Abstract
Retroviral RNA encapsidation involves a recognition event between genomic RNA (gRNA) and one or more domains in Gag. In HIV-1, the nucleocapsid (NC) domain is involved in gRNA packaging and displays robust nucleic acid (NA) binding and chaperone functions. In comparison, NC of human T-cell leukemia virus type 1 (HTLV-1), a deltaretrovirus, displays weaker NA binding and chaperone activity. Mutation of conserved charged residues in the deltaretrovirus bovine leukemia virus (BLV) matrix (MA) and NC domains affects virus replication and gRNA packaging efficiency. Based on these observations, we hypothesized that the MA domain may generally contribute to NA binding and genome encapsidation in deltaretroviruses. Here, we examined the interaction between HTLV-2 and HIV-1 MA proteins and various NAs in vitro. HTLV-2 MA displays higher NA binding affinity and better chaperone activity than HIV-1 MA. HTLV-2 MA also binds NAs with higher affinity than HTLV-2 NC and displays more robust chaperone function. Mutation of two basic residues in HTLV-2 MA α-helix II, previously implicated in BLV gRNA packaging, reduces NA binding affinity. HTLV-2 MA binds with high affinity and specificity to RNA derived from the putative packaging signal of HTLV-2 relative to nonspecific NA. Furthermore, an HIV-1 MA triple mutant designed to mimic the basic character of HTLV-2 MA α-helix II dramatically improves binding affinity and chaperone activity of HIV-1 MA in vitro and restores RNA packaging to a ΔNC HIV-1 variant in cell-based assays. Taken together, these results are consistent with a role for deltaretrovirus MA proteins in viral RNA packaging.
INTRODUCTION
When retroviruses assemble in infected cells, two copies of full-length genomic RNA (gRNA) are selected for packaging. Although gRNA constitutes only a very small portion of total RNA in the cytoplasm, it is selectively packaged into virions (1). The specific packaging process is believed to involve recognition of gRNA packaging signals by the Gag polyprotein (1–4). In HIV-1 assembly, the nucleocapsid (NC) domain of Gag is the dominant nucleic acid (NA) binding region and is essential for specific incorporation of gRNA. HIV-1 NC is also a robust chaperone protein, wherein it remodels NAs to their most thermodynamically stable state through duplex destabilization, aggregation, and rapid binding kinetics (5–8). The solution structures of HIV-1 NC bound to stem-loop 2 (SL2) and SL3 derived from the psi (Ψ) packaging signal have been studied by nuclear magnetic resonance (NMR) spectroscopy. In these structures, the two zinc finger motifs of NC specifically bind to guanosines in the G-rich RNA tetraloops (9, 10).
The matrix (MA) domain of Gag also has several established functions in retroviral replication (11). HIV-1 MA is required for targeting of Gag to the plasma membrane of infected cells via its myristoyl moiety (12, 13). Basic residues of HIV-1 MA also contribute to membrane binding (14–16). In the absence of NC and protease activity, the HIV-1 MA domain has been shown to bind RNA and facilitate immature virus particle formation (17, 18). A number of additional studies have supported the NA binding properties of HIV-1 MA (19–21), yet how this capability contributes to virus replication has not been elucidated. A more recent study showed that HIV-1 MA-RNA interactions can be outcompeted by phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]-containing liposomes but not other liposomes (22). It has been proposed that RNA binding to MA negatively regulates membrane binding both by preventing nonspecific interactions between basic residues and acidic lipids and by suppressing myristate-dependent hydrophobic interactions (23).
An early study suggested that the MA domain of bovine leukemia virus (BLV), a deltaretrovirus, plays a more significant role in specific RNA binding than NC (24). BLV MA was reported to form a specific complex with the dimeric 5′ end of the gRNA sequence but not with other RNAs in vitro, whereas BLV NCp12 bound randomly and nonspecifically (24). More recent studies showed that both the BLV MA and NC domains are involved in gRNA packaging and that conserved charged residues in MA are critical for this function. When two conserved residues, K41 and H45, were individually mutated to alanine, RNA packaging efficiency was significantly reduced (by 66 and 92%, respectively). In contrast, mutation of these same residues does not affect Gag membrane localization (25). The MA domain of the deltaretrovirus human T-cell leukemia virus type 2 (HTLV-2) contains 11 basic residues scattered throughout the primary sequence, which form a cluster of exposed positive charges at the surface of the protein; 8 of the 11 residues are also present in HTLV-1 MA (26, 27). Surprisingly, replacement of the basic residues of HTLV-1 MA with Leu/Ile did not affect intracellular targeting of Gag, even though most of the mutations completely abolished viral infectivity and dramatically reduced viral particle production (27). Based on these data, it has been suggested that, as for BLV MA, HTLV-1 MA may also play a role in gRNA packaging (25).
Interestingly, HTLV-1 NC displays reduced NA binding affinity and chaperone function relative to those of HIV-1 NC yet has robust duplex-destabilizing capabilities (28, 29). Our laboratory has previously explored the mechanistic basis for the poor NA binding and chaperone properties of HTLV-1 NC, and our studies show that removal of HTLV-1 NC's anionic C-terminal domain (CTD) improves the chaperone function to a level comparable to those of other retroviral NCs (30). An intramolecular N-terminal domain (NTD)-CTD interaction reduces the kinetics of association with NAs in the unbound state, whereas an NTD-CTD interaction between neighboring molecules reduces the NC-NA dissociation in the bound state. These properties inhibit both NA aggregation and rapid protein dissociation from single-stranded DNA (ssDNA), which are required for chaperone function (6). The amino acid sequences of HTLV-1 and HTLV-2 NCs are 72% identical and have similar isoelectric points, close to neutral. Therefore, HTLV-2 NC is likely to possess structural and biochemical properties similar to those of HTLV-1 NC.
Sequence alignment of HTLV-1, HTLV-2, BLV, and HIV-1 MA proteins shows high homology among the three deltaretroviruses, especially HTLV-1 and HTLV-2, which share 58% identity. These data suggest that there is likely to be conserved function among deltaretroviral MA proteins. In contrast, HIV-1 and HTLV-2 MA proteins share only ∼10% sequence identity. Nevertheless, they adopt quite similar secondary structures, with N-terminal basic residues exposed in similar positions on one side of α-helix II, as shown by NMR spectroscopy (26). The functional similarity between these two MA proteins is unknown.
The gRNA packaging signal of BLV is a bipartite RNA motif consisting of a primary (SL1 and SL2) region and a secondary region containing a single stem-loop (31). It has been shown that replacement of the BLV packaging signal with a similar region from either HTLV-1 or HTLV-2 leads to only a partial BLV replication defect (32, 33). These data support at least some level of conserved function in deltaretroviral RNA packaging signals.
Based on the available data, we hypothesized that in deltaretroviruses, MA plays an equally important role in gRNA recognition and packaging as that of NC. To test this hypothesis, we compared the capabilities of HTLV-2 MA and HIV-1 MA to bind and aggregate NAs and to chaperone the annealing of complementary structures. We chose HTLV-2 MA as a representative deltaretrovirus for these studies due to the availability of a high-resolution NMR structure (26). Comparisons between HTLV-2 MA and NC were also made. In addition, to probe the NA binding specificity, HTLV-2 MA α-helix II variants were prepared and compared to the wild-type (WT) protein (Fig. 1). Finally, an HIV-1 MA variant designed to mimic HTLV-2 MA (Fig. 1) was prepared and tested in vitro as well as in cell-based studies. Taken together, our results support an important role for HTLV-2 MA in NA binding and chaperone activities and support the conclusion that MA may generally function in deltaretrovirus gRNA packaging. The results highlight retrovirus-specific differences in protein-RNA interactions that play critical roles in the retrovirus life cycle.
FIG 1.
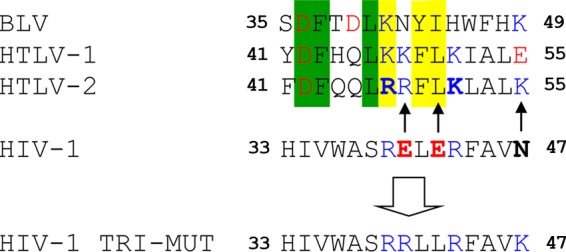
Structure-based sequence alignment of MA α-helix II from HTLV-2, HTLV-1, and HIV-1 (26) along with BLV MA. Residues are boxed as follows: green, identical in HTLV-1 and -2 and BLV; yellow, conserved in HTLV-1 and -2 and BLV. Basic residues are shown in blue, and acidic residues are shown in red. Residues in boldface type indicate residues changed to generate HTLV-2 and HIV-1 MA variants. Arrows show the changes made to generate the HIV-1 MA TRI-MUT variant to mimic HTLV-2 MA.
MATERIALS AND METHODS
Plasmid construction.
Plasmids containing the genes encoding histidine-tagged HTLV-2 MA in a pET-11a vector and histidine-tagged HIV-1 MA in a pET-16b vector were constructed by using standard methods. The R47A/K51A HTLV-2 MA and E40R/E42L/N47K HIV-1 MA (TRI-MUT) variants were constructed by using the QuikChange mutagenesis kit from Stratagene (La Jolla, CA). BL21-CodonPlus(DE3)-RP competent cells (Stratagene, La Jolla, CA) were transformed with plasmids encoding the WT and mutant MA proteins, and mutations were confirmed by sequencing of the entire gene. The WT/ΔNC HIV-1 proviral plasmid used in this study was the delNC construct (34), a gift from David Ott, AIDS and Cancer Virus Program. The MAE40R/E42L/N47K/ΔNC HIV-1 proviral plasmid was generated by mutating nucleotides (nt) 907 to 909 from GAG to CGC, nt 913 to 914 from GA to TT, and nt 930 from G to T (nt positions refer to those from HIV-1 pNL4-3 [GenBank accession no. AF324493]).
Protein preparation.
All MA proteins were purified according to a previously reported protocol (26), except that Talon metal affinity resin (Clontech Laboratories, Inc., Mountain View, CA) was used and the purified proteins were dialyzed into a solution containing 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, 5 mM 2-mercaptoethanol (β-ME), and 1 mM dithiothreitol (DTT). The concentrations of purified proteins were determined by using the Bradford assay (35).
The HTLV-2 NC protein was prepared essentially as described previously (28, 36, 37). NC was stored in a lyophilized form at −80°C. Prior to use, NC was resuspended in NC storage buffer containing 20 mM HEPES (pH 7.5), 5 mM β-ME, and 0.1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) (pH 7.5). The concentration of NC was determined by measuring its absorbance at 280 nm and using the extinction coefficient 11,740 M−1 cm−1.
Circular dichroism spectroscopy.
Circular dichroism (CD) spectra were measured at room temperature by using an Aviv 202 CD spectrometer (Aviv Biomedical, Lakewood, NJ) with a 0.1-cm-path-length cuvette. Prior to analysis, proteins were dialyzed into 10 mM sodium phosphate (pH 7.5) and diluted to a concentration of 0.2 mg/ml. Spectra were accumulated over three scans.
Nucleic acid preparation.
NA oligonucleotides used in this work are shown in Fig. 2. The 6-carboxyfluorescein (FAM)-labeled 20-nt ssDNA oligonucleotide (5′-FAM DNA20) was obtained from TriLink Biotechnologies (San Diego, CA). The following high-performance liquid chromatography (HPLC)-purified FAM- and fluorescein (Fl)-labeled HTLV-2 RNA oligonucleotides and unlabeled HTLV-2 SL1 RNA were purchased from Dharmacon RNA Technologies (Lafayette, CO): 5′-Fl-UUAUGGGACAAAUCCA-3′ (5′-Fl-SL1) and 5′-Fl-UUGGGCUUUCCCCAACUCCAAUACCCAAAGCCC-3′ (5′-Fl-SL2) (note that the two U's in italics are not encoded by HTLV-2). RNAs derived from HIV-1 (3′-FAM-MicroTAR and 3′-FAM-SL3) and 3′-FAM-minihelixLys,3, derived from the acceptor-TΨC stem-loop of human tRNALys,3, were also purchased from Dharmacon RNA Technologies (Lafayette, CO).
FIG 2.
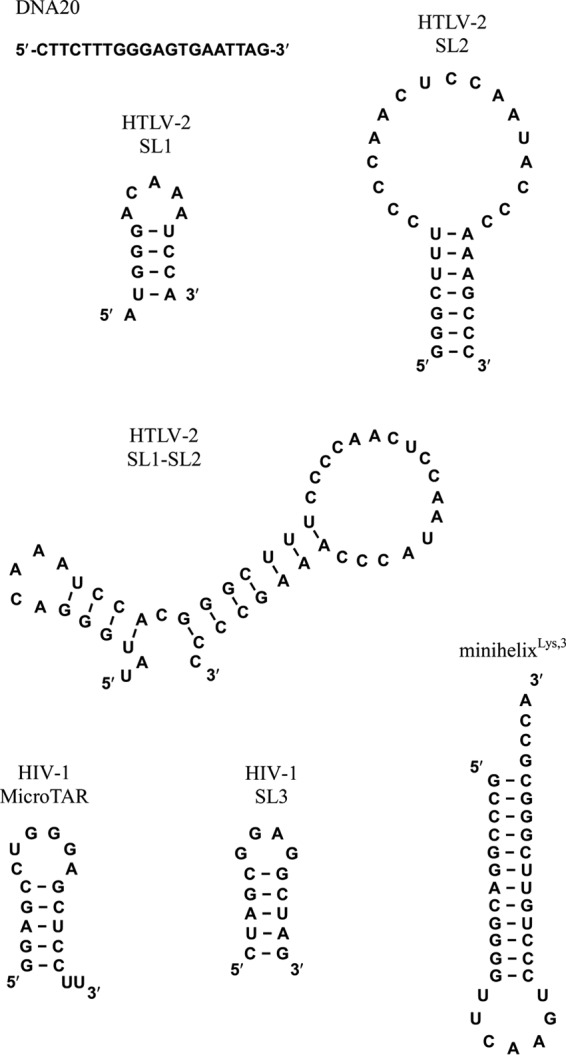
Oligonucleotide constructs used in this work.
HIV-1 trans-activation response element (TAR) DNA (38) was obtained from Integrated DNA Technologies (Coralville, IA), purified on a 12% (wt/vol) denaturing polyacrylamide gel, and stored at −20°C. Unlabeled HTLV-2-derived SL2 and SL1-SL2, human tRNALys,3 (39), and HIV-1 TAR RNA (38) were in vitro transcribed with T7 RNA polymerase as described previously (40). For gel shift annealing assays, TAR RNA was internally radiolabeled with [α-32P]GTP during in vitro transcription with T7 RNA polymerase by using standard protocols, followed by gel purification. All RNA oligonucleotides were dissolved in diethyl pyrocarbonate-treated water and stored at −20°C.
The concentrations of RNA and DNA oligonucleotides were determined by measuring the absorbance at 260 nm with the following extinction coefficients: 6.04 × 105 M−1 cm−1 for human tRNALys,3 (76-mer), 5.648 × 105 M−1 cm−1 for TAR DNA (59-mer), 5.337 × 105 M−1 cm−1 for TAR RNA (59-mer), 4.586 × 105 M−1 cm−1 for SL1-SL2 (50-mer), 3.499 × 105 M−1 cm−1 for 3′-FAM-minihelixLys,3 (37-mer), 3.242 × 105 M−1 cm−1 for 5′-Fl-SL2 (33-mer), 2.139 × 105 M−1 cm−1 for 5′-FAM DNA20, 1.893 × 105 M−1 cm−1 for 3′-FAM-MicroTAR (18-mer), 1.88 × 105 M−1 cm−1 for 5′-Fl-SL1 (16-mer), and 1.506 × 105 M−1 cm−1 for 3′-FAM-SL3 (16-mer).
Prior to use, all oligonucleotides except 5′-FAM DNA20 were refolded in a solution containing 25 mM HEPES (pH 7.5) and 100 mM NaCl by heating at 80°C for 2 min and cooling to 60°C for 2 min, followed by the addition of MgCl2 to a final concentration of 10 mM and placement on ice.
Fluorescence anisotropy measurements.
Equilibrium dissociation constants were determined by measuring the fluorescence anisotropy (FA) of 20 nM fluorescently labeled NA as a function of increasing concentrations of unlabeled proteins. The labeled NAs were incubated with various amounts of the unlabeled proteins for 30 min at room temperature in a solution containing 20 mM HEPES (pH 7.5) and 50 mM NaCl. Anisotropy measurements were carried out on a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA). The excitation and emission wavelengths were 485 and 525 nm, respectively. All measurements were performed at least three times, and the data were averaged. The titration curves were fit to the following equation, which assumes a 1:1 binding stoichiometry (41–43): A = Amin + {(Y + S + Kd) − [(Y + S + Kd)2 − (4YS)]1/2} · (Amax − Amin)/(2Y), where A is the measured anisotropy at a particular total concentration of the protein (S) and the labeled NAs (Y), Amin is the minimum anisotropy, Amax is the final maximum anisotropy, and Kd is the dissociation constant. For FA competition assays, fluorescently labeled NAs were prebound to proteins of interest at saturating concentrations (5 μM for HTLV-2 MA, 5 μM for HTLV-2 NC, and 7 μM for HIV-1 MA) for 10 min under the same conditions as those described above for direct FA measurements, followed by incubation with increasing amounts of unlabeled SL1, SL2, SL1-SL2, tRNA, or inositol hexaphosphate (IP6) for 30 min prior to taking FA readings.
Sedimentation assays.
Sedimentation assays were carried out essentially as described previously (44). Briefly, refolded 32P-labeled TAR RNA (15 nM) was combined with TAR DNA (90 nM) in a solution containing 20 mM HEPES (pH 7.5), 20 mM NaCl, and 5 mM DTT. Upon addition of proteins to a final concentration of 1, 5, or 10 μM, reaction mixtures (30 μl) were incubated at 37°C for 30 min. Solutions were then centrifuged at 12,000 rpm in a microcentrifuge for 20 min. The supernatant (2 μl) was collected and analyzed by scintillation counting. The percent radioactivity remaining in the supernatant, relative to the RNA-only sample (set to 100%), was plotted as a function of the protein concentration.
Annealing assays.
TAR DNA/RNA annealing assays were performed essentially as described previously (44). Briefly, refolded 32P-labeled TAR RNA was combined with unlabeled complementary TAR DNA in a solution containing 20 mM HEPES (pH 7.5), 20 mM NaCl, and 5 mM DTT at 37°C. Reactions were initiated by adding 90 nM DNA and MA or NC proteins (5 μM) to 15 nM RNA, followed by incubation in the reaction buffer for the indicated times. Reactions were quenched by placing solutions on ice, followed by the addition of SDS to a 1% (vol/vol) final concentration. Samples were extracted twice with a 4:1 mixture of phenol-chloroform and loaded onto SDS–12% polyacrylamide gels (375 mM Tris-HCl [pH 8.8], 0.1% [wt/vol] SDS, and a 19:1 mixture of acrylamide-bisacrylamide [wt/vol]) run at 25°C in Tris-glycine (25 mM Tris, 250 mM glycine [pH 8.3]) running buffer. Gels were visualized by using a Typhoon Trio Imager and quantified with Bio-Rad Quantity One software.
Determination of RNA packaging efficiency.
HIV-1 proviral plasmid DNA (10 μg) was transfected into 2 million 293T cells by using a calcium phosphate-mediated method in 10-cm plates (45). Cells and supernatant were collected at 48 h posttransfection for preparation of RNAs and lysates. For quantification of viral RNA, virus-like particles (VLPs) were harvested by filtering the supernatant through a 0.2-μm syringe filter prior to ultracentrifugation in a Beckman 50.2 Ti rotor at 25,000 rpm for 2 h at 4°C. Viral RNA was extracted from VLP pellets with a High Pure Viral RNA kit (Roche Applied Science, Indianapolis, IN). Total cellular RNAs were prepared by using an RNeasy kit and QIAshredder (Qiagen, Inc., Valencia, CA). Total viral and cellular RNAs were then treated with DNase by using a DNA-free kit (Ambion, Inc., Austin, TX) to remove any contaminating genomic DNA. Reverse transcription (RT) and quantitative PCR (qPCR) were performed individually for both viral and cellular RNAs with the Transcriptor High Fidelity cDNA synthesis kit (Roche) and the SYBR green qPCR reagent kit (Invitrogen Corp., Carlsbad, CA), respectively. Primers used for quantitative PCR of HIV-1 gag RNA were 5′-ACATCAAGCAGCCATGCAAAT and 5′-ATGTCACTTCCCCTTGGTTCTCT. A serial dilution of an HIV-1 vector (10−6 to 10 ng) containing the target sequence was prepared as a standard for quantitative PCR analysis. The quantity of Gag can be determined from viral and cellular samples based on the defined standard. Experiments were performed with a MyiQ single-color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA), and data were analyzed with MyiQ software (version 1.0; Bio-Rad). For quantification of VLP production, the supernatant was filtered through a 0.2-μm syringe filter prior to ultracentrifugation in a Beckman 50.2 Ti rotor at 25,000 rpm for 2 h at 4°C. Transfected cells were also harvested and washed with Dulbecco's phosphate-buffered saline (Invitrogen). VLPs and cell pellets were resuspended individually in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA). Lysates were electrophoresed on 12.5% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Bio-Rad). HIV-1 Gag was detected with a primary rabbit anti-HIV-1 p24 antiserum (Advanced Biotechnologies, Inc., Columbia, MD) at a 1:1,500 dilution, followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific, Inc., Rockford, IL) at a 1:10,000 dilution. Band intensities were quantified with the ChemiDoc XRS system (Bio-Rad). The RNA packaging efficiency was determined by using methods described previously (25).
RESULTS
Nucleic acid binding of HTLV-2 MA and NC.
Previous studies have shown that HIV-1 NC is an excellent RNA binding and chaperone protein and plays an essential role in gRNA packaging, while HIV-1 MA is not required for specific RNA packaging (46). To test whether HTLV-2 NC and MA proteins play roles similar to those of their HIV-1 counterparts, FA assays were used to determine equilibrium dissociation constants (Kd) for HTLV-2 NC and MA binding to a nonspecific 20-mer ssDNA (5′-FAM DNA20) and to HTLV-2 SL1 and SL2 sequences derived from the putative gRNA packaging signal (Fig. 2) (32). Representative binding curves for 5′-FAM DNA20 are shown in Fig. 3. The data were fit to a binding model that assumes a 1:1 binding stoichiometry, and apparent Kd values of 215 nM and 138 nM were determined for the HTLV-2 NC and MA proteins, respectively. The Kd values for HTLV-2 genome-derived SL1 and SL2 sequences are summarized in Table 1. Under the relatively low-ionic-strength conditions of 50 mM NaCl, the binding of NC to ssDNA and SL2 was ∼2-fold and ∼6-fold weaker than the binding of MA, respectively. Under these conditions, neither HTLV-2 MA or NC has a strong preference for binding to SL1 or SL2 relative to ssDNA, although MA binds with a ∼2-fold-higher affinity to SL2 than to ssDNA.
FIG 3.
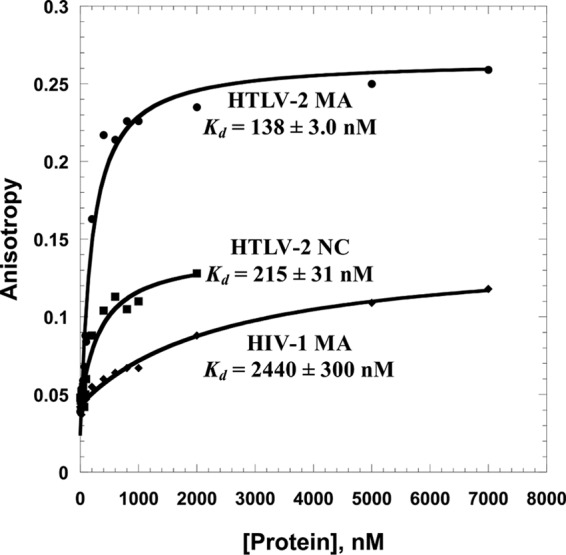
Representative FA binding assays wherein 20 nM 5′-FAM DNA20 was titrated with HTLV-2 MA, HTLV-2 NC, and HIV-1 MA. The curves are single exponential fits of the data.
TABLE 1.
Binding parameters of HTLV-2 proteinsa
| HTLV-2 protein (NaCl concn [mM]) | Mean Kd (nM) ± SD |
||
|---|---|---|---|
| ssDNA | SL1 | SL2 | |
| NC (50) | 215 ± 31 | 614 ± 120 | 419 ± 170 |
| MA (50) | 138 ± 3.0 | 341 ± 170 | 74 ± 18 |
| MA (100) | 945 ± 200 | — | 186 ± 80 |
| MA (150) | 6,200 ± 1,600 | — | 493 ± 97 |
| R47A/K51A MA (50) | 864 ± 320 | — | 633 ± 200 |
| R47A/K51A MA (100) | NB | — | NB |
| R47A/K51A MA (150) | NB | — | NB |
Shown are apparent equilibrium dissociation constants (Kd) obtained from FA measurements performed at various ionic strengths. NB indicates that no binding was detected with up to 7 μM protein. A dash indicates that the value was not determined.
Although MA does not display a strong preference for binding to SL2 at low ionic strength, the binding to SL2 is less sensitive to increasing salt concentrations than the binding to ssDNA (Table 1). At 100 mM NaCl, SL2 binding was 5-fold stronger than ssDNA binding, whereas at 150 mM NaCl, binding was ∼13-fold stronger. Thus, under physiological conditions, HTLV-2 MA may play a role in selective binding to gRNA.
FA competition assays were performed to further test the specificity of HTLV-2 MA binding. In these experiments, MA was prebound to fluorescently labeled minihelixLys,3, derived from the acceptor-TΨC sequence of human tRNALys,3, or HIV-1 SL3 (Fig. 2). The complexes were titrated with unlabeled HTLV-2-derived SL1, SL2, or SL1-SL2 or human tRNALys,3. As shown in Fig. 4, SL1 was unable to compete effectively for MA binding to the prebound RNAs. In contrast, SL2 and SL1-SL2 readily competed off the nonspecific RNAs. Surprisingly, tRNALys,3 was even more effective than SL1-SL2 at competing for binding, and the same result was obtained with another tRNA (tRNAAla) tested (data not shown).
FIG 4.
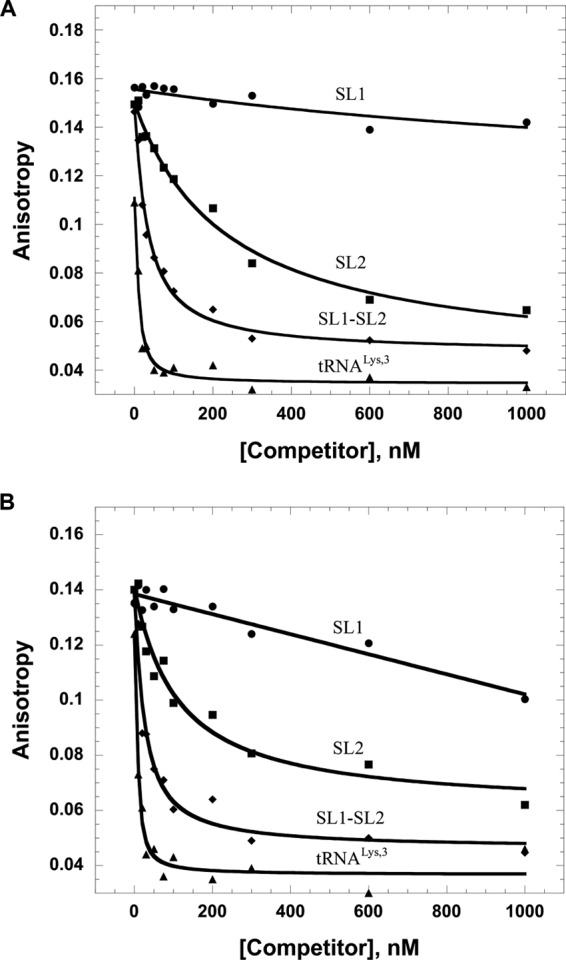
FA competition assays wherein 20 nM fluorescently labeled HIV-1 SL3 (A) or minihelixLys,3 (B) was preincubated with 5 μM HTLV-2 MA, followed by titration with unlabeled HTLV-2-derived SL1, SL2, and SL1-SL2 RNAs or tRNALys,3.
Nucleic acid chaperone activity of HTLV-2 MA and NC.
Annealing of TAR DNA hairpin to a complementary TAR RNA hairpin, which mimics the annealing step of minus-strand transfer in reverse transcription, was performed as a model assay to study the chaperone function of the HTLV-2 NC and MA proteins (38). A comparison of the annealing of TAR RNA/DNA hairpins in the presence of saturating concentrations of NC and MA is shown in Fig. 5. These data suggest that HTLV-2 MA exhibits much better chaperone activity than NC. The effective annealing rates and final percentages of RNA annealed in the presence of MA were significantly higher (∼15-fold and ∼12-fold, respectively) than those in the presence of NC. In contrast to HIV-1, in which NC is highly basic, HTLV-2 NC has an acidic C-terminal domain (overall pI ∼7.0), similar to HTLV-1 NC, which also lacks strong NA binding and chaperone activities (28). However, HTLV-2 MA is a relatively basic protein (overall pI ∼9.6), which may explain its relatively robust NA binding and chaperone functions.
FIG 5.
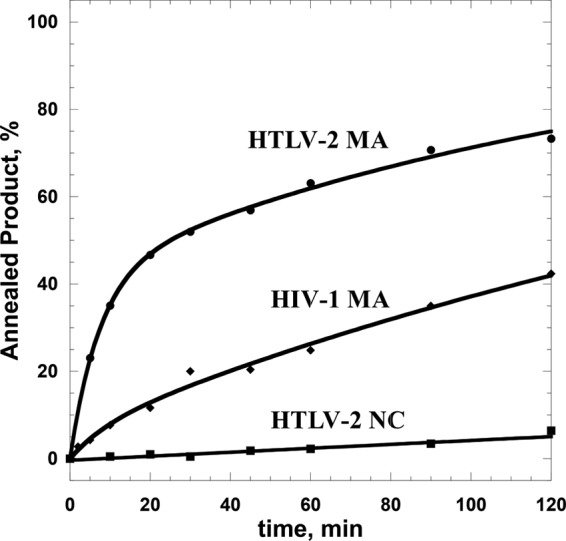
Annealing time courses of 15 nM TAR RNA and 90 nM TAR DNA at 37°C in the presence of 5 μM proteins.
Role of basic amino acid residues in α-helix II of HTLV-2 MA.
The sequence alignment reveals high homology between deltaretroviral MA proteins, including the presence of charged residues in α-helix II (Fig. 1), which is predicted to have an overall positive charge. In contrast, HIV-1 MA possesses a helix II that is neutral overall. A previous study showed that mutation of BLV MA K41 and H45 to Ala significantly reduced the gRNA packaging efficiency (25). To test whether these conserved residues influence RNA binding of HTLV-2 MA in vitro, the two corresponding basic residues, R47 and K51, were mutated to alanine. Binding to nonspecific 5′-FAM DNA20 and HTLV-2 SL2 RNA was tested by using FA. Table 1 shows that when assays were conducted with 50 mM NaCl, the simultaneous mutation of two residues to generate the R47A/K51A MA variant led to ∼6-fold and ∼8-fold reductions in ssDNA and SL2 binding, respectively. The effect on SL2 binding was even more dramatic under conditions of higher ionic strength (Table 1). At ≥100 mM NaCl, binding to ssDNA and SL2 was undetectable, whereas the WT protein still bound SL2 with a relatively high affinity (186 nM and 493 nM at 100 mM and 150 mM NaCl, respectively). WT MA binding to ssDNA was much more salt sensitive, with apparent Kd values of 945 nM and 6,200 nM measured at 100 mM and 150 mM NaCl, respectively. Thus, R47 and K51 may interact with SL2 through primarily nonelectrostatic forces.
Comparison of HTLV-2 and HIV-1 MA proteins.
Superposition of the HTLV-2 and HIV-1 MA structures reveals a similar three-dimensional fold despite limited primary sequence identity (26). To gain further insights into functional differences between these proteins, we conducted FA assays to compare their binding to 5′-FAM DNA20. This fairly random ssDNA sequence was used to minimize sequence-specific binding effects. The Kd value obtained for HTLV-2 MA (138 nM) is ∼17-fold lower than that measured for HIV-1 MA (2,440 nM), showing that HTLV-2 MA binds NAs with a higher affinity than HIV-1 MA (Fig. 3). Furthermore, SL2 binding of HIV-1 MA was 6-fold weaker and more sensitive to increasing salt concentrations than HTLV-2 MA (Tables 1 and 2).
TABLE 2.
Binding parameters of WT HIV-1 MA and variantsa
| HIV-1 MA protein (NaCl concn [mM]) | Mean Kd (nM) ± SD |
||
|---|---|---|---|
| ssDNA | MicroTAR | SL2 | |
| WT (50) | 2,440 ± 300 | 1,360 ± 280 | 471 ± 150 |
| WT (100) | — | — | 1,320 ± 140 |
| WT (150) | — | — | 5,190 ± 700 |
| E40R (50) | 154 ± 6.0 | 254 ± 76 | — |
| E40R/E42L (50) | 461 ± 190 | 271 ± 170 | — |
| E40R/E42L/N47K (50) | 230 ± 31 | 185 ± 16 | — |
Shown are apparent equilibrium dissociation constants (Kd) obtained from FA measurements performed at various ionic strengths. A dash indicates that the value was not determined.
Figure 5 compares the annealing of TAR DNA/RNA hairpins in the presence of saturating concentrations of HTLV-2 MA and HIV-1 MA. HTLV-2 MA facilitates the annealing reaction more effectively than HIV-1 MA, with an ∼2-fold-higher kobs. The ability to aggregate NAs is another critical component of a chaperone protein, and a sedimentation assay was used to measure this property (data not shown). The higher percentage of aggregated RNA observed at all protein concentrations tested shows that HTLV-2 MA is a much more effective NA-aggregating agent than HIV-1 MA. HTLV-2 MA achieved 80% aggregation at all concentrations tested (1 to 10 μM), whereas HIV-1 MA aggregated only ∼36% of the NAs at the lowest concentration tested (1 μM) and aggregated even smaller amounts at 5 μM (25%) and 10 μM (6%). Although it is not known why less aggregation was observed with increasing HIV-1 MA concentrations, it may be due to the increase in the salt concentration as more protein was added.
Electrostatic potential surfaces of HTLV-2 and HIV-1 MA proteins obtained by using Swiss-PdbViewer reveal a major difference between the two proteins in the N terminus. In particular, four basic residues (R47, R48, K51, and K55) located in HTLV-2 MA helix II form a large patch of positive charge (data not shown). In contrast, HIV-1 MA lacks a cluster of basic residues on helix II. Instead, two basic (R39 and R43) and two acidic (E40 and E42) residues form a neutral surface on one face of the N-terminal domain. In order to test whether the basic character of helix II specifically affects the NA binding properties of HIV-1 MA, an E40R/E42L/N47K chimeric triple mutant (TRI-MUT) was designed to mimic the more basic HTLV-2 MA helix II domain. The HIV-1 MA TRI-MUT variant was overexpressed, purified, and shown to be well folded by CD spectroscopy (data not shown). Binding of this HIV-1 MA variant to both nonspecific DNA and HIV-1-derived RNA was investigated next. FA binding assays showed that the TRI-MUT variant showed a ∼10-fold-higher ssDNA binding affinity than WT HIV-1 MA (Fig. 6A and Table 2) as well as significantly improved chaperone function (Fig. 6B). Binding to an HIV-1 TAR RNA-derived sequence (MicroTAR) was also ∼7-fold tighter for the TRI-MUT variant. The binding and chaperone properties of the TRI-MUT variant are similar to those of HTLV-2 MA (Fig. 3, 5, and 6). Surprisingly, even the double mutant variant (E40R/E42L) and a single point mutant (E40R) demonstrated significantly improved binding properties relative to those of WT HIV-1 MA when measured using both ssDNA and MicroTAR RNA (Table 2). Taken together, the data support a critical function for the conserved basic residues located in helix II of HTLV-2 MA on NA binding and chaperone properties.
FIG 6.
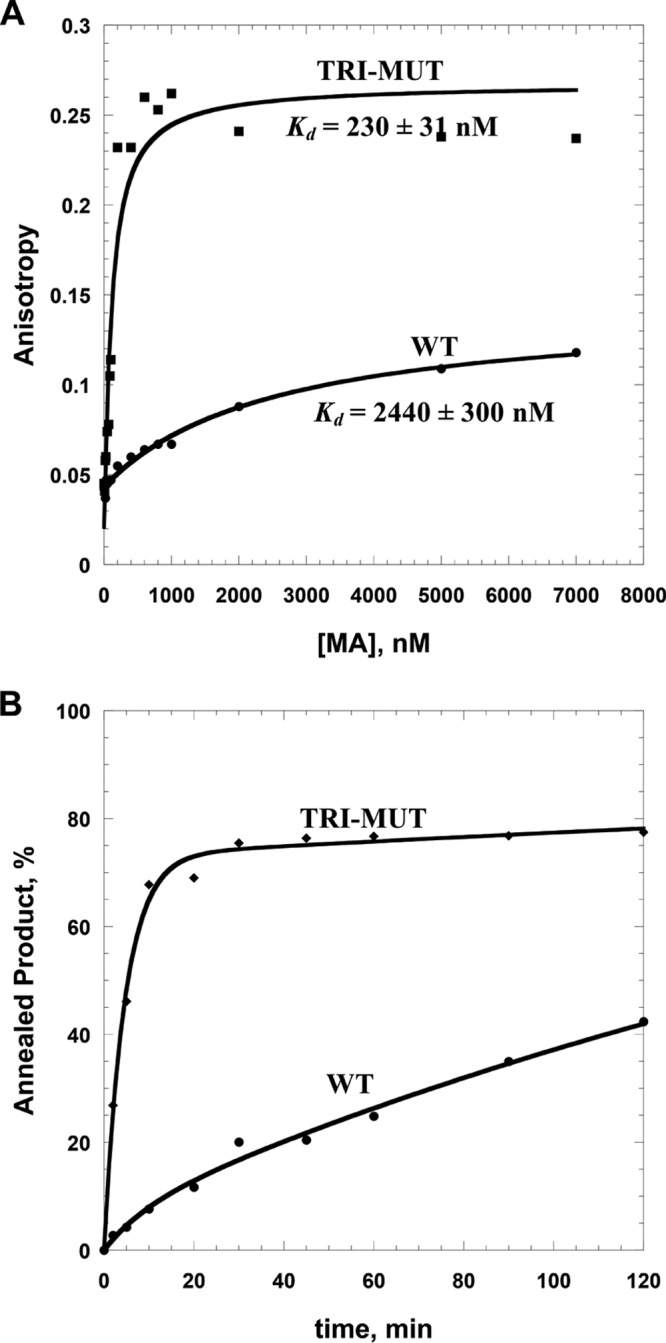
Comparison of WT HIV-1 MA and TRI-MUT NA binding and chaperone functions. (A) Representative FA binding assays using 20 nM 5′-FAM DNA20 in the presence of increasing amounts of protein. Lines are fits to the data (see equation in the text). (B) Annealing time courses of 15 nM TAR RNA and 90 nM TAR DNA in the presence of 5 μM protein.
IP6 competes for MA-nucleic acid binding.
Since a primary function of all retroviral MA domains involves membrane binding to phospholipids, we tested the NA binding of HTLV-2 MA, HTLV-2 NC, and HIV-1 MA in the presence of increasing concentrations of IP6, which has been shown to have an effect on HIV-1 Gag assembly in vitro (47) (Fig. 7). Proteins were prebound to HTLV-2 SL2 at saturating concentrations. As expected, addition of IP6 did not compete for HTLV-2 NC-SL2 interactions, consistent with the fact that NC does not participate in membrane binding. In contrast, both HTLV-2 and HIV-1 MAs were displaced from Fl-SL2 by IP6. The final anisotropy value of ∼0.05 at 20 μM IP6 suggests that Fl-SL2 is completely displaced. These results suggest that IP6 interacts with both HTLV-2 and HIV-1 MA proteins but not with HTLV-2 NC in the presence of RNA.
FIG 7.
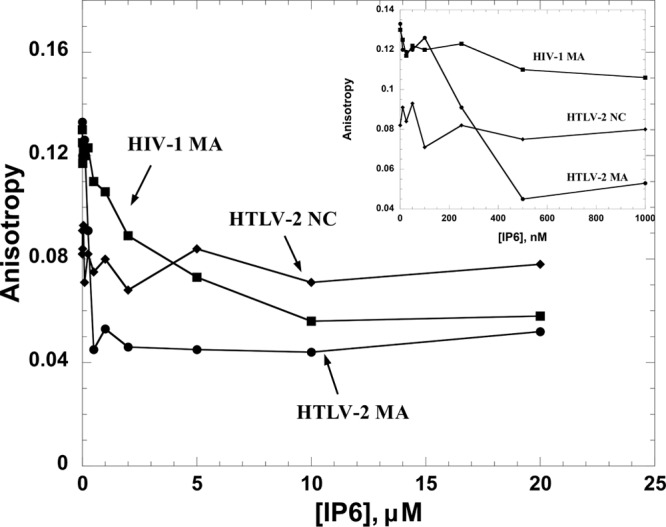
FA competition assays wherein 20 nM 5′-Fl-SL2 was preincubated with HTLV-2 MA, HIV-1 MA, or HTLV-2 NC at saturating protein concentrations (5 μM for HTLV-2 MA, 5 μM for HTLV-2 NC, and 7 μM for HIV-1 MA), followed by titration with IP6. The inset shows an expanded view of the low-IP6-concentration region of the titration.
Cell-based assays to probe the role of MA helix II in HIV-1 RNA packaging.
Although NC is the key player in HIV-1 gRNA packaging (4, 48–51), MA has also been shown to participate in BLV gRNA packaging (25). Furthermore, data provided in this study and elsewhere (17, 19–21, 34) suggest that HIV-1 MA interacts with gRNA. We have shown that the HIV-1 MA TRI-MUT variant exhibits higher NA binding affinity in vitro than WT HIV-1 MA. To test the effect of the triple mutation on RNA packaging in HIV-1, we transfected WT/ΔNC and MAE40R/E42L/N47K/ΔNC HIV-1 clones individually into 293T cells and harvested VLPs. The amount of Gag polypeptide (Pr48) was detected and quantified from VLPs as well as from virus-producing cells (Fig. 8A and B). Total RNAs from VLPs and virus-producing cells were used in a two-step, real-time RT-PCR analysis to quantify the expression level of the HIV-1 gag gene (Fig. 8C). The viral RNA packaging efficiency was determined as described previously (25). The results shown in Fig. 8D indicate that HIV-1 RNA packaging is about 5 times more efficient for MAE40R/E42L/N47K/ΔNC virus than for WT/ΔNC virus (n = 3). (The packaging efficiency of the WT virus with NC, as determined in independent experiments, was ∼17-fold higher than that of WT/ΔNC virus [data not shown].) These cell-based data support the in vitro results and show that the introduction of basic charges into helix II of HIV-1 MA enhances the RNA binding and packaging ability of HIV-1 MA in the absence of NC.
FIG 8.
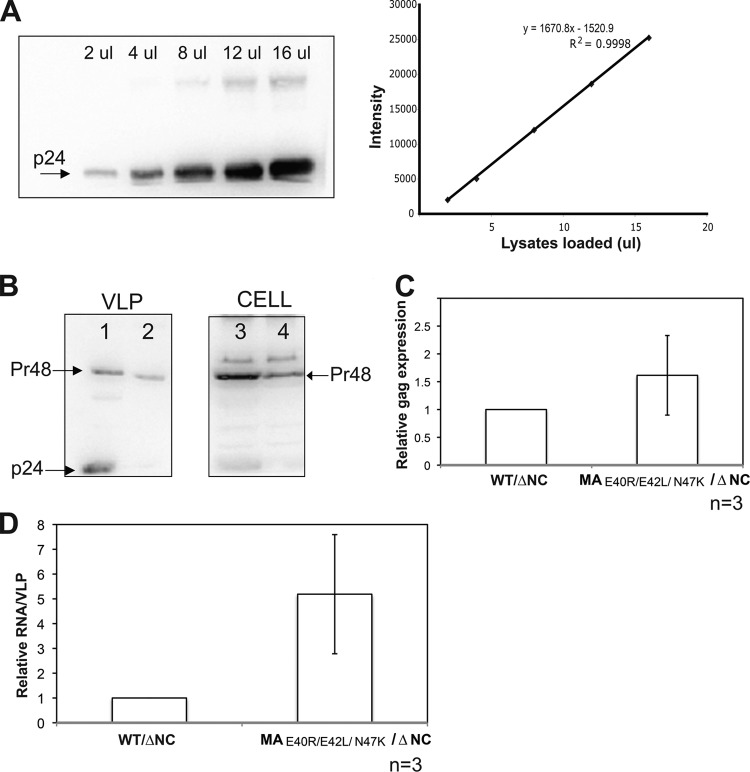
HIV-1 gRNA packaging efficiency of the HIV-1 MA TRI-MUT variant in an NC deletion background. (A) Serial dilution of WT HIV-1 p24 for protein quantification. (Left) Immunoblot displaying a serial dilution (2 μl to 16 μl lysate per loading) of WT HIV-1 p24. (Right) Band intensities (arbitrary units) (y axis) plotted against the volume of lysates loaded (x axis) to evaluate the linearity and accuracy of protein quantification. The linear equation and an R2 value are indicated. (B) Immunoblots were probed with antisera against HIV-1 p24. The amount of protein was determined based on the linear standard shown in panel A. Lanes 1 and 2 show VLP lysates collected from WT/ΔNC (lane 1) and MAE40R/E42L/N47K/ΔNC (lane 2). Lanes 3 and 4 show lysates from producing cells expressing the WT/ΔNC clone (lane 3) and the MAE40R/E42L/N47K/ΔNC clone (lane 4). (C) Relative expression level of gag detected by two-step quantitative RT-PCR. The expression level of gag from MAE40R/E42L/N47K/ΔNC is normalized to that from WT/ΔNC. Error bars represent standard deviations from three independent experiments (n = 3). (D) Relative fold change of RNA packaging efficiency. The RNA packaging efficiency (y axis) was calculated as previously described (25). The RNA packaging efficiency of the MAE40R/E42L/N47K/ΔNC mutant was normalized to that of WT/ΔNC. Error bars represent standard deviations from three individual experiments (n = 3).
DISCUSSION
In this work, the NA binding and chaperone properties of two different MA proteins, HTLV-2 and HIV-1, were compared in vitro. FA binding studies demonstrated that in the deltaretrovirus HTLV-2, MA binds NAs with higher affinity than NC, which is in contrast to HIV-1. Furthermore, salt-dependent binding assays support specific binding of HTLV-2 MA to the SL2 stem-loop structure derived from the putative HTLV-2 RNA packaging signal and suggest that conserved basic residues in helix II contribute to binding. Competition binding studies also supported preferential binding to SL2 or the combined SL1-SL2 over SL1 alone, but surprisingly, tRNA was found to be even more effective than SL1-SL2 at competing for HTLV-2 MA binding. This may be due to nonspecific binding interactions of MA with longer nucleic acid sequences. A recent study of BLV MA showed high-affinity (10 to 20 nM) binding to RNAs derived from the BLV genome, supporting a conservation of function among deltaretroviral MA proteins (52). In contrast, previous work has shown that HIV-1 MA is not required for specific gRNA binding (46).
The chaperone activities of HTLV-2 MA and NC were tested by using gel shift annealing assays and compared with those of HIV-1 proteins. Our data suggest that HTLV-2 MA facilitates TAR RNA/DNA annealing more effectively than NC, which is also in contrast to HIV-1, where NC is a potent chaperone protein. The different behaviors of the two retroviral genera may be due to the different distributions of local electrostatic potential in the respective Gag proteins. HIV-1 NC is well known for its highly basic character (pI 9.86), whereas HTLV-2 NC is neutral overall (pI 7.68); in contrast, HTLV-2 MA (pI 9.51) exhibits a more basic character than HIV-1 MA (pI 9.02). A basic cluster is located in the N-terminal domain of HTLV-2 MA, forming a highly positively charged surface that is absent from HIV-1 MA. We have previously shown that HTLV-1 NC, which shares high sequence identity with HTLV-2 NC, lacks NA-aggregating capabilities and displays relatively poor chaperone activity despite the fact that it is a strong duplex destabilizer compared to other retroviral NCs (28). Combined with previous cell-based studies, which demonstrated that basic residue variants of BLV MA are defective in RNA packaging (25), these data suggest that deltaretroviral MA proteins are major players in the initial stage of gRNA selection and packaging, whereas NC plays a more minor role at this stage of the life cycle. Further functional analyses will be needed to map the MA-genome interactions at the molecular level.
The basic residues of MA also play a role in directing membrane targeting of Gag. In HIV-1, a highly basic region spanning residues in the N terminus forms an interface with acidic phospholipids and, along with the N-terminal myristoyl group, facilitates membrane binding of Gag to PI(4,5)P2-containing liposomes (23, 53, 54). Interestingly, Gag targeting and membrane binding mediated by HTLV-1 MA do not appear to require PI(4,5)P2 (55). Instead, HTLV-1 Gag binding to liposomes is driven largely by electrostatic interactions instead of specific interactions with PI(4,5)P2. In contrast to HIV-1, HTLV-1 Gag membrane binding in vitro is not suppressed by RNA. These data suggested that HIV-1 and HTLV-1 use different mechanisms to regulate membrane targeting (55). Previous studies have also indicated that HTLV-1 and HIV-1 Gag proteins target different plasma membrane microdomains in Jurkat T cells (56). Our competition assays with IP6 demonstrate that HTLV-2 MA is competed off NAs even more effectively than HIV-1 MA and suggest that the MA-IP6-interacting surface likely overlaps the MA-NA binding site. Similar results were recently reported for BLV MA (52). IP6 possesses more negative-charge density than the head group of PI(4,5)P2. Therefore, these results are in agreement with previous studies showing that electrostatic interactions are the major driving force for HTLV-1 Gag-liposome binding (55), although it is still unclear which specific phospholipid is required for HTLV-1 and -2 membrane targeting. More detailed liposome binding studies are needed to determine the specific factor that contributes to deltaretroviral MA membrane targeting and subcellular localization.
The chaperone activity of retroviral NC proteins plays a critical role in facilitating NA remodeling events throughout reverse transcription. With a 228-nt gRNA R region, which is predicted to fold into a complex secondary structure (57), HTLV-1 would be expected to require a robust chaperone to facilitate the minus-strand transfer steps of reverse transcription. Surprisingly, HTLV-1 and -2 NC proteins display relatively poor NA binding and overall chaperone activity; however, once bound, HTLV-1 NC is a strong duplex destabilizer (28, 29). We therefore suggest that in deltaretroviruses, MA is involved in key early steps involving genome recognition and packaging; however, once the local concentration of Gag is high enough, MA preferentially binds to the membrane, and NC domain binding to NAs occurs, allowing NC to carry out the chaperone function required for reverse transcription (Fig. 9).
FIG 9.
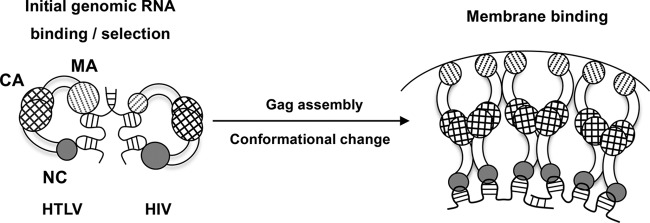
Proposed mechanism of the Gag-genome interaction. Larger circles indicate a major role in NA binding, and smaller circles indicate a more minor role. In initial genome selection (left), Gag binds to gRNA with both NC and MA domains. In HIV, the NC domain has specific binding interactions with the psi packaging signal, with only minor contributions from MA. In HTLV, the MA domain contributes significantly more to initial binding and specific interactions with gRNA than NC. Upon reaching the plasma membrane, the conformation of Gag changes, as the MA domain preferentially binds to membrane lipids. In HTLV, the high local concentration of Gag allows the NC domain to remain bound to the genome despite its relatively weak affinity.
The fact that we can increase RNA packaging efficiency (and perhaps specificity as well) of a ΔNC HIV-1 virus by increasing the basic character of the helix II domain of HIV-1 MA is remarkable. This result is consistent with previous studies showing that HIV-1 MA and NC have redundant roles in virus assembly (17, 34). Mutations introduced into NA binding regions of either NC or MA do not severely affect RNA incorporation; however, mutation of RNA binding areas of both domains results in particles without gRNA packaged (17, 34). Although the cluster of basic amino acids located in helix II of BLV MA is known to contribute to gRNA incorporation (25), additional studies are needed to confirm that the basic character of MA in other retroviruses, such as HTLV-1 and -2, is involved in genome packaging.
In summary, based on previous studies and the new results reported here, we propose a model (Fig. 9) in which initial gRNA binding and selection involve a folded conformation of Gag with both MA and NC domains binding to RNA. In HIV-1, NC plays a major role at this stage and binds the genome specifically. We have recently shown that HIV-1 Gag binds to psi-specific RNAs differently than to non-psi RNAs; our data suggest that psi binding involves primarily the NC domain, whereas non-psi binding involves both the NC and MA domains (58). In contrast, we propose that in deltaretroviruses, MA plays a larger role in recognizing gRNA at the initial stage. When Gag assembles at the plasma membrane, it triggers a conformational change (59, 60), wherein phosphorylated phosphatidylinositides or other membrane microdomains bind to the MA domain while NC remains bound to RNA. Furthermore, once a high local concentration is achieved, the slow NA dissociation kinetics of HTLV NC (28) allow it to remain bound to the RNA. Taken together, these studies suggest that deltaretroviruses use distinct protein-RNA interactions for gRNA packaging. The implications of these findings for specific viral RNA recognition by HTLV MA and MA's role in the context of Gag remain to be explored. Additional studies to map HTLV MA-RNA interactions are under way to gain further insights into MA's diverse roles in the viral life cycle.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by NIH grants GM065056 (to K.M.-F.) and GM098500 (to L.M.M.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with Leidos Biomedical Research, Inc. (R.J.G.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We thank members of the AIDS and Cancer Virus Program, Frederick National Laboratory for Cancer Research, David E. Ott for providing the delNC HIV-1 proviral plasmid, and Donald G. Johnson and Catherine V. Hixson for assistance with preparation of the HTLV-2 NCp15 protein.
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177–218. 10.1007/978-3-642-80145-7_6 [DOI] [PubMed] [Google Scholar]
- 2.Jewell NA, Mansky LM. 2000. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 81:1889–1899 http://vir.sgmjournals.org/content/81/8/1889.long [DOI] [PubMed] [Google Scholar]
- 3.Rein A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513–522 [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286. 10.1016/S0079-6603(05)80006-6 [DOI] [PubMed] [Google Scholar]
- 6.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. 2006. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 363:867–877. 10.1016/j.jmb.2006.08.070 [DOI] [PubMed] [Google Scholar]
- 7.Narayanan N, Gorelick RJ, DeStefano JJ. 2006. Structure/function mapping of amino acids in the N-terminal zinc finger of the human immunodeficiency virus type 1 nucleocapsid protein: residues responsible for nucleic acid helix destabilizing activity. Biochemistry 45:12617–12628. 10.1021/bi060925c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlix JL, Garrido JL, Morellet N, Mely Y, de Rocquigny H. 2007. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 55:299–346. 10.1016/S1054-3589(07)55009-X [DOI] [PubMed] [Google Scholar]
- 9.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491–511. 10.1006/jmbi.2000.3979 [DOI] [PubMed] [Google Scholar]
- 10.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388. 10.1126/science.279.5349.384 [DOI] [PubMed] [Google Scholar]
- 11.Parent LJ, Gudleski N. 2011. Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J. Mol. Biol. 410:553–564. 10.1016/j.jmb.2011.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. U. S. A. 87:523–527. 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spearman P, Wang JJ, Vander Heyden N, Ratner L. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 82:2405–2417. 10.1128/JVI.01614-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Caballero D, Hatziioannou T, Martin-Serrano J, Bieniasz PD. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on gag precursor-membrane interactions. J. Virol. 78:9560–9563. 10.1128/JVI.78.17.9560-9563.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott DE, Coren LV, Gagliardi TD. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 79:13839–13847. 10.1128/JVI.79.22.13839-13847.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ott DE, Coren LV, Shatzer T. 2009. The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly. J. Virol. 83:7718–7727. 10.1128/JVI.00099-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hearps AC, Wagstaff KM, Piller SC, Jans DA. 2008. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 47:2199–2210. 10.1021/bi701360j [DOI] [PubMed] [Google Scholar]
- 20.Lochrie MA, Waugh S, Pratt DG, Jr, Clever J, Parslow TG, Polisky B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 25:2902–2910. 10.1093/nar/25.14.2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purohit P, Dupont S, Stevenson M, Green MR. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584. 10.1017/S1355838201002023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83:12196–12203. 10.1128/JVI.01197-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107:1600–1605. 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh I, Kyushiki H, Sakamoto Y, Ikawa Y, Yoshinaka Y. 1991. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic RNA fragment. J. Virol. 65:6845–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Norris KM, Mansky LM. 2003. Involvement of the matrix and nucleocapsid domains of the bovine leukemia virus Gag polyprotein precursor in viral RNA packaging. J. Virol. 77:9431–9438. 10.1128/JVI.77.17.9431-9438.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen AM, Massiah MA, Turner BG, Sundquist WI, Summers MF. 1996. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J. Mol. Biol. 264:1117–1131. 10.1006/jmbi.1996.0700 [DOI] [PubMed] [Google Scholar]
- 27.Le Blanc I, Rosenberg AR, Dokhelar MC. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 82:10129–10142. 10.1128/JVI.01169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darugar Q, Kim H, Gorelick RJ, Landes C. 2008. Human T-cell lymphotropic virus type 1 nucleocapsid protein-induced structural changes in transactivation response DNA hairpin measured by single-molecule fluorescence resonance energy transfer. J. Virol. 82:12164–12171. 10.1128/JVI.01158-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K. 2010. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J. Biol. Chem. 285:295–307. 10.1074/jbc.M109.051334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansky LM, Krueger AE, Temin HM. 1995. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J. Virol. 69:3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansky LM, Wisniewski RM. 1998. The bovine leukemia virus encapsidation signal is composed of RNA secondary structures. J. Virol. 72:3196–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansky LM, Gajary LC. 2002. The primary nucleotide sequence of the bovine leukemia virus RNA packaging signal can influence efficient RNA packaging and virus replication. Virology 301:272–280. 10.1006/viro.2002.1578 [DOI] [PubMed] [Google Scholar]
- 34.Ott DE, Coren LV, Chertova EN, Gagliardi TD, Nagashima K, Sowder RC, II, Poon DT, Gorelick RJ. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77:5547–5556. 10.1128/JVI.77.10.5547-5556.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 36.Carteau S, Gorelick RJ, Bushman FD. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980–8988. 10.1128/JVI.74.19.8980-8988.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2009. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper.” J. Mol. Biol. 386:789–801. 10.1016/j.jmb.2008.12.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stello T, Hong M, Musier-Forsyth K. 1999. Efficient aminoacylation of tRNA(Lys,3) by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res. 27:4823–4829. 10.1093/nar/27.24.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milligan JF, Uhlenbeck OC. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51–62. 10.1016/0076-6879(89)80091-6 [DOI] [PubMed] [Google Scholar]
- 41.Lundblad JR, Laurance M, Goodman RH. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607–612. 10.1210/me.10.6.607 [DOI] [PubMed] [Google Scholar]
- 42.Reid SL, Parry D, Liu HH, Connolly BA. 2001. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry 40:2484–2494. 10.1021/bi001956p [DOI] [PubMed] [Google Scholar]
- 43.Muller B, Restle T, Reinstein J, Goody RS. 1991. Interaction of fluorescently labeled dideoxynucleotides with HIV-1 reverse transcriptase. Biochemistry 30:3709–3715. 10.1021/bi00229a017 [DOI] [PubMed] [Google Scholar]
- 44.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. 2006. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 363:244–261. 10.1016/j.jmb.2006.08.039 [DOI] [PubMed] [Google Scholar]
- 45.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. 10.1016/0042-6822(73)90341-3 [DOI] [PubMed] [Google Scholar]
- 46.Poon DT, Li G, Aldovini A. 1998. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol. 72:1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta SA, Zhao Z, Clark PK, Tarasov S, Alexandratos JN, Campbell SJ, Kvaratskhelia M, Lebowitz J, Rein A. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365:799–811. 10.1016/j.jmb.2006.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimarelli A, Sandin S, Hoglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046–3057. 10.1128/JVI.74.7.3046-3057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poon DT, Wu J, Aldovini A. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375:592–610. 10.1016/j.virol.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 52.Qualley DF, Lackey CM, Paterson JP. 2013. Inositol phosphates compete with nucleic acids for binding to bovine leukemia virus matrix protein: implications for deltaretroviral assembly. Proteins 81:1377–1385. 10.1002/prot.24281 [DOI] [PubMed] [Google Scholar]
- 53.Ono A, Orenstein JM, Freed EO. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855–2866. 10.1128/JVI.74.6.2855-2866.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099–3104. 10.1073/pnas.93.7.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inlora J, Chukkapalli V, Derse D, Ono A. 2011. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 85:3802–3810. 10.1128/JVI.02383-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazurov D, Heidecker G, Derse D. 2006. HTLV-1 Gag protein associates with CD82 tetraspanin microdomains at the plasma membrane. Virology 346:194–204. 10.1016/j.virol.2005.10.033 [DOI] [PubMed] [Google Scholar]
- 57.Askjaer P, Kjems J. 1998. Mapping of multiple RNA binding sites of human T-cell lymphotropic virus type I Rex protein within 5′- and 3′-Rex response elements. J. Biol. Chem. 273:11463–11471. 10.1074/jbc.273.19.11463 [DOI] [PubMed] [Google Scholar]
- 58.Webb JA, Jones CP, Parent LJ, Rouzina I, Musier-Forsyth K. 2013. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA 19:1078–1088. 10.1261/rna.038869.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. 2007. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365:812–824. 10.1016/j.jmb.2006.10.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rein A, Datta SA, Jones CP, Musier-Forsyth K. 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 36:373–380. 10.1016/j.tibs.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]