Abstract
HLA-C-restricted T cells have been shown to play an important role in HIV control, but their impact on protection or pathogenesis in other viral infections remains elusive. Here, we characterized the hierarchy of HLA class I-restricted hepatitis B virus (HBV) epitopes targeted by CD8 T cells in HBV-infected subjects. The frequency of CD8 T cells specific for a panel of 18 HBV epitopes (restricted by HLA-A∗0201/03/07 [hereinafter HLA-A0201/03/07], -A1101, -A2402/07, -B5801, -B4001, -B1301, and -Cw0801) was quantified in a total of 59 subjects who resolved HBV infection. We found that the HLA-Cw0801-restricted epitope comprised of Env residues 171 to 180 (Env171–180) is immunoprevalent in the Southeast Asian subjects (10/17 HLA-Cw0801-positive subjects) and immunodominant in the majority of HLA-Cw0801-positive subjects able to control HBV infection. HLA-Cw0801-restricted Env171–180-specific CD8 T cells recognized endogenously produced HBV surface antigen (HBsAg) and tolerated amino acid variations within the epitope detected in HBV genotypes B and C. In conclusion, we demonstrate that the HLA-Cw0801-restricted Env171–180 T cell response is an important component of the HBV-specific adaptive T cell immunity in Asians infected with HBV. Thus, HLA-C restricted T cells might play an important role in various viral infections.
INTRODUCTION
During viral infections, CD8 T cells clear virus-infected cells due to their ability to recognize viral proteins presented, in the form of short peptides, by different major histocompatibility complex (MHC) class I molecules on the surface of the cells. Two allelic forms of MHC class I proteins coded by three distinct genes, HLA-A, -B, and -C, are expressed in human nucleated cells. Virus-specific CD8 T cells recognizing HLA-A/B viral peptide complexes have been amply characterized in humans, with HLA-B-restricted CD8 T cells often associated with superior antiviral activity (1, 2). In contrast, since HLA-C molecules seem to be expressed at levels 10% lower than HLA-A and HLA-B molecules, CD8 T cells specific for viral peptides presented by HLA-C molecules have been thought to be rare and characterized by weak antiviral activity (3).
Seminal data obtained in HIV infection has, however, challenged this concept. The first observation was derived from a genome-wide association study that identified a strong association between a dimorphism 35 kb upstream of the HLA-C gene promoter and levels of HIV viremia (4). Such results were complemented by the finding that the HLA-C variant −35C, associated with lower viremia, was linked with higher expression of HLA-C molecules in European/American populations, showing that higher expression of HLA-C molecules confers protection from HIV (5, 6). Recently, the protective value of HLA-C-restricted T cell responses in HIV infections was extended to Asian populations, where it was shown that the high expression of HLA-C molecules results in a stronger HLA-C-restricted HIV-specific immune response and an increased frequency of viral mutations on targeted epitopes (7).
However, the protective impact of HLA-C-restricted T cells might be an exclusive feature of HIV infection since during HIV replication, the HIV negative replication factor protein (nef) selectively downregulates HLA-A and HLA-B expression without interfering with HLA-C expression levels (8). Even though HLA-C-restricted CD8 T cells have been detected in other viral infections (human cytomegalovirus [HCMV], Epstein-Barr virus [EBV], and hepatitis C virus [HCV]) (3), the role played by HLA-C-restricted T cells in viral protection or pathogenesis remains elusive.
We recently characterized in a Han Chinese patient with acute hepatitis B virus (HBV) infection an HLA-Cw∗0801 (hereinafter HLA-Cw0801)-restricted T cell response specific for the conserved region of residues 171 to 180 of the envelope protein (Env171–180) of HBV genotypes A, C, D, and F (9). Whether this epitope is frequently targeted by HBV-specific T cells in HLA-Cw08-positive (HLA-Cw08+) patients and whether such HLA-C08-restricted T cells represent a dominant or subdominant response within an infected individual are not known.
Such information is important since knowledge of the repertoire of HBV-specific CD8 T cells is very limited in patients infected with HBV genotypes B and C (the dominant HBV strains in Northeast/Southeast Asia), and an understanding of the prevalence of the CD8 response against defined HBV epitopes in South Asian patients carrying specific HLA class I (horizontal immunodominance or immunoprevalence) is needed. At the moment, HBV-specific CD8 T cell characterization in this patient population is limited to few epitopes (10, 11) or is based on theoretical epitopes predicted by the presence of mutations associated with selected HLA alleles (10, 12). Furthermore, we have demonstrated that CD8 T cell epitopes frequently recognized and immunodominant in HLA-A02 Caucasian patients infected by HBV genotypes A and D are rarely detected in HLA-A02 South Asian patients (13).
Thus, to have a better understanding of the HBV epitopes targeted by CD8 T cells in subjects infected with HBV genotypes B and C, we interrogated the HBV-specific T cell repertoire of 59 patients of South Asian origin who control HBV infection (with or without an episode of acute hepatitis) and found that the Env171–180 epitope was recognized in most of the HBV-infected subjects carrying the HLA-Cw0801 allele. The vertical immunodominance hierarchy, defined as the hierarchy of dominant or subdominant CD8 T cell responses within a subject, of the HBV-specific cytotoxic T lymphocyte (CTL) responses in these patients was also determined. This additional parameter helps in the definition of possible protective epitopes since often, within an infected individual, the dominant CD8 T cell response should exert more antiviral activity than subdominant ones.
By reporting that the HLA-Cw0801-restricted Env171–180 epitope represents a horizontal and vertical immunodominant HBV epitope in South Asian populations, we provide valuable information about the important antigenic region targeted by the immune system during HBV infection and also show that the substantial role of HLA-C-restricted T cells in viral infection might not be restricted only to HIV infection.
MATERIALS AND METHODS
Patient population.
HBV-infected patients of Southeast Asian origin were enrolled at the National University Hospital of Singapore or at Chulalongkorn University, Bangkok, Thailand. This study was approved by the ethics committees of both Chulalongkorn University and the National University Hospital of Singapore. Eight patients had clinical, biochemical, and virological evidence of acute HBV infection (alanine aminotransferase [ALT] levels of >10 times the upper limit of normal, detection of HBV surface antigen [HBsAg] and serum anti-HBV core protein [HBc] IgM, and HBsAg clearance within 2 months from clinical onset of hepatitis). Fifty-one patients had only serological evidence of HBV contact (HBsAg negative and IgG anti-HBc and anti-HBV surface protein [HBs] positive) but no history of acute hepatitis. All patients were HLA typed (BGI, Hong Kong, China), gave written informed consent, and were serologically negative for HIV and HCV. We also selected three patients with chronic HBV infection (HbsAg positive for over 6 months) that were positive for the HLA-Cw0801 allele.
Synthetic peptides.
A library of 313 synthetic 15-mer peptides overlapping by 10 amino acid (aa) residues covering the whole HBV genotype C proteome sequence were purchased from Chiron Mimotopes (Victoria, Australia). The pools of core and X peptides were made into a 9-by-8 matrix, containing eight or nine peptides/pool, respectively, whereas envelope peptides were pooled in a 9-by-9 matrix containing nine peptides/pool and polymerase peptides formed a 14-by-12 matrix containing 12 or 14 peptides/pool, as described previously (13). The known HBV epitopes were purchased from Proimmune (Oxford, United Kingdom) and from GenScript (Piscataway, NJ).
Stimulation of PBMCs.
Peripheral blood mononuclear cells (PBMCs) from patients were isolated by Ficoll-Hypaque density gradient centrifugation (Sigma Chemical Co., St. Louis, MO) and resuspended in AIM-V medium (Gibco-BRL Laboratories, Gaithersburg, MD) with 2% human AB serum. PBMCs were in vitro expanded with peptides for 10 days before assays were performed. For full proteome screening, 20% of PBMCs were pulsed with 10 μg/ml of each overlapping peptide for 1 h at 37°C and then washed and cocultured with the remaining PBMCs (80%) in AIM-V medium with 2% human AB serum and 20 U/ml of interleukin-2 (IL-2) (R&D Systems, Abingdon, United Kingdom). For single peptide expansion, HBV peptides were added directly at 5 μg/ml for 15-mer peptides and at 1 μg/ml for 9- to 10-mer peptides.
Intracellular cytokine staining (ICS) and degranulation assays.
In vitro-expanded PBMCs were incubated in medium alone (control) or with viral peptides (5 μg/ml) for 5 h in the presence of brefeldin A (10 mg/ml). After being washed, the cells were stained with anti-CD8 phycoerythrin (PE)-Cy7 and anti-CD3 peridinin chlorophyll protein (PerCp)-Cy5.5 monoclonal antibodies (MAbs) for 30 min at 4°C and then fixed and permeabilized using Cytofix/Cytoperm Fixation/Permeabilization solution (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Cells were then stained with anti-gamma interferon (IFN-γ) PE for 30 min on ice, washed, and analyzed by flow cytometry. To assess degranulation activity, CD107a PE antibody (BD Pharmingen, San Diego) was added to all wells at the beginning of the 5 h of incubation. Following the incubation, cells were washed and labeled with CD8 PE-Cy7 and CD3 PerCp-Cy5.5 as described above.
IFN-γ ELISPOT assay.
IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assays were performed as previously described (13) using a panel of 313 overlapping peptides covering the full proteome sequence of HBV genotype C pooled in the described mixtures. HBV-specific T cell responses were analyzed in IFN-γ ELISPOT assays either ex vivo using fresh or frozen PBMCs or after short-term peptide-specific polyclonal T cell expansion (10 days). Briefly, 96-well plates (Multiscreen-HTS; Millipore, Billerica, MA) were coated overnight at 4°C with 5 μg/ml capture mouse anti-human IFN-γ monoclonal antibody (1DIK; Mabtech, Sweden). Plates were then blocked with AIM-V medium supplemented with 10% heat-inactivated fetal calf serum (FCS) for 30 min at room temperature. A total of 1 × 105 PBMCs or 5 × 104 cells from short-term polyclonal T cell lines were seeded per well, in duplicates for each individual peptide mixture. Plates were incubated for 18 h at 37°C in the absence or presence of peptides (at a final concentration of 5 μg/ml). After the incubation, plates were developed using the alkaline phosphatase substrate 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium chloride (BCIP/NBT; KPL, MD) according to the recommended protocol from Mabtech. The colorimetric reaction was stopped after 10 to 15 min by washing the plates with distilled water. Plates were air dried, and spots were counted using an automated ELISPOT reader (ImmunoSpot reader; CTL Technologies, OH). The number of peptide-specific IFN-γ-secreting cells was calculated by subtracting the nonstimulated control value from that of the stimulated sample. Positive controls consisted of PBMCs stimulated with phorbol myristate acetate (10 ng/ml) and ionomycin (100 ng/ml). In the direct ex vivo assays, wells were considered positive when the number of spot-forming units (SFU) was above 5 and at least three times the mean value of the unstimulated control wells (three wells/patient). The positivity criteria for in vitro ELISPOT assays is less stringent, requiring that wells that have SFU counts above 5 and at least two times the mean value of unstimulated control wells, but the responses were reconfirmed using IFN-γ ICS.
HLA restriction, fine specificity, and epitope avidity assay.
The HLA restriction of the T cell responses was deduced by coculturing the short-term T cell lines for 5 h with a panel of HLA class I-matched Epstein-Barr virus (EBV)-transformed B cells pulsed with the specific peptide. After T cells were cocultured with EBV B cells, IFN-γ- and CD107a-expressing CD8+ cells were quantified by flow cytometry. To determine the minimal epitope, short peptides (8- to 10-mer) were designed based on the responding 15-mer peptide and tested using IFN-γ ICS. The functional avidity of the minimal epitopes was determined by pulsing EBV B cell lines carrying the appropriate HLA class I molecules with serially diluted concentrations of the corresponding peptides (1 μM, 100 nM, 1 nM, 100 pM, and 1 pM), followed by coculturing with the corresponding short-term T cell lines (derived from the PBMCs of HBV peptide-responding patients) and quantification of the IFN-γ and CD107a response by flow cytometry. We then calculated the percentage of activated T cells and interpolated the corresponding 50% effective concentrations (EC50s) from the data.
Endogenous processing and presentation of the Env171–180 epitope.
SNU-423, an HLA-Cw0801-expressing hepatocellular carcinoma cell line, was transduced with the full HBV envelope (genotype C) using a Lenti-X HTX packaging system (Clontech, Mountain View, CA). A total of 50,000 SNU-423 cells expressing Env (SNU-423Env) and parental SNU-423 cells were plated on a 96-well flat-bottom plate overnight. The next day, 50,000 cells from a short-term in vitro-expanded T cell line specific for Env171–180 were added to both SNU-423 targets and cultured for 6 h in the presence of the protein transport inhibitor brefeldin A. After the culture, the cells were stained for CD8, IFN-γ, tumor necrosis factor alpha (TNF-α), and IL-2, according to the ICS protocol detailed above, and subsequently acquired on a FACSCanto instrument. As a positive control, the same number of HLA-Cw0801-expressing Epstein-Barr virus (EBV)-transformed B cells was pulsed with 1 μM Env171–180 peptide for 1 h and used as targets.
RESULTS
Horizontal immunodominance of HLA-Cw0801-restricted T cells.
The horizontal immunodominance (immunoprevalence) of 18 distinct HBV-specific CD8 T cell epitopes restricted by HLA-A0201/03/07, HLA-A1101, HLA-A2402/07, HLA-B5801, HLA-B4001, HLA-B1301, and HLA-Cw0801 (Table 1) were tested in HBV patients of Southeast Asian ethnicities (Chinese Han, Thai, Malay, and Indonesian Javanese). The amino acid sequences of different epitopes and their HLA-class I restrictions are displayed in Table 1. These epitopes were chosen as they are restricted by HLA alleles commonly present in the Southeast Asian population. To analyze the frequency of subjects carrying the respective HLA-class I allele who are able to respond to the corresponding HBV epitopes, the HLA-class I profile (four-digit resolution) of 59 subjects (HBsAg negative and anti-HBc and anti-HBs positive) with or without a reported episode of acute hepatitis was characterized.
TABLE 1.
List of known HBV epitopes tested
| Allele | Protein and amino acid position | HBV genotype Ca |
|---|---|---|
| HLA-A alleles | ||
| A0201/07 | Core 8–16 | EFGASVELL |
| A0201/07 | Core 18–27 | FLPSDFFPSI |
| A0201/07 | Env 61–70 | NLLGWSPQA |
| A0201/07 | Env 183–191 | FLLTRILTI |
| A0201/07 | Env 335–343 | WLSLLVPFV |
| A0201/07 | Env 338–347 | LLVPFVQWFV |
| A0201/07/03 | Env 348–357 | GLSPTVWLSV |
| A0201/07/03 | Env 370–379 | NILNPFLPLL |
| A0201/07 | Pol 455–463 | GLPRYVARL |
| A1101 | Core 88–96 | YVNVNMGLK |
| A1101 | Core 141–151 | TLPETTVVRR |
| A2402/07 | Core 117–125 | EYLVSFGVW |
| A2402/07 | Pol 756–764 | KYTSFPWLL |
| HLA-B alleles | ||
| B5801 | Env 356–364 | SVIWMMWYW |
| B5801 | Pol 165–174 | ASFCGSPYSW |
| B4001b | Core 75–83 | LEDPASREL |
| B1301 | Pol 386–395 | SRLVVDFSQF |
| HLA-C allele | ||
| Cw0801 | Env 171–180 | FLGPLLVLQA |
Amino acid positions with genotype-specific mutations are highlighted in bold.
Predicted epitope (12).
Selected subjects were positive for the following alleles: 24 for HLA-A02 (8 A0201, 9 A0203, and 9 A0207), 13 for HLA-A24 (9 A2402 and 4 A2407), 17 for Cw0801, 24 for A1101, 8 for B1301, 8 for B5801, and 7 for B4001. PBMCs of each subject were stimulated with the respective HLA class I-restricted epitopes in vitro for 10 days. The presence of epitope-specific CD8 T cells was determined with ICS and ELISPOT assays. Figure 1 shows a summary of the results obtained.
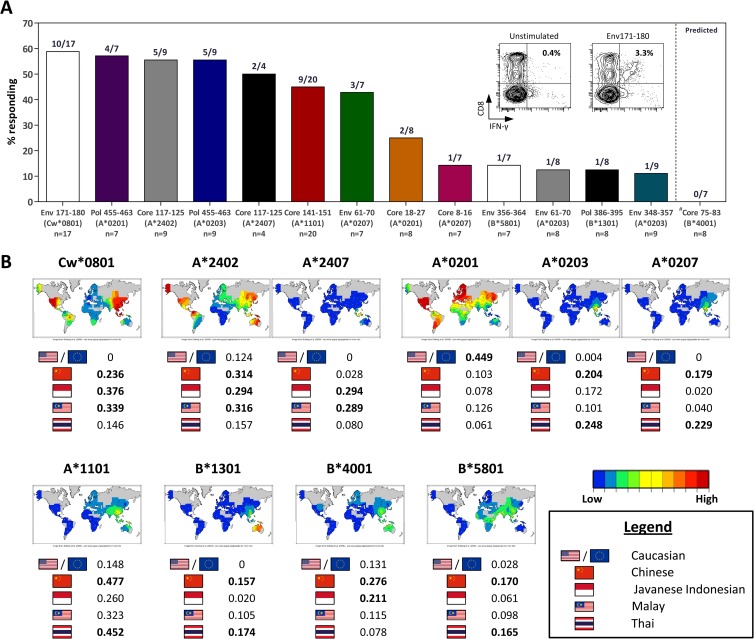
FIG 1.
Immunoprevalence of HLA-Cw0801-restricted Env171–180-specific T cell response in Southeast Asian subjects who resolved HBV infection. (A) Bars show the prevalence of the HLA-Cw0801/Env171–180 T cell response in Southeast Asian subjects who resolved HBV infection in comparison to previously characterized HBV epitopes restricted by HLA alleles commonly found in the Southeast Asian population. The inset shows the HLA-Cw0801/Env171–180 T cell response from a representative subject. Cells were gated for CD3 positivity, and the epitope-specific CD8 T cell response is shown as the frequency in the total T cell population. The number of subjects tested and the HLA restriction for each epitope are indicated below the bars. (B) Phenotypic frequency and the geographical distribution of HLA alleles commonly found in the Southeast Asian population. Heat maps and data were obtained from NCBI MHC database (dbMHC [http://www.ncbi.nlm.nih.gov/gv/mhc]).
As described previously by Tan et al., the immunoprevalence profile of CD8 T cell responses varies between the different populations. Unlike the situation in HLA-A0201-positive Caucasian HBV patients, where the response against the core protein epitope consisting of residues 18 to 27 (core18–27) was highly common, the prevalence of this response was low in our HLA-A0201-positive Southeast Asian cohort (two positive subjects out of eight tested) (Fig. 1A). Conversely, the response to the polymerase epitope consisting of residues 455 to 463 (Pol455–463) that was less frequently found in HLA-A0201-positive Caucasians was one of the most prevalent responses observed in our HLA-A0201-positive Southeast Asian subjects (four positive subjects out of seven tested) (Fig. 1A).
In our Southeast Asian cohort, four responses were the most frequently detected among various CD8 T cell epitopes tested. In descending order, the first was against the HLA-Cw0801-restricted Env171–180 epitope, which was detected in 10 of the 17 HLA-Cw0801 subjects; the second was against the Pol455–463 epitope restricted to HLA-A0201 (4 out of 7 subjects); third was the HLA-A2402-restricted core117–125 epitope found in 5 out of 9 subjects; and the fourth response was again directed at the Pol455–463 epitope but restricted to HLA-A0203 (Fig. 1A). Taking into account the greater prevalence of HLA-Cw0801 in Southeast Asia (Fig. 1B), our data show that the HLA-Cw0801-restricted Env171–180 CD8 T cell response represents a highly immunoprevalent CTL response in Southeast Asian patients with self-limiting HBV infection.
Vertical immunodominance of the HLA-Cw0801-restricted T cell response.
We then evaluated the vertical immunodominance of the HLA-Cw0801/Env171–180 response. For this purpose, the whole HBV-specific T cell repertoire was studied in two HLA-Cw0801+ patients with acute HBV infection. PBMCs of the two patients, obtained 4 weeks after the onset of acute hepatitis symptoms (patient 1) or after resolution of disease and HBsAg seroconversion (patient 2), were stimulated with 61 mixtures of overlapping peptides covering the whole proteome of HBV genotype C. ELISPOT assays to detect IFN-γ-producing cells were performed directly ex vivo and after 10 days of in vitro expansion. In accordance with previous results, direct ex vivo analysis of IFN-γ-producing cells did not generate results that could be used with confidence to elucidate an immunodominance hierarchy (data not shown). At best, 8 to 6 spots/105 cells were visualized by stimulation with different peptide pools. We have recently demonstrated, directly ex vivo, that the lack of detectable functional HBV-specific T cells during the acute phase of HBV infection is the result of the combined action of T cell exhaustion and the suppressive effect of arginase (14). On the other hand, in patient 2, the few HBV-specific IFN-γ spots detected ex vivo are compatible with the low frequency of memory HBV-specific CD8 T cells present after resolution of infection.
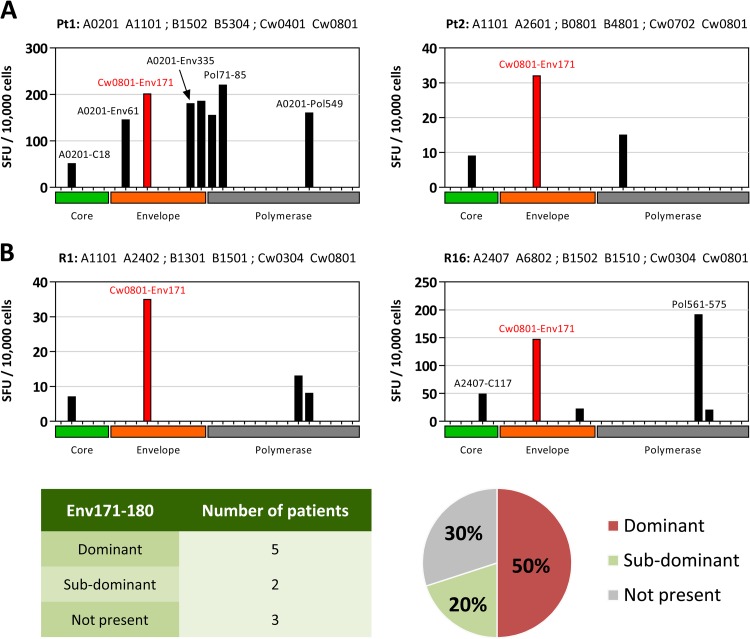
In contrast to the ex vivo data, a clear population of HBV peptide-responsive T cells was detected with ELISPOT assays in both patients after in vitro expansion. Deconvolution of the single peptide responsible for the T cell activation and definition of their CD4 or CD8 phenotype was performed on the individual T cell lines with intracellular cytokine staining. Figure 2A summarizes the results of these consecutive experiments, displaying only the spot counts that were derived from mixtures containing peptides that were confirmed to activate HBV-specific CD8 T cells. CD8 T cells responding to the Env171–180 HLA-Cw0801-restricted epitope clearly represent a numerically important response in both acute patients. In patient 1, even though the response to a Pol71–85 epitope was slightly bigger, Env171–80 was able to stimulate the strongest CD8 T cell response specific to the envelope protein. Remarkably, Env171–80-specific CD8 T cells were dominant over the response to the HLA-A201-restricted CD8 T cell epitope core18-27, one of the most studied HBV-specific CD8 T cell epitopes. In patient 2, the memory CD8 T cell response to Env171–180 was dominant in comparison to responses against polymerase- and core-specific CD8 T cell responses.
FIG 2.
Vertical immunodominance of the HLA-Cw0801/Env171–180 T cell response. (A) PBMCs from two representative subjects with a documented history of acute hepatitis B were expanded in vitro with the HBV genotype C peptide library (15-mer peptides covering the full HBV core, envelope, and polymerase proteins) and screened using IFN-γ ELISPOT assays. Each bar shows the response against a pool of 15-mer peptides covering the corresponding HBV protein, and the single epitope responsible for the ELISPOT hits was confirmed by ICS and annotated accordingly. SFU counts shown were background subtracted and normalized to 10,000 cells. Pt1, patient 1; Pt2, patient 2. (B) PBMCs from 15 subjects with serological evidence of prior HBV infection (anti-HBs and anti-HBc positive) were expanded in vitro and screened as described above. Detailed responses from two representative subjects (R1 and R16) are shown (top). The vertical immunodominance of the HLA-Cw0801/Env171–180 T cell response is summarized in the table and pie chart (bottom).
The vertical immunodominance of the Cw0801-restricted CD8 T cell response detected in the two patients with acute hepatitis B was then measured in an additional 15 HLA-Cw0801+ subjects with serological profiles indicative of previous HBV infection (anti-HBs and anti-HBc positive). In 10 out of these 15 HLA-Cw0801+ subjects, HBV-specific T cells directed against peptides covering the whole HBV proteome were detectable after in vitro expansion. Seven out of the 10 HLA-Cw0801+ subjects with detectable expanded HBV-specific T cells showed an Env171–180-specific CD8 T cell response. As visually displayed in Fig. 2B, the HLA-Cw0801/Env171–180-specific CD8 T cells represent the dominant response in 50% of HLA-Cw0801+ HBV-infected subjects with detectable CTL responses, in addition to being the most abundant CD8 T cell response in five out of the seven Env171–180-responsive subjects.
Since the Cw0801-restricted CD8 T cell response against Env171–180 sequence is immunodominant in the majority of HLA-Cw08+ subjects able to control HBV infection, we wanted to determine whether the Env171–180 response could also be detected in patients with chronic HBV infection. Three HLA-Cw0801+ chronic HBV patients were tested. Not surprisingly, we were unable to detect the Env171–180 response even after 10 days of in vitro expansion (data not shown), consistent with the established view of dysfunctional/deleted HBV-specific T cell responses in chronic HBV patients.
Functional affinity of HBV-specific CD8 T cells.
In a previous analysis of the relative magnitude of several different HIV- and EBV-specific CD8 T cell responses, the vertical and horizontal immunodominance of the epitope-specific CD8 T cell response was directly correlated with its functional affinity (2). We therefore analyzed the functional affinity of CD8 T cells specific for HLA-Cw0801/Env171–180, -A0201/core18–27, -A0201/Env370–379, -A0201/Pol455–463, -B5801/Env356–364, and -B1301/Pol386–395 and correlated such results with their immunodominance hierarchy in the HBV-infected subjects.
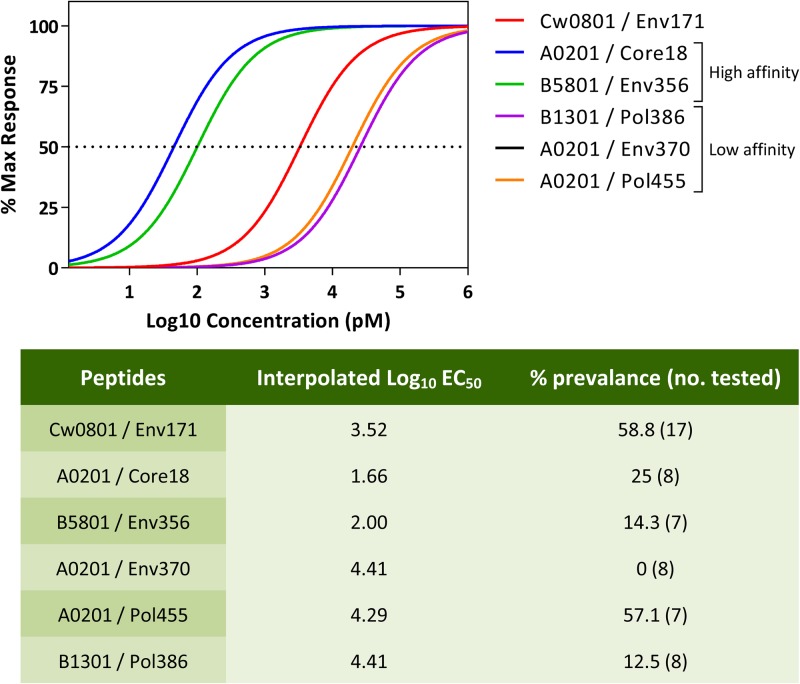
T cells of all specificities reached maximal activation when cultured together with target cells pulsed with a 1 μM concentration of the corresponding peptides (Fig. 3). Functional affinity of the different cell lines can be divided into two main categories: high functional affinity was observed for CD8 T cells specific for HLA-A0201/core18–27 and HLA-B5801/Env356–364 that were able to recognize target cells pulsed with peptides at concentrations as low as 1 to 10 pM. Other CD8 T cell specificities displayed lower functional affinity (HLA-B1301/Pol386–395, -A0201/Pol455–463, and -A0201/Env370–379), being activated only by targets pulsed with at least 1 to 10 nM concentrations of the different peptides and with EC50s that were 2 logs higher. HLA-Cw0801/Env171–180-specific CD8 T cells displayed a functional affinity curve that was between these two categories. Note that the high functional affinity of the HLA-A0201/core18–27 CD8 T cells was related to the response of a CD8 T cell line selected in Caucasian HBV patients infected with HBV genotype A/D virus, one that is dominant in HLA-A201+ Caucasian HBV patients.
FIG 3.
Comparative functional affinity of the HLA-Cw0801/Env171–180 epitope against known HBV epitopes. EBV-transformed B cells expressing specific HLA alleles were pulsed with different concentrations of the corresponding HBV epitopes and used as targets to stimulate in vitro expanded epitope-specific T cells. The dose-response curve was generated using IFN-γ production as a measurement of activation. The table below shows the interpolated EC50s of the various epitopes and the corresponding immunoprevalence of the response. Max, maximum.
We also constructed a table (Fig. 3) where the hierarchy of horizontal immunodominance and functional affinity was compared. Unfortunately, based on our limited sample size, we did not see an increased prevalence of T cell responses with higher functional affinity.
Influence of HBV mutations on Env171–180-specific CD8 T cell response.
Having demonstrated the horizontal and vertical immunodominance of the HLA-Cw0801-restricted Env171–180-specific T cell response and its high functional affinity, we analyzed whether amino acid mutations naturally present in the Env171–180 region can alter T cell recognition.
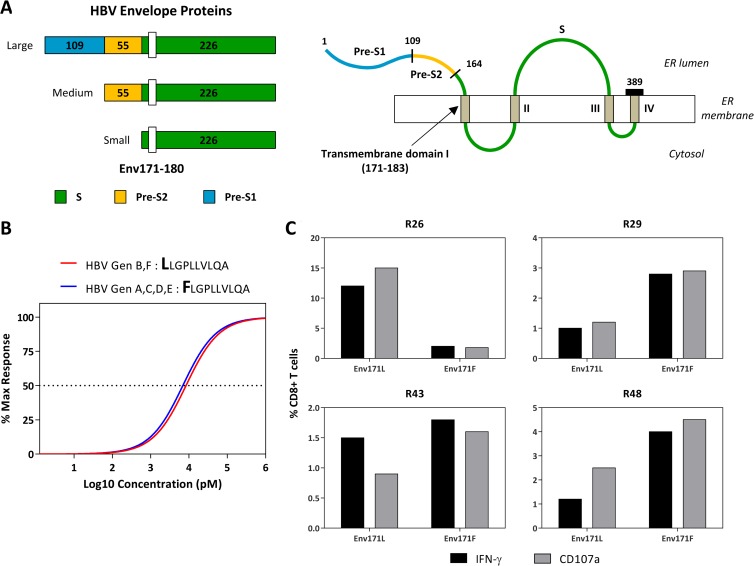
The amino acid sequence FLGPLLVLQA corresponding to the HLA-Cw0801/Env171–180 epitope is located within the first transmembrane domain of the S region of HBV envelope proteins (a schematic representation is present in Fig. 4A). Importantly, recent experimental analysis of the role of the transmembrane domains in the production of HBV surface (S) protein have shown that mutations introduced within this region (at positions 172 and 178) are able to completely block S synthesis and subviral particle production. Such a constraint can explain why the region of residues 171 to 180 appears remarkably conserved between the different HBV isolates, with only a single amino acid substitution of Leu to Phe at position 171 (171L) present in isolates of HBV genotype B. We therefore analyzed whether HLA-Cw0801-specific CD8 T cells similarly recognized the sequence LLGPLLVLQA (substitution in boldface), corresponding to the envelope region of residues 171 to 180 of HBV genotype B, and the sequence FLGPLLVLQA, present in HBV genotypes A, C, and D. Two experimental systems were used. The first, illustrated in Fig. 4B, shows that the amino acid substitution at position 171 did not alter the ability of Env171–180 CD8 T cells to recognize target cells pulsed with the peptides LLGPLLVLQA (HBV genotypes B and F) and FLGPLLVLQA (HBV genotypes A, C, D, and E). We also tested whether the two peptides have similar in vitro abilities to expand Env171–180-specific CD8 T cells. PBMCs of HLA-Cw0801+ subjects were stimulated with the two peptides, and the expansion of Env171–180 CD8 T cells was tested. Figure 4C shows that both peptides were able to expand in vitro HLA-Cw0801/Env171–180-restricted CD8 T cells. In different subjects the expansion of Env171–180 CD8 T cells specific for the two peptides differs, perhaps because of primary infection with different HBV genotypes. Overall, in all of the subjects the variations at aa 171 did not abolish the immunogenicity of the Env171–180 region.
FIG 4.
Impact of genotype-specific amino acid mutations on the HLA-Cw0801/Env171–180 T cell response. (A) Schematic diagram showing the location of the Env171–180 epitope within the full-length HBV envelope protein. (B) Functional affinity of genotype-specific Env171–180 variants. The dose-response curve was generated as described in the legend of Fig. 3. (C) Frequencies of Env 171L- or Env 171F-specific T cells from four representative subjects (R26, R29, R43, and R48) with serological evidence of prior HBV infection. The frequencies were quantified after 10 days of in vitro expansion with the respective epitopes.
Alternative HLA restriction of the Env171–180 epitope.
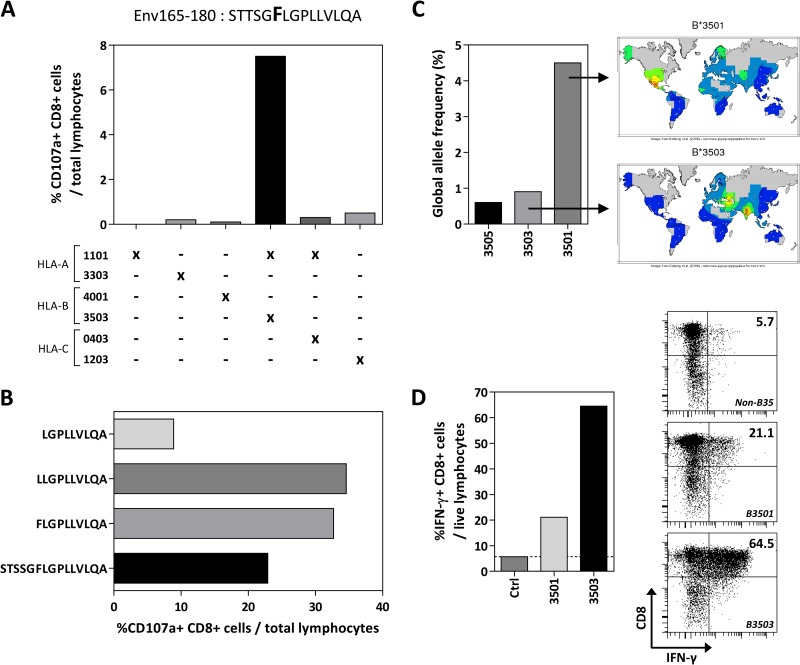
Interestingly, by analyzing the T cell responses against the HBV envelope in HLA-Cw0801-negative HBV-infected patients, we detected one subject (HLA-A1101, -A3303, -B3503, -B4001, -C0403, and -C1203) that mounted a specific CD8 T cell response to the region of residues 165 to 180, containing the HLA-Cw0801/Env171–180 epitope. Env165–180-specific CD8 T cell lines were expanded, and HLA class I restriction and fine specificity were determined (Fig. 5A and B). We were able to demonstrate that this patient presented an HLA-B3503-restricted CD8 T cell response against the same Env171–180 region, which appears, therefore, to be a promiscuous epitope capable of being presented by at least two different HLA class I molecules, HLA-B3503 and HLA-Cw0801. Similar to the result with the HLA-Cw0801/Env171–180 epitope, the amino acid mutation at position 171 did not alter the immunogenicity of this HLA-B3503-restricted epitope.
FIG 5.
Alternate HLA restriction of the Env171–180 epitope. (A) The HLA restriction of the Env165–180 CD8 T cell response found in a non-HLA-Cw0801 subject was deduced using a panel of six EBV-transformed B cell lines. The EBV B cell lines were pulsed with the Env165–180 peptide and used to stimulate short-term T cell lines derived from the subject. HLA alleles that were common between the subject and the respective EBV B cell lines are indicated by an X. (B) The optimal epitope of the HLA-B3503-restricted Env165–180 response was identified by stimulating Env165–180-specific T cell lines with truncated peptides spanning Env165–180. (C) The allele frequencies of the top three most common HLA-B35 subtypes are shown in bars while the heat map shows their geographical distributions. Heat maps and data were obtained from the NCBI dbMHC website (http://www.ncbi.nlm.nih.gov/gv/mhc). (D) EBV B cells expressing HLA-B3501 or B3503 were pulsed with Env165–180 peptide and used as targets to activate the Env165–180-specific short-term T cell lines derived from the subject described above. An EBV B cell line not expressing HLA-B35 was used as a control.
Among the HLA-B35 alleles, the HLA-B3501 subtype is the most frequently detected HLA-B35 allele globally (the top three most frequent HLA-B35 alleles are shown in Fig. 5C). Also, while HLA-B3503 is rarely found in South Asian populations, the incidence of the HLA-B3501 subtype is moderately frequent in Caucasians in the United States (∼10% phenotype frequency) (Fig. 5C). Hence, we tested whether HLA-B3503-restricted CD8 T cells can also recognize Env171–180 when it is presented by HLA-B3501. Using a short-term T cell line specific for Env171–180 derived from the HLA-B3503-expressing subject, we were able to show that HLA-B3501 can present the Env171–180 peptide to HLA-B3503/Env171–180-specific T cells (Fig. 5D), suggesting that the same T cell response could also be present in the Caucasian population, which frequently expresses the HLA-B3501 allele.
Endogenous processing and presentation of Env171–180.
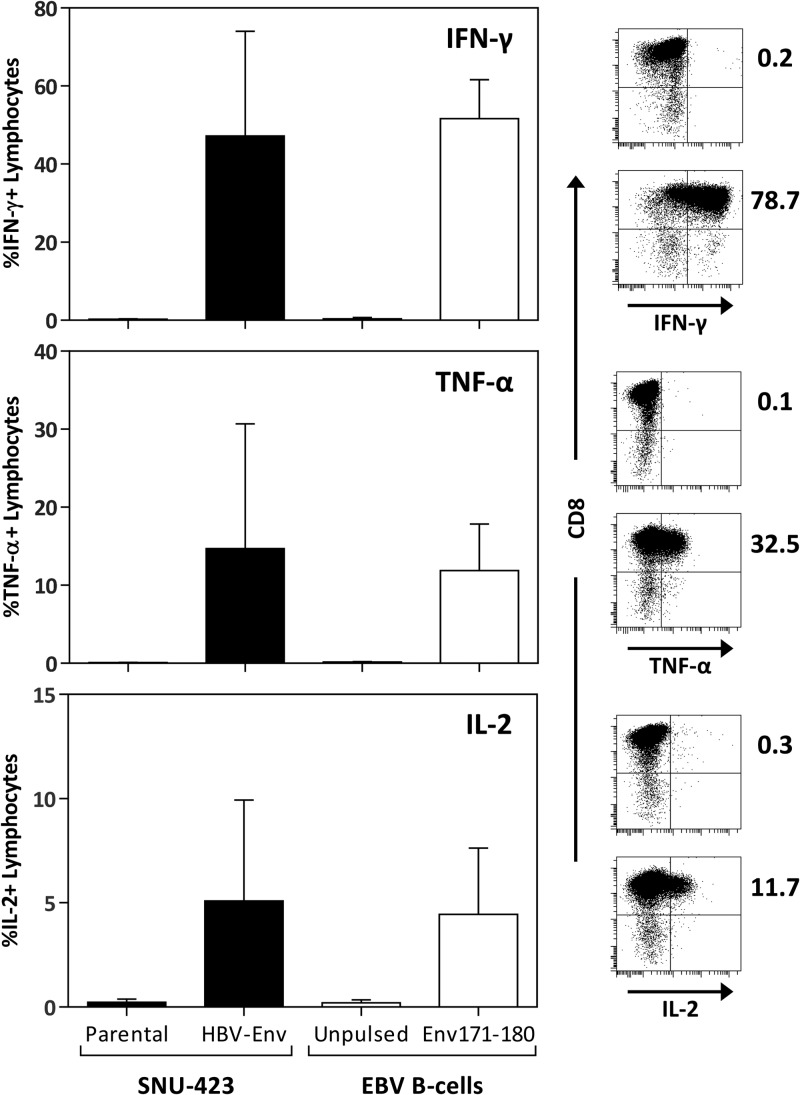
Finally, we sought to determine whether this epitope could be processed and presented in a physiological manner, a crucial point for T cell responses characterized using synthetic peptides. A HLA-Cw0801-expressing hepatocellular carcinoma cell line, SNU-423, was transduced with the full-length HBV genotype C envelope protein (SNU-423Env). The cell line was then used as a target to stimulate an in vitro-expanded short-term T cell line specific for Env171–180 derived from a patient with self-limiting HBV infection. As shown in Fig. 6, SNU-423Env efficiently stimulated T cells specific for Env171–180 to produce IFN-γ, TNF-α, and IL-2, with frequencies comparable to those obtained using EBV-transformed B cells pulsed with the corresponding peptides. This confirms that the Env171–180 epitope could be naturally processed, presented, and subsequently activate specific T cells in HLA-Cw0801 patients infected with HBV.
FIG 6.
Endogenous processing of the Env171–180 epitope from full-length HBV envelope protein. The HLA-Cw0801-expressing hepatocellular carcinoma cell line SNU-423 was transduced with the full-length HBV genotype C envelope gene. The transduced line was then used as a target to stimulate HLA-Cw0801/Env171–180-specific short-term T cell lines, and activation was determined by intracellular cytokine staining for IFN-γ, TNF-α, and IL-2. The frequency of epitope-specific cytokine-producing lymphocytes was compared to that obtained using HLA-Cw0801-expressing EBV B cells pulsed with the Env171–180 peptide as targets.
DISCUSSION
During HBV infection, CD8 T cells classically target multiple epitopes and establish a hierarchy of dominant and subdominant HBV-specific CD8 T cell responses within the infected subject. Measurement of the presence of a defined CD8 T cell response against a single epitope in subjects sharing the same HLA class I molecule is called horizontal immunodominance and is important to understanding the prevalence of a defined T cell response in a given population. The analysis of the hierarchy of dominant and subdominant responses within a single individual, defined as vertical immunodominance, represents, in contrast, a functional parameter that helps in the definition of possible protective epitopes (15). Often, within an infected individual, the dominant CD8 T cell response suppresses viral replication more efficiently than subdominant responses, and the dominant response is also associated with better functional avidity.
By analyzing the responses against several previously characterized epitopes, we showed that the HLA-Cw0801-restricted CD8 T cell response specific for the Env171–180 epitope is both horizontally and vertically immunodominant in HBV-infected patients of South Asian origin. The HLA-Cw0801 molecule is expressed in <5% of European/American populations, but it is, in contrast, present in 18 to 20% of the Southeast Asian population, with peaks of about 35 to 40% in specific large ethnic groups (Malay and Indonesian Javanese). Based on these observations, we think that this response might play an important role in the control or pathogenesis of HBV infection in Asian patients, who are the primary world population most affected by HBV persistence. Importantly, by demonstrating that HLA-C-restricted T cells are frequently detectable in HBV infection, our data demonstrate that HLA-C-restricted T cells are not an exclusive feature of HIV infection but are a robust component of antiviral immunity in infections with viruses, like HBV, that are not known to interfere with HLA-A and HLA-B molecule expression.
The immunodominance of the HLA-Cw0801/Env171–180 T cell response in comparison to other HBV-specific CTL responses can be explained by virological and immunological features. First, the Env171–180 epitope resides in the first transmembrane region of HBV envelope that is critical for infectivity (16), suggesting that this region has a high level of conservation among various HBV genotypes and, hence, the frequent T cell response against it. In addition, we demonstrated that the Env171–180-specific CD8 T cells were able to tolerate the genotype-specific amino acid mutation present at position 171 and be activated by both variants of the epitope. This is unlike the HLA-A0201-restricted core18–27 response (found mainly in subjects infected with HBV genotypes A and D) where amino acid mutation (isoleucine in genotypes B and C; valine in genotypes A and D) at position 27 has a clear negative impact on functional recognition by T cells (13). Therefore, the Env171–180 epitope could potentially be found in HLA-Cw0801+ subjects infected by all HBV isolates. Indeed, our immunoprevalence data derived from the study of patients of South Asian origin likely infected by either HBV genotype B or C support this hypothesis.
The other important factor that likely facilitates the horizontal and vertical immunodominance of the HLA-Cw0801/Env171–180 CD8 T cell response is the expression levels of HLA-Cw0801 molecules on nucleated cells. HLA-Cw08 alleles (both HLA-Cw0801 and -Cw0802) have been shown to be in strong linkage disequilibrium with the −35C polymorphism that was associated with high HLA-C cell surface expression (6). More importantly, the higher surface expression of HLA-C correlates with a stronger HLA-C-restricted CD8 T cell response in HIV-infected patients (7). Hence, the elevated expression of HLA-Cw0801 is also likely to contribute to the immunodominance of our Env171–180 T cell response in HBV patients.
Note that our data do not directly demonstrate that Env171–180-specific CD8 T cells have a protective or pathogenic role during HBV infection. Such conclusions might be better supported by large population studies associating HLA-class I and/or class II profiles of HBV-infected patients with normal subjects. However, recently the superior antiviral activity of vertical immunodominant epitopes has been demonstrated in a work that investigated the impact of HIV-specific CTL responses on HIV control and showed that vertical immunodominant epitopes exerted more pressure on HIV variability (15). Therefore, the vertical immunodominance of the HLA-Cw0801/Env171–180-specific CD8 T cells, in addition to their exclusive detection in subjects that controlled the infection, supports the important antiviral activity of such responses during HBV infection.
Other than the immunodominance of the Env171–180 epitope, its location within transmembrane domain I of HBV envelope further substantiated its significant antiviral potential. Experimental analysis of the role of the transmembrane domains in the production of HBV surface protein has clearly shown that mutations introduced within the first transmembrane domain (at positions 172 and 178) completely blocked S protein synthesis and subviral particle production (17). Together with the demonstration that this domain is also crucial for infectivity (16), this means that it is unlikely that HBV will escape CTL pressure that targets this region due to the major inherent viral fitness cost, making the Env171–180 epitope a T cell response with important antiviral potential.
On a different note, the demonstration that an important CD8 T cell epitope can target a conserved viral region also exposed the limitation of using viral mutations associated with HLA-class I profile as a method of identifying new CTL epitopes. Such a method has been used extensively in past years and is driven by a sound rationale, but this approach will never be able to pinpoint the CTL epitopes against conserved regions of the virus that might represent responses crucial for protection. This caveat will have to be properly considered in works employing such an approach to CTL epitope characterization.
Another interesting observation of the Env171–180 response was also the ability of the epitope to be presented by a different HLA-class I. The Env171–180 epitope can induce both HLA-Cw0801- and HLA-B3502-restricted responses in patients carrying these two different HLA class I molecules, showing the strong immunogenicity of this epitope. Perhaps the conserved nature of this region might facilitate the detection of CD8 T cell epitopes since our method still relies on peptides that were synthesized based on published sequences and might not be identical to the sequence of the infecting virus. One other possibility is that the transmembrane region might actually be presented more efficiently by HLA class I molecules since their embedded nature in the endoplasmic reticulum (ER) membrane might facilitate HLA class I loading. More detailed analysis of different CD8 T cell epitopes will need to be performed to understand whether epitopes located in transmembrane regions of envelope proteins might have a selective advantage to be presented by HLA class I molecules.
Overall, in addition to showing that an HLA-Cw-restricted CD8 T cell response can be easily triggered in not only HIV but also HBV infection, we also provide information on the hierarchy of new HBV-specific CD8 T cell epitopes that will facilitate pathogenetic studies of HBV infection in patients of Asian ethnicities.
ACKNOWLEDGMENTS
P.S. was supported by the Royal Golden Jubilee Ph.D. Program and the 90th Anniversary of Chulalongkorn University Fund (Ratchadapisek-somphot Endowment Fund). The study was supported by a research grant from the Thailand Research Fund (RMU5180051), core funding from the Agency for Science, Technology and Research (A∗STAR), and a Singapore Translational Research (STaR) Investigator award given to Antonio Bertoletti (NMRC/STaR/013/2012).
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Berger CT, Frahm N, Price DA, Mothe B, Ghebremichael M, Hartman KL, Henry LM, Brenchley JM, Ruff LE, Venturi V, Pereyra F, Sidney J, Sette A, Douek DC, Walker BD, Kaufmann DE, Brander C. 2011. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J. Virol. 85:9334–9345. 10.1128/JVI.00460-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bihl F, Frahm N, Di Giammarino L, Sidney J, John M, Yusim K, Woodberry T, Sango K, Hewitt HS, Henry L, Linde CH, Chisholm JV, III, Zaman TM, Pae E, Mallal S, Walker BD, Sette A, Korber BT, Heckerman D, Brander C. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094–4101 [DOI] [PubMed] [Google Scholar]
- 3.Blais ME, Dong T, Rowland-Jones S. 2011. HLA-C as a mediator of natural killer and T-cell activation: spectator or key player? Immunology 133:1–7. 10.1111/j.1365-2567.2011.03422.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947. 10.1126/science.1143767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M. 2013. Influence of HLA-C expression level on HIV control. Science 340:87–91. 10.1126/science.1232685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas R, Apps R, Qi Y, Gao X, Male V, O'hUigin C, O'Connor G, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ, Buchbinder S, Kirk GD, Martin MP, Telenti A, Deeks SG, Walker BD, Goldstein D, McVicar DW, Moffett A, Carrington M. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290–1294. 10.1038/ng.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blais ME, Zhang Y, Rostron T, Griffin H, Taylor S, Xu K, Yan H, Wu H, James I, John M, Dong T, Rowland-Jones SL. 2012. High frequency of HIV mutations associated with HLA-C suggests enhanced HLA-C-restricted CTL selective pressure associated with an AIDS-protective polymorphism. J. Immunol. 188:4663–4670. 10.4049/jimmunol.1103472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671. 10.1016/S1074-7613(00)80065-5 [DOI] [PubMed] [Google Scholar]
- 9.Chang CX, Tan AT, Or MY, Toh KY, Lim PY, Chia AS, Froesig TM, Nadua KD, Oh HL, Leong HN, Hadrup SR, Gehring AJ, Tan YJ, Bertoletti A, Grotenbreg GM. 2013. Conditional ligands for Asian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur. J. Immunol. 43:1109–1120. 10.1002/eji.201243088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott WG, Tsai P, Leung E, Trevarton A, Ofanoa M, Hornell J, Gane EJ, Munn SR, Rodrigo AG. 2010. Associations between HLA class I alleles and escape mutations in the hepatitis B virus core gene in New Zealand-resident Tongans. J. Virol. 84:621–629. 10.1128/JVI.01471-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai SL, Chen MH, Yeh CT, Chu CM, Lin AN, Chiou FH, Chang TH, Liaw YF. 1996. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD8+ cytotoxic T lymphocytes. J. Clin. Invest. 97:577–584. 10.1172/JCI118450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desmond CP, Gaudieri S, James IR, Pfafferott K, Chopra A, Lau GK, Audsley J, Day C, Chivers S, Gordon A, Revill PA, Bowden S, Ayres A, Desmond PV, Thompson AJ, Roberts SK, Locarnini SA, Mallal SA, Lewin SR. 2012. Viral adaptation to host immune responses occurs in chronic hepatitis B virus (HBV) infection, and adaptation is greatest in HBV e antigen-negative disease. J. Virol. 86:1181–1192. 10.1128/JVI.05308-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan AT, Loggi E, Boni C, Chia A, Gehring AJ, Sastry KS, Goh V, Fisicaro P, Andreone P, Brander C, Lim SG, Ferrari C, Bihl F, Bertoletti A. 2008. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J. Virol. 82:10986–10997. 10.1128/JVI.01124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandalova E, Laccabue D, Boni C, Watanabe T, Tan A, Zong HZ, Ferrari C, Bertoletti A. 2012. Increased levels of arginase in patients with acute hepatitis B suppress antiviral T cells. Gastroenterology 143:78–87 e73. 10.1053/j.gastro.2012.03.041 [DOI] [PubMed] [Google Scholar]
- 15.Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, Li H, Pavlicek JW, Cai F, Rose-Abrahams M, Treurnicht F, Hraber P, Riou C, Gray C, Ferrari G, Tanner R, Ping LH, Anderson JA, Swanstrom R, Cohen M, Karim SS, Haynes B, Borrow P, Perelson AS, Shaw GM, Hahn BH, Williamson C, Korber BT, Gao F, Self S, McMichael A, Goonetilleke N. 2013. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J. Clin. Invest. 123:380–393. 10.1172/JCI65330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepere-Douard C, Trotard M, Le Seyec J, Gripon P. 2009. The first transmembrane domain of the hepatitis B virus large envelope protein is crucial for infectivity. J. Virol. 83:11819–11829. 10.1128/JVI.01026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegler VD, Bruss V. 2013. Role of transmembrane domains of hepatitis B virus small surface proteins in subviral-particle biogenesis. J. Virol. 87:1491–1496. 10.1128/JVI.02500-12 [DOI] [PMC free article] [PubMed] [Google Scholar]