Abstract
Viral coat proteins function in virion assembly and virus biology in a tightly coordinated manner with a role for virtually every amino acid. In this study, we demonstrated that the coat protein (CP) of Wheat streak mosaic virus (WSMV; genus Tritimovirus, family Potyviridae) is unusually tolerant of extensive deletions, with continued virion assembly and/or systemic infection found after extensive deletions are made. A series of deletion and point mutations was created in the CP cistron of wild-type and/or green fluorescent protein-tagged WSMV, and the effects of these mutations on cell-to-cell and systemic transport and virion assembly of WSMV were examined. Mutants with overlapping deletions comprising N-terminal amino acids 6 to 27, 36 to 84, 85 to 100, 48 to 100, and 36 to 100 or the C-terminal 14 or 17 amino acids systemically infected wheat with different efficiencies. However, mutation of conserved amino acids in the core domain, which may be involved in a salt bridge, abolished virion assembly and cell-to-cell movement. N-terminal amino acids 6 to 27 and 85 to 100 are required for efficient virion assembly and cell-to-cell movement, while the C-terminal 65 amino acids are dispensable for virion assembly but are required for cell-to-cell movement, suggesting that the C terminus of CP functions as a dedicated cell-to-cell movement determinant. In contrast, amino acids 36 to 84 are expendable, with their deletion causing no obvious effects on systemic infection or virion assembly. In total, 152 amino acids (amino acids 6 to 27 and 36 to 100 and the 65 amino acids at the C-terminal end) of 349 amino acids of CP are dispensable for systemic infection and/or virion assembly, which is rare for multifunctional viral CPs.
INTRODUCTION
Viral coat proteins (CPs) are multifunctional, with roles in almost every aspect of the virus life cycle (1, 2). In addition to virion assembly and disassembly, multiple nonstructural functions in virus biology have been attributed to the CPs, such as symptom modulation (e.g., see references 3 and 4), vector transmission (1), replication (5), viral RNA translation (6), virus translocation (1, 7), suppression of host RNA silencing (8, 9), and activation of R-gene-mediated host defenses (10). To facilitate these multiple tasks, the CPs should function in a tightly coordinated manner with a role for virtually every amino acid (1, 2). In contrast to other virus-encoded proteins, the CPs possess several determinants with distinct and/or overlapping functions in virus biology (11). Hence, the CPs tolerate minimal or no mutations without having deleterious effects on virus biology (e.g., see references 4 and 11 to 14).
Successful systemic infection of plants by viruses requires the virus to spread beyond the initially infected cells through specialized intercellular connections termed plasmodesmata (PD) until the virus comes in contact with the vascular system. The virus is then passively transported through phloem-associated cells and egressed at a distal place, followed by further cell-to-cell movement, allowing the virus to spread systemically (15). Cell-to-cell movement of plant viruses is mediated by virus-encoded movement proteins (MPs) through interactions with host factors which allow the virus to move through PD to adjacent cells in virion or nucleoprotein complex form (16, 17). Plant viruses can be divided into at least three types on the basis of the characteristics of cell-to-cell movement. The first group of viruses encodes a single dedicated Tobamovirus 30K-like MP, which increases the size exclusion limit of PD to allow virions or ribonucleoprotein complexes to pass through modified PD channels (15, 18). The second group of viruses includes icosahedral viruses, such as the Secoviridae, Bromoviridae, and Caulimoviridae, which mediate cell-to-cell movement with the involvement of both MPs and CPs through tubule-like structures (19, 20). The third group of viruses includes potex-, carla-, and hordeiviruses and some furo-like viruses, which encode the triple-gene-block proteins that function collectively without forming tubule-like structures (21).
The family Potyviridae comprises the largest number of positive-stranded RNA viruses infecting a wide range of plant species (22). The cell-to-cell movement mechanism of potyvirids does not fall into any of the three categories described above. Instead, several virus-encoded proteins have been reported to be involved in virus movement, with no dedicated MP. The proteins involved in the movement function of potyvirids also play at least one additional role in virus biology (23). Potyviral helper component proteinase (HC-Pro) (24), genome-linked protein (25), cylindrical inclusion protein (26), and CP (11, 13, 24) were reported to have a function in the cell-to-cell movement of potyviruses. Recently, P3N-PIPO, which probably translates from the P3-coding region at a conserved shifty sequence at the 5′ end of PIPO (27), was also implicated in cell-to-cell movement (28–31). Among potyvirid species, the role of the CP in virus biology has been well studied for members of the Potyvirus genus. The CP has three distinct domains: the variable N- and C-terminal domains that are exposed on the virion surface and susceptible to mild trypsin treatment and the more conserved central core domain that forms the core subunit structure of the virion (32, 33). The N-terminal Asp-Ala-Gly (DAG) motif and its adjacent amino acids in the CP are required for aphid transmission of potyviruses by a specific interaction between DAG and the PTK motif located in the HC-Pro (34–36). The elegant work of Dolja et al. (11, 13) identified distinct functions of CP in virion assembly and cell-to-cell movement of Tobacco etch potyvirus (TEV).
Wheat streak mosaic virus (WSMV), an economically important wheat virus, is the type species of the genus Tritimovirus of the family Potyviridae with a 9,384-nucleotide (nt) [excluding the 3′ poly(A) tail] single-stranded plus-sense RNA genome encapsidated in flexuous filamentous particles of 690 to 700 nm by 11 to 15 nm (37). WSMV is transmitted by the wheat curl mite (Aceria tosichella Keifer) in a semipersistent manner, and HC-Pro has been implicated as a viral determinant for wheat curl mite transmission (38–40). The WSMV genome contains a single large open reading frame encoding a polyprotein that is translated from the genomic RNA (37). The polyprotein is cleaved cotranslationally and in trans by three virus-encoded proteinases, P1, HC-Pro, and NIa proteinase, into 10 mature proteins. In contrast to the multifunctional potyviral HC-Pro (23), WSMV HC-Pro is dispensable for systemic infection (41). P1 of WSMV, and not HC-Pro as in potyviruses, was identified as the suppressor of RNA silencing (42).
The WSMV CP is 349 amino acid (aa) residues long, whereas the CPs of TEV and Potato virus Y are 263 and 267 amino acids long, respectively (32, 37, 43). The CP determinants in virion assembly, virus transport, pathogenicity, and vector transmission of WSMV are not known, except that the N-terminal region of CP was identified as a host- and strain-specific long-distance transport factor (44). Recently, we demonstrated that the C-terminal aspartic acid residues of CP are involved in host-specific virus movement and have a role in efficient cell-to-cell movement in wheat and a long-distance transport function in maize inbred line SDp2 (45). In the present study, we identified a region between amino acids 155 and 285 of CP to be the core domain through predictive structural modeling. Additionally, we explored the roles of CP in WSMV biology by introducing point mutations in the central core domain and a series of deletions at the N- and C-terminal regions and examining the effects of these mutations on cell-to-cell and long-distance movement and virion assembly. In total, 152 amino acids (amino acids 6 to 27 and 36 to 100 and the 65 amino acids at the C-terminal end) of 349 amino acids of CP are dispensable for virion assembly and/or systemic infection of WSMV, albeit at reduced levels. Remarkably, 49 amino acid residues comprising amino acids 36 to 84 of CP are expendable, with their deletion having no obvious effects on WSMV virion assembly or systemic infection of wheat.
MATERIALS AND METHODS
Construction of CP mutants.
An infectious cDNA clone of WSMV Sidney 81, pSP6-WSMV-S81 (46), was the basis for all the mutants generated in this study. Previously, we modified pSP6-WSMV-S81 to express green fluorescent protein (GFP) as a marker protein in a pUC-based construct, pSP6-WSMV-S81-GFP-6K1/CI(7aa), and in a T-DNA-based pCAMBIA construct, pCAM-WSMV-S81-GFP-6K1/CI(7aa) (47). Point mutations and in-frame deletions in the CP cistron were created in pSP6-WSMV-S81, pSP6-WSMV-S81-GFP-6K1/CI(7aa), and pCAM-WSMV-S81-GFP-6K1/CI(7aa) using mutagenic oligonucleotides, followed by overlap extension PCR (48). The overlap extension PCR was performed with oligonucleotides W-3 and W-89 (44), and the PCR products were ligated into pSP6-WSMV-S81 or pSP6-WSMV-S81-GFP-6K1/CI(7aa) between the BstEII (at nt 6319) and SpeI (at the 3′ end) restriction endonuclease sites, as described previously (44). Point or deletion mutations in the CP cistron in pCAM-WSMV-S81-GFP-6K1/CI(7aa) were created by replacing a PstI-NotI restriction fragment (nt 4816 at the 3′ end) with the corresponding fragment from pSP6-WSMV-S81 or pSP6-WSMV-S81-GFP-6K1/CI(7aa) containing mutations in the CP cistron.
Standard molecular biology methods were used for PCR, overlap extension PCR, ligations, and transformations, as described by Sambrook and Russell (49). Escherichia coli strain JM109 was used to transform cDNA clones of WSMV, and plasmid DNA was prepared from 40 ml of a culture grown overnight using a Bio-Rad plasmid midiprep kit (Bio-Rad, Hercules, CA). The presence of point or deletion mutations in the cDNA clones was confirmed by nucleotide sequencing, and 3 independent clones per mutant were tested in phenotypic studies. Each mutant was examined in 2 to 3 independent experiments, and the results presented are from one independent clone.
Inoculation of wheat with in vitro transcripts and virion stability assay.
One microgram of NotI- or SpeI-linearized plasmid was used to prepare in vitro transcripts in a 40-μl mixture reaction as described by Tatineni et al. (47). Freshly prepared in vitro transcripts were mixed with an equal volume of 2% sodium pyrophosphate, pH 9.0, containing 1.0% baked Celite and inoculated onto 13 to 20 wheat cv. Tomahawk seedlings at the single-leaf stage. Wheat seedlings were washed with distilled water 5 min after inoculation and incubated in a greenhouse at 20 to 27°C for symptom development. Wheat seedlings were observed for symptom development at 7 to 25 days postinoculation (dpi).
The virion stability of the WSMV CP deletion mutants was examined by preparing crude extract from symptomatic leaves of transcript-inoculated plants in 20 mM sodium phosphate buffer, pH 7.0, at a 1:20 dilution. The extracts were incubated at room temperature for 45 min, inoculated onto wheat seedlings at the single-leaf stage, and observed in a greenhouse for symptom development at 20 to 27°C.
RT-PCR assay.
Total RNA extracted from upper fully expanded symptomatic wheat leaves of CP deletion mutant-infected plants (50) was used to synthesize the first-strand cDNA in a 10-μl reaction volume with random primers as described previously (51). One microliter of first-strand cDNA was used for PCR in a 25-μl reaction volume with plus- and minus-sense CP-specific primers with the following PCR program: 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, 54°C for 30 s, and 72°C for 90 s and one cycle at 72°C for 10 min. The reverse transcription-PCR (RT-PCR) products were analyzed on 1.0% agarose gels in TAE (Tris-acetate-EDTA) buffer.
Cell-to-cell movement of WSMV CP mutants.
In vitro transcripts of GFP-tagged WSMV CP point/deletion mutants were inoculated onto wheat seedlings at the single-leaf stage as described above. The cell-to-cell movement of GFP-tagged wild-type and mutant viruses was monitored by examining the formation of fluorescent foci on inoculated wheat leaves under a Zeiss Stereo Discovery V12 fluorescence microscope (Carl Zeiss MicroImaging, Inc., New York, NY) using a GFP narrow-band filter at 4 and 14 dpi. The GFP fluorescence pictures were taken using an AxioCam MRc5 camera attached to the V12 fluorescence microscope, and the sizes of the foci were measured using a program provided with the AxioCam MRc5 camera.
Agroinfiltration assays.
pCAM-WSMV-S81-GFP-6K1/CI(7aa) constructs with point or deletion mutations in the CP cistron were transformed into Agrobacterium tumefaciens strain EHA105. Agrosuspensions containing WSMV constructs (optical density at 600 nm, 1.0) were mixed with an equal volume of Agrobacterium harboring Tomato bushy stunt virus p19, a suppressor of RNA silencing (52), and infiltrated into the abaxial side of Nicotiana benthamiana leaves using needleless 3-ml syringes. The agroinfiltrated plants were incubated in a growth chamber at a 24°C maximum temperature and a 20°C minimum temperature with a 14-h photoperiod. N. benthamiana leaves were collected at 7 days postagroinfiltration (dpa) for GFP fluorescence observation, followed by virion purification.
Purification of virions and electron microscopy.
Virions were partially purified from 4 to 6 g of symptomatic wheat leaves or 7 g of agroinfiltrated N. benthamiana leaves expressing GFP, as described by Tatineni et al. (53). Briefly, frozen infected tissue was ground in 0.1 M sodium citrate buffer (SCB), pH 6.5, containing 0.1% β-mercaptoethanol (3 ml/g tissue). The extract was filtered through four layers of muslin cloth and clarified by centrifugation at 8,000 × g for 10 min. The supernatant was treated with 2% Triton X-100 at 4°C for 15 to 20 min and layered on 5 ml of 20% (wt/vol) sucrose in SCB, followed by centrifugation at 118,000 × g for 1.5 h in a Beckman 50.2 Ti rotor. The virus pellet was suspended in 300 μl of 40 mM sodium phosphate buffer, pH 7.0, containing 5% sucrose at 4°C overnight. The purified virus was clarified by centrifugation at 3,000 × g for 5 min, 20 μl of the purified virus preparation was used to prepare 400-mesh carbon-coated copper grids, and purified virus was observed under a Hitachi H-7500 transmission electron microscope. For Western blots, purified virus was mixed with an equal volume of 2× sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue), and the mixture was incubated in a boiling water bath for 3 min.
Assays of WSMV CP mutant virion assembly.
The ability to form virions by the CP mutants was examined by purifying virions from systemically infected wheat (movement-dependent virion assembly [MDVA] assay) and from agroinfiltrated N. benthamiana leaves (movement-independent virion assembly [MIVA] assay). The MDVA assay can provide information on virion assembly only if a mutant efficiently infects wheat in a manner similar to that of a wild-type virus and not if mutants fail to infect or inefficiently infect wheat due to defects in cell-to-cell movement but not in virion assembly. In contrast, the MIVA assay with N. benthamiana facilitates examination of virion assembly independently of virus movement (see below).
Recently, we found that Agrobacterium harboring a GFP-tagged variant of WSMV in a binary vector [pCAM-WSMV-S81-GFP-6K1/CI(7aa)] replicated weakly and formed infectious virions in agroinfiltrated N. benthamiana leaves without cell-to-cell and long-distance movement (47) (data not shown), which facilitates examination of virion assembly independently of virus movement. Purification of virions from agroinfiltrated leaves through a 20% sucrose cushion separates encapsidated virions from nonencapsidated free CP. We utilized the MDVA and MIVA assays to examine the ability to form virions and the efficiency of virion formation by CP mutants. Partially purified virions from wheat and N. benthamiana leaves were observed under an electron microscope and/or subjected to Western blotting using an anti-WSMV serum.
Western blot assay.
Partially purified denatured virions were subjected to SDS-PAGE on 4 to 20% gels (Invitrogen, Carlsbad, CA), followed by transfer onto a polyvinylidene difluoride (PVDF) membrane using an iBlot apparatus (Invitrogen). The blots were developed using a 1:15,000 dilution of anti-WSMV serum as a primary antibody and horseradish peroxidase-labeled goat anti-rabbit IgG (at 1:50,000) as a secondary antibody. The PVDF membranes were developed using Immobilon Western blot substrate (Millipore, Billerica, MA), and images of immunoreactive protein bands were captured using a Molecular Imager ChemiDoc XRS+ imaging system with Image Lab software (Bio-Rad).
Predictive structural modeling.
Since there is no three-dimensional structure of the CP or a closely related protein, the I-TASSER (iterative threading assembly refinement) server, a widely used integrated platform for structure prediction, was used to model the structure of the WSMV CP both with and without user-supplied restraints (54). The crystal structure of Papaya mosaic virus (PMV) CP (chain A; Protein Data Bank [PDB] accession number 4DOX) (55) was used as a user restraint for generating models of the full-length version and a number of truncated versions of CP. The confidence (C) score, based on the quality of threading template alignments and structural assembly simulation convergence parameters, along with the TM score, a measurement of structural similarity between two structures, were used to assess the reliability of the predicted models.
RESULTS
The core domain of CP is estimated to extend from R155 to M285 on the basis of predictive structural modeling.
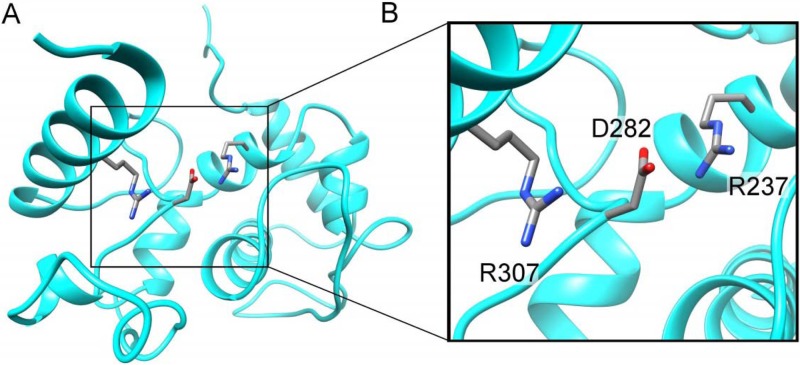
Initial models generated by the I-TASSER server using the full CP sequence with and without user restraint had very low C scores (−4.2 to −3.5) and TM scores (0.27 to 0.33). For a predictive model to be considered reliable, it should have C scores of >−1.5 and TM scores of >0.5 (54). These models failed on both scores. However, the models generated using the structure with PDB accession number 4DOX as the user restraint (55) did provide insight into the possibility of modeling a C-terminal core domain. The PMV CP crystal structure displays a C-terminal core domain with a short N-terminal domain consisting of a loop with a single helix that extends away from the core (55). This loop contains a Phe residue that is inserted into a hydrophobic pocket of an adjacent core domain. The entire sequence of the protein with PDB accession number 4DOX contains only 173 amino acid residues and therefore cannot be used to model the full WSMV CP sequence of 349 amino acids. However, models generated using the protein with PDB accession number 4DOX as a restraint showed sequence and structural alignment of the CP C-terminal domain with the C-terminal core domain structure of the PMV CP crystal structure. After recognition of this, models were generated using truncated CP starting at R155 (C-term-R155) that had acceptable C (−1.53) and TM (0.53) scores. Models generated for C-term-R155-Delta17 (which is missing the last 17 amino acids), C-term-R155-Delta35 (Fig. 1), and C-term-R155-Delta65 had C scores of −0.82, −0.25, and 0.04, respectively, and TM scores of 0.61, 0.68, and 0.72, respectively. These observations suggest that CP with progressive deletions from the C terminus yielded better structural models for the core domain. Thus, we predict that the core domain extends approximately from amino acid R155 to amino acid M285, which suggests that the remaining amino acids are part of a separate C-terminal domain. Since all of these models were generated using the protein with PDB accession number 4DOX, they are similar in fold and are all an α helix.
FIG 1.
Predicted structural model of the WSMV CP core domain. (A) Predicted ribbon structure of the core domain of WSMV CP showing a 3-amino-acid salt bridge between R237, D282, and R307. This structural model was generated with I-TASSER using WSMV CP sequence C-term-R155-Delta35aa together with the PMV CP structural model (chain A; PDB accession number 4DOX) (55) as the user-supplied restraint. (B) Enlarged view of the predicted 3-amino-acid salt bridge between R237, D282, and R307 in the core domain.
All of the models revealed two salt bridges between the critical amino acid residues R237, D282, and R307, with D282 being in the middle (Fig. 1), which is experimentally supported by mutational analysis of R237 and D282 (see below). These residues are highly conserved in related viruses, and residue R307 aligns with residue R223 in TEV CP. Previously, a salt bridge was predicted between the R154 and D198 residues in the TEV CP (13).
The N-terminal domain (residues 1 to 155) was also modeled using I-TASSER; reliable models for this domain were not found. The C and TM scores for all of the N-terminal deletions were quite low. The best scores obtained were for the full-length N-terminal domain (C score = −1.79, TM score = 0.5), and they were still below the cutoff for what is considered trustworthy.
The core domain of WSMV CP is required for cell-to-cell movement and virion formation.
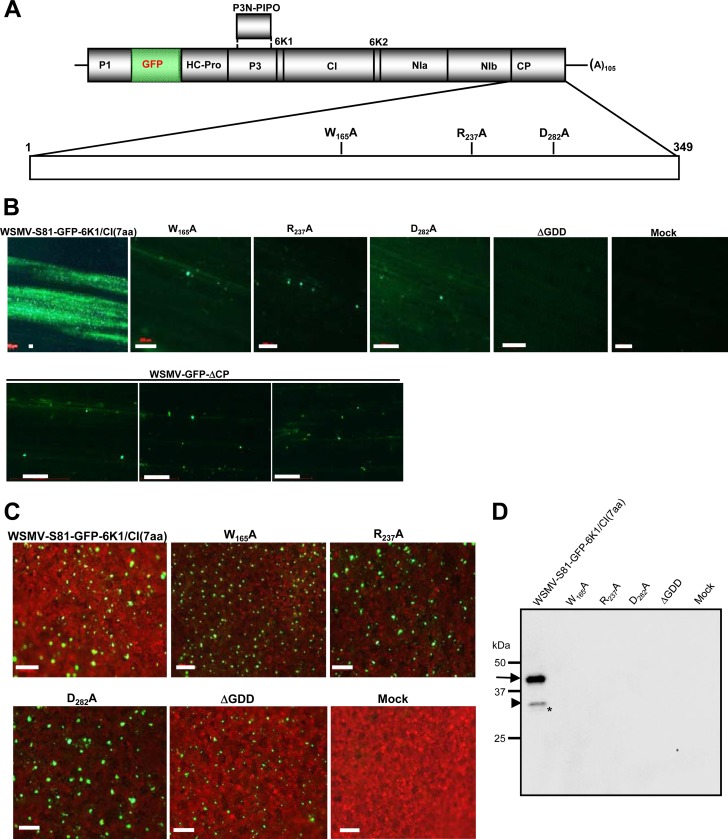
The conserved tryptophan, arginine, and aspartic acid residues at positions 165, 237, and 282, respectively, in the CP were individually mutated to alanine residues in pSP6-WSMV-S81-GFP-6K1/CI(7aa) to obtain W165A, R237A, and D282A (Fig. 2A). These three amino acid residues are conserved among the CPs of a majority of potyvirid species, and R237 and D282 are conserved among the CPs of plant filamentous viruses and are implicated in cell-to-cell movement and virion assembly of TEV (13, 56). In vitro transcripts of GFP-tagged wild-type virus and CP mutants were inoculated onto wheat seedlings at the single-leaf stage. The wild-type virus WSMV-S81-GFP-6K1/CI(7aa) systemically infected wheat at 7 dpi, while mutant viruses failed to infect wheat even at 25 dpi (Table 1). At 9 dpi, wild-type virus induced large foci spreading throughout most of the inoculated leaf lamina; in contrast, mutant viruses restricted spreading to 1 to 3 cells (Fig. 2B), indicating that these mutations debilitated the cell-to-cell movement of WSMV.
FIG 2.
The core domain of WSMV CP is required for cell-to-cell movement and virion assembly. (A) Genomic organization of WSMV-S81-GFP-6K1/CI(7aa) showing the locations of individual proteins encoded by the polyprotein. The vertical lines between the encoded proteins are sequences that code for proteolytic processing sites. An enlarged view of the CP cistron showing the locations of point mutations W165A, R237A, and D282A is shown below the genomic organization. (B) Infection foci of in vitro transcripts of GFP-tagged wild-type virus and the point, ΔGDD, and WSMV-GFP-ΔCP mutants on wheat at 9 dpi. The ΔGDD mutant was created by deleting the GDD motif and five amino acids on either side of the GDD motif in the NIb cistron. WSMV-GFP-ΔCP was created by deleting codons encoding CP amino acid residues, except for 5 amino acids each at the N and C termini. Note that viruses with point mutations in the core domain or WSMV-GFP-ΔCP were restricted to 1 to 3 cells and the ΔGDD mutant failed to form fluorescent foci on inoculated wheat leaves. The wild-type virus formed large foci. Bars, 200 μm. (C) Expression of GFP in agroinfiltrated leaves of N. benthamiana at 7 dpa. Bars, 200 μm. (D) Western blot of partially purified virions from agroinfiltrated N. benthamiana leaves at 7 dpa. The immunoblot was probed with an anti-WSMV serum. The locations of the 45-, 32-, and 29-kDa protein bands are indicated with an arrow, an arrowhead, and an asterisk, respectively. Note that only the wild-type virus formed virions. The protein size markers used in SDS-PAGE are indicated on the left.
TABLE 1.
Infectivity of in vitro RNA transcripts generated from cloned DNA copies of WSMV CP deletion mutants on wheat at 21 dpi
| Mutant | No. of plants infected/total no. inoculated (% infected) |
|
|---|---|---|
| Expt 1 | Expt 2 | |
| WSMV-S81 (wild type)a | 13/15 (87) | 17/19 (89) |
| WSMV-CPΔ36-84aaa | 17/18 (94) | 18/19 (95) |
| WSMV-CPΔ85-100aab | 15/17 (88) | 19/20 (95) |
| WSMV-CPΔ48-100aac | 12/16 (75) | 25/26 (96) |
| WSMV-CPΔ36-100aac | 10/15 (67) | 16/20 (80) |
| WSMV-CPΔC14aad | 10/13 (77) | 16/19 (84) |
| WSMV-CPΔC17aae | 4/16 (25) | 2/18 (11) |
| WSMV-CPΔN6-27aae | 7/15 (47) | 10/18 (56) |
| WSMV-GFP-W165Af | 0/16 (0) | 0/18 (0) |
| WSMV-GFP-R237Af | 0/14 (0) | 0/17 (0) |
| WSMV-GFP-D282Af | 0/17 (0) | 0/16 (0) |
| WSMV-GFP-ΔGDDf | 0/13 (0) | 0/17 (0) |
| WSMV-GFP-ΔCPf | 0/16 (0) | 0/14 (0) |
| Mock infected | 0/15 (0) | 0/17 (0) |
The first visible symptoms developed at 6 to 9 dpi.
The first visible symptoms developed at 9 to 12 dpi.
The first visible symptoms developed at 11 to 15 dpi.
The first visible symptoms developed at 14 to 18 dpi.
The first visible symptoms developed at 15 to 21 dpi.
The mutant failed to infect wheat systemically at 25 dpi.
The virion assembly competency of these mutants was examined in an MIVA assay by transferring CP point mutations to pCAM-WSMV-S81-GFP-6K1/CI(7aa) (47). As a control, a mutant with a GDD deletion (ΔGDD mutant) was created in pSP6-WSMV-S81-GFP-6K1/CI(7aa) and pCAM-WSMV-S81-GFP-6K1/CI(7aa) by deleting the GDD (Gly-Asp-Asp) motif plus five amino acids on either side of the GDD motif in the NIb cistron. As expected, in vitro transcripts of pSP6-WSMV-S81-GFP-6K1/CI(7aa) with the ΔGDD mutation failed to form detectable fluorescent foci in wheat at 9 dpi and failed to infect wheat systemically at 25 dpi (Table 1; Fig. 2B). The point and ΔGDD mutants of pCAM-WSMV-S81-GFP-6K1/CI(7aa) were transformed into A. tumefaciens strain EHA105, and agrosuspensions harboring CP or GDD mutants were infiltrated into N. benthamiana leaves. At 7 dpa, GFP fluorescence was similarly observed in point mutants and wild-type virus (Fig. 2C). Surprisingly, the GFP fluorescence was also observed in ΔGDD mutant-infiltrated leaves (Fig. 2C), suggesting that constitutively produced viral transcripts under the 35S promoter most likely translated and the GFP protein might have been released from the polyprotein by viral proteinases. Moreover, GFP expression in agroinfiltrated leaves served as a good marker for the expression of polyprotein and also as an indication of the virus-encoded RNA-silencing suppressor protein effectively counteracting the host defense system.
Partially purified virions obtained from agroinfiltrated N. benthamiana leaves at 7 dpa were subjected to Western blotting using antibodies against WSMV. A WSMV CP of 45 kDa and truncated 31- and 29-kDa proteins were found in partially purified virion preparations of wild-type virus (Fig. 2D). Previously, it has been reported that three CP bands of 45, 31, and 29 kDa were found in purified virion preparations of WSMV, and these were attributed to in vivo proteolysis of CP due to leaf senescence (57). However, the exact nature of multiple CP bands in purified virion preparations is not known. In contrast to wild-type virus, CP did not accumulate at detectable levels in purified virion preparations of mutant-infiltrated N. benthamiana leaves (Fig. 2D), suggesting that W165, R237, and D282 in the CP are critical for virion assembly. Though the GFP fluorescence was observed in ΔGDD mutant-infiltrated leaves, CP did not accumulate at detectable levels in partially purified virion preparations (Fig. 2D). This result suggests that failure to form virions by a replication-deficient ΔGDD mutant could be due to a lack of free CP at a threshold level.
WSMV CP is dispensable for replication.
The requirement of CP for WSMV replication was examined by deleting codons encoding the CP amino acid residues except for 5 amino acids each at the N and C termini in pSP6-WSMV-S81-GFP-6K1/CI(7aa) to obtain pSP6-WSMV-GFP-ΔCP. In vitro transcripts of pSP6-WSMV-GFP-ΔCP elicited fluorescent foci that were restricted to single cells in wheat and that were similar to those of the R237A and D282A mutants (Fig. 2B). As expected, no fluorescent foci were observed in ΔGDD mutant-inoculated wheat seedlings (Fig. 2B). The development of foci by WSMV-GFP-ΔCP similar to those of the R237A and D282A mutants, together with the results obtained with analogous CP mutants of TEV with mutants R237A and D282A that are dispensable for replication (13), suggests that deletion of CP does not appreciably affect WSMV replication in wheat but that CP is required for cell-to-cell movement.
Amino acids 36 to 84 in CP are dispensable for virion assembly and systemic infection of wheat.
The requirement of the N-terminal region of WSMV CP for virion assembly and systemic infection of wheat was examined by deleting codons encoding amino acid residues 36 to 84 in pSP6-WSMV-S81 to obtain pSP6-WSMV-CPΔ36-84aa (Fig. 3A). Wheat seedlings inoculated with in vitro transcripts of this mutant developed systemic chlorotic streaks and mosaic symptoms at 9 dpi similar to those of seedling inoculated with wild-type virus (Fig. 3B; Table 1), demonstrating that amino acid residues 36 to 84 of CP are dispensable for systemic infection of wheat. Systemic infection of wheat by WSMV-CPΔ36-84aa was further confirmed by RT-PCR amplification of the CP cistron from deletion mutant-infected plants (Fig. 3C), followed by nucleotide sequencing of PCR amplicons.
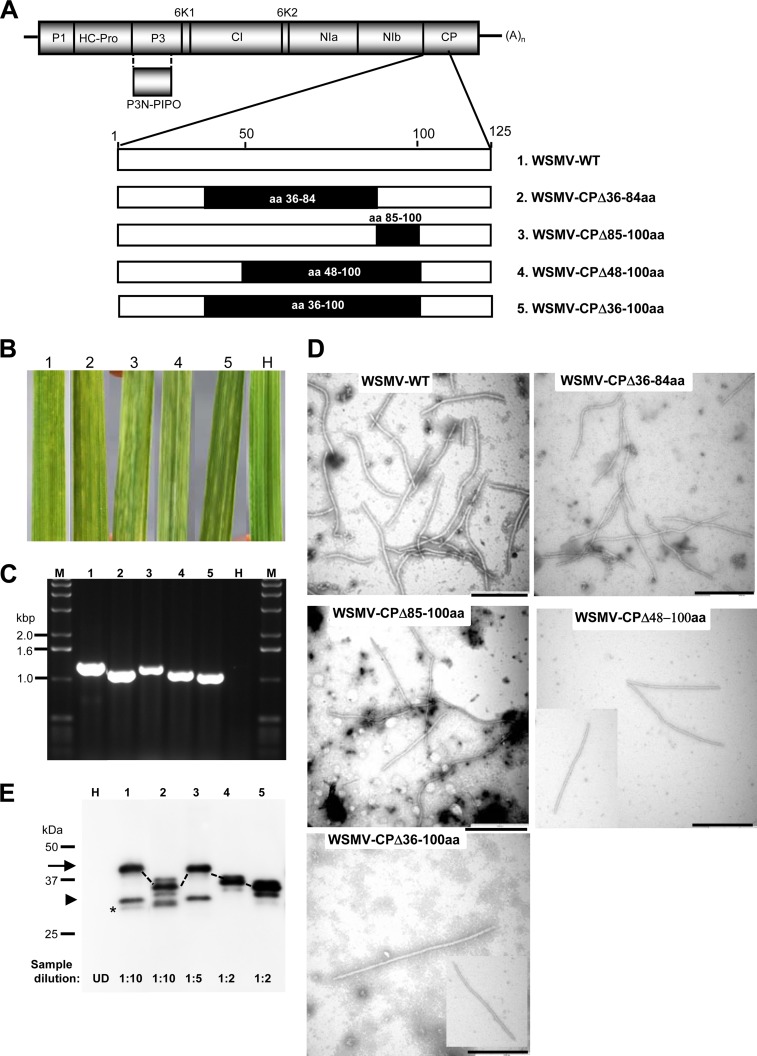
FIG 3.
WSMV with deletion of amino acids 36 to 100 in the CP systemically infects wheat. (A) Genomic organization of WSMV with proteins encoded by the polyprotein. An enlarged view of the N-terminal 125 amino acids is shown at the bottom of the schematic representation of the genome. The positions of the amino acids deleted from the CP are indicated with solid boxes, and the respective mutants are indicated at the right. WT, wild type. (B) Systemic symptoms induced by in vitro transcripts of WSMV CP deletion mutants on wheat. The numbers above the leaves correspond to the mutants indicated by number in panel A. Symptoms were induced by mutants 1 and 2 and mutants 3 to 5 at 9 and 16 dpi, respectively. Leaf H, a buffer-inoculated healthy wheat leaf. (C) RT-PCR amplification of the CP cistron from deletion mutant-infected wheat. The RT-PCR products were electrophoresed through a 1.0% agarose gel, and the numbers above the gel correspond to the mutants indicated by number in panel A. The sizes of the bands in the DNA ladder (lanes M) are indicated on the left. (D) Electron micrographs of virions of the wild-type virus and deletion mutants from systemically infected wheat. Bars, 500 nm. (E) Western blot analysis of partially purified virions from wheat leaves infected with the wild-type virus and CP deletion mutants. The numbers above the immunoblot correspond to the mutants indicated by number in panel A. The location of CP is indicated with an arrow, and the corresponding CP of the deletion mutants is marked with dotted lines. The positions of CP and the truncated 31- and 29-kDa CPs are indicated with an arrow, an arrowhead, and an asterisk, respectively. The protein size markers used in SDS-PAGE are indicated on the left. The dilution of the partially purified virions used for immunoblotting is indicated at the bottom of the blot. UD, undiluted.
The ability to form virions by WSMV-CPΔ36-84aa was examined by partially purifying virions from systemically infected wheat at 14 dpi, and virus particles with no obvious morphological differences from wild-type virus particles were found under an electron microscope (Fig. 3D). Western blot analysis using an anti-WSMV serum suggested that the CP of 35 kDa was detected in virion preparations of WSMV-CPΔ36-84aa in amounts similar to those detected in virion preparations of wild-type virus (Fig. 3E; compare lane 2 with lane 1). In addition to full-length CP, this mutant virus also accumulated a truncated form of the 29-kDa protein but lacked a 31-kDa protein (Fig. 3E).
Amino acids 85 to 100 of CP are required for efficient systemic infection of wheat.
We next examined the requirement of amino acid residues 85 to 100 for systemic infection by deleting the corresponding codons in pSP6-WSMV-S81 to obtain pSP6-WSMV-CPΔ85-100aa (Fig. 3A). Wheat seedlings inoculated with in vitro transcripts of this mutant developed mild chlorotic streaks and chlorotic spots at 9 to 12 dpi, whereas wheat seedlings inoculated with in vitro transcripts of wild-type virus developed these symptoms at 6 to 8 dpi (Table 1; Fig. 3B), suggesting that deletion of amino acid residues 85 to 100 in CP delayed the onset of systemic symptoms, followed by moderate symptoms at 18 to 21 dpi. We next examined possible cumulative effects of deletions comprising amino acid residues 48 to 100 and 36 to 100 on systemic infection of wheat by creating pSP6-WSMV-CPΔ48-100aa and pSP6-WSMV-CPΔ36-100aa, respectively (Fig. 3A). Wheat inoculated with in vitro transcripts of these two mutants developed mild systemic chlorotic streaks on wheat at 11 to 15 dpi, whereas wheat inoculated with in vitro transcripts of WSMV-CPΔ85-100aa and wild-type virus developed systemic chlorotic streaks at 9 to 12 and 6 to 9 dpi, respectively (Table 1; Fig. 3B). Systemic infection of wheat by WSMV with deletions encompassing amino acids 85 to 100 was further confirmed by detecting RT-PCR amplicons from deletion mutant-infected plants smaller than those from wild-type virus-infected plants (Fig. 3C) and nucleotide sequencing of PCR amplicons.
WSMV with overlapping deletions comprising amino acids 85 to 100 in the CP formed virions in wheat that appeared to be morphologically similar to those of wild-type virus (Fig. 3D). Western blot analysis of partially purified virions with an anti-WSMV serum revealed that virions of WSMV-CPΔ85-100aa, WSMV-CPΔ48-100aa, and WSMV-CPΔ36-100aa accumulated in wheat at levels 2- to 5-fold less than those of virions of wild-type virus (Fig. 3E). Taken together, these data suggest that WSMV with deletion of amino acid residues 36 to 100 in the CP is capable of virion assembly and systemic infection of wheat, though at reduced efficiencies.
N-proximal amino acids 6 to 27 and the C-proximal 17 amino acids of CP are dispensable for long-distance transport in wheat.
The requirement of the N- and C-proximal amino acids for systemic infection of wheat and virion assembly was examined by deleting codons encoding amino acid residues 6 to 27 at the N terminus as well as both 14 and 17 amino acid residues from the C terminus in WSMV-CPΔ6-27aa, WSMV-CPΔC14aa, and WSMV-CPΔC17aa, respectively (Fig. 4A). In vitro transcripts of WSMV-CPΔN6-27aa infected 47 to 56% of wheat at 15 to 21 dpi, with a few chlorotic streaks being detected in each leaf, and WSMV-CPΔC14aa infected 77 to 84% of wheat seedlings at 14 to 18 dpi, with mild to moderate symptoms being detected (Table 1; Fig. 4B). In contrast, deletion of 17 amino acids from the C terminus had a deleterious effect on systemic infection, with only 11 to 25% of plants being infected at 15 to 21 dpi and a few chlorotic streaks being detected in each leaf (Table 1; Fig. 4B). Infection of wheat by the N- and C-terminal deletion mutants was further confirmed by RT-PCR amplification of the CP cistron from systemically infected wheat (Fig. 4C), followed by nucleotide sequencing of PCR amplicons. These data demonstrated that amino acid residues 6 to 27 at the N terminus and the 17 amino acids from the C terminus are dispensable for systemic infection of wheat.
FIG 4.
The N- and C-proximal regions of WSMV CP are dispensable for systemic infection in wheat. (A) Schematic diagram of the organization of the WSMV genome and the locations of deletions (bold and underlined) in the N- and C-terminal regions of CP, which are indicated below the genomic organization. (B) Symptoms elicited by in vitro transcripts of the wild-type virus and N- and C-proximal deletion mutants on wheat at 18 days postinoculation. The numbers above the leaves correspond to the mutants indicated by number in panel A. Leaf H, a buffer-inoculated healthy wheat leaf. (C) RT-PCR analysis of the CP cistron from wheat plants infected with the wild-type virus and mutants with N- or C-terminal deletions in the CP. The sizes of the DNA markers (lanes M) are indicated at the left. The numbers above the gel correspond to the mutants indicated by number in panel A. (D) Electron micrographs of the wild-type virus and N- or C-terminal deletion mutants of WSMV in wheat. Bars, 500 nm. (E) Western blot analysis of partially purified virions from wheat leaves infected with the wild-type virus and N- or C-terminal deletion mutants. The positions of CP and truncated CPs of 31 and 29 kDa are indicated with an arrow, an arrowhead, and an asterisk, respectively. The protein size markers used in SDS-PAGE are indicated on the left. The dilution of the partially purified virions used for immunoblotting is indicated at the bottom of the blot. UD, undiluted.
The N- and C-proximal deletion mutants formed virions with no obvious morphological differences from wild-type virus virions (Fig. 4D), though these mutants accumulated virions at levels 2- and 10-fold less than those of virions of wild-type virus (Fig. 4E). Taken together, these data revealed that N-proximal amino acid residues 6 to 27 and the C-proximal 14 and 17 amino acids are dispensable for virion formation and systemic infection, though the presence of these amino acids enhanced the fitness of the virus.
Amino acids 6 to 27 and 85 to 100 and the C-terminal region of CP are required for efficient cell-to-cell movement.
Deletion of amino acids 6 to 27 or 85 to 100 or the C-terminal 14 or 17 amino acids in the CP delayed the onset of systemic infection and caused mild symptoms in wheat, suggesting that these mutants are defective in movement. The movement of CP deletion mutants in wheat was monitored by transferring deletions into pSP6-WSMV-S81-GFP-6K1/CI(7aa) (47). The GFP-tagged variant of WSMV has successfully been used as an excellent marker virus to monitor virus movement and distribution in wheat and SDp2 maize (44, 45, 47).
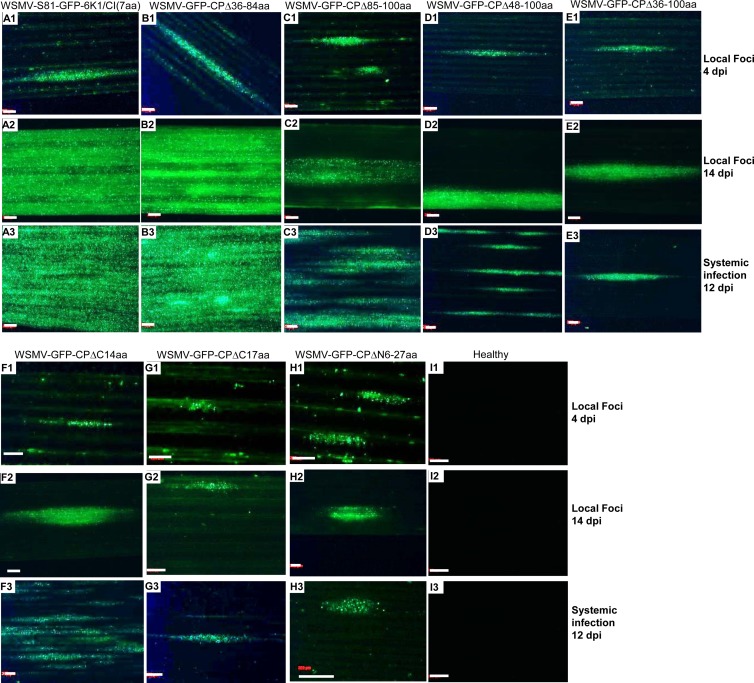
In vitro transcripts of GFP-tagged variants of WSMV CP deletion mutants were inoculated onto two sets of wheat seedlings at the single-leaf stage. One set of inoculated plants was observed for GFP fluorescent foci on inoculated leaves at 4 and 14 dpi, and the other set of plants was observed for systemic fluorescent foci at 12 or 21 dpi (Fig. 5). WSMV with deletion of amino acids 36 to 84 in the CP formed foci slightly larger than those formed by wild-type virus (Table 2; Fig. 5A1 and B1), suggesting that amino acids 36 to 84 are not required for cell-to-cell movement. In contrast, WSMV with deletions comprising amino acids 85 to 100, 48 to 100, or 36 to 100 formed smaller foci of 1.73, 1.45, and 1.36 mm2, respectively, whereas wild-type virus formed foci of 2.31 mm2 (Table 2; Fig. 5C1 to E1). WSMV with deletion of amino acids 6 to 27 or the C-terminal 14 or 17 amino acids formed substantially smaller foci of 0.53, 0.64, and 0.40 mm2, respectively, than the 2.31 mm2 foci formed by wild-type virus (Table 2; Fig. 5F1 to H1). These data revealed that amino acids 85 to 100 and 6 to 27 and the C-terminal 17 amino acids are required for efficient cell-to-cell movement in wheat. Furthermore, at 14 dpi, GFP fluorescence was spread throughout most of the inoculated leaves of GFP-tagged wild-type virus or a mutant with a deletion comprising amino acids 36 to 84 (Fig. 5A2 and B2). In contrast, fluorescence was restricted to individual foci in leaves inoculated with WSMV with deletions comprising amino acids 6 to 27 or 85 to 100 or the C-terminal 14 or 17 amino acids (Fig. 5C2 to H2), indicating that these amino acids are required for efficient cell-to-cell movement in wheat.
FIG 5.
The N- and C-proximal regions and amino acids 85 to 100 of WSMV CP are required for efficient cell-to-cell movement in wheat. In vitro transcripts of GFP-tagged wild-type virus and CP deletion mutants were inoculated onto wheat seedlings at the single-leaf stage. The fluorescent foci developed by the wild-type virus and CP deletion mutants at 4 and 14 dpi are presented in the top (A1 to I1) and middle (A2 to I2) rows, respectively, and the GFP fluorescence in the upper noninoculated leaves at 12 dpi (21 dpi for WSMV-CPΔC17aa and WSMV-CPΔN6-27aa mutant viruses) is presented in the bottom rows (A3 to I3). Bars, 500 μm. Note that WSMV with a deletion of amino acids 36 to 84 in the CP elicited local and systemic infection foci similar to those found in wild-type virus; in contrast, deletions comprising amino acids 6 to 27 or 85 to 100 or the C-terminal 14 or 17 amino acids affected the efficiency of cell-to-cell movement in wheat.
TABLE 2.
Infection focus sizes on wheat seedlings produced by in vitro RNA transcripts generated from cloned cDNA copies of GFP-tagged WSMV CP deletion mutantsa
| Mutant | Focus size (mm2) |
|---|---|
| WSMV-S81-GFP-6K1/CI(7aa) (wild type) | 2.31 ± 0.44 |
| WSMV-GFP-CPΔ36-84aa | 2.68 ± 0.38 |
| WSMV-GFP-CPΔ85-100aa | 1.73 ± 0.36 |
| WSMV-GFP-CPΔ48-100aa | 1.45 ± 0.37 |
| WSMV-GFP-CPΔ36-100aa | 1.36 ± 0.30 |
| WSMV-GFP-CPΔC14aa | 0.64 ± 0.17 |
| WSMV-GFP-CPΔC17aa | 0.40 ± 0.21 |
| WSMV-GFP-CPΔN6-27aa | 0.53 ± 0.19 |
| Mock | 0 |
Wheat seedlings were incubated in a greenhouse at 20 to 27°C. Focus sizes were estimated from 20 to 30 individual foci at 4 dpi.
At 12 dpi, GFP fluorescence covered most of the upper noninoculated leaves of wheat infected with wild-type virus or the WSMV-GFP-CPΔ36-84aa mutant (Fig. 5A3 and B3). In contrast, at 12 to 21 dpi, GFP fluorescence was restricted to several foci without spreading throughout the leaf lamina of plants inoculated with mutants with deletions comprising amino acids 85 to 100 or 6 to 27 or the C-terminal 14 or 17 amino acids (Fig. 5C3 to H3), which was consistent with the defective cell-to-cell movement nature of these mutants.
Amino acids 6 to 27 and 85 to 100 of CP are required for efficient virion assembly.
The requirement of CP amino acids 6 to 27 and 85 to 100 for efficient virus translocation in wheat prompted an examination of the virion assembly competency of these mutants in an MIVA assay. Deletions of codons encoding amino acid residues 6 to 27, 85 to 100, or 36 to 100 were introduced into the CP cistron in pCAM-WSMV-S81-GFP-6K1/CI(7aa) (47). Wild-type virus and a CP mutant with deletion of amino acids 36 to 84 were included as positive controls. Agrobacterium harboring pCAM-WSMV-S81-GFP-6K1/CI(7aa) and the deletion mutants was infiltrated into N. benthamiana leaves.
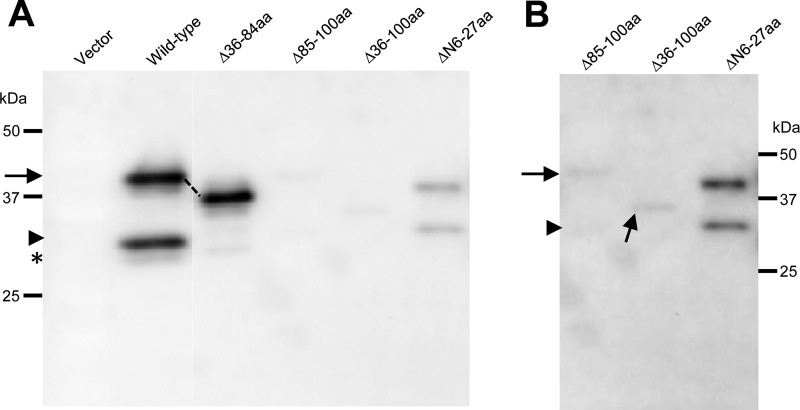
Virions were partially purified from agroinfiltrated leaves at 7 dpa and analyzed by Western blotting using an anti-WSMV serum. WSMV with a deletion of amino acid residues 36 to 84 in the CP accumulated virions in amounts similar to those for wild-type virus, while deletion of amino acids 85 to 100 or 36 to 100 drastically reduced virion formation, as CP accumulated in partially purified virions at levels much reduced from those in the virions of wild-type virus (Fig. 6A and B). Deletion of amino acids 6 to 27 also affected virion formation substantially compared to that for wild-type virus, but WSMV with deletion of 6 to 27 amino acids formed virions at slightly higher levels than those for virions formed by mutants with deletions comprising amino acids 85 to 100 (Fig. 6A and B). These results, together with the detection of virions in systemically infected wheat, suggested that amino acids 6 to 27 and 85 to 100 are dispensable for virion formation but that the presence of these amino acids enhances the fitness of virion assembly.
FIG 6.
WSMV CP amino acids 6 to 27 and 85 to 100 are required for efficient virion assembly. (A) MIVA assay of wild-type virus and CP deletion mutants in agroinfiltrated Nicotiana benthamiana leaves at 7 dpa. Western blot analysis of partially purified virions from agroinfiltrated N. benthamiana leaves was performed with an anti-WSMV serum. Note that the deletion of amino acids 36 to 84 resulted in levels of accumulation of virions approximately similar to those for wild-type virus; in contrast, deletion of amino acids 85 to 100, 36 to 100, or 6 to 27 in the CP significantly affected virion assembly in WSMV. (B) Longer exposure of immunoblot lanes consisting of partially purified virions of WSMV CP mutants with deletions comprising amino acids 85 to 100, 36 to 100, or 6 to 27. The positions of CP and truncated CPs of 31 and 29 kDa are indicated with arrows, arrowheads, and an asterisk, respectively. The protein size markers used in SDS-PAGE are indicated.
The C terminus of CP is required for cell-to-cell movement but is dispensable for virion assembly.
WSMV with deletion of 14 amino acids from the C terminus of CP had a delayed onset of systemic symptoms, and an additional 3-amino-acid deletion substantially affected cell-to-cell movement and the ability to infect wheat systemically (Table 1; Fig. 5 and 7A). We next introduced a deletion of 22 and 27 amino acids from the C terminus of CP in pSP6-WSMV-S81-GFP-6K1/CI(7aa), and in vitro transcripts of these mutants formed fluorescent foci confined to 3 to 10 cells at 14 dpi (Fig. 7A) but failed to establish systemic infection in wheat at 25 dpi. These results suggest that the C-terminal region of CP is involved in the cell-to-cell movement of WSMV, but it is not known whether these deletions affected virion encapsidation. To examine virion assembly of these mutants in an MIVA assay, a deletion of 14, 17, 22, or 27 amino acids from the C terminus of CP was introduced into pCAM-WSMV-S81-GFP-6K1/CI(7aa), and the mutants were agroinfiltrated into N. benthamiana leaves. Western blotting of partially purified virions from agroinfiltrated leaves indicated that CP accumulated in the purified virions of the deletion mutants in amounts similar to those in the purified virions of wild-type virus (Fig. 7B). Moreover, mutants with a deletion of 14, 17, 22, or 27 amino acids from the C terminus formed virions similar to those of the wild-type virus (Fig. 7C; data not shown). These data demonstrate that the 27 amino acids from the C terminus of CP are dispensable for virion formation but are crucial for cell-to-cell movement in wheat.
FIG 7.
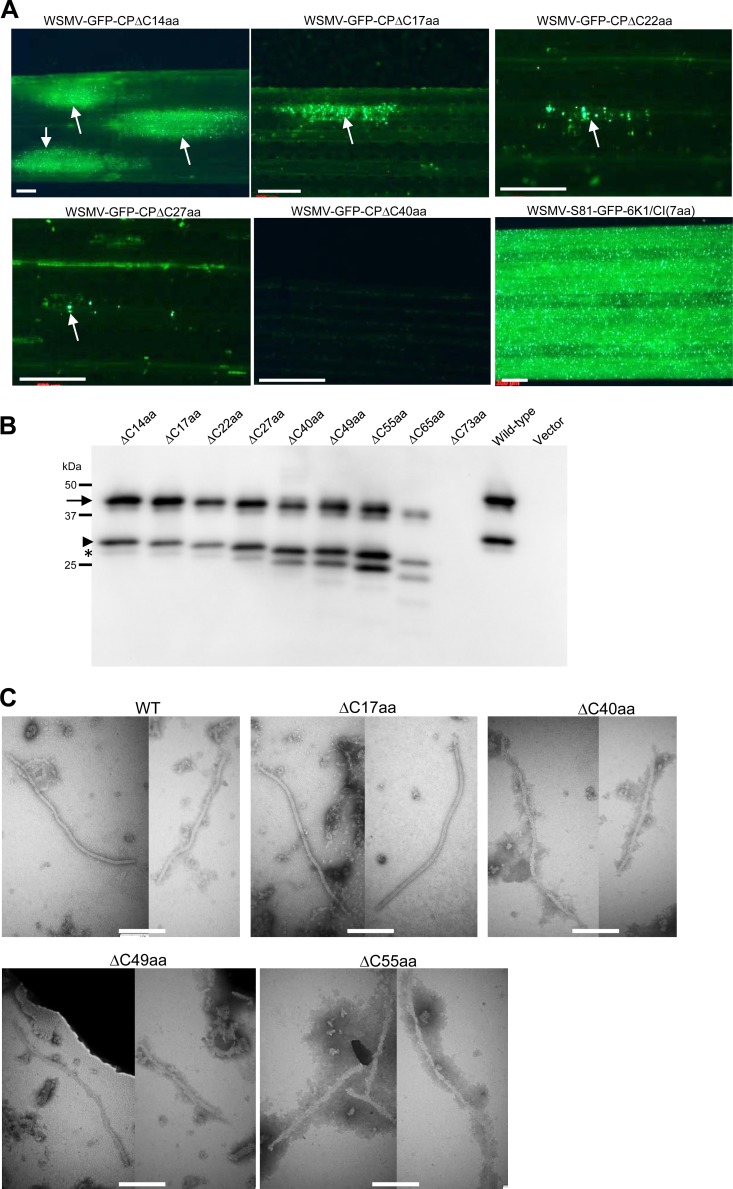
The C-terminal 65 amino acids of WSMV CP are dispensable for virion assembly but are required for cell-to-cell movement. (A) Progressive deletions at the C terminus of CP increasingly debilitated the cell-to-cell movement of WSMV in wheat. GFP fluorescent foci induced by GFP-tagged WSMV mutants with progressive deletions at the C terminus of CP on inoculated wheat leaves at 14 days postinoculation are indicated with arrows. Bars, 500 μm. (B) MIVA assay of WSMV mutants with progressive deletions from the C terminus of CP in agroinfiltrated N. benthamiana leaves at 7 dpa. Partially purified virions from agroinfiltrated N. benthamiana leaves were analyzed by immunoblotting at 7 dpa with an anti-WSMV serum. Note that WSMV with progressive deletions of up to 65 amino acids at the C terminus of CP is capable of virion assembly. The protein size markers used in SDS-PAGE are indicated on the left. (C) Electron micrographs of virions (wild type, WSMV-CPΔC17aa) and virus-like particles (noninfectious, WSMV-CPΔC40aa, WSMV-CPΔC49aa, and WSMV-CPΔC55aa) in partially purified virions of GFP-tagged WSMV (wild type) and the C-terminal deletion mutants from agroinfiltrated N. benthamiana leaves at 7 dpa. Bars, 200 nm.
We next introduced a deletion of 40, 49, 55, 65, or 73 amino acids from the C terminus of CP in pSP6-WSMV-S81-GFP-6K1/CI(7aa) and pCAM-WSMV-S81-GFP-6K1/CI(7aa). In vitro transcripts of these mutants failed to form detectable fluorescent foci in wheat at 15 dpi (Fig. 7A), suggesting that these deletions debilitated the cell-to-cell movement function of WSMV. Western blotting of partially purified virions from agroinfiltrated N. benthamiana leaves revealed that mutants with a deletion of 40, 49, or 55 amino acids from the C terminus accumulated CP in partially purified virions in amounts similar to those in partially purified virions of wild-type virus (Fig. 7B). In contrast, deletion of 65 amino acids at the C-terminal region resulted in the assembly of virions at levels ∼15 to 20% of those of virions of wild-type virus, while deletion of 73 amino acids completely abolished virion assembly (Fig. 7B). Mutants with a deletion of 40, 49, or 55 amino acids from the C terminus formed flexuous filamentous virus-like particles in an MIVA assay (Fig. 7C). These results indicate that the C-terminal 65 amino acids of CP are dispensable for virion assembly but are required for the cell-to-cell movement of WSMV.
DISCUSSION
Viral CPs are involved in most aspects of the virus life cycle and have overlapping structural and nonstructural functions in virion assembly and virus biology, respectively (1, 2). Thus, point mutations or small deletions comprising a few amino acids in the CP can elicit negative effects on CP functions (e.g., see references 4, 11 to 14, and 58). Previously, we mapped differential infection of SDp2 maize and wheat by WSMV to five amino acids at the N terminus and three aspartic acid residues at the C terminus of CP (44, 45). In this study, in contrast to other viruses, we found that WSMV CP is unusually tolerant of extensive deletions and is able to continue systemic infection with these deletions, which facilitated identifying the CP determinants involved in cell-to-cell movement and virion assembly.
Three point mutations, W165A, R237A, and D282A, targeting conserved amino acids in the core domain of CPs of potyvirid species (56) restricted the virus mostly to a few cells, and the mutants failed to form virions at detectable levels, suggesting that these mutations might have affected the protein-protein or protein-RNA interactions required for virion assembly and/or cell-to-cell movement. Though the CP cistron is not required for WSMV replication, mutants with mutations in the core domain failed to form virions at detectable levels, suggesting a functional role for these amino acids in virion assembly and/or stability. Our predictive structural model of the core indicates that R237 and D282 are likely involved in a salt bridge that confers structural stability to the CP. The analogous mutations in potyviral CPs also abolished cell-to-cell transport and virion assembly (11, 13, 24). Taken together, the core domains of poty- and tritimoviral CPs possess similar functions in virion assembly and possibly in cell-to-cell movement.
WSMV CP is unusual in that 49 amino acid residues comprising amino acids 36 to 84 are dispensable for normal virion assembly and systemic infection. What are the roles of these amino acids in WSMV biology? Why is the virus keeping amino acids that are not required for virion assembly and systemic infection? The ClustalW alignment of CP sequences from representative species of each genus of the Potyviridae family revealed no conserved or similar amino acids located at the N-terminal region. Additionally, the CPs of members of the Tritimovirus genus contain fewer conserved amino acids in the N-terminal region than in the central core domain and C-terminal region. The variable nature of the N-terminal region of CPs of the potyvirid species, together with amino acids 36 to 84, which are expendable for systemic infection, suggests that these amino acids may possess virus-specific specialized functions, such as vector transmission, host range, and cross-protection functions. The N-terminal DAG motif of potyviral CPs was reported to be involved in aphid transmission (34); in contrast, WSMV CP does not contain such a motif (37). However, mite transmission studies of CP deletion mutants may decipher CP's role, if any, in vector transmission. Moreover, efficient infection of wheat by WSMV with a 49-amino-acid deletion (amino acids 36 to 84) in the CP could facilitate expression and display of specialty epitopes/peptides embedded in virions.
Additionally, WSMV mutants with overlapping deletions encompassing amino acids 85 to 100 in the CP were capable of systemic infection, albeit with delayed and milder symptoms, yet it is rare for a virus with a deletion of as large as 65 amino acid residues (amino acids 36 to 100) in the CP to systemically infect plants. These mutants displayed restrictive cell-to-cell movement and delayed the onset of systemic infection in wheat compared to the time of onset for wild-type virus. The delayed systemic infection phenotype of these mutants could be due to the requirement of amino acids 85 to 100 for virion assembly and/or cell-to-cell transport. WSMV mutants with deletions comprising amino acids 85 to 100 formed virions at levels 2- to 5-fold less than those for wild-type virus in wheat, but the levels were substantially reduced in an MIVA assay, suggesting that these amino acids are required for efficient virion assembly. The differences in the levels of virion assembly by CP deletion mutants in MDVA (in wheat) and MIVA (in N. benthamiana) assays might be due to limited cell-to-cell and long-distance movement in wheat, whereas it is due to virus restriction exclusively to infiltrated mesophyll cells in N. benthamiana. Our data suggest that amino acids 85 to 100 are required for efficient virion assembly, but we cannot exclude the possible role of these amino acids in cell-to-cell movement independently of virion assembly. It is also possible that deletion of amino acids 85 to 100 might have affected virion stability. However, mutants with deletions in crude sap are highly infectious in wheat, suggesting that virions lacking amino acids 85 to 100 are stable and are capable of protecting viral genomic RNA.
In contrast to potyviral CPs, we found several differences with regard to the role of the N- and C-proximal regions in virus biology. The potyviral N-proximal region (amino acids 5 to 29) was dispensable for virion assembly but has been found to be required for long-distance movement (13). In contrast, we found that N-terminal amino acids 6 to 27 of WSMV CP are required for efficient virion assembly but are dispensable for long-distance transport. It is possible that the observed differences between TEV and WSMV might be because the deletion of the N-proximal region of TEV CP is slightly longer than the corresponding deletion in WSMV CP (amino acids 5 to 29 versus amino acids 6 to 27). Nonetheless, the N-terminal regions of poty- and tritimoviral CPs have clearly different requirements for virion assembly. Progressive deletions of up to 65 amino acids from the C terminus of CP increasingly affected the cell-to-cell movement of WSMV, though virions or virus-like particles were formed. Two plausible reasons can be deduced from these results: the C-terminal 65 amino acids are involved in a dedicated cell-to-cell movement function, and virions with deletions from the C-terminal end are defective in virion stability/disassembly. Because progressive deletions from the C-terminal region increasingly affected cell-to-cell movement, it is likely that the C terminus of CP functions as a determinant for cell-to-cell movement. However, we cannot exclude a possible role for these amino acids in virion stability and/or disassembly. In contrast to systemic infection by WSMV with a 17-amino-acid deletion from the C terminus, the analogous mutant of TEV failed to infect tobacco systemically (11), suggesting that either a differential requirement for the C-terminal region of these two viruses or differences in long-distance transport of viruses in eudicot and monocot plants could account for these variations.
The systemic infection of wheat by cell-to-cell movement-defective N- and C-terminal deletion mutants suggests that these mutants entered the vascular system and successfully egressed at distal regions. The delayed systemic infection by these mutants is most likely due to defects in cell-to-cell movement, which might have caused delayed virus entry into the vascular system. Since the CP of WSMV has not been demonstrated to have suppressor activity against host RNA silencing (42), defects in combating the host defense system are not likely causing delayed systemic infection by CP deletion mutants. Though it is not known for WSMV, the N and C termini of potyviral CPs are exposed outside the virion (32, 33), and it is possible that these amino acids are potential targets for interactions with host factors. Thus, deletion of these amino acids might have prevented such interactions, consequently affecting the cell-to-cell movement of virus. We observed a correlation between virion assembly and cell-to-cell transport of WSMV: mutants that affected virion assembly also affected cell-to-cell transport, and no mutant with a cell-to-cell movement function without virion formation was found. These results suggest that the cell-to-cell movement function of WSMV is facilitated as the virion form, as reported for como- and closteroviruses (59, 60). However, virion formation is not the only requirement for cell-to-cell transport, as deletions from the C terminus of CP affected cell-to-cell movement but not virion assembly. The CPs of several plant viruses were also implicated in virus translocation (1, 4, 12, 61).
The availability of a structural model of CP would be helpful to understand the structural basis for the results of this study. However, currently there are no high-resolution structural models for CPs of the Potyviridae family that can be used for homology modeling of the WSMV CP. The recent crystal structure of the CP of the distantly related PMV (55), which is also a flexuous filamentous plant virus, was used in this study to identify the likely core domain region of WSMV CP using the predictive modeling server I-TASSER. Although this core domain model does not aid in the interpretation of the N terminus deletion data, it does provide insight into the effects of the single-site mutations R237 and D282, which are predicted to form a salt bridge as well as the C-terminal deletions. The salt bridge is critical in maintaining a stably folded domain, and removal of the salt bridge via mutagenesis disrupts the stable fold. In the CP core model, there is the presence of a second conserved Arg (R307) on the opposite side of the D282 residue from R237 that likely helps to further stabilize the folded structure. Figure 1 shows that the predicted core domain contains two salt bridges, with D282 being in the middle. Swapping these residues would keep one salt bridge but disrupt the other by placing two positively charged Arg residues in close proximity, which could explain why swapping of two salt bridge residues in TEV CP failed to restore virion assembly (13). As for the C-terminal deletions, it was observed that deletions of up to 65 amino acids from the C terminus are predicted by the model to have very little effect on the folding of the core. This fits well with the C-terminal deletion data that indicate that mutants with deletions of up to 65 amino acids from the C terminus form virions but that the mutant with a 73-amino-acid deletion does not. Additionally, this model also predicts that the N-terminal region is likely a separate domain possibly connected to the core by a short loop and may have a domain-swapping interaction similar to that of PMV CP (55). Deletions that do not disrupt this interaction would not be expected to have an effect on virion assembly.
Point and deletion mutations in the CP cistron of WSMV facilitated the identification of determinants for virion assembly and/or cell-to-cell movement. The N-terminal amino acids 6 to 27 and 85 to 100 are required for cell-to-cell movement and efficient virion assembly, and the C-terminal 65 amino acids are required for cell-to-cell movement but are dispensable for virion assembly. Additionally, amino acid residues 36 to 84 are not required for virus translocation and virion assembly. The ability of WSMV to infect wheat systemically, despite extensive deletions in the CP, may have serious consequences on the development of CP-based resistance against WSMV. Our results indicate that resistance based on CP antagonism may be overcome by WSMV. Additionally, the availability of viable CP deletion mutants will facilitate defining the roles of CP in the transmission of WSMV by the wheat curl mite and disease development.
ACKNOWLEDGMENTS
We thank T. Jack Morris for discussions during early stages of this investigation, Stephen N. Wegulo for editorial comments, and Jonathan Horrell and Melissa Bartels for technical assistance.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 13 November 2013
REFERENCES
- 1.Callaway A, Giesman-Cookmeyer D, Gillock ET, Sit TL, Lommel SA. 2001. The multifunctional capsid protein of plant RNA viruses. Annu. Rev. Phytopathol. 39:419–460. 10.1146/annurev.phyto.39.1.419 [DOI] [PubMed] [Google Scholar]
- 2.Ivanov KI, Makine K. 2012. Coat protein, host factors, and plant viral replication. Curr. Opin. Virol. 2:712–718. 10.1016/j.coviro.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 3.Dawson WO. 1992. Tobamovirus-plant interactions. Virology 186:359–367. 10.1016/0042-6822(92)90001-6 [DOI] [PubMed] [Google Scholar]
- 4.Rao ALN, Grantham GL. 1996. Molecular studies on Bromovirus capsid protein II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 226:294–305 [DOI] [PubMed] [Google Scholar]
- 5.Bol JF. 2005. Replication of alfamo- and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43:39–62. 10.1146/annurev.phyto.43.101804.120505 [DOI] [PubMed] [Google Scholar]
- 6.Dreher TW, Miller WA. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185–197. 10.1016/j.virol.2005.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington JC, Kasschau KD, Mahajan SK, Schaad MC. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu F, Ren T, Morris TJ. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511–522. 10.1128/JVI.77.1.511-522.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang MB, Metzlaff M. 2005. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8:216–222. 10.1016/j.pbi.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 10.Soosaar JLM, Burch-Smith TM, Dinesh-Kumar S. 2005. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 3:789–798. 10.1038/nrmicro1239 [DOI] [PubMed] [Google Scholar]
- 11.Dolja VV, Haldeman-Cahill R, Montgomery AE, Vandenbosch KA, Carrington JC. 1995. Capsid protein determinants involved in cell-to-cell and long distance movement of Tobacco etch Potyvirus. Virology 206:1007–1016. 10.1006/viro.1995.1023 [DOI] [PubMed] [Google Scholar]
- 12.Dawson WO, Bubrick P, Grantham GL. 1988. Modification of the Tobacco mosaic virus coat protein gene affecting replication, movement and symptomatology. Phytopathology 78:783–789. 10.1094/Phyto-78-783 [DOI] [Google Scholar]
- 13.Dolja VV, Haldeman R, Robertson NL, Dougherty WG, Carrington JC. 1994. Distinct functions of capsid protein in assembly and movement of Tobacco etch Potyvirus in plants. EMBO J. 13:1482–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SH, Sit TL, Kim KH, Lommel SA. 2012. The Red clover necrotic mosaic virus capsid protein N-terminal lysine-rich motif is a determinant of symptomatology and virion accumulation. Mol. Plant Pathol. 13:744–754. 10.1111/j.1364-3703.2011.00784.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueki S, Citovsky V. 2011. To gate, or not to gate: Regulatory mechanisms for intercellular protein transport and virus movement in plants. Mol. Plant 4:782–793. 10.1093/mp/ssr060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benitez-Alfonso Y, Faulkner C, Ritzenthaler C, Maule AJ. 2010. Plasmodesmata: gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23:1403–1412. 10.1094/MPMI-05-10-0116 [DOI] [PubMed] [Google Scholar]
- 17.Harries PA, Schoelz JE, Nelson RS. 2010. Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact. 23:1381–1393. 10.1094/MPMI-05-10-0121 [DOI] [PubMed] [Google Scholar]
- 18.Kawakami S, Watanabe Y, Beachy RN. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complex. Proc. Natl. Acad. Sci. U. S. A. 101:6291–6296. 10.1073/pnas.0401221101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laporte C, Vetter G, Loudes AM, Robinson DG, Hillmer S, Stussi-Garaud C, Ritzenthaler C. 2003. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15:2058–2075. 10.1105/tpc.013896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouwels J, van der Velden T, Willemse J, Borst JW, van Lent J, Bisseling T, Wellink J. 2004. Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. J. Gen. Virol. 85:3787–3796. 10.1099/vir.0.80497-0 [DOI] [PubMed] [Google Scholar]
- 21.Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. 2010. Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23:1231–1247. 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- 22.Berger PH, Adams MJ, Barnett OW, Brunt AA, Hammond J, Hill JH, Jordan RL, Kashiwazaki S, Rybicki E, Spence N, Stenger DC, Ohki ST, Uyeda I, van Zaayen A, Valkonen J, Vetten HJ. 2005. Family Potyviridae, p 819–841 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy: classification and nomenclature of viruses, 8th report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 23.Urcuqui-Inchima S, Haenni AL, Candresse T, Bernardi F. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157–175. 10.1016/S0168-1702(01)00220-9 [DOI] [PubMed] [Google Scholar]
- 24.Rojas MR, Zerbini FM, Allison RF, Gilbertson RL, Lucas WJ. 1997. Capsid protein and helper component-proteinase function as Potyvirus cell-to-cell movement proteins. Virology 237:283–295. 10.1006/viro.1997.8777 [DOI] [PubMed] [Google Scholar]
- 25.Schaad MC, Haldeman-Cahill R, Cronin S, Carrington JC. 1996. Analysis of the VPg-proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70:7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrington JC, Jensen PE, Schaad MC. 1998. Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14:393–400. 10.1046/j.1365-313X.1998.00120.x [DOI] [PubMed] [Google Scholar]
- 27.Chung BY-W, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105:5897–5902. 10.1073/pnas.0800468105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi IR, Horken KM, Stenger DC, French R. 2005. An internal RNA element in the P3 cistron of Wheat streak mosaic virus revealed by synonymous mutations that affect both movement and replication. J. Gen. Virol. 86:2605–2614. 10.1099/vir.0.81081-0 [DOI] [PubMed] [Google Scholar]
- 29.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang W. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6:e1000962. 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen RH, Hajimorad MR. 2010. Mutational analysis of the putative PIPO of Soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400:1–7. 10.1016/j.virol.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 31.Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. 2012. Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8:e1002639. 10.1371/journal.ppat.1002639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allison RF, Dougherty WG, Parks TD, Willis L, Johnston RF, Kelly M, Armstrong FB. 1985. Biochemical analysis of the capsid protein gene and capsid protein of Tobacco etch virus: N-terminal amino acids located on the virion's surface. Virology 147:309–316. 10.1016/0042-6822(85)90133-3 [DOI] [PubMed] [Google Scholar]
- 33.Shukla DD, Strike PM, Tracy SL, Gough KH, Ward CW. 1988. The N- and C-termini of the coat proteins of potyviruses are surface-located and the N-terminus contains the major virus-specific epitopes. J. Gen. Virol. 69:1497–1508. 10.1099/0022-1317-69-7-1497 [DOI] [Google Scholar]
- 34.Atreya PL, Atreya CD, Pirone TP. 1991. Amino acid substitutions in the coat protein result in loss of insect transmissibility of a plant virus. Proc. Natl. Acad. Sci. U. S. A. 88:7887–7891. 10.1073/pnas.88.17.7887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanc S, Lopez-Moya JJ, Wang R, Garcia-Lampasona S, Thornbury D, Pirone TP. 1997. A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 231:141–147. 10.1006/viro.1997.8521 [DOI] [PubMed] [Google Scholar]
- 36.Pirone TP, Blanc S. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227–247. 10.1146/annurev.phyto.34.1.227 [DOI] [PubMed] [Google Scholar]
- 37.Stenger DC, Hall JS, Choi I-R, French R. 1998. Phylogenetic relationships within the family Potyviridae: wheat streak mosaic virus and brome streak mosaic virus are not members of the genus Rymovirus. Phytopathology 88:782–787. 10.1094/PHYTO.1998.88.8.782 [DOI] [PubMed] [Google Scholar]
- 38.Slykhuis JT. 1955. Aceria tulipae Keifer (Acarina: Eriophyidae) in relation to spread of wheat streak mosaic virus. Phytopathology 45:116–128 [Google Scholar]
- 39.Stenger DC, Hein GL, Gildow FE, Horken KM, French R. 2005. Plant virus HC-Pro is a determinant of eriophyid mite transmission. J. Virol. 79:9054–9061. 10.1128/JVI.79.14.9054-9061.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenger DC, Hein GL, French R. 2006. Nested deletion analysis of Wheat streak mosaic virus HC-Pro: mapping of domains affecting polyprotein processing and eriophyid mite transmission. Virology 350:465–474. 10.1016/j.virol.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 41.Stenger DC, French R, Gildow FE. 2005. Complete deletion of Wheat streak mosaic virus HC-Pro: a null mutant is viable for systemic infection. J. Virol. 79:12077–12080. 10.1128/JVI.79.18.12077-12080.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young BA, Stenger DC, Qu F, Morris TJ, Tatineni S, French R. 2012. Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res. 163:672–677. 10.1016/j.virusres.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 43.Robaglia C, Durand-Tardif M, Tronchet M, Boudazin G, Astier-Manifacier S, Casse-Delbart F. 1989. Nucleotide sequence of Potato virus Y (N strain). J. Gen. Virol. 70:935–947. 10.1099/0022-1317-70-4-935 [DOI] [PubMed] [Google Scholar]
- 44.Tatineni S, Van Winkle DH, French R. 2011. The N-terminal region of Wheat streak mosaic virus coat protein is a host- and strain-specific long-distance transport factor. J. Virol. 85:1718–1731. 10.1128/JVI.02044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatineni S, French R. 10 October 2013. The C-terminus of Wheat streak mosaic virus coat protein is involved in differential infection of wheat and maize through host-specific long-distance transport. Mol. Plant Microbe Interact. [Epub ahead of print.] 10.1094/MPMI-09-13-0272-R [DOI] [PubMed] [Google Scholar]
- 46.Choi IR, French R, Hein GL, Stenger DC. 1999. Fully biologically active in vitro transcripts of the eriophyid mite-transmitted Wheat streak mosaic tritimovirus. Phytopathology 89:1182–1185. 10.1094/PHYTO.1999.89.12.1182 [DOI] [PubMed] [Google Scholar]
- 47.Tatineni S, McMechan JA, Hein GL, French R. 2011. Efficient and stable expression of GFP through Wheat streak mosaic virus-based vectors in cereal hosts using a range of cleavage sites: formation of dense fluorescent aggregates for sensitive virus tracking. Virology 410:268–281. 10.1016/j.virol.2010.10.043 [DOI] [PubMed] [Google Scholar]
- 48.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, p 2344 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 50.McNeil JF, French R, Hein GL, Baenziger PS, Askridge KM. 1996. Characterization of genetic variability among natural populations of Wheat streak mosaic virus. Phytopathology 86:122–127 [Google Scholar]
- 51.Tatineni S, Graybosch RA, Hein GL, Wegulo SN, French R. 2010. Wheat cultivar-specific disease synergism and alteration of virus accumulation during co-infection with Wheat streak mosaic virus and Triticum mosaic virus. Phytopathology 100:230–238. 10.1094/PHYTO-100-3-0230 [DOI] [PubMed] [Google Scholar]
- 52.Qu F, Morris TJ. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant Microbe Interact. 15:193–202. 10.1094/MPMI.2002.15.3.193 [DOI] [PubMed] [Google Scholar]
- 53.Tatineni S, Ziems AD, Wegulo SN, French R. 2009. Triticum mosaic virus: a distinct member of the family Potyviridae with an unusually long leader sequence. Phytopathology 99:943–950. 10.1094/PHYTO-99-8-0943 [DOI] [PubMed] [Google Scholar]
- 54.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang S, Wang T, Bohon J, Gagné M-EL, Bolduc M, Leclerc D, Li H. 2012. Crystal structure of the coat protein of the flexible filamentous Papaya mosaic virus. J. Mol. Biol. 422:263–273. 10.1016/j.jmb.2012.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dolja VV, Boyko VP, Agranovsky AA, Koonin EV. 1991. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology 184:79–86. 10.1016/0042-6822(91)90823-T [DOI] [PubMed] [Google Scholar]
- 57.Brakke MK, Skopp RN, Lane LC. 1990. Degradation of Wheat streak mosaic virus capsid protein during leaf senescence. Phytopathology 80:1401–1405. 10.1094/Phyto-80-1401 [DOI] [Google Scholar]
- 58.Choi CW, Qu F, Ren T, Ye X, Morris TJ. 2004. RNA silencing-suppressor function of Turnip crinkle virus coat protein cannot be attributed to its interaction with the Arabidopsis protein TIP. J. Gen. Virol. 85:3415–3420. 10.1099/vir.0.80326-0 [DOI] [PubMed] [Google Scholar]
- 59.Wellink J, van Kammen A. 1989. Cell-to-cell transport of cowpea mosaic virus requires both the 58/48K proteins and the capsid proteins. J. Gen. Virol. 70:2279–2286. 10.1099/0022-1317-70-9-2279 [DOI] [Google Scholar]
- 60.Alzhanova DV, Napuli AJ, Creamer R, Dolja VV. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70homolog. EMBO J. 20:6997–7007. 10.1093/emboj/20.24.6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saito T, Yamanaka K, Okada Y. 1990. Long-distance movement and viral assembly of Tobacco mosaic virus mutants. Virology 176:329–336. 10.1016/0042-6822(90)90002-9 [DOI] [PubMed] [Google Scholar]