Abstract
Herpesvirus nucleocapsids traverse the nuclear envelope into the cytoplasm in a process called nuclear egress that includes disruption of the nuclear lamina. In several herpesviruses, a key player in nuclear egress is a complex of two proteins, whose homologs in human cytomegalovirus (HCMV) are UL50 and UL53. However, their roles in nuclear egress during HCMV infection have not been shown. Based largely on transfection studies, UL50 and UL53 have been proposed to facilitate disruption of the nuclear lamina by recruiting cellular protein kinase C (PKC), as occurs with certain other herpesviruses, and/or the viral protein kinase UL97 to phosphorylate lamins. To investigate these issues during HCMV infection, we generated viral mutants null for UL50 or UL53. Correlative light electron microscopic analysis of null mutant-infected cells showed the presence of intranuclear nucleocapsids and the absence of cytoplasmic nucleocapsids. Confocal immunofluorescence microscopy revealed that UL50 and UL53 are required for disruption of the nuclear lamina. A subpopulation of UL97 colocalized with the nuclear rim, and this was dependent on UL50 and, to a lesser extent, UL53. However, PKC was not recruited to the nuclear rim, and its localization was not affected by the absence of UL50 or UL53. Immunoprecipitation from cells infected with HCMV expressing tagged UL53 detected UL97 but not PKC. In summary, HCMV UL50 and UL53 are required for nuclear egress and disruption of nuclear lamina during HCMV infection, and they recruit UL97, not PKC, for these processes. Thus, despite the strong conservation of herpesvirus nuclear egress complexes, a key function can differ among them.
INTRODUCTION
Viruses that replicate and package their genomes in the nucleus must have a mechanism by which they escape from the nucleus during their exit from the cell. Herpesvirus nucleocapsids are transported out of the nucleus through a process of envelopment and de-envelopment across the nuclear membranes. This unusual process is called nuclear egress (reviewed in references 1 to 3). Although the molecular details of nuclear egress are not completely understood, all known alpha-, beta-, and gammaherpesviruses encode two highly conserved viral proteins—one a nuclear membrane protein and the other a nucleoplasmic protein—that form what is termed the nuclear egress complex (NEC) (3, 4). Herpesviruses differ in their requirement for the viral NEC. Based on studies using null mutant viruses, productive replication and nuclear egress of alphaherpesviruses such as herpes simplex virus 1 (HSV-1) and pseudorabies virus (PrV) and of gammaherpesviruses such as Epstein-Barr virus (EBV) are impaired in the absence of the NEC, but not completely abolished (5–10). In the betaherpesvirus human cytomegalovirus (HCMV), which is an important human pathogen with limited treatment options (11–16), null mutants for the homologous proteins (UL50 and UL53) are replication incompetent (17, 18). Such mutants have been difficult to study due to a lack of complementing cell lines. For murine cytomegalovirus (MCMV), elegant studies showed that conditionally expressed dominant negative mutants of either of the homologous proteins, i.e., M50 or M53, resulted in defects in nuclear egress (19, 20). However, to our knowledge, the nuclear egress phenotypes of either HCMV or MCMV null mutants for these proteins have not been examined.
A critical step in nuclear egress is disruption of the nuclear lamina, which permits nucleocapsids to gain access to the inner nuclear membrane. In alphaherpesviruses, the NEC is required for this disruption (21, 22). However, whether this is true for HCMV has yet to be determined. This disruption resembles the phosphorylation-mediated depolymerization of nuclear lamina that accompanies dissolution of the nuclear envelope during mitosis. Cellular kinases that participate in this cell cycle process include protein kinase C (PKC)—a family of 15 isoforms divided into three subclasses, i.e., conventional (or classical), novel, and atypical (23), some of which are primarily involved in lamin B phosphorylation (24–26)—and cyclin-dependent kinase 1 (Cdk-1), whose phosphorylation of lamin A/C on critical residues, including serine 22, is crucial for lamina disassembly during mitosis (27, 28). In an influential publication, it was reported that conventional PKC is recruited to the nuclear lamina during MCMV infection and colocalizes with the NEC in transfected cells (29). Subsequently, it was shown that in HSV-1-infected cells, at least two PKC isoforms (belonging to the conventional and novel subclasses) are recruited to the nuclear rim and that this is dependent on the presence of the NEC components (30). Thus, a common view is that PKC recruitment by the NEC is a crucial step in lamina dissolution during herpesvirus nuclear egress, which informed a study implicating atypical PKC in a cellular process akin to herpesvirus nuclear egress in Drosophila melanogaster muscle cells (31).
For HCMV, it has been proposed, as is the case for other herpesviruses, that UL50 and UL53 recruit PKC for disruption of the nuclear lamina, based primarily on immunoprecipitation studies with transiently expressed UL50 and UL53 and on yeast two-hybrid studies (32–34). In transfection studies, coexpression of HCMV UL50 and UL53 in the absence of other viral proteins is sufficient to cause disruptions in the nuclear lamina resembling those observed during HCMV infection (35). However, during HCMV infection, an HCMV protein kinase, UL97, mimics Cdk-1 for phosphorylation of lamin A/C on serine 22 and is required for disruption of the nuclear lamina and efficient nuclear egress (36). Accordingly, pharmacological or genetic ablation of UL97 prevents nuclear lamina disruption in infected cells, despite the presence of UL50 and UL53. These observations have led to questions about the role(s) of UL50 and UL53 in the presence or absence of UL97 and during HCMV nuclear egress. Moreover, recruitment of PKC to the nuclear lamina and interactions between UL50 and UL53 and viral and/or cellular kinases during the authentic context of HCMV infection have remained largely unexplored. Whether UL97 is recruited to the nuclear lamina during lamina disruption and nuclear egress has also not been determined.
To investigate the role of HCMV UL50 and UL53 in nuclear egress, we generated bacterial artificial chromosome (BAC) constructs carrying null mutations for UL50 or UL53 and examined the effects of these mutations on nuclear egress, disruption of the nuclear lamina during infection, the subcellular distribution of cellular and viral protein kinases during infection, and the possible interaction of UL50 and UL53 with the cellular kinases and/or the viral kinase UL97. The results reveal important differences between HCMV and other systems.
MATERIALS AND METHODS
Generation of recombinant and mutant viruses. (i) Null constructs and rescued derivatives.
The UL50 and UL53 null BACs were generated by replacing methionine residues with stop codons and introducing frameshift mutations within the respective coding sequences (Table 1). These changes were engineered into pBADGFP (37), an HCMV BAC containing the green fluorescent protein (GFP) cassette under the control of the major immediate early promoter of HCMV, using the “en passant” recombination method of Tischer and coworkers (38, 39). Briefly, PCR primers (see the supplemental material at https://coen.med.harvard.edu) were used to amplify an I-Sce–AphAI (Kanr) DNA sequence from plasmid pEP-KanaS (38). The PCR product was gel purified and electroporated into GS1783 cells harboring the bacmid pBADGFP. Kanamycin-resistant integrates were resolved by heat shock and l-(+)-arabinose induction of I-SceI, and the resulting BACs, 50N pBADGFP and 53N pBADGFP, were sequenced to confirm the introduced changes. The rescued derivatives of 50N pBADGFP and 53N pBADGFP, termed 50NR pBADGFP and 53NR pBADGFP, respectively, were generated by mutagenesis of the stop codons back to the original residues in the UL50 and UL53 coding sequences. The bacmids carrying the rescued sequences were electroporated into human foreskin fibroblast (HFF) cells (Hs27; ATCC CRL-1684) (American Type Culture Collection, Manassas, VA) as described previously, with plasmids pCGN71 (40, 41), expressing the viral transcriptional transactivator pp71, and pBRep-Cre, to generate the rescue viruses 50NR BADGFP and 53NR BADGFP.
TABLE 1.
Summary of HCMV AD169 BAC constructs
| Construct | Genetic background | Reference for background construct | Change(s) introduced |
|---|---|---|---|
| 50N pBADGFP | pBADGFP | 37 | UL50 residues Ala45, Met46, and Leu47 were mutated to stop codons, and an additional nucleotide was added after the third stop codon to introduce a translational frameshift |
| 50NR pBADGFP | UL50 Null pBADGFP | This study | Restored the WT UL50 coding sequence to the UL50 null construct |
| 53N pBADGFP | pBADGFP | 37 | UL53 residues Met81, Met82, and Met84 were mutated to stop codons, and nucleotide 252 was deleted to introduce a translational frameshift |
| 53NR pBADGFP | UL53 Null pBADGFP | This study | Restored the WT UL53 coding sequence to the UL53 null construct |
| 53-F AD169-RV | AD169-RV | 42 | FLAG sequence at C terminus of UL53 |
| 53-F pBADGFP | pBADGFP | 37 | FLAG sequence at C terminus of UL53 |
| 50N 53-F pBADGFP | UL50 Null pBADGFP | This study | FLAG sequence at C terminus of UL53 |
| FLAG-97 pBADGFP | pBADGFP | 37 | FLAG sequence at N terminus of UL97 |
| 50N FLAG-97 pBADGFP | UL50 Null pBADGFP | This study | FLAG sequence at N terminus of UL97 |
| 53N FLAG-97 pBADGFP | UL53 Null pBADGFP | This study | FLAG sequence at N terminus of UL97 |
| 50NR FLAG-97 pBADGFP | UL50 Null Rescue pBADGFP | This study | FLAG sequence at N terminus of UL97 |
| 53NR FLAG-97 pBADGFP | UL53 Null Rescue pBADGFP | This study | FLAG sequence at N terminus of UL97 |
(ii) UL53-FLAG AD169 BACs.
AD169-RV, a BAC clone of HCMV strain AD169 (42), was used in the generation of UL53-FLAG virus (53-F). Sequences encoding a single FLAG epitope tag (DYKDDDDK) were fused to sequences encoding the C terminus of UL53 within the context of the HCMV genome, and additional nucleotides were incorporated to remove the overlap between the UL53 and UL54 open reading frames (ORFs) to prevent any changes in protein coding content of the UL54 gene. The two-step Red recombination method was used to introduce these mutations (38, 39). Briefly, PCR primers 53CTFa1Fw (5′-3′) and 53CTFa1Rv (5′-3′) were used to amplify an I-Sce–AphAI (Kanr) DNA sequence from plasmid pEP-KanaS (38). The PCR product was gel purified and used as a template for a second PCR, using primers 53CTFp2Fw and 53CTFp2Rv. The resulting PCR product was gel purified and electroporated into GS1783 cells harboring the AD169-RV bacmid, and the procedure described above was followed to obtain the bacmid UL53-FLAG AD169-RV. This bacmid was electroporated into HFF cells to generate the virus 53-FLAG AD169-RV. The same strategy was then used to fuse sequences encoding the FLAG sequence to the C terminus of UL53 in the wild-type (WT) pBADGFP and 50N pBADGFP backgrounds, generating the constructs 53-F pBADGFP and 50N 53-F pBADGFP, respectively.
(iii) FLAG-UL97 BACs.
The FLAG sequence was introduced at the N terminus of the UL97 coding sequence by using the primers listed in the supplemental material at our website (https://coen.med.harvard.edu) and a previously described strategy (41) on the WT, 50N, and 53N pBADGFP constructs, as well as the rescued derivatives, 50NR pBADGFP and 53NR pBADGFP, to generate the constructs FLAG-97 pBADGFP, 50N FLAG-97 pBADGFP, 53N FLAG-97 pBADGFP, 50NR FLAG-97 pBADGFP, and 53NR FLAG-97 pBADGFP, respectively. These bacmids were electroporated into HFF cells as described above to generate the respective BADGFP viruses.
Virus replication assays.
To assess HCMV replication, 1 × 105 HFF cells per well were seeded into 24-well plates 24 h before infection. At the time of infection, the indicated virus was added to each well at the multiplicity of infection (MOI) indicated in the text. After incubation for 1 h at 37°C, the inoculum was removed and replaced with 1 ml of complete Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 5% fetal bovine serum (FBS) (Gibco). At the indicated time points, the medium from each well (virus supernatant) was taken from the cells and stored at −80°C until required. Dilutions of each virus supernatant were titrated simultaneously onto fresh monolayers of HFF cells to determine virus titers.
CLEM.
Correlative light electron microscopy (CLEM) was used to analyze GFP-positive cells after electroporations with either WT pBADGFP, 50N pBADGFP, or 53N pBADGFP. On day 6 postelectroporation, the cells were reseeded onto no. 2 glass-bottom P35G-2-14-C-grid dishes (MatTek). On day 7, medium was aspirated from dishes and replaced with phosphate-buffered saline (PBS). A Nikon Ti inverted fluorescence microscope (w/Perfect Focus model; HC PL APO ×10/0.7 CS air objective) was used to image the transfected cells with fluorescence as well as in the reflection mode to visualize the cells and the grid. Samples were then prepared for electron microscopy (EM) by fixing with 3% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.0, for 20 min and rinsing 3 times with the same buffer. Samples were postfixed with 1% osmium with 0.8% potassium ferricyanide in buffer for 15 min on ice in a chemical hood and then rinsed thrice with buffer and twice with distilled water. They were stained with 1% aqueous uranyl acetate overnight at 4°C in the dark. Samples were then rinsed with water and the gridded coverslips removed from the dishes with “glass bottom fluid” (DCF-OS-30; MatTek) according to the manufacturer's directions. The samples were then dehydrated in a graded ethanol series, using progressive lowering of the temperature (43). After a final dip in fresh 100% ethanol followed by a dip in 100% propylene oxide, they were infiltrated with a 2:1 propylene oxide-Epon araldite solution followed by treatment with a 1:2 propylene oxide-Epon araldite solution at room temperature for 30 min each. This was followed by treatment with 100% Epon araldite for 1 h. The samples were then mounted for polymerization at 65°C for 48 h. The glass coverslips were removed from the embedded samples by scoring the glass with a diamond pencil and then applying an ice cube until chips of glass could be removed. GFP-positive cells and their grid coordinates previously imaged using phase microscopy were identified, excised, and remounted for serial sectioning at 75 nm on a Reichert Ultracut S ultramicrotome. Sections were stained with uranyl acetate and then lead citrate before being viewed on a TecnaiG2 Spirit BioTWIN microscope. Images were taken with an AMT 2k charge-coupled device (CCD) camera. Transmission EM was performed in the Harvard Cell Biology EM Core Facility.
Immunofluorescence.
A total of 1 × 105 HFF cells were seeded on glass coverslips in a 24-well plate. Cells were mock infected or infected with AD169-RV (MOI of 1) and were fixed at 72 h postinfection (p.i.). Cells electroporated with the pBADGFP constructs were trypsinized and reseeded on glass coverslips at 1 × 105 cells/well in a 24-well plate at 6 days postelectroporation and were fixed and/or permeabilized on day 7. Methanol fixation and permeabilization (for anti-UL50 or anti-UL53 staining) were performed using ice-cold methanol for 20 min. Formaldehyde fixation was accomplished at room temperature with freshly made 4% formaldehyde in Dulbecco's phosphate-buffered saline (DPBS). After washing in DPBS, formaldehyde-fixed samples were permeabilized with 0.1% Triton X-100 dissolved in DPBS at room temperature (RT) for 10 min. The cells were washed again with DPBS and incubated in 1% bovine serum albumin (BSA) dissolved in DPBS for 1 h at RT. Mouse anti-FLAG antibody (Sigma) at 1:1,000, anti-lamin A/C (1:10) (N-18; Santa Cruz), anti-pan-PKC, anti-PKC-α, or anti-PKC-δ (1:10) (Santa Cruz), anti-Cdk-1 (1:100) (P0H1; Cell Signaling), or anti-UL44 (1:100) (Virusys) antiserum was applied as indicated in the text and incubated for 1 h at RT. For detection of UL50 and UL53, custom rabbit antisera against UL50 (amino acids 1 to 169) and UL53 (amino acids 50 to 292), purified from Escherichia coli as described previously (44), were prepared and affinity purified commercially (Open Biosystems, Huntsville, AL). These antibodies were used at a dilution of a 1:100. Antiserum was removed by washing cells three times with DPBS for 5 min each time, with rocking. This procedure was repeated with the appropriate fluorescently labeled secondary antibodies (anti-rabbit, anti-mouse, or anti-goat labeled with Alexa Fluor 488, 568, or 647; Molecular Probes). DAPI (4′,6-diamidino-2-phenylindole) was applied in the last 10 min of the secondary antibody incubation. After three washes with DPBS, the prepared coverslips were mounted on microscope slides with ProLong antifade reagent (Invitrogen-Molecular Probes). All imaging experiments were done at the Nikon Imaging Center at Harvard Medical School, using a Nikon Ti microscope with a spinning disk confocal laser at a magnification of ×100. Images shown were obtained by acquiring sequential optical planes in the z axis, using the MetaMorph program. Statistical tests were performed using GraphPad Prism, version 6.
Immunoprecipitation of UL53-FLAG from HFF cells for Western blotting.
Two 150-mm plates of HFF cells (7 × 107 cells) were infected with UL53-FLAG AD169-RV or, as a negative control, with AD169-RV, at an MOI of 1 PFU/cell. At 72 h p.i., the cells were washed once with DPBS and harvested into 3 ml of hypotonic buffer (10 mM HEPES [pH 7.4], 1 mM dithiothreitol [DTT], 1 mM MgCl2, 10 mM KCl, and protease inhibitor cocktail). After incubation on ice at 4°C for 15 min, the cells were Dounce homogenized with 40 strokes. The homogenate was centrifuged at 1,000 × g for 10 min. The supernatant was removed, and the nuclear pellet was resuspended in 3 ml of lysis buffer (50 mM HEPES, pH 7.4, 1 mM DTT, 200 mM NaCl, 5 mM EDTA, 50 mM arginine, 0.5% Triton X-100, EDTA-free protease inhibitor cocktail [Roche Applied Sciences, Indianapolis, IN]). This was followed by end-over-end rotation at 4°C for 15 min. The homogenate was Dounce homogenized again with 40 strokes and then centrifuged at 18,000 × g for 25 min. The supernatant was precleared with 500 μl of anti-IgG resin (Sigma) for 2 h and then applied to 300 μl of settled EZ-View anti-FLAG M2 affinity resin (Sigma) for 4 h at 4°C with rotation. Beads were then washed 4 times with 3 ml of lysis buffer for 15 min with rotation at 4°C. The protein was then eluted from beads by low-pH elution using 270 μl of 0.1 M glycine, pH 2.5, and incubation at room temperature for 15 min. This was followed by addition of 30 μl of neutralization buffer (0.5 M Tris, pH 7.4, 1.5 M NaCl) and 10% glycerol to the final volume of 300 μl. The eluate was stored in 100-μl aliquots at −80°C until further analysis.
For Western blotting, 10 μl of a 1:1 mixture of the protein eluate and 2× Laemmli buffer and 30 μl of a 1:1 mixture of the flowthrough from the beads were separated in a 10% gradient SDS-PAGE gel (Bio-Rad). The proteins were transferred to a nitrocellulose membrane, blocked with 1% milk in PBST (PBS with 1% Tween 20), and probed with the various antibodies. The antibody dilutions were as follows: rabbit anti-UL50 (this study), 1:100; rabbit anti-UL53 (this study), 1:100; rabbit anti-UL97 (41), 1:1,000; goat anti-lamin A/C (N-18; Santa Cruz), 1:200; rabbit or mouse anti-pan PKC or rabbit anti-PKC-α or anti-PKC-δ (Santa Cruz), 1:200; and mouse anti-Cdk-1 (Cell Signaling), 1:800. Goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody, goat anti-rabbit HRP-conjugated antibody, or chicken anti-goat HRP-conjugated antibody (Southern Biotech) and chemiluminescence solution (Pierce) were used to detect primary antibodies, and the images were obtained using film.
RESULTS
Construction and characterization of HCMV UL50 null and UL53 null BACs.
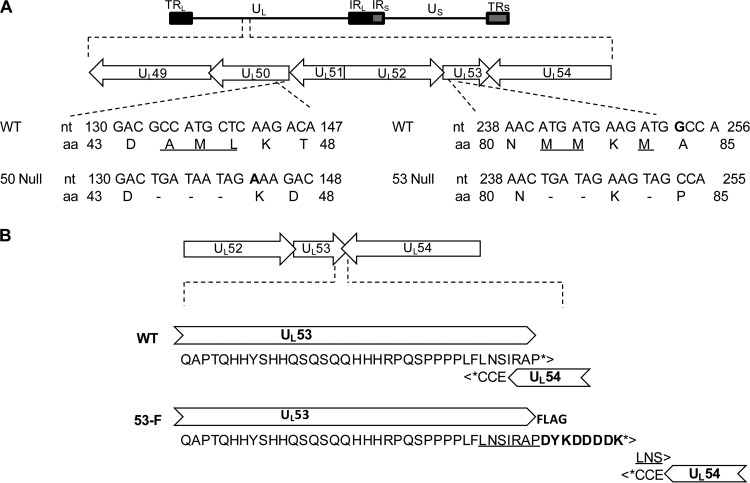
We wished to test whether UL50 and UL53 are required for nuclear egress and lamina disruption in HCMV-infected cells. Deletion of the entire coding region for either protein without effects on other viral functions is not feasible, because both UL50 and UL53 overlap neighboring essential genes (the C-terminal coding region for UL50 with that for UL49 and the N- and C-terminal coding regions for UL53 with UL52 and UL54, respectively) (Fig. 1A). Therefore, we engineered mutants in which the second methionine and neighboring codons within UL50 and UL53 were changed to stop codons and a nucleotide was either added or deleted to shift the reading frame and lead to generation of drastically truncated forms of UL50 (expressing amino acids 1 to 43) and UL53 (expressing amino acids 1 to 80) during infection (Fig. 1A and Table 1). Neither of these truncated proteins is known to be sufficient for efficient binding to any potential interaction partner. The mutations were introduced into a GFP-encoding BAC derived from strain AD169, namely, pBADGFP (37), using two-step Red recombination to generate the UL50 null (50N pBADGFP) and UL53 null (53N pBADGFP) bacmids. These two bacmids were separately electroporated, alongside WT parental BAC DNA, into HFF cells. In cells electroporated with wild-type pBADGFP, spread of the GFP signal in the cell monolayer was observed, which was followed by cytopathic effect (CPE) and release of detectable levels of infectious virus. In contrast, there was no spread of the GFP signal, CPE, or release of detectable infectious virus in cells electroporated with either the 50N or 53N pBADGFP. Thus, the 50N and 53N BADGFP viruses are nonviable.
FIG 1.
Construction of HCMV UL50 and UL53 null mutants. (A) Organization of the HCMV genome. TRL, terminal repeat long; UL, unique long; IRL/S, internal repeat long and neighboring internal repeat short; US, unique short; TRS, terminal repeat short. Below the top schematic, the region of the viral genome from UL49 to UL54 is expanded, showing the overlaps between UL49 and UL50 and between UL52, UL53, and UL54. Below this, the mutations introduced into the UL50 and UL53 coding sequences for generation of 50N and 53N pBADGFP are shown. Amino acids that were mutated to stop codons (dashes) are underlined. Extra nucleotides are shown in bold. nt, nucleotide; aa, amino acid. (B) Construction of HCMV UL53-FLAG AD169-RV. Fusion of sequences encoding the FLAG tag to sequences encoding the C terminus of UL53 in a BAC of AD169 HCMV was performed. The UL52-UL53-UL54 region of the viral genome is expanded to show the details of the UL53 C-terminal sequence overlap with UL54 in the original wild-type (WT) genome. The stop codons for UL53 and UL54 are indicated by asterisks. The duplicated sequence is underlined, and the FLAG sequence is indicated in bold.
To test whether these lethal phenotypes were due to the engineered mutations, we generated rescued derivatives of the 50N and 53N bacmids by restoring wild-type sequences to the UL50 or UL53 coding region, resulting in bacmids designated 50NR pBADGFP and 53NR pBADGFP (Table 1). Electroporation of these BACs into HFF cells resulted in increasing numbers of cells expressing GFP and in the development and spread of CPE. The resulting viruses, 50NR BADGFP, and 53NR BADGFP, showed replication kinetics comparable to those of the WT virus in a multiple-cycle growth curve (MOI = 0.1) (see Fig. S1A in the supplemental material at https://coen.med.harvard.edu). Thus, the engineered UL50 and UL53 null mutations are lethal, consistent with studies of UL50 and UL53 insertion mutants (17, 18).
Effect of null mutations on nuclear egress.
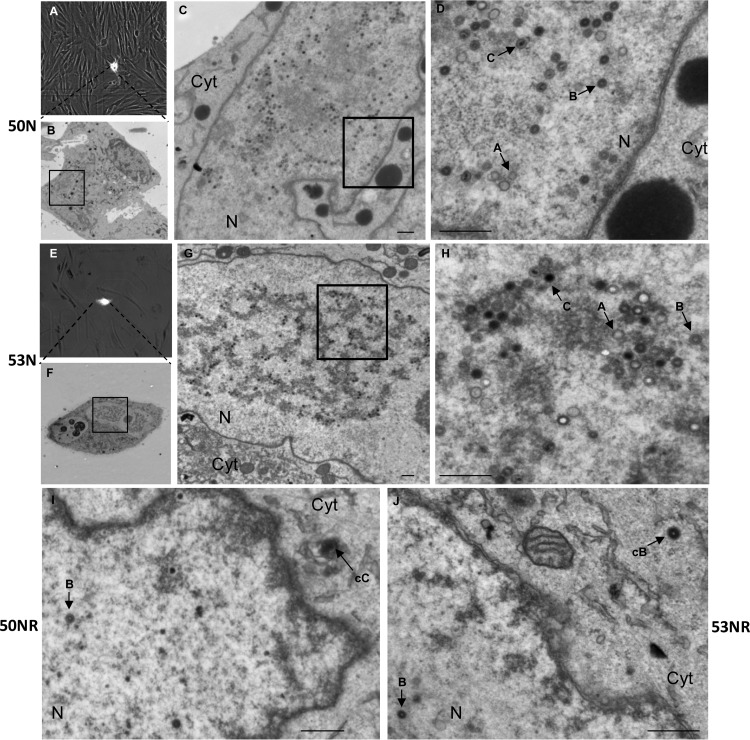
We wanted to determine if the 50N and 53N viruses exhibited defects in nuclear egress. We first asked whether virus infection could progress to the stage of replication compartment formation in the absence of UL50 or UL53, as determined by electroporating cells with either WT pBADGFP or the 50N or 53N pBADGFP construct and staining for a viral DNA polymerase subunit (UL44) and lamin A/C at 7 days postelectroporation (see Fig. S1B at https://coen.med.harvard.edu). The sizes of the replication compartments demarcated by UL44 staining were similar in all three infections, suggesting that the UL50 and UL53 null mutant viruses are defective at a stage after DNA synthesis. We next utilized CLEM, which allows analysis of cells by EM based on localization of cells expressing a fluorophore (45). HFF cells electroporated with either 50N or 53N pBADGFP were imaged using phase and fluorescence microscopy on day 7 and then fixed for transmission EM. We then traced back EM serial sections (60 μm) to the GFP-positive cells from the phase-fluorescence microscopy images (Fig. 2A and E; a color version is available in Fig. S2 at https://coen.med.harvard.edu), as detailed in Materials and Methods. In the absence of either UL50 or UL53, abundant nucleocapsids representing all three forms (A, B, and C) could be detected in the nucleus (Fig. 2D and H), but no capsids were detected in the cytoplasm in any of the sections. In contrast, cells electroporated with the 50NR or 53NR pBADGFP-rescued derivatives consistently displayed the presence of both nuclear and cytoplasmic capsids (Fig. 2I and J). Although the number of cytoplasmic capsids in cells electroporated with rescued derivative bacmids was not high (as is also the case in WT-infected cells [46, 47]) and the number of cells analyzed was limited, these results indicate that both UL50 and UL53 are critical for efficient nuclear egress of HCMV.
FIG 2.
CLEM analysis of HCMV UL50 and UL53 null mutants. HFF cells were electroporated with UL50 null (50N) (A to D) or UL53 null (53N) (E to H) pBADGFP or the rescued derivative 50NR (I) or 53NR pBADGFP (J) and were seeded on gridded coverslips. Phase and fluorescence microscopy was performed on electroporated cells (A and E) on day 7 postelectroporation, and the cells were then processed for EM (B to D and F to J). The original GFP-positive cells (A and E) were traced back, and serial sections were scanned for nuclear and cytoplasmic nucleocapsids, for each sample. The insets in panels B, C, F, and G indicate regions magnified in the next panels (C, D, G, and H, respectively). Bars, 500 nm. N, nucleus; Cyt, cytoplasm; cB and cC, cytoplasmic B and C capsids, respectively. Arrows A, B, and C point to examples of nuclear A, B, and C capsids.
Effect of null mutations on the subcellular distribution of UL50 or UL53.
HCMV UL50 and UL53 colocalize at the nuclear rim both in transient-expression studies and during infection, and localization of UL53 to the nuclear rim requires UL50 in transfection assays (35, 44, 48). To investigate the effect of the absence of one of these proteins on the subcellular distribution of the other during HCMV infection, we generated antisera by immunizing rabbits with bacterially expressed versions of these proteins. We also generated a virus expressing UL53 tagged with a FLAG epitope (Fig. 1B). Sequences encoding a FLAG-epitope tag were added to the C terminus of the UL53 coding sequence on the AD169-RV BAC by employing a strategy similar to that used to construct an HSV-1 UL31 null mutant (9). Briefly, we introduced additional nucleotides to remove the overlap between the UL53 and UL54 ORFs, such that the FLAG tag would not alter the protein coding content of the UL54 gene (Fig. 1B). The replication kinetics of the corresponding virus, 53-F AD169-RV (53-F), were indistinguishable from those of untagged WT virus (MOI = 1) (see Fig. S3A at https://coen.med.harvard.edu). To determine the subcellular distribution of FLAG-tagged UL53, we mock infected cells or infected them with 53-F AD169-RV (53-F). At 72 h postinfection, cells were fixed and stained with DAPI to visualize nuclei and with FLAG antibody. We observed FLAG staining at the nuclear rim, consistent with the FLAG tag not altering the subcellular distribution of UL53 (see Fig. S3B at https://coen.med.harvard.edu).
Subsequently, we fused a sequence encoding the FLAG tag with the sequence encoding the C terminus of UL53 in WT pBADGFP and 50N pBADGFP, to generate 53-F pBADGFP and 50N 53-F pBADGFP, respectively (Table 1). We then electroporated HFF cells with either the WT 53-F, 50N 53-F, or 53N pBADGFP bacmid. At 7 days postelectroporation, the cells were fixed and stained with DAPI to visualize nuclei and with the anti-FLAG or anti-UL50 antibody to visualize UL53-FLAG or UL50, respectively. We identified electroporated cells by the GFP signal and analyzed them using confocal microscopy (see Fig. S4 at https://coen.med.harvard.edu). In 53-F pBADGFP-electroporated cells, UL53-FLAG was localized predominantly at the nuclear rim (see Fig. S4F and H at the above URL). However, in the absence of UL50 (50N), UL53-FLAG was distributed throughout the nucleus, with no preferential localization at the nuclear rim (see Fig. S4J and L at the above URL). In contrast, in both WT and 53N pBADGFP-electroporated cells, UL50 was comparably localized to the nuclear rim (see Fig. S5M and N at the above URL), a result later confirmed by costaining with UL50 and lamin B antisera (data not shown). Infection with the 50N-rescued derivative, 50 NR BADGFP, showed WT localization of UL53 at the nuclear rim (see Fig. S6G to I at the above URL). Additionally, we consistently detected bright staining in a region outside the nucleus, not observed in mock-infected cells, with the affinity-purified rabbit anti-UL50 and anti-UL53 antisera (see Fig. S5 and Fig. S6 at the above URL). This could be due to the proteins accumulating in the assembly compartment (49), although we note that mouse anti-FLAG antibodies detected only faint staining in this region in cells infected with UL53-FLAG-expressing viruses (see Fig. S3 and S4 at the above URL). Thus, during HCMV infection, as has been observed in transfected cells (35, 44), UL50 was necessary for the localization of UL53 to the nuclear rim, but not vice versa.
UL50 and UL53 are required for disruption of the nuclear lamina during HCMV infection.
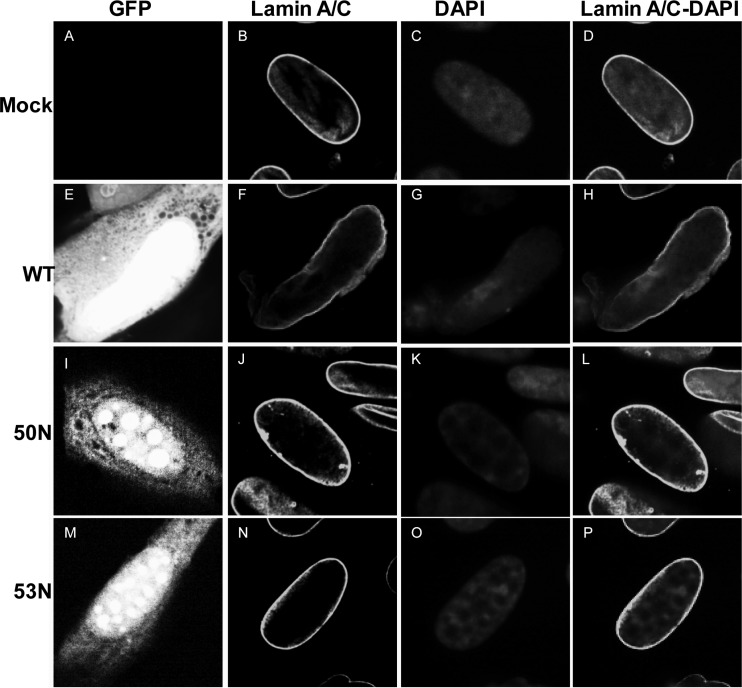
Infection with HCMV induces remodeling of the nuclear lamina, characterized by ruffling, thinning, and generation of gaps (35, 36, 50). Although transfection studies have shown that HCMV UL50 and UL53 are sufficient to cause structural changes to the nuclear lamina (35), these proteins are not sufficient to induce disruption of the nuclear lamina in infected cells when UL97 is genetically or pharmacologically ablated (36). Thus, to test the roles of these proteins in nuclear lamina disruption in infected cells, we electroporated cells with WT, 50N, or 53N pBADGFP bacmid, stained cells with antibodies against lamin A/C at day 7 postelectroporation, and visualized GFP-positive cells by confocal microscopy (Fig. 3; a color version is available as Fig. S7 at https://coen.med.harvard.edu). Electroporation of the WT BAC led to deformation of the nuclear shape, characteristic ruffling and thinning of the nuclear lamina, and generation of gaps visible by light microscopy (Fig. 3F and H). In contrast, electroporation with either the UL50 or UL53 mutant BAC (50N or 53N, respectively) resulted in cells with oval nuclei and an almost intact nuclear lamina, similar in appearance to the lamin A/C staining in mock-electroporated cells (Fig. 3B, J, and N). Cells infected with the rescued derivatives of the null mutant viruses showed nuclear lamina modifications similar to those of WT virus-infected cells (see Fig. S6E, H, and K at https://coen.med.harvard.edu). Thus, UL50 and UL53 are each required for disruption of the nuclear lamina in infected cells.
FIG 3.
Nuclear lamina structure in the absence of HCMV UL50 or UL53. HFFs were mock electroporated (A to D) or electroporated with WT (E to H), UL50 null (50N) (I to L), or UL53 null (53N) (M to P) pBADGFP. Cells were fixed on day 7 and stained with antibody against lamin A/C (B, F, J, and N), the nucleus was stained with DAPI (C, G, K, and O), and cells were visualized using confocal microscopy, with GFP-positive cells (E, I, and M) identified by green fluorescence.
Distribution of cellular kinases PKC and Cdk-1 during HCMV infection.
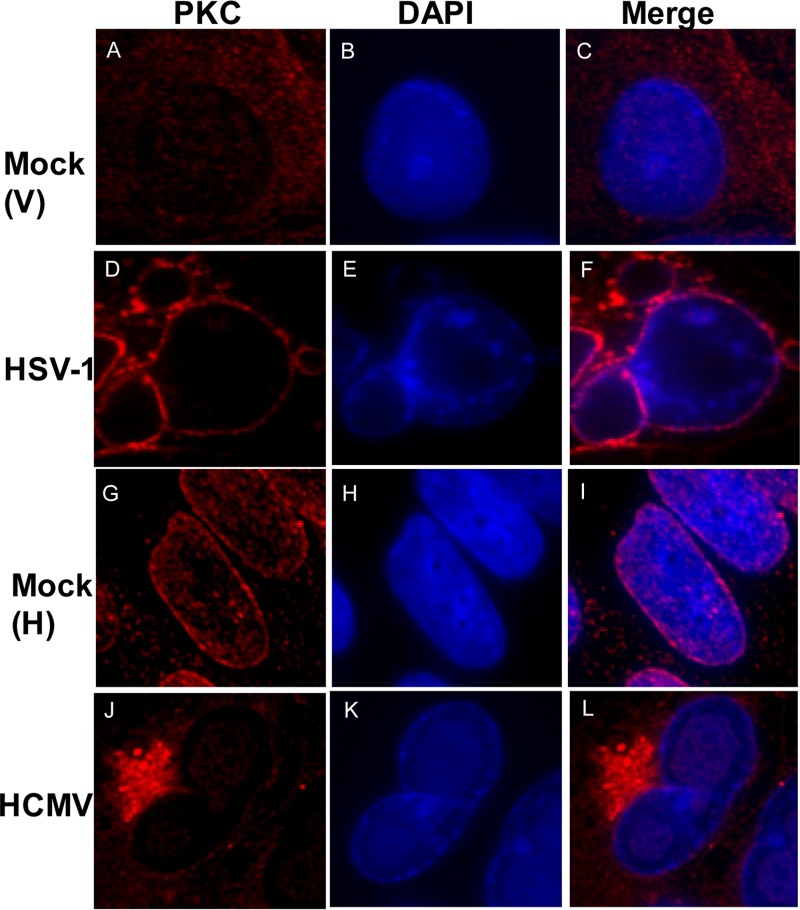
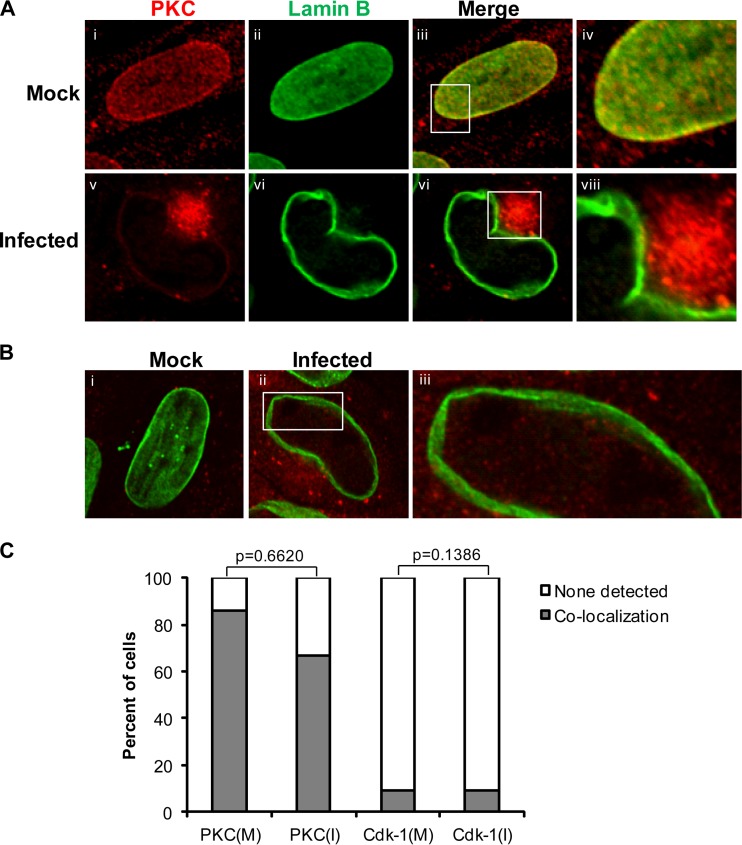
Recruitment of PKC to the nuclear lamina by UL50 and UL53 and their homologs has been proposed to be required for disruption of the nuclear lamina during HCMV, HSV-1, and MCMV infections (29, 30, 32, 33). However, it has not been demonstrated that such recruitment occurs during HCMV infection. We compared the subcellular distribution of PKC during HSV-1 and HCMV infections. We infected Vero cells with HSV-1 (or mock infected them), as in the previous HSV-1 study (30), and stained these cells with an antibody that recognizes all PKC isoforms (as used in the previous HSV-1 study [30]). We also infected HFF cells with wild-type HCMV (or mock infected them) and stained the cells with the same pan-PKC antibody. In both cases, we used DAPI to stain nuclei. In the HSV-infected cells, PKC localized predominantly to the nuclear rim (Fig. 4D and F), which differs from the diffuse distribution in mock-infected cells (Fig. 4A and C), as has been reported previously (30). Similar results were found in HSV-1-infected HFF cells (data not shown). In contrast, in HCMV-infected cells compared to mock-infected cells, PKC localized primarily to a perinuclear region (Fig. 4J and L). We observed staining of PKC in this region by using both mouse and rabbit antibodies (see Fig. S8A and B at https://coen.med.harvard.edu) and costaining of PKC staining with a viral tegument protein, pp28 (see Fig. S8B at the above URL), consistent with this region being the assembly compartment (49). Importantly, there was no evident recruitment of PKC to the nuclear rim in HCMV-infected cells above that seen in mock-infected cells (compare Fig. 4G and J). In fact, if anything, there appeared to be less PKC at the nuclear rim in HCMV-infected cells than in mock-infected cells. We repeated these experiments with antisera that recognized PKC isoforms α (classical PKC) and δ (novel PKC), which are recruited to the nuclear rim during HSV-1 infection (30), and again saw no recruitment of these isoforms to the nuclear rim during HCMV infection above that observed in mock-infected controls (data not shown).
FIG 4.
PKC distribution in HSV-1- or HCMV-infected cells. Vero cells were mock infected [Mock (V)] (A to C) or infected with HSV-1 (D to F) at an MOI of 1, and HFFs were mock infected [Mock (H)] (G to I) or infected with HCMV (J to L) at an MOI of 1. Vero cells were fixed at 16 h, and HFFs were fixed at 72 h postinfection. Samples were stained with a pan-PKC antibody (red) and DAPI (blue) and visualized by confocal microscopy.
Given the differences between our results for HCMV-infected cells and those for other systems, we next compared the distributions of PKC and another cellular kinase, Cdk-1, which plays an important role in lamin A/C phosphorylation during mitosis (28, 51, 52), between HCMV-infected and mock-infected cells. This time we costained for lamin B to demarcate the nuclear lamina (Fig. 5). For both kinases, we found points of colocalization with lamin B staining, although there were very few such points of Cdk-1 colocalization with lamin B in either mock-infected or infected cells, and only in a small fraction of cells (Fig. 5B, panels ii and iii, and C). On the other hand, consistent with previous results showing overexpression of Cdk-1 in HCMV-infected cells (53, 54), the Cdk-1 signal was stronger in the cytoplasm of infected cells than in mock-infected cells (Fig. 5B, panels i and ii). Comparing 21 infected cells with 21 mock-infected cells, again, if anything, we found fewer infected cells than mock-infected cells showing colocalization of PKC with nuclear lamina, although the difference was not statistically significant (Fig. 5C). Additionally, there was generally less intense staining for PKC at the nuclear rim in infected cells than in mock-infected cells (Fig. 5A, compare panels i and iii with panels iv and vi). We also found no statistically significant difference between mock-infected cells and infected cells in colocalization of Cdk-1 to the nuclear lamina (Fig. 5C). Thus, we found no evidence of recruitment of either PKC or Cdk-1 to the nuclear lamina in HCMV-infected cells. Interestingly, in HCMV-infected cells, there was generally less disruption of lamin B staining in terms of thinning and gaps than of lamin A/C staining (compare Fig. 5A, panel viii, B, panel iii, and Fig. S8A at https://coen.med.harvard.edu with Fig. 3F and Fig. S6 and S7 at the same URL).
FIG 5.
Cellular distribution and nuclear rim localization of cellular kinases PKC and Cdk-1 during HCMV infection. (A) HFFs were mock infected (i to iv) or infected with HCMV (v to viii) at an MOI of 1. At 72 h p.i., cells were fixed and stained for lamin B (green) and PKC (red). The samples were visualized using confocal microscopy. (B) HFFs were mock infected (i) or infected with HCMV (ii and iii) at an MOI of 1. At 72 h p.i., cells were fixed and stained for lamin B (green) and Cdk-1 (red). The insets indicate magnified (×3) sections of the respective images. (C) Cells showing colocalization (shaded bars) or no colocalization (unshaded bars) for the cellular kinases and lamin B were counted among the infected (I) and mock-infected (M) samples. The data were analyzed using two-tailed Fisher's exact test, and the P values for the differences between the mock-infected and infected samples are shown.
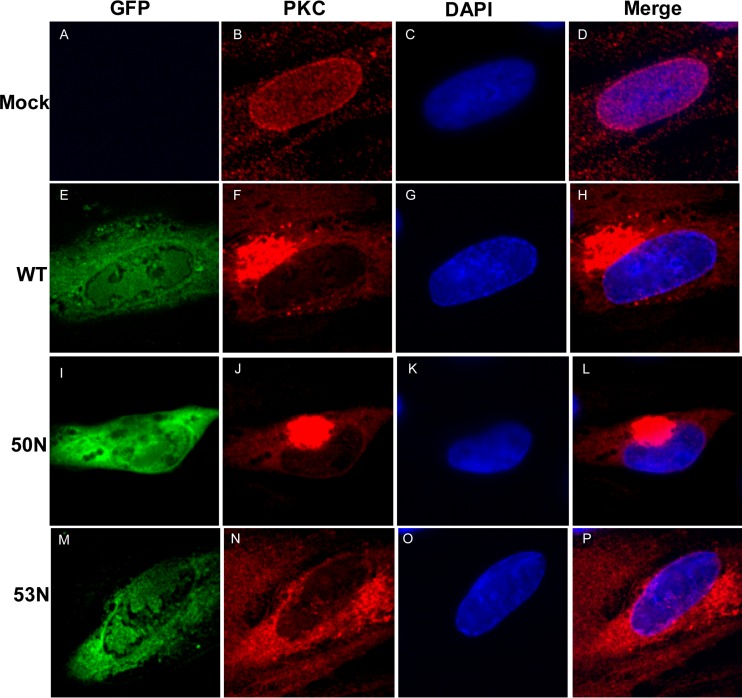
Based on transfection experiments, PKC has been suggested to be recruited to the nuclear lamina through interaction with UL50 (32). To ascertain if the absence of UL50 or UL53 affects the distribution of PKC in infected cells, HFF cells electroporated with WT pBADGFP, 50N pBADGFP, or 53N pBADGFP were compared for pan-PKC staining. The absence of either UL50 or UL53 did not lead to any observable differences in the distribution of PKC at the nuclear rim in the GFP-positive cells (Fig. 6). Thus, we found no evidence for UL50- or UL53-dependent recruitment of PKC in HCMV-infected cells.
FIG 6.
Cellular distribution of PKC in the absence of HCMV UL50 or UL53. HFFs were mock electroporated (A to D) or electroporated with WT (E to H), UL50 null (50N) (I to L), or UL53 null (53N) (M to P) pBADGFP. Cells were fixed on day 7 and stained with antibody against PKC (red), and the nucleus was stained with DAPI (blue). Cells positive for GFP (green) were visualized by confocal microscopy.
A subpopulation of UL97 localizes to the nuclear rim, depending on UL50 and UL53, during HCMV infection.
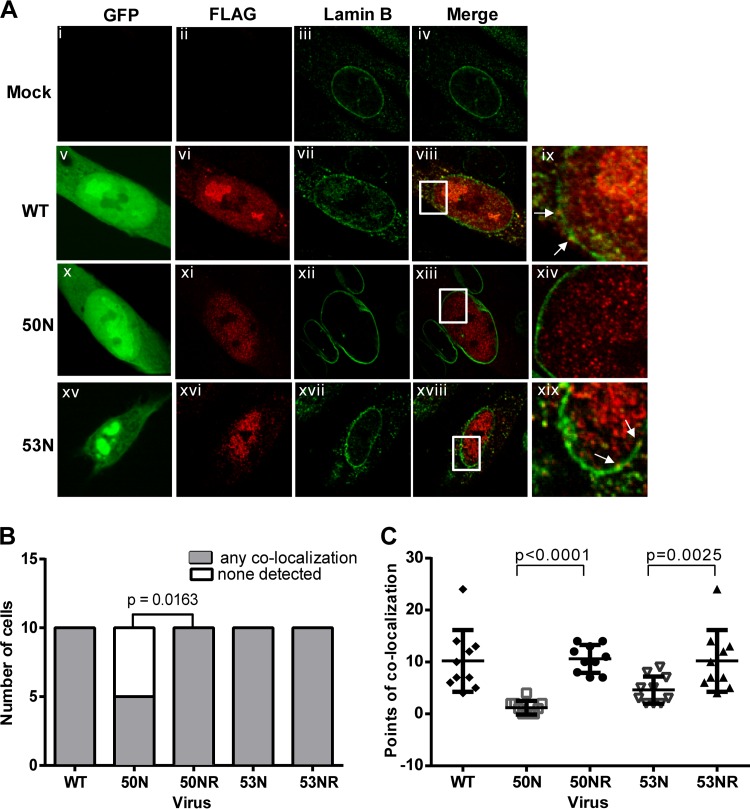
Since we found that neither PKC nor Cdk-1 is recruited to the nuclear rim during HCMV infection, we investigated whether the viral protein kinase UL97 is. Because various antibodies against UL97 did not work well in preliminary immunofluorescence assays (data not shown), we fused a sequence encoding a FLAG epitope to sequences encoding the N terminus of UL97 in the WT, 50N, and 53N pBADGFP bacmids, as well as in their rescued derivatives (Table 1). We electroporated HFF cells with these bacmids, and 7 days later, we fixed the cells, stained them with anti-FLAG and anti-lamin B antibodies, and analyzed the GFP-expressing cells by confocal microscopy. As previously shown for UL97 during HCMV infection (50), FLAG-UL97 was distributed mainly throughout the nucleus in cells electroporated with WT pBADGFP or the rescued derivatives, with some staining in the cytoplasm as well. However, a small portion of the FLAG staining colocalized with lamin B in discrete foci (Fig. 7A, panels vi, viii, and ix, and data not shown). Notably, in a number of cells electroporated with 50N pBADGFP, all of the FLAG staining was interior to the lamin B staining (Fig. 7A, panel xiv). To quantify the results, FLAG and lamin B colocalization in optical sections from 10 GFP-positive cells from electroporation with either the 50N or 53N pBADGFP construct were compared to those from 10 GFP-positive cells electroporated with 50NR or 53NR pBADGFP, respectively, and were assessed in terms of the percentage of cells showing such colocalization (Fig. 7B). The absence of UL50 led to significant decreases in the percentage of cells showing any observable colocalization of FLAG-UL97 with the lamina (P = 0.0163) (Fig. 7B). We also counted the number of foci of FLAG-UL97 and lamin B colocalization in these optical sections. The 50N-infected cells either showed no colocalization or had fewer foci of colocalization than any of the WT-infected or 50NR-infected cells (Fig. 7C), and the difference between the 50N mutant and its rescued derivative (and WT) was highly significant (P < 0.0001).
FIG 7.
Cellular distribution of UL97 in the absence of HCMV UL50 or UL53. (A) HFFs were mock electroporated (i to iv) or electroporated with WT (v to ix), UL50 null (50N) (x to xiv), or UL53 null (53N) (xv to xix) pBADGFP expressing FLAG epitope-tagged UL97. HFFs were also electroporated with rescued derivatives 50NR and 53NR. Cells were fixed on day 7 and stained with antibodies against FLAG (red) and lamin B (green). GFP-positive cells were located by confocal microscopy. Panels ix, xiv, and xix represent magnified (×3) sections of the insets in the panels to their left. White arrows point at regions of colocalization. (B) For statistical analysis, 10 infected (GFP-positive) cells from each electroporation were analyzed for whether they showed any points of colocalization (shaded bars) between FLAG-UL97 and lamin B or no observable colocalization (unshaded bars). The difference between the data for cells infected with 50N versus 50NR was analyzed using one-tailed Fisher's exact test, and the P value is shown. (C) The number of foci of FLAG-UL97 and lamin B colocalization was counted in representative optical sections of 10 cells/sample. Differences in these data between cells infected with a mutant versus its rescued derivative were analyzed using one-way analysis of variance (ANOVA) followed by Sidak's multiple-comparison test, and P values are shown.
In cells electroporated with 53N pBADGFP, all cells exhibited colocalization of FLAG-UL97 and lamin B (Fig. 7A, panels xvi, xviii, and xix, and B). However, there were generally fewer such points of colocalization between UL97 and lamin B in optical sections of 53N-infected cells compared with those in cells infected with WT or 53NR (Fig. 7A and C). Although these differences were less dramatic than those for 50N-infected cells versus WT- or 50NR-infected cells, they were still highly significant (P = 0.0025) (Fig. 7C). These data suggest that a subpopulation of UL97 is recruited to the nuclear lamina, dependent on UL50 and, to a lesser extent, UL53 in infected cells.
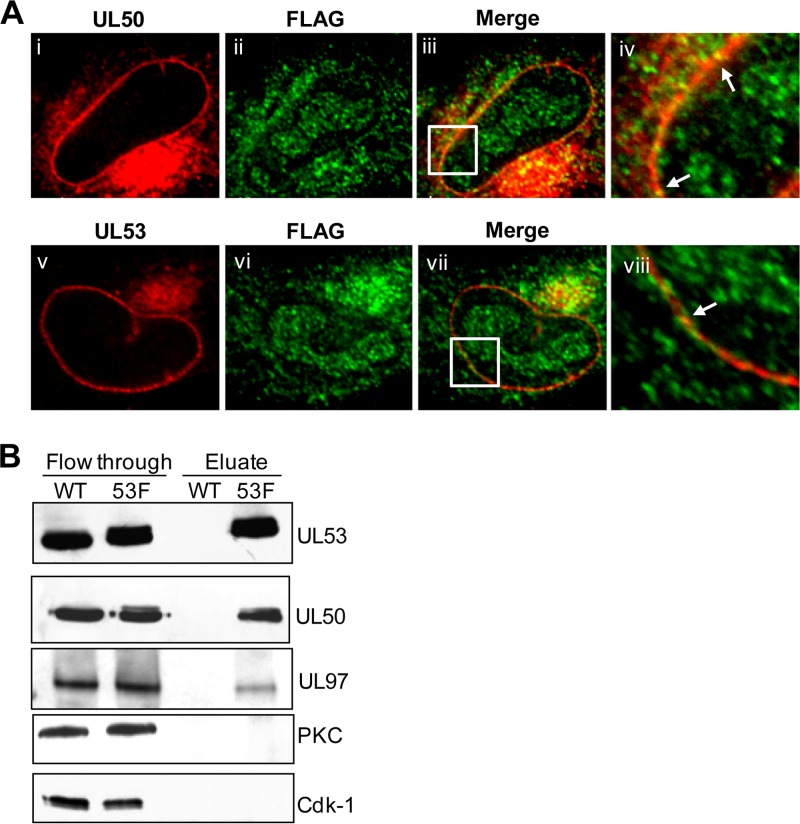
We also investigated whether UL97 colocalizes with UL50 and UL53 in infected cells. We observed foci of colocalization of a subpopulation of UL97 with both UL50 (Fig. 8A, panel iii) and UL53 (Fig. 8A, panel vii) at the nuclear rim in cells infected with FLAG-UL97 AD169-RV. Thus, localization of UL97 to the nuclear rim is associated with colocalization with UL50 and UL53.
FIG 8.
Association of UL97 with UL50 and UL53. (A) HFFs were infected with FLAG-UL97 BADGFP at an MOI of 1. At 72 h p.i., cells were fixed and stained for FLAG (green) and either UL50 (red) (i to iv) or UL53 (red) (v to viii). The samples were visualized using confocal microscopy. The insets indicate magnified (×3) sections of the respective images. Arrows show areas of colocalization. (B) Nuclear lysates were obtained at 72 h p.i. from HFF cells infected with HCMV AD169-RV (WT) or UL53-FLAG AD169-RV (53F) at an MOI of 1. Lysates were precleared and incubated with anti-FLAG M2 monoclonal antibody-conjugated agarose beads. Bound proteins were eluted using low pH and analyzed by Western blotting using antibodies against the proteins indicated to the right of the panel.
UL97, but neither PKC nor Cdk-1, detectably coimmunoprecipitates with HCMV UL50-UL53.
We next investigated whether the localization of UL97 to the nuclear rim and its colocalization with UL50 and UL53 correlated with a physical association of the UL50 and UL53 complex with UL97. To this end, we used the UL53-FLAG AD169-RV virus (53-F) in immunoprecipitation experiments. Nuclear extracts from HFF cells infected with either 53-F or the untagged WT virus were immunoprecipitated using an antibody recognizing FLAG bound to beads. Immunoprecipitated proteins were eluted from beads by using low pH, and the eluate and unbound proteins (flowthrough) were separated by SDS-PAGE and examined by Western blotting using antibodies against UL50, UL53, UL97, PKC, and Cdk-1 (Fig. 8B). Immunoreactive bands corresponding to UL50 and UL53-FLAG were detected in eluates from 53-F-infected cells but not the WT virus-infected cells. UL97 protein kinase was also detected in the eluate from cells infected with the tagged but not untagged virus. Neither Cdk-1 nor PKC was found in either eluate, although both were readily detected in the unbound fraction (Fig. 8B). For PKC detection, we again used antisera recognizing all PKC isoforms (30), as well as the specific anti-PKC-α and anti-PKC-δ antibodies described above for the immunofluorescence studies (Fig. 8B and data not shown). Thus, these results are consistent with our immunofluorescence data, suggesting that the UL50-UL53 complex preferentially associates with the viral kinase UL97 rather than with PKC or Cdk-1.
DISCUSSION
HCMV UL50 and UL53 are homologs of proteins from other herpesviruses that form a complex, the NEC, involved in nuclear egress (reviewed in references 3 and 55). Despite the medical importance of HCMV, whether UL50 and UL53 actually function during nuclear egress in HCMV-infected cells has not been reported previously. In this study, using null mutant viruses and CLEM, we showed that these two proteins are indeed crucial for efficient nuclear egress during infection, and thus deserve the name HCMV NEC. We further demonstrated that UL50 is required for localization of UL53 to the nuclear rim and that both UL50 and UL53 are required for disruption of the nuclear lamina. We then found that a subpopulation of the viral protein kinase, UL97, colocalizes with the nuclear lamina, dependent on UL50 and, to a lesser extent, UL53, and associates with the NEC in immunofluorescence and coimmunoprecipitation experiments. UL97 is required for proper phosphorylation of lamin A/C, disruption of the nuclear lamina, and efficient nuclear egress in infected cells (36, 46, 47, 50). Thus, our results support a model in which one function of the HCMV NEC is to recruit UL97 to the nuclear rim, where it phosphorylates lamin A/C to disrupt the nuclear lamina so that viral nucleocapsids can gain access to the inner nuclear membrane for budding into the perinuclear space.
We were initially surprised to find no evidence for recruitment of PKC to the nuclear lamina during HCMV infection given reports demonstrating such recruitment during nuclear egress of MCMV and HSV-1 (29, 30) and studies in support of such recruitment for lamina disruption by the HCMV NEC (29, 32, 33, 50). Reviewing the latter studies, an early report found that a PKC inhibitor present throughout infection reduced incorporation of phosphate into lamins during HCMV infection (29). However, that inhibitor is cytotoxic (data not shown) and/or could have affected UL97 expression or activity. In a second study, yeast two-hybrid assays detected interactions of PKC isoforms ε and ζ with UL50, but puzzlingly, coimmunoprecipitation and immunofluorescence assays of transfected cells showed associations with an overexpressed GFP fusion of a different PKC isoform, PKC-α (33). Regardless, these studies did not examine HCMV-infected cells. A third study did report a coimmunoprecipitation experiment (using a PKC-α monoclonal antibody) with HCMV-infected cells that detected low levels of an immunoreactive species roughly the size of UL53 in the immunoprecipitate (32). However, this study did not document the presence of PKC in the immunoprecipitate or the use of a standard isotype control and did not report whether UL50 was present (i.e., whether any association was with the NEC rather than UL53 alone) (32). A fourth study suggested an important role for PKC in infected cells based on residual levels of lamina disruption in some cells infected with a UL97 mutant HCMV that appeared to be reduced further by treatment with an inhibitor with activity against both UL97 and PKC (50). However, this compound inhibits serine-threonine kinases generally (56), and the quantitative comparison was performed at 3 weeks postinfection; lacked a control of mock-infected cells, some of which exhibit lamina disruption (data not shown); and was not subjected to a statistical analysis. Thus, although these previous results were all consistent with the possibility that the HCMV NEC could recruit PKC for lamin phosphorylation, they neither demonstrated such recruitment in infected cells nor showed that PKC was important for disruption of the nuclear lamina during HCMV infection.
Furthermore, we found no evidence for the recruitment of Cdk-1 to the nuclear lamina during HCMV infection. Our failure to detect recruitment of PKC or Cdk-1 is consistent with our observations here of minimal effects of HCMV infection on the thickness of lamin B staining and with previous results, including the failure to detect phosphorylation of lamin A/C on sites phosphorylated by PKC (36), the failure of a Cdk inhibitor to decrease lamin A/C phosphorylation in HCMV-infected cells (36), and the failure of a dominant negative Cdk-1 to decrease HCMV replication (57). We note that although phosphorylation of specific residues on lamins—most notably serine 22 of lamin A/C (which is phosphorylated by UL97 [36])—is sufficient for lamina depolymerization (51), there is evidence that PKC phosphorylation of lamins is not (27, 28). We also note that the anti-PKC antibody that we used detects all PKC isoforms (30) and that we readily detected PKC recruitment to the nuclear lamina in HSV-infected cells by using the same antibody. In summary, we conclude that the HCMV NEC mainly recruits and interacts with UL97, not PKC, during infection.
Requirement for HCMV NEC during nuclear egress as revealed by CLEM.
To our knowledge, CLEM has not previously been used to study cells infected with a nonviable virus or, for that matter, any virus, although it has been used in transfected cells to study activities of RNA virus proteins (58, 59). By using CLEM to analyze null mutants, we avoided using conditional knockdown of viral protein expression or dominant negative mutants, with their potential drawbacks of leakiness or activities that would not occur with null mutants. In this regard, it is interesting that the UL53 null mutant did not exhibit any obvious overrepresentation of capsids lacking DNA, in contrast to dominant negative MCMV M53 mutants (60). On the other hand, HSV and EBV mutants null for the UL53 homolog also showed overrepresentation of empty capsids (9, 61). Thus, in contrast to what is seen with these other herpesviruses, HCMV UL53 does not appear to be crucial for genome packaging, although we cannot exclude a subtler role. Although using CLEM to analyze null mutants has important limitations, particularly in terms of the number of cells that can readily be examined, it should be valuable for characterization of other nonviable mutant viruses.
Specific roles of UL50 and UL53 in disruption of nuclear lamina.
The UL50-dependent colocalization of UL97 with the nuclear lamina suggests that UL50's role in lamina disruption might simply be to recruit UL97. This would be consistent with a report that coexpression of UL50 and UL97 following transfection resulted in increased numbers of cells with altered nuclear lamina relative to cells in which UL97 was transfected alone (32), although in this previous analysis, lamina alterations were not quantified in vector-transfected cells or cells expressing UL50 alone. Perhaps UL50 and UL97 interact directly. However, such an interaction was not detected in coimmunoprecipitates by use of a virus expressing affinity-tagged UL97 (41) or in transient-transfection and yeast two-hybrid studies (33). UL50 and UL97 were found to interact with the host protein p32 in yeast two-hybrid assays, leading to a model in which UL50 recruits UL97 via p32 (32, 33). However, p32 is notorious for promiscuously interacting with many diverse proteins (62, 63). The precise mechanism of recruitment of UL97 to the nuclear lamina deserves further study.
While UL53 was not strongly required for UL97 colocalization with the nuclear lamina, its absence modestly but significantly decreased such colocalization. It is possible that this modest effect nevertheless explains the role of UL53 in lamina disruption. It is also possible that UL53 somehow positively regulates the function of UL97. Regardless, the roles of the HCMV NEC subunits during nuclear egress, including recruitment of UL97 to the nuclear lamina, appear to have no cellular substitutes.
Modifications to the nuclear lamina in the absence of UL97.
Expression of UL50 and UL53, like that of their counterparts in HSV-1, PrV, Kaposi's sarcoma-associated herpesvirus (KSHV), and MCMV, in the absence of any other herpesvirus protein, induces modifications to the nuclear lamina in transfected cells (7, 21, 29, 35, 64), while in HCMV-infected cells, in the absence of UL97 activity, there are no visible disruptions of the nuclear lamina above those seen in mock-infected cells (36, 47). Thus, UL50 and UL53 are sufficient for modification of the nuclear lamina in transfected cells, whereas they are necessary but not sufficient in infected cells. We caution that the modifications to the nuclear lamina in transfected cells (7, 21, 29, 35) may differ qualitatively and quantitatively from those in infected cells. Alternatively, overexpression of the HCMV NEC might drive an otherwise inefficient process in transfected cells. This process might also operate in HCMV-infected cells when UL97 is inactivated, as nuclear egress and viral replication do continue, albeit inefficiently. Although this process might conceivably involve PKCs, Cdk-1 may be a better candidate for modification of the lamina in these settings. Consistent with this idea, UL97-independent increases in phosphorylation of lamin A/C on serine 22 have been detected in HCMV-infected cells, which may help to explain low-level nuclear egress in the absence of UL97 (36, 47). In addition, we found that a Cdk inhibitor reduced HCMV-induced deformation of the nucleus, which is an indirect indication of nuclear lamina modification, while a pan-PKC inhibitor had little, if any, effect (unpublished results).
PKC and nuclear egress in other systems.
At the risk of being provocative, our failure to find evidence for recruitment of PKC to the nuclear lamina during HCMV infection led us to consider the evidence for NEC-dependent recruitment of PKC in cells infected with HSV-1 and MCMV. In both cases, clear recruitment of certain PKCs to the nuclear rim during infection has been shown (29, 30), with strong evidence for NEC-dependent recruitment of PKC during HSV-1 infection (30). To our knowledge, no studies (e.g., using conditional dominant negative mutants) have examined whether the NEC is required for recruitment of PKC in MCMV-infected cells. However, there is evidence linking PKCs to nuclear egress, especially for HSV-1 (30, 65), and, more recently, to export of ribonucleoprotein (RNP) complexes from muscle cells during synaptic development in Drosophila (31). Interestingly, in the Drosophila system, alterations in the nuclear lamina do not appear to entail gaps like those seen in HSV-1- and HCMV-infected cells (21, 36, 50, 66–68) but, rather, remodeling of the lamina so that it surrounds the RNPs (31). Thus, important differences can be discerned among various systems of nuclear egress. We speculate that despite the sequence and functional conservation of the HSV-1 and HCMV NECs, the HSV-1 NEC evolved to recruit one or more PKCs, while the HCMV NEC evolved to recruit the viral protein kinase UL97 for lamina disruption during nuclear egress.
ACKNOWLEDGMENTS
We thank Jean Pesola for help with statistical analysis, Kirsi Hellström and Tero Ahola for advice in designing the CLEM experiments, Maria Ericsson of the Harvard Medical School Electron Microscope Facility for suggesting the use of CLEM, Marty Fernandez and Brian Bender for assistance, Tim Mitchison for support and encouragement, and Tim Mitchison, Blair Strang, and Adrian Wilkie for critical readings of the manuscript. We are also grateful for the assistance of staff and the availability of equipment at the Nikon Imaging Center at Harvard Medical School for acquisition and analysis of immunofluorescence data and at the EM Core Facility for acquisition and analysis of EM data.
This work was supported by NIH grants RO1 AI026077 (to D.M.C.), F32 AI075766 (to J.P.K.), and P01 CA139980 and R01 GM039565 (to Tim Mitchison).
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Mettenleiter TC, Klupp BG, Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423–429. 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Minson T. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610–1611. 10.1128/JVI.80.3.1610-1612.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547. 10.1128/JVI.76.4.1537-1547.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnee M, Ruzsics Z, Bubeck A, Koszinowski UH. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80:11658–11666. 10.1128/JVI.01662-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378. 10.1128/JVI.76.1.364-378.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073. 10.1128/JVI.74.21.10063-10073.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241–7246. 10.1073/pnas.0701757104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye GJ, Roizman B. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. U. S. A. 97:11002–11007. 10.1073/pnas.97.20.11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YE, Van Sant C, Krug PW, Sears AE, Roizman B. 1997. The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117–129. 10.1128/JVI.74.1.117-129.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katlama C. 1993. Cytomegalovirus infection in acquired immune-deficiency syndrome. J. Med. Virol. 1993(Suppl 1):128–133 [DOI] [PubMed] [Google Scholar]
- 12.Winston DJ, Ho WG, Champlin RE. 1990. Cytomegalovirus infections after allogeneic bone marrow transplantation. Rev. Infect. Dis. 12(Suppl 7):S776–S792 [DOI] [PubMed] [Google Scholar]
- 13.Pultoo A, Jankee H, Meetoo G, Pyndiah MN, Khittoo G. 2000. Detection of cytomegalovirus in urine of hearing-impaired and mentally retarded children by PCR and cell culture. J. Commun. Dis. 32:101–108 [PubMed] [Google Scholar]
- 14.Tokumoto JI, Hollander H. 1993. Cytomegalovirus polyradiculopathy caused by a ganciclovir-resistant strain. Clin. Infect. Dis. 17:854–856. 10.1093/clinids/17.5.854 [DOI] [PubMed] [Google Scholar]
- 15.Erice A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biron KK. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 71:154–163. 10.1016/j.antiviral.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 17.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401. 10.1073/pnas.1635160100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228. 10.1073/pnas.2334032100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popa M, Ruzsics Z, Lotzerich M, Dolken L, Buser C, Walther P, Koszinowski UH. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J. Virol. 84:9035–9046. 10.1128/JVI.00681-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupp B, Ruzsics Z, Buser C, Adler B, Walther P, Koszinowski UH. 2007. Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50. J. Virol. 81:5508–5517. 10.1128/JVI.02796-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds AE, Liang L, Baines JD. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78:5564–5575. 10.1128/JVI.78.11.5564-5575.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mou F, Wills E, Baines JD. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83:5181–5191. 10.1128/JVI.00090-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellor H, Parker PJ. 1998. The extended protein kinase C superfamily. Biochem. J. 332:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson LJ, Fields AP. 1996. bII protein kinase C is required for the G2/M phase transition of cell cycle. J. Biol. Chem. 271:15045–15053. 10.1074/jbc.271.25.15045 [DOI] [PubMed] [Google Scholar]
- 25.Goss VL, Hocevar BA, Thompson LJ, Stratton CA, Burns DJ, Fields AP. 1994. Identification of nuclear bII protein kinase C as a mitotic lamin kinase. J. Biol. Chem. 269:19074–19080 [PubMed] [Google Scholar]
- 26.Collas P, Thompson L, Fields AP, Poccia DL, Courvalin JC. 1997. Protein kinase C-mediated interphase lamin B phosphorylation and solubilization. J. Biol. Chem. 272:21274–21280. 10.1074/jbc.272.34.21274 [DOI] [PubMed] [Google Scholar]
- 27.Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. 1990. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61:591–602. 10.1016/0092-8674(90)90471-P [DOI] [PubMed] [Google Scholar]
- 28.Eggert M, Radomski N, Linder D, Tripier D, Traub P, Jost E. 1993. Identification of novel phosphorylation sites in murine A-type lamins. Eur. J. Biochem. 213:659–671. 10.1111/j.1432-1033.1993.tb17806.x [DOI] [PubMed] [Google Scholar]
- 29.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857. 10.1126/science.1071506 [DOI] [PubMed] [Google Scholar]
- 30.Park R, Baines JD. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494–504. 10.1128/JVI.80.1.494-504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, Moore MJ, Budnik V. 2012. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149:832–846. 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milbradt J, Auerochs S, Sticht H, Marschall M. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90:579–590. 10.1099/vir.0.005231-0 [DOI] [PubMed] [Google Scholar]
- 33.Milbradt J, Auerochs S, Marschall M. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88:2642–2650. 10.1099/vir.0.82924-0 [DOI] [PubMed] [Google Scholar]
- 34.Marschall M, Marzi A, aus dem Siepen P, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357–33367. 10.1074/jbc.M502672200 [DOI] [PubMed] [Google Scholar]
- 35.Camozzi D, Pignatelli S, Valvo C, Lattanzi G, Capanni C, Dal Monte P, Landini MP. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89:731–740. 10.1099/vir.0.83377-0 [DOI] [PubMed] [Google Scholar]
- 36.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. 10.1371/journal.ppat.1000275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strang BL, Bender BJ, Sharma M, Pesola JM, Sanders RL, Spector DH, Coen DM. 2012. A mutation deleting sequences encoding the amino terminus of human cytomegalovirus UL84 impairs interaction with UL44 and capsid localization. J. Virol. 86:11066–11077. 10.1128/JVI.01379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 39.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30 [DOI] [PubMed] [Google Scholar]
- 40.Baldick CJ, Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamil JP, Coen DM. 2007. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 81:10659–10668. 10.1128/JVI.00497-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729. 10.1128/JVI.74.17.7720-7729.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carleman E, Villiger W, Actaraub JD, Kellenberger E. 1986. Low temperature embedding, p 24–59 In Müller M, Becker RP, Boyde A, Wolsewick JJ. (ed), Science of biological specimen preparation. AMF O'Hare, Chicago, IL [Google Scholar]
- 44.Sam MD, Evans BT, Coen DM, Hogle JM. 2009. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83:2996–3006. 10.1128/JVI.02441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vicidomini G, Gagliani MC, Cortese K, Krieger J, Buescher P, Bianchini P, Boccacci P, Tacchetti C, Diaspro A. 2010. A novel approach for correlative light electron microscopy analysis. Microsc. Res. Tech. 73:215–224. 10.1002/jemt.20777 [DOI] [PubMed] [Google Scholar]
- 46.Krosky PM, Baek MC, Coen DM. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905–914. 10.1128/JVI.77.2.905-914.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reim NI, Kamil JP, Wang D, Lin A, Sharma M, Ericsson M, Pesola JM, Golan DE, Coen DM. 2013. Inactivation of retinoblastoma protein does not overcome the requirement for human cytomegalovirus UL97 in lamina disruption and nuclear egress. J. Virol. 87:5019–5027. 10.1128/JVI.00007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dal Monte P, Pignatelli S, Zini N, Maraldi NM, Perret E, Prevost MC, Landini MP. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005–1012 [DOI] [PubMed] [Google Scholar]
- 49.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986. 10.1128/JVI.74.2.975-986.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milbradt J, Webel R, Auerochs S, Sticht H, Marschall M. 2010. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285:13979–13989. 10.1074/jbc.M109.063628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heald R, McKeon F. 1990. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61:579–589. 10.1016/0092-8674(90)90470-Y [DOI] [PubMed] [Google Scholar]
- 52.Ward GE, Kirschner MW. 1990. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61:561–577. 10.1016/0092-8674(90)90469-U [DOI] [PubMed] [Google Scholar]
- 53.Jault FM, Jault JM, Ruchti F, Fortunato EA, Clark C, Corbeil J, Richman DD, Spector DH. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez V, McElroy AK, Spector DH. 2003. Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J. Virol. 77:13214–13224. 10.1128/JVI.77.24.13214-13224.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee CP, Chen MR. 2010. Escape of herpesviruses from the nucleus. Rev. Med. Virol. 20:214–230. 10.1002/rmv.643 [DOI] [PubMed] [Google Scholar]
- 56.Marschall M, Stein-Gerlach M, Freitag M, Kupfer R, van Den Bogaard M, Stamminger T. 2001. Inhibitors of human cytomegalovirus replication drastically reduce the activity of the viral protein kinase pUL97. J. Gen. Virol. 82:1439–1450 [DOI] [PubMed] [Google Scholar]
- 57.Hertel L, Chou S, Mocarski ES. 2007. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 3:e6. 10.1371/journal.ppat.0030006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spuul P, Balistreri G, Hellstrom K, Golubtsov AV, Jokitalo E, Ahola T. 2011. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 85:4739–4751. 10.1128/JVI.00085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polishchuk RS, Polishchuk EV, Marra P, Alberti S, Buccione R, Luini A, Mironov AA. 2000. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J. Cell Biol. 148:45–58. 10.1083/jcb.148.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pogoda M, Bosse JB, Wagner FM, Schauflinger M, Walther P, Koszinowski UH, Ruzsics Z. 2012. Characterization of conserved region 2-deficient mutants of the cytomegalovirus egress protein pM53. J. Virol. 86:12512–12524. 10.1128/JVI.00471-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granato M, Feederle R, Farina A, Gonnella R, Santarelli R, Hub B, Faggioni A, Delecluse HJ. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82:4042–4051. 10.1128/JVI.02436-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363–24370. 10.1074/jbc.272.39.24363 [DOI] [PubMed] [Google Scholar]
- 63.van Leeuwen HC, O'Hare P. 2001. Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J. Cell Sci. 114:2115–2123 [DOI] [PubMed] [Google Scholar]
- 64.Desai PJ, Pryce EN, Henson BW, Luitweiler EM, Cothran J. 2012. Reconstitution of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 86:594–598. 10.1128/JVI.05988-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leach NR, Roller RJ. 2010. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology 406:127–137. 10.1016/j.virol.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bjerke SL, Roller RJ. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261–276. 10.1016/j.virol.2005.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simpson-Holley M, Baines J, Roller R, Knipe DM. 2004. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591–5600. 10.1128/JVI.78.11.5591-5600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buser C, Walther P, Mertens T, Michel D. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042–3048. 10.1128/JVI.01564-06 [DOI] [PMC free article] [PubMed] [Google Scholar]