (Fig. 1). LC-MS/MS analysis covered both ends of the protein, giving a molecular mass of 19.3 kDa. It was not possible to estimate the copy number of T5p144 due to the comigration with the prevalent pb10 protein (120 copies per phage). Functional assignment of T5p144 was based on two observations: (i) its gene is located downstream of the head protein gene pb8 and precedes the tail gene module, a position typical for head completion protein genes, and (ii) T5p144 was detected in T5 ghosts and mature capsids but not in empty proheads (Fig. 1), suggesting that the protein binds to the capsid after DNA packaging. Similarity searches related T5p144 to a family of proteins that make up the head-to-tail connector by binding to the portal protein and are represented by the gp6 protein of phage HK97 (Pfam PF05135) and the gp15 protein of phage SPP1. T5p144 most closely resembles a subfamily of gp6-like head-to-tail connectors (TIGR02215), which show strong similarity at both N- and C-terminal ends and include a large insertion (60 to 90 amino acids) located in the central region of the protein (37). Remarkably, closure of the T5 capsid is ensured solely by T5p144, as no other protein was detected in the mature capsid and no other gene encoding a head completion protein was found.
T5 DNA packaging proteins and maturation endonucleases. (i) Small subunit of phage T5 terminase.
The DNA packaging molecular motor consists most frequently of two subunits: the DNA-binding small subunit (TerS), which recognizes the phage DNA produced by replication, and the large catalytic subunit (TerL) that powers the ATP-dependent translocation of the DNA into the capsid and cuts the substrate DNA concatemers to generate the mature genome (38). We previously characterized the T5 TerL (39). Here, we assigned the DNA-binding subunit to ORF T5p152, taking into account the following features shared by the TerS described so far: (i) ORF T5p152 is located upstream of the gene encoding TerL, (ii) the protein size—160 residues—matches that of other TerS proteins (184 residues for phage SPP1 and 164 residues for phage T4), and (iii) similarity searches through HHpred related T5p152 to many proteins sharing a helix-turn-helix (HTH) DNA-binding motif that resembles the HTH motif found in the TerS of other bacteriophages as in the G1P protein of the SPP1-like phage SF6 (40).
(ii) Formation of single-chain interruptions in phage T5 DNA.
The head morphogenesis module includes the two unknown genes T5p150 and T5p149 inserted between the terminase and capsid structural genes. By similarity searches through HHBlits, we linked both proteins to endonuclease families. T5p150 was related to the superfamily of GIY-YIG endonucleases (IPR000305) that includes phage-encoded intron endonucleases (41). Notably, T5p150 is close to T5p137, a second putative endonuclease inserted in the tail morphogenesis module (Fig. 2). Both genes are optional among the T5 relatives since they are absent in the genomes of EPS7 (3), H8 (2), SPC35 (4), and pVp-1 (6). This feature, together with their position in intragenic regions of the morphogenesis module, suggests that T5p150 and T5p137 may be mobile freestanding homing endonucleases like the previously identified Seg or I-TevI nucleases of phage T4 (41). On the other hand, T5p149, which we linked to the family of His-metal H-N-H endonucleases (IPR003615) (42), is conserved in all T5 relatives. A striking feature of T5 is the presence of single-chain interruptions or “nicks” located at specific positions in the minus strand of the DNA molecule. In order to investigate the relation between the nicks and the presence of the endonucleases among the morphogenesis genes, we sequenced the late genes of T5amHA911, a mutant that produces a full nick-less genome under nonsuppressive growth conditions (43). We found a unique nonsense mutation in the T5p149 gene (Table 1), strongly suggesting that it is involved in generating the single-strand interruptions. Two unknown genes, SciA and SciB, whose functions are required for the formation of the nicks, were previously mapped at the 5′ end of the late gene region (43). Our results suggest that T5p149 is one of them. Krauel and Heller previously attributed the SciB function to T5p152 (44) that we identified as the terminase small subunit in this work (see above). T5 TerS has no endonuclease activity per se; however, we cannot exclude that it contributes to regulating the formation of the single-chain interruptions during the packaging process.
Structure of T5 tail.
Only four of the 11 tail proteins have a known function and localization (pb1, pb2, pb5, and pb6; see the introduction). The lack of demonstrable similarity between tail proteins of different phages often prevents their direct identification. However, the tail gene order is conserved in the Siphoviridae and Myoviridae genomes (18), which guides prediction of gene function. In order to unambiguously determine the topology of uncharacterized T5 tail proteins, we undertook their localization by electron microcopy (EM) immuno-cross-linking and gold labeling.
(i) Tail termination and tail completion: T5p142 and T5p143 proteins.
Specific termination and completion proteins make up the proximal end of the tail that is connected to the portal vertex of the capsid. Two proteins, T5p142 (18.4 kDa) and T5p143 (27.8 kDa), were found in T5 tails, which most likely correspond to the above functions (Fig. 1; Table 2). Both genes are predicted to be transcribed from the same short operon located downstream from the P-39 promoter and are located between the head module and the TTP pb6 (Fig. 2). This is the usual position for the tail completion proteins. Similarity searches through HHpred linked T5p142 to the structure gpU, the tail terminator of phage λ (PDB 3FZB) (45) and, more generally, the sequence family of gpU (PF06141) with a probability of >95%. These hits, as well as the genomic position, strongly support structural and functional similarity, even though the sequences are highly diverse. We confirmed the position of T5p142 at the proximal end of the T5 tail in EM by cross-linking with anti-p142 IgGs and gold labeling (Fig. 3B).
FIG 3.
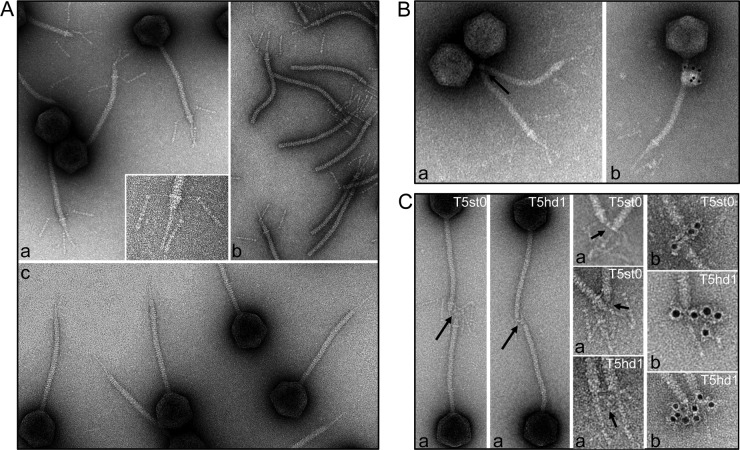
(A) Analysis of the tail tip morphology in phage T5st0 in wide-field and blowup views (a), in isolated tails (b), and in the T5hd1 mutant lacking the L-shaped fibers (c). (B) Localization of T5p142 in T5st0 and (C) of pb3 in T5st0 and T5hd1: the position of the proteins was identified by IgG cross-linking (a, black arrows) or by visualization of the IgG molecules associated with goat anti-rabbit IgG–gold conjugate (b). For pb3, blowups of the tail tips are shown to highlight immunolocalization. The diameter of the tail tube is 12 nm.
T5p143 could not be related to other phage tail proteins by similarity searches. However, the gene location two ORFs upstream of the TTP gene suggests that T5p143 may be a homologue of the highly conserved family of tail completion proteins (PF06763), of which phage λ gpZ is the prototype (18). Genetic and biochemical data indicate that gpZ proteins of λ and its relatives in myophages P2 and Mu are essential for infectivity; however, their function and organization within the tail are unknown.
(ii) Tail tube proteins and assembly chaperones.
The central tail operon encodes the TTP pb6, the T5p140 to T5p137 proteins, and the TMP pb2. The originality of pb6 is its trimeric organization (9). Similarity searches found no homologues of pb6, apart from the TTPs from T5 relatives. The pb6 sequence (464 residues) includes a C-terminal IgG-like domain (type Big2), which is conserved in many TTPs (46). A representative of this extra domain is the C-terminal domain of the TTP gpV of phage λ that is not required for TTP subunit polymerization and tail tube formation and is exposed to the surface of the tail tube (47). The pb6 N-terminal region (370 residues), predicted to form the core of the 3-fold symmetric tail tube, is about twice as long as other known TTP domains. Secondary structure predictions indicate that it may include a duplicate domain mainly arranged in β-strands, but structural investigation will be required to determine whether the trimeric organization of pb6 could mask a pseudohexameric structure similar to the 6-fold symmetry shared by the tail tube proteins of most bacteriophages (48).
The 34.5-kDa T5p140 protein (Fig. 1 and 2; Table 2) is a minor tail component conserved among all T5 relatives. Neither database searches nor its location downstream from the TTP matched any established tail protein or specific function of other known phage families, suggesting that T5p140 is a landmark in the tail tube operon of T5-like phages.
The products of the neighboring genes T5p139 and T5p138, which are conserved in all T5 relatives, were not found in the T5 particles, indicating that they are not structural proteins. Similarity searches yielded no conclusive clues as to their function, but the location of these two genes, between the TTP and TMP genes, is reminiscent of tail assembly chaperones seen in other Siphoviridae and Myoviridae (18, 49). It has been shown that production of these chaperones is controlled by a conserved programmed translational frameshift, which leads to alternative transcript products, as exemplified by gpG and gpGT proteins of phage λ (50). Such a mechanism for T5p139/p138 genes seems unlikely, as several required sequence features are absent in the T5 genome. Most importantly, T5p139/p138 are separated by a 62-nucleotide intervening sequence and no known frameshifting signals for the +1 (or −1) mechanisms are found within the relevant chromosome region.
(iii) Tail tip components.
The third tail operon encodes pb9, pb3, and pb4; the newly identified 15-kDa protein T5p132; and the LTF protein pb1 (Fig. 2; Table 2). These genes are located in the region dedicated to the tail tip complex that initiates tail assembly as described for phage λ (18, 51).
(a) L-shaped fibers and their attachment base.
pb1 forms the three LTF of T5 that bind to the O-antigen domains of the E. coli lipopolysaccharide (LPS) (52). Negative-staining EM of phage particles and isolated tails showed that the LTF are attached to a collar structure at the junction between the tail tube and the cone (Fig. 3A, panels a and b). Each fiber consists of a thin proximal rod (≈30 nm) connected by a hinge to a thicker distal part (≈47 nm). The distal region exhibits a series of globular domains that may represent secondary-structure elements, as observed in long tail fibers of T4 (53) or in the short kinked fibers of T7 (54, 55). In the fiberless T5hd1 mutant previously isolated by Saigo (13), the lack of fibers is correlated with the lack of the collar (Fig. 3A, panel c), suggesting that the two structures belong to the same building block. Interestingly, the Western blot detection of the protein T5p132 in T5hd1 showed that T5p132 was absent in the fiberless particles (Fig. 1B). We checked the sequence of both T5p132 and pb1 (lft) genes in the T5hd1 genome and found only one mutation in the lft gene, which yields truncated protein pb1 (Table 1). These observations suggest that T5p132 and pb1 coassemble to form the collar and the L-fibers. Notably, immunolocalization of T5p132 failed, as we did not observe any cross-linking or immunogold labeling using anti-T5p132 IgGs. We thus surmised that T5p132 might be shielded by interaction with pb1 and/or other tail proteins in the collar structure. Similarity searches did not hit meaningful homologues of T5p132 in other phages.
(b) Cone and straight fiber proteins.
We used antibodies directed against the pb9 and pb3 proteins to localize these proteins within the tail tip. We visualized the sites of cross-links and gold labeling with anti-pb9 in the upper part of the cone, right under the collar onto which are attached the L-fibers (see accompanying paper by Flayhan et al. [56]) and in the lower part of the cone with anti-pb3 IgGs (Fig. 3C). This indicated that both proteins are the major components of the cone structure.
The gene position of pb9, downstream of the TMP gene, is the landmark of the Distal tail proteins (Dit), which form a hexameric ring located at the tail tube end of several siphophages infecting Gram-positive bacteria. Together with the localization of pb9, this strongly suggests that pb9 is the Dit protein of phage T5. This was recently confirmed by the resolution of the crystal structure of pb9, which reveals a two-domain protein, one domain of which shows structural similarity with the N-terminal domain of the Dit proteins of phages SPP1, p2, and TP901-1, infecting Gram-positive bacteria (56).
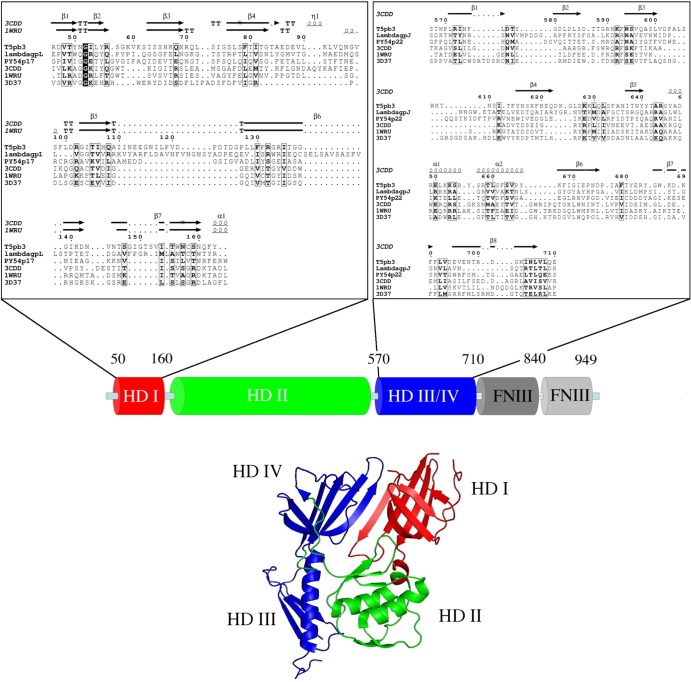
PSI-BLAST searches with the pb3 protein (103 kDa) found two families of phage proteins that are homologues of two distinct regions of the pb3 sequence. Iterative searches linked the N-terminal region (residues 1 to 175) to diverse prophage proteins and to the gp17 protein of Yersinia phage PY54. All these sequences were related to the conserved domain Pfam DUF2163 that describes the N-terminal region of the tail tip protein gpL of phage λ (57) (high scores and same position in the genome map). The pb3 central and C-terminal regions (residues 400 to 850) were linked to a family of prophage and phage host recognition proteins (Pfam 13550), including the gp22 protein of phage PY54. The typical representative of this family is the gpJ protein of phage λ, of which the trimer constitutes a portion of the conical tip and the central tail fiber (58). Using the fold recognition @tome2 server, we linked pb3 and related proteins to the structure of trimeric gp27 baseplate hub proteins (BHPs) of the T4-like Myoviridae that make a cylinder connection between the tail tube and the central cell-puncturing device (Fig. 4) (59). BHPs are four-domain proteins: the first (HDI) and third (HDIII) domains consist of seven- or eight-stranded antiparallel β-barrels and form the region located in contact with the tail tube. The second domain (HDII) together with the fourth domain (HDIV) forms the distal region of the hub cylinder, in contact with the central needle protein gp5. Figure 4 shows the @tome2 sequence-structure alignments of pb3 with the BHP p44 of phage Mu (PDB 1WRU and close structures 3CDD and 3D37) (60). The pairwise alignments of phage PY54 p17 with the BHPs suggest that the gpL-like N-terminal region folds as HDI (probability 94%), while the alignments of the central regions of pb3 (residues 566 to 710), λgpJ (residues 325 to 509), and PY54p22 (residues 289 to 433) hint at their folding as HDIII-HDIV domains (probability, 98%). HDII is the only domain of p44 that was not found in pb3. The corresponding region is much larger in pb3 than in known BHPs (∼300 versus 100 residues), suggesting that its fold is different in T5. The only homologue of the T4-like BHP that has been identified in a siphophage is the trimeric ORF16 protein of phage p2 (PDB 2WZP) (61), whose gene is located in the same position as that of pb3 and gpL, i.e., downstream of the Dit genes ORF15, pb9, and gpM, respectively. These structural predictions and the gene position, together with the location of pb3 at the interface between the cone and the central tail fiber, strongly suggest that pb3 would trimerize to form the T5 BHP. Finally, the C-terminal region of pb3 (residues 730 to 934) was predicted to fold as two successive FNIII fibronectin domains. Three similar domains were also found after the BHP domain in the central region of gpJ.
FIG 4.
Sequence analysis and structure prediction of pb3. (Top) Multiple sequence alignment of protein pb3 and related phage proteins. The pairwise alignments of pb3 with protein p17 of Yersinia phage PY54 and gpL of phage λ were generated with clustalW. The pairwise alignments of pb3, λgpJ, and PY54p22 with the baseplate hub proteins from the PDB data bank (1WRU, phage Mu protein p44; 3D37, tail protein from Neisseria meningitidis MC58; 3CDD, 43-kDa tail protein from prophage MuSO2) were obtained from the @tome2 metaserver (http://atome.cbs.cnrs.fr). Sequence-structure alignments were manually refined with the help of the program ViTO (77). Secondary structure assignments were performed using DSSP. Similarity calculations were performed using the Risler matrix (30). (Bottom) Domain architecture of pb3. The putative hub domains (HDs) (HDI in red, HDII in green, and HDIII/HDIV in blue) are related to the structure of MuSO2 BHP (PBD 3CDD). The C-terminal end of pb3 was related to two successive fibronectin type III domains (PDB 1FNF) through HHsearch with 99.6% probability.
We could not find full-length homologues of the 75-kDa protein pb4 except in other T5-like genomes. However, using PSI-BLAST, we could link the 180 residues L406 to F591 of pb4 to the sequence of the central tail fiber/host recognition protein gp54 of the Streptococcus pneumoniae phage Dp-1. This sequence matched a collagen triple helix repeat (Pfam 01391) that is the signature of fiber structures. Moreover, as the pb4 gene is located downstream of the Dit and BHP genes, we propose that pb4 assembles to pb3 to constitute the main part of the central fiber.
To further explore the central fiber composition, we investigated the positions of the TMP pb2 by immunolocalization. We did not observe any cross-linking or significant gold labeling with anti-pb2 IgG, arguing that pb2 is not accessible to antibodies in T5 particles. These observations challenge the previous assumption that pb2 is the major component of the central tail fiber (17) but rather support the idea that pb2 fills the tail tube with its long coiled-coil TMP domain and has its C-terminal hydrophobic domain sequestered within the tail tip and not exposed to the external medium (16).
(iv) The receptor binding protein pb5 is located at the tip of the central tail fiber.
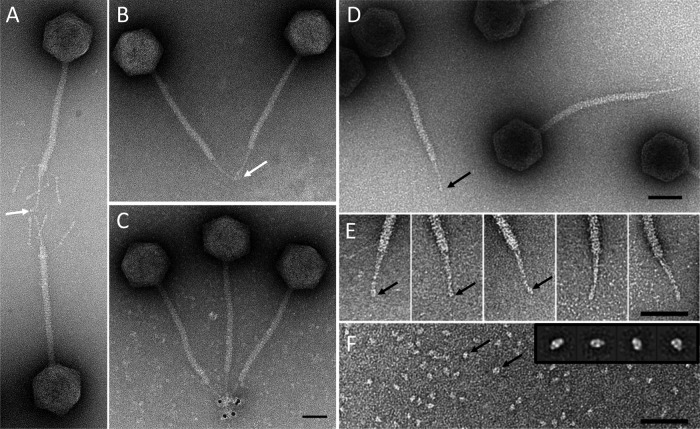
IgG cross-linking and gold labeling located pb5 in the rod-shaped extremity of the central tail fiber. Immunolocalization of pb5 was shown in T5st0 and T5hd1 (Fig. 5A, B, and C). Purified pb5 is a monomer, which forms, in vitro, a highly stable 1/1 stoichiometric complex with its receptor FhuA (21, 22). In either small angle neutron scattering and EM and single-particle analysis of purified monomeric pb5, side views revealed a prolate molecule 7 nm long and 5 nm wide (Fig. 5F) (23). These dimensions and shape fit very well in the volume of the central fiber extremity, arguing for the presence of a single copy of pb5 in the T5 tail tip. This observation strongly suggests that the pb5-FhuA complex formed in vitro is relevant to the physiological binding of T5 to its receptor. The position of pb5 is fully consistent with its RBP function and contradicts the previous localization of pb5 at the base of the tail tip cone (17).
FIG 5.
Localization of pb5 in the tail tip. Immunolocalization of pb5 by IgG cross-linking (white arrows) on phages T5st0 (A) and T5hd1 (B) or by visualization of the IgG molecules associated with anti-rabbit IgG–5-nm gold conjugate on T5hd1 (C). Panel E shows a gallery of tail tips selected from high-resolution images of T5hd1 (D). Panel F shows an image of purified pb5 (black arrows) with a gallery of the major class averages obtained by processing 659 selected single particles of pb5 in the inset (23). Bars, 50 nm.
Conformational changes of the tail tip after binding of T5 to its receptor FhuA.
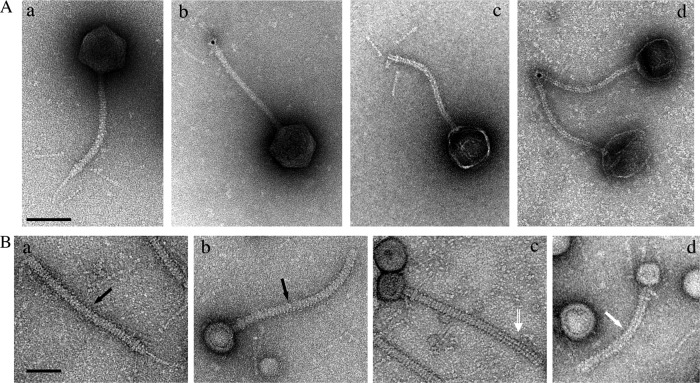
Incubation of phage T5 with its purified receptor FhuA promotes DNA ejection in the external medium (33) or into liposomes reconstituted with FhuA (62). We visualized the structural rearrangements occurring within the tail tip upon DNA ejection and investigated the position of pb2. Empty phages were imaged after digestion of ejected DNA (see Materials and Methods). While the collar and L-fibers did not appear significantly modified, the cone was open, giving access to the hollow tail tube, and the central fiber was lost (Fig. 6A, panels c and d). The presence of pb2 at the end of the cone, at the position of the original central fiber, was attested by labeling with anti-pb2 IgG, indicating that destabilization of the fiber was associated with unmasking of pb2 (Fig. 6A, panel d). Structural modification of the fiber together with pb2 exposure to the outside might be the immediate consequence of binding of pb5 to FhuA, preceding DNA egress, as witnessed by T5 particles full of DNA that have lost their tail tips (Fig. 6A, panel b). Incubation of isolated T5 tails with FhuA led to major aggregation of the tail tips, hindering the analysis of the conformational changes of the tail tips (data not shown). However, most of the tails exhibited an empty tail tube, confirming that pb2 exit is promoted by binding to FhuA, regardless of DNA transfer. An interesting observation came from partially purified tail preparations that contained membrane vesicles resulting from host cell lysis (see Materials and Methods). Upon lysis, some tails bind to FhuA receptors that are present in outer membrane vesicles. All these tails appeared to be buried in the vesicles by their tail fiber, leaving the cone rising above the surface of the membrane (Fig. 6B). The presence of uranyl acetate staining in the tail lumen indicates whether the tails are empty or not, i.e., whether the TMP domain of pb2 was expelled. We observed different states: filled tails, tails exhibiting partial ejection of pb2, and completely empty tails. These pictures suggest that anchoring of the central fiber into the host envelope does not require the full ejection of pb2. It raises the question of which tail proteins physically constitute the channel through which DNA is transported and of the fate of pb2 after the cell wall perforation.
FIG 6.
Modification of the tail tip after binding of phage T5 and tails to FhuA. (A) T5st0 particles incubated with purified FhuA and treated with Benzonase before EM (see Materials and Methods). A control image of T5 particles treated in the absence of FhuA is shown in subpanel a. The loss of the central tail fiber was visible in all phages with an empty capsid (c and d) and also in some phages with filled heads (b). The position of pb2 was visualized with gold-conjugated anti-pb2 IgG only in phage particles whose central fiber was affected (b and d). (B) EM images of partially purified tail preparations (addition of LDAO was omitted): free tails (a) and tails bound to membrane vesicles (b, c, and d). Bound tails include filled (b) and intermediate (c) tails and empty ones from pb2 (d), as indicated with black arrows, an open white arrow, and a solid white arrow, respectively. Upper bar, 100 nm; bottom bar, 50 nm.
DISCUSSION
We report the first comprehensive description of T5 morphogenesis genes and the topology of their products in viral particles and assigned functions, and we highlight major new structural features of T5, which is the prototype of a large family of lytic siphophages infecting Gram-negative hosts (4–7). New similarities between T5 and several families of Siphoviridae and Myoviridae are also identified, providing additional examples of the evolutionary connections between related viruses.
T5 capsid assembly and maturation.
The T5 prohead is assembled from the major head pb8, the portal protein pb7, and the protease pb11. The role of pb11 in the specific cleavage of the N-terminal scaffolding domain of pb8 has been previously demonstrated in vitro on tube structures assembled from the purified full-length precursor of pb8 (10). Here, we show that the three procapsid proteins are processed prior to DNA packaging, suggesting that pb11 is responsible for these multiple proteolytic events, which likely control the formation of T5 procapsids. Head proteases are essential for capsid assembly and maturation in other phages. In phage HK97, the gp4 uncleaved protease stabilizes the initial procapsid prior to autoproteolysis and digestion of the scaffolding domain of the major head protein gp5 (63). In phage λ, the gpC protease degrades the scaffolding protein gpNu3 and cleaves the N-terminal end of the portal protein gpB (64). For both phages, the digested proteases escape the capsid before or upon DNA packaging. Remarkably, in T5, a low copy number of processed pb11 remains associated with the mature capsid (9 ± 3 per T5 particle, as estimated by densitometry measurements on SDS-PAGE). Thus, pb11, like other proteases from the myophages T4 (65), P2 (66), and phiKZ (67), might not be just a morphogenetic enzyme but also a structural component of the mature capsid. In all these phages, it is not known whether the remaining processed proteases serve an additional function in packaging, organization, or ejection of the DNA.
The analysis of T5 morphogenesis genes reasonably indicates that T5p144 is the only head completion protein, which closes the portal vertex after DNA packaging, to form the connector and provide the attachment site for the tail. This is a striking difference from other tailed phages described so far (see reference 68 for a recent review). In the siphophages λ, SPP1, and HK97, the connector includes two ring-layers of head completion proteins: the upper one is formed by the adaptor protein λgpW, SPP1gp15, or HK97gp6 that seals the portal gate, and the bottom one is formed by the stopper protein λgpFII, SPP1gp16, or HK97gp7. In phage T4, capsid assembly is ended by successive addition of a dodecamer of gp13 and a hexamer of gp14 (69). T5p144 is the first characterized example of a single head-tail connector protein, which might be a variant of the adaptor-stopper system. Further structural studies should shed light on how T5p144 connects the dodecamer of the portal pb7 to the putative hexameric T5p142 tail terminator and provide information on this family of atypical head completion proteins.
These data provide a comprehensive view of the protein composition of the T5 capsid. The head morphogenesis module includes, in addition to the terminase subunits, a conserved endonuclease (T5p149), which might generate the nicks found in the T5 genome. Whether and how these proteins are involved in T5 assembly or in DNA packaging will be the subject of future investigation.
T5 tail structure and architecture of T5 tail tip complex.
By combining gene map analysis with sequence similarities and immunolocalization of unknown tail proteins, we obtained an almost complete view of the tail components. While the proteins forming the top of the tail—tail terminator T5p142 and tail completion T5p143—are likely organized like their homologous proteins in most other siphophages, the proteins forming the tail tip architecture show an unusual topology. The conical structure of the tail tip is formed by two proteins: pb9, the Dit protein located in the upper part (56), and pb3, located in the lower part in contact with the central straight fiber. The Dit protein pb9 would serve as the docking site for the long L-shaped side fibers and their collar base (formed by pb1 and the newly identified T5p132 protein). When these fibers are absent—as seen in the T5hd1 mutant—we can observe the intact conical structure. We propose that pb3 is the BHP of T5 on the basis of its similarity with the gp27-like BHPs of the T4 myophage. Thus, pb3 would form the gate at the end of the tail tube, which opens to allow DNA ejection in response to binding of T5 to its receptor. This hypothesis is supported by EM images, showing that once T5 has ejected its DNA after in vitro interaction with FhuA or when isolated tails are inserted into vesicles, the cone is open and forms a hollow tube in continuity with the tail tube. Interestingly, pb3 was linked to two distinct tail proteins of phages λ and PY54. The N-terminal end of pb3 forming the putative BHP-HDI domain was related to gpL homologues, and the central region forming the putative BHP-HDIII/IV domains was related to the host specificity/straight fiber gpJ homologues. While genes gpL and gpJ are separated by the two genes gpK and gpI in phages λ and PY54, the corresponding sequences are fused in the same pb3 ORF in T5. gpL, which is a two-domain protein (57), probably does not form the BHP of phage λ but includes the HD1 fold that is shared by many tail proteins (see reference 56 and references therein). gpJ is long enough to encode the BHP function together with the central fiber and RBP functions. Similarly, the gp21 protein of the Gram-positive bacterium-infecting phage SPP1 bears these three functions (70, 71). Our data argue in favor of pb4 forming the fiber (probably a trimer as suggested by SDS-PAGE densitometry analysis) that is bound to pb3 at the lower part of the cone. However, pb3 might also form part of this fiber as it is significantly larger than the known BHPs and includes two fibronectin domains that are found in many tail fiber proteins (Fig. 4). Finally, the extremity of the straight fiber is formed by a monomer of the RBP pb5. Figure 7 shows a schematic representation of the tail architecture and illustrates how the same functions are distributed into separate genes to form similar tail tip complexes in T5, λ, and SPP1. The main difference is the repartition of the Dit, BHP, central fiber, and RBP genes. Only four proteins (pb9, pb3, pb4, and pb5) likely form the T5 tail tip complex, while additional proteins are required in phage λ (gpL, -K, and -I) (51) and possibly in phage SPP1 (gp22, gp23, gp23.1, gp24, and gp24.1) (71).
FIG 7.
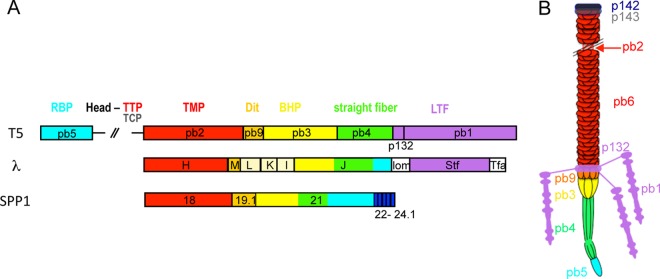
(A) Arrangement of the tail tip genes in the siphophages T5, λ, and SPP1. Genes or part of them predicted to encode the same functions are depicted in the same color: TMP, red; Dit, orange; BHP, yellow; central straight fiber, green; RBP, cyan; LTF or Stf side fibers, violet. The proteins L, K, and I of λ (beige) are known to be part of the tail tip complex, but their location is unknown. Genes 22, 23, 23.1, 24, and 24.1 of SPP1, whose function and location are unknown, might encode proteins of the tail tip distal end. (B) Schematic representation of T5 tail architecture based on the description of its structural proteins.
The fate of the tail tip proteins upon DNA transfer remains an open question. In the podovirus T7, proteins forming the capsid internal core are ejected to form a conduit through the whole infected cell envelope (72). In T4, the proteins forming the internal tail tube and the cell-puncturing device, and which are evolutionarily related to the type VI secretion apparatus, very likely perforate the host cell (73). In T5, EM images and immunolocalization analysis indicate that the TMP pb2, which is initially buried in the tail tube and tail tip complex, is released after binding of T5 to FhuA. Exit of pb2 from the tail tube is associated with the destabilization of the straight fiber. The same observations—TMP release and fiber disruption—were made with phage SPP1 as it binds its receptor YueB (70, 74). EM images of the T5 tails inserted into membrane vesicles (this study) together with previous cryo-electron tomography images showing that T5 can inject its DNA into liposomes containing FhuA (75) suggest that it is the straight fiber that forms a channel across the host envelope, while the cone opens and remains in close contact with the outer surface of the membrane. We have previously shown that pb2 is a pore-forming protein that is very likely involved in the host envelope perforation (16); however, further investigation is required to reveal how pb2, together with other tail straight fiber proteins, channel T5 DNA across the host membranes.
Concluding remarks.
Several structural and genetic features of phage T5 have been revealed that lead to a deeper understanding of the phage structure and of its assembly. T5 properties described in this work and previously are both unique in some details and generally applicable to the huge family of dsDNA tailed phages. The study of T5 and its relatives will thus help to provide an increasingly detailed picture of the variable strategies used by bacteriophages to infect their hosts.
ACKNOWLEDGMENTS
Special thanks go to L. Letellier for support at the early stages of this study and P. Tavares and J. F. Conway for fruitful discussions and critical reading of the manuscript. We thank I. Molineux for providing T5D17am34d, T5D20am30d, and T5amHA911 mutants and K. Heller for providing the T5hd1 mutant. We are grateful to M. Argentini and D. Cornu for the use of the mass spectrometry facilities from the SiCaps platform at ImaGif, CNRS Gif sur Yvette; to J. Drouin for devoted technical assistance; and to C. Pereira and F. Grégoire for help with the purification of proteins for immunization. We thank H. Stahlberg (C-CINA, Biozentrum, University of Basel, Switzerland) for support.
We used the RoBioMol platform of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. This work was supported by funding from the French National Research Agency (ANR-06-PCVI-0002), the Institut Fédératif de Recherche 115, and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement 211800. The EM work was supported in part by the Swiss National Science Foundation (SystemsX.ch RTD CINA).
Footnotes
Published ahead of print 6 November 2013
REFERENCES
- 1.Wang J, Jiang Y, Vincent M, Sun Y, Yu H, Bao Q, Kong H, Hu S. 2005. Complete genome sequence of bacteriophage T5. Virology 332:45–65. 10.1016/j.virol.2004.10.049 [DOI] [PubMed] [Google Scholar]
- 2.Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, Schuerch DW, Klebba JE, Roe BA, Gomez JAL, Schallmey M, Newton SMC, Klebba PE. 2007. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J. Bacteriol. 189:5658–5674. 10.1128/JB.00437-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong J, Kim KP, Heu S, Lee SJ, Adhya S, Ryu S. 2008. Identification of host receptor and receptor-binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202–209. 10.1111/j.1574-6968.2008.01397.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050. 10.1128/AEM.02504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu YD, Stanford K, Kropinski AM, Ackermann HW, Johnson RP, She YM, Ahmed R, Villegas A, McAllister TA. 2012. Genomic, proteomic and physiological characterization of a T5-like bacteriophage for control of Shiga toxin-producing Escherichia coli O157:H7. PLoS One 7:e34585. 10.1371/journal.pone.0034585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JH, Jun JW, Choresca CH, Shin SP, Han JE, Park SC. 2012. Complete genome sequence of a novel marine siphovirus, pVp-1, infecting Vibrio parahaemolyticus. J. Virol. 86:7013–7014. 10.1128/JVI.00742-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DH, Lee JH, Shin H, Ji S, Roh E, Jung K, Ryu S, Choi J, Heu S. 2012. Complete genome sequence of Pectobacterium carotovorum subsp. carotovorum bacteriophage My1. J. Virol. 86:11410–11411. 10.1128/JVI.01987-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zweig M, Cummings DJ. 1973. Structural proteins of bacteriophage T5. Virology 51:443–453. 10.1016/0042-6822(73)90443-1 [DOI] [PubMed] [Google Scholar]
- 9.Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. 2006. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 361:993–1002. 10.1016/j.jmb.2006.06.081 [DOI] [PubMed] [Google Scholar]
- 10.Huet A, Conway JF, Letellier L, Boulanger P. 2010. In vitro assembly of the t=13 procapsid of bacteriophage T5 with its scaffolding domain. J. Virol. 84:9350–9358. 10.1128/JVI.00942-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preux O, Durand D, Huet A, Conway JF, Bertin A, Boulogne C, Drouin-Wahbi J, Trevarin D, Perez J, Vachette P, Boulanger P. 2013. A two-state cooperative expansion converts the procapsid shell of bacteriophage T5 into a highly stable capsid isomorphous to the final virion head. J. Mol. Biol. 425:1999–2014. 10.1016/j.jmb.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 12.Kaliman AV, Kulshin VE, Shlyapnikov MG, Ksenzenko VN, Kryukov VM. 1995. The nucleotide sequence of the bacteriophage T5 ltf gene. FEBS Lett. 366:46–48. 10.1016/0014-5793(95)00482-O [DOI] [PubMed] [Google Scholar]
- 13.Saigo K. 1978. Isolation of high-density mutants and identification of nonessential structural proteins in bacteriophage T5; dispensability of L-shaped tail fibers and a secondary major head protein. Virology 85:422–433. 10.1016/0042-6822(78)90449-X [DOI] [PubMed] [Google Scholar]
- 14.Schwarzer D, Stummeyer K, Gerardy-Schahn R, Muhlenhoff M. 2007. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J. Biol. Chem. 282:2821–2831. 10.1074/jbc.M609543200 [DOI] [PubMed] [Google Scholar]
- 15.Feucht A, Schmid A, Benz R, Schwarz H, Heller KJ. 1990. Pore formation associated with the tail-tip protein pb2 of bacteriophage T5. J. Biol. Chem. 265:18561–18567 [PubMed] [Google Scholar]
- 16.Boulanger P, Jacquot P, Plancon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. 2008. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 283:13556–13564. 10.1074/jbc.M800052200 [DOI] [PubMed] [Google Scholar]
- 17.Heller KJ, Schwarz H. 1985. Irreversible binding to the receptor of bacteriophages T5 and BF23 does not occur with the tip of the tail. J. Bacteriol. 162:621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. 2012. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 726:115–142. 10.1007/978-1-4614-0980-9_6 [DOI] [PubMed] [Google Scholar]
- 19.Heller KJ, Bryniok D. 1984. O antigen-dependent mutant of bacteriophage T5. J. Virol. 49:20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondigler M, Holz T, Heller KJ. 1996. Identification of the receptor-binding regions of pb5 proteins of bacteriophages T5 and BF23. Virology 219:19–28. 10.1006/viro.1996.0218 [DOI] [PubMed] [Google Scholar]
- 21.Plançon L, Janmot C, Le Maire M, Desmadril M, Bonhivers M, Letellier L, Boulanger P. 2002. Characterization of a high-affinity complex between the bacterial outer membrane protein FhuA and the phage T5 protein pb5. J. Mol. Biol. 318:557–569. 10.1016/S0022-2836(02)00089-X [DOI] [PubMed] [Google Scholar]
- 22.Flayhan A, Wien F, Paternostre M, Boulanger P, Breyton C. 2012. New insights into pb5, the receptor binding protein of bacteriophage T5, and its interaction with its Escherichia coli receptor FhuA. Biochimie 94:1982–1989. 10.1016/j.biochi.2012.05.021 [DOI] [PubMed] [Google Scholar]
- 23.Breyton C, Flayhan A, Gabel F, Lethier M, Durand G, Boulanger P, Chami M, Ebel C. 2013. Assessing the conformational changes of pb5, the receptor-binding protein of phage T5, upon binding to its Escherichia coli receptor FhuA. J. Biol. Chem. 288:30763–30772. 10.1074/jbc.M113.501536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheible PP, Rhoades M. 1975. Heteroduplex mapping of heat-resistant deletion mutants of bacteriophage T5. J. Virol. 15:1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanni YT. 1958. Lysis inhibition with a mutant of bacteriophage T5. Virology 5:481–501. 10.1016/0042-6822(58)90041-2 [DOI] [PubMed] [Google Scholar]
- 26.Boulanger P. 2009. Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. Methods Mol. Biol. 502:227–238. 10.1007/978-1-60327-565-1_13 [DOI] [PubMed] [Google Scholar]
- 27.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentz R, Bujard H. 1985. Promoters recognized by Escherichia coli RNA polymerase selected by function: highly efficient promoters from bacteriophage T5. J. Bacteriol. 164:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noirclerc-Savoye M, Gallet B, Bernaudat F, Vernet T. 2010. Large scale purification of linear plasmid DNA for efficient high throughput cloning. Biotechnol. J. 5:978–985. 10.1002/biot.201000132 [DOI] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. 2009. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U. S. A. 106:20240–20245. 10.1073/pnas.0911500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 33.Boulanger P, le Maire M, Bonhivers M, Dubois S, Desmadril M, Letellier L. 1996. Purification and structural and functional characterization of FhuA, a transporter of the Escherichia coli outer membrane. Biochemistry 35:14216–14224. 10.1021/bi9608673 [DOI] [PubMed] [Google Scholar]
- 34.Hendrickson HE, McCorquodale DJ. 1971. Genetic and physiological studies of bacteriophage T5. I. An expanded genetic map of T5. J. Virol. 7:612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng H, Shen N, Pei J, Grishin NV. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13:2260–2269. 10.1110/ps.04726004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Mushegian A. 2004. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J. Bacteriol. 186:4369–4375. 10.1128/JB.186.13.4369-4375.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. 2010. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J. Mol. Biol. 395:754–768. 10.1016/j.jmb.2009.10.067 [DOI] [PubMed] [Google Scholar]
- 38.Casjens SR. 2011. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9:647–657. 10.1038/nrmicro2632 [DOI] [PubMed] [Google Scholar]
- 39.Ponchon L, Boulanger P, Labesse G, Letellier L. 2006. The endonuclease domain of bacteriophage terminases belongs to the resolvase/integrase/ribonuclease H superfamily: a bioinformatics analysis validated by a functional study on bacteriophage T5. J. Biol. Chem. 281:5829–5836. 10.1074/jbc.M511817200 [DOI] [PubMed] [Google Scholar]
- 40.Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA. 2012. Structural basis for DNA recognition and loading into a viral packaging motor. Proc. Natl. Acad. Sci. U. S. A. 109:811–816. 10.1073/pnas.1110270109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunin-Horkawicz S, Feder M, Bujnicki JM. 2006. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics 7:98. 10.1186/1471-2164-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta P, Katta K, Krishnaswamy S. 2004. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 13:295–300. 10.1110/ps.03115604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers SG, Godwin EA, Shinosky ES, Rhoades M. 1979. Interruption-deficient mutants of bacteriophage T5. I. Isolation and general properties. J. Virol. 29:716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krauel V, Heller KJ. 1991. Cloning, sequencing, and recombinational analysis with bacteriophage BF23 of the bacteriophage T5 oad gene encoding the receptor-binding protein. J. Bacteriol. 173:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pell LG, Liu A, Edmonds L, Donaldson LW, Howell PL, Davidson AR. 2009. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J. Mol. Biol. 389:938–951. 10.1016/j.jmb.2009.04.072 [DOI] [PubMed] [Google Scholar]
- 46.Auzat I, Dröge A, Weise F, Lurz R, Tavares P. 2008. Origin and function of the two major tail proteins of bacteriophage SPP1. Mol. Microbiol. 70:557–569. 10.1111/j.1365-2958.2008.06435.x [DOI] [PubMed] [Google Scholar]
- 47.Pell LG, Gasmi-Seabrook GM, Morais M, Neudecker P, Kanelis V, Bona D, Donaldson LW, Edwards AM, Howell PL, Davidson AR, Maxwell KL. 2010. The solution structure of the C-terminal Ig-like domain of the bacteriophage lambda tail tube protein. J. Mol. Biol. 403:468–479. 10.1016/j.jmb.2010.08.044 [DOI] [PubMed] [Google Scholar]
- 48.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160–4165. 10.1073/pnas.0900044106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu J, Hendrix RW, Duda RL. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11–21. 10.1016/j.molcel.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 50.Xu J, Hendrix RW, Duda RL. 2013. A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. J. Mol. Biol. 425:3476–3487. 10.1016/j.jmb.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsura I, Kuhl PW. 1975. Morphogenesis of the tail of bacteriophage lambda. III. Morphogenetic pathway. J. Mol. Biol. 91:257–273 [DOI] [PubMed] [Google Scholar]
- 52.Heller K, Braun V. 1982. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 41:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ. 2010. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl. Acad. Sci. U. S. A. 107:20287–20292. 10.1073/pnas.1011218107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Doval C, van Raaij MJ. 2012. Crystallization of the C-terminal domain of the bacteriophage T7 fibre protein gp17. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68:166–171. 10.1107/S1744309111051049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, Hainfeld JF, Studier FW. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351–365 [DOI] [PubMed] [Google Scholar]
- 56.Flayhan A, Vellieux FMD, Lurz R, Maury O, Contreras-Martel C, Girard E, Boulanger P, Breyton C. 2014. Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages. J. Virol. 88:820–828. 10.1128/JVI.02135-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tam W, Pell LG, Bona D, Tsai A, Dai XX, Edwards AM, Hendrix RW, Maxwell KL, Davidson AR. 2013. Tail tip proteins related to bacteriophage lambda gpL coordinate an iron-sulfur cluster. J. Mol. Biol. 425:2450–2462. 10.1016/j.jmb.2013.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendrix RW, Casjens S. 2006. Bacteriophage lambda and its genetic neighborhood, p 409–447 In Calendar R. (ed), The bacteriophages. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 59.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553–557. 10.1038/415553a [DOI] [PubMed] [Google Scholar]
- 60.Kondou Y, Kitazawa D, Takeda S, Tsuchiya Y, Yamashita E, Mizuguchi M, Kawano K, Tsukihara T. 2005. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J. Mol. Biol. 352:976–985. 10.1016/j.jmb.2005.07.044 [DOI] [PubMed] [Google Scholar]
- 61.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichiere J, van Heel M, Campanacci V, Moineau S, Cambillau C. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 107:6852–6857. 10.1073/pnas.1000232107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambert O, Plançon L, Rigaud JL, Letellier L. 1998. Protein-mediated DNA transfer into liposomes. Mol. Microbiol. 30:761–765. 10.1046/j.1365-2958.1998.01107.x [DOI] [PubMed] [Google Scholar]
- 63.Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, Johnson JE. 2011. The prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J. Mol. Biol. 408:541–554. 10.1016/j.jmb.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Medina E, Wieczorek D, Medina EM, Yang Q, Feiss M, Catalano CE. 2010. Assembly and maturation of the bacteriophage lambda procapsid: gpC is the viral protease. J. Mol. Biol. 401:813–830. 10.1016/j.jmb.2010.06.060 [DOI] [PubMed] [Google Scholar]
- 65.Black LW, Showe MK. 1993. Morphogenesis of T4 head, p 218–258 In Karam JD. (ed), Molecular biology of bacteriophage T4. ASM Press, Washington, DC [Google Scholar]
- 66.Marvik OJ, Sharma P, Dokland T, Lindqvist BH. 1994. Bacteriophage P2 and P4 assembly: alternative scaffolding proteins regulate capsid size. Virology 200:702–714. 10.1006/viro.1994.1234 [DOI] [PubMed] [Google Scholar]
- 67.Thomas JA, Weintraub ST, Wu W, Winkler DC, Cheng N, Steven AC, Black LW. 2012. Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage phiKZ. Mol. Microbiol. 84:324–339. 10.1111/j.1365-2958.2012.08025.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tavares P, Zinn-Justin S, Orlova EV. 2012. Genome gating in tailed bacteriophage capsids. Adv. Exp. Med. Biol. 726:585–600. 10.1007/978-1-4614-0980-9_25 [DOI] [PubMed] [Google Scholar]
- 69.Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. 2013. The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol. 425:1731–1744. 10.1016/j.jmb.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goulet A, Lai-Kee-Him J, Veesler D, Auzat I, Robin G, Shepherd DA, Ashcroft AE, Richard E, Lichiere J, Tavares P, Cambillau C, Bron P. 2011. The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J. Biol. Chem. 286:25397–25405. 10.1074/jbc.M111.243360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, Santos MA, Sao-Jose C. 2012. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol. Microbiol. 83:289–303. 10.1111/j.1365-2958.2011.07931.x [DOI] [PubMed] [Google Scholar]
- 72.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. 10.1126/science.1231887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159. 10.1073/pnas.0813360106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sao-Jose C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. 2006. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J. Biol. Chem. 281:11464–11470. 10.1074/jbc.M513625200 [DOI] [PubMed] [Google Scholar]
- 75.Böhm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL. 2001. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11:1168–1175. 10.1016/S0960-9822(01)00349-9 [DOI] [PubMed] [Google Scholar]
- 76.McCorquodale JD, Warner HR. 1988. Bacteriophages T5 and related phages, p 439–476 In Calendar R. (ed), The viruses, vol 1 Plenum Press, New York, NY [Google Scholar]
- 77.Catherinot V, Labesse G. 2004. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics 20:3694–3696. 10.1093/bioinformatics/bth429 [DOI] [PubMed] [Google Scholar]