Abstract
MLN4924 is a first-in-class cancer drug that inhibits the Nedd8-activating enzyme (NAE). Herein, we report that MLN4924 inhibits Vpx/Vpr-induced SAMHD1 degradation by inhibiting the neddylation of E3 ubiquitin ligase and blocks macaque simian immunodeficiency virus (SIVmac) replication in myeloid cells. SAMHD1 is required for MLN4924-mediated SIVmac inhibition. Our findings indicate the potential efficacy of inhibiting neddylation as an antiretroviral strategy and identify the readily available anticancer drug MLN4924 as a candidate agent for that purpose.
TEXT
Cullin-Ring E3 ubiquitin ligases (CRLs), the most prominent class of E3 ubiquitin ligases, are essential for the turnover of a large number of proteins in mammalian cells and are critical for maintaining cellular protein homeostasis (1). The ubiqutin-like protein NEDD8 has a dedicated E1-activating enzyme (NAE) and E2-conjugating enzymes (UBE2M and UBE2F), and it plays an important role in the enzymatic activity of the CRL E3 ligase family through direct conjugation to the Cullin scaffold. MLN4924, a novel small-molecule NAE inhibitor (2), has entered phase 1 trials and has demonstrated significant therapeutic benefit in cancer therapy (Fig. 1A). Preclinical studies in a range of cancer models have demonstrated that MLN4924 inhibits diverse CRL E3 functions in cancer cells and causes an accumulation of their substrates, thus inducing cell cycle arrest and apoptosis (2).
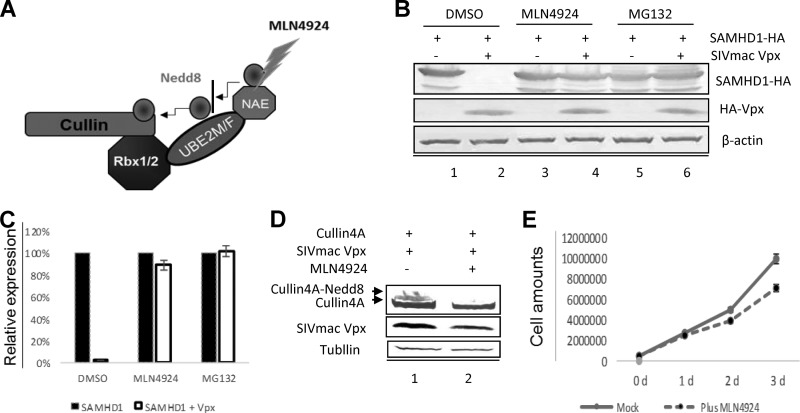
FIG 1.
MLN4924 inhibits Vpx-induced SAMHD1 degradation by blocking neddylation activation of Vpx-CRL4(DCAF1) E3 ligase. (A) Schematic diagram indicating where MLN4924 inhibits NAE at the initial neddylation step. (B) Effects of MLN4924 on Vpx-mediated degradation of SAMHD1. HEK293T cells were co-transfected with pSAMHD1-HA plus a control vector or Vpx-HFA expression vector. At 36 h after transfection, the cells were DMSO-treated or treated with MG132/MLN4924 for 24 h. Cell lysates were harvested and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA, with β-actin as the loading control. (C) Effects of MLN4924 on relative expression of SAMHD1 (100%) with or without Vpx expression. Error bars represent the standard deviation from three experiments. (D) MLN4924 inhibits the neddylation of Cullin4A in the presence of SIV Vpx. HEK293T cells were first transfected with pMyc-Cullin4A and pHA-SIVmac Vpx, and 24 h later, cells were treated with DMSO or MLN4924, respectively, for 24 h. The cells were then harvested for immunoblot analysis with the indicated antibodies. (E) MLN4924 treatment was feeble cytotoxic to the HEK293T cells. HEK293T cells were treated with MLN4924, or with DMSO as a negative control. Viable cells were counted daily from Day 0 to Day 3.
The HIV-1 and HIV-2 infections that have led to a worldwide epidemic remain without a cure or broadly effective prophylaxis. In recent years, studies of host-virus interactions have revealed the presence of naturally occurring restriction factors that could potentially inhibit the replication of HIV and the closely related simian immunodeficiency virus (SIV). HIV has overcome the potential effects of these restriction factors by expressing virally encoded accessory proteins, such as Vif, Vpu, and Vpx, which interact, respectively, with the restriction factors APOBEC3, tetherin, and the recently identified SAMHD1 (3–8). Interestingly, all these viral proteins load their targets onto hijacked CRL E3 ligases for ubiquitination-dependent degradation. Drugs and mutations of viral proteins restore the accumulation of these restriction factors and dramatically inhibit viral infection and replication (3–6, 9). Because of the role of neddylation in the enzymatic activity of the CRL E3 ligases, it is important to understand what role this process plays in the degradation of CRL ligases that have been hijacked by viral proteins. A better understanding of this role might lead to the identification of new targets for antiretroviral drug and vaccine development.
MLN4924 impairs the macaque SIVmac239 Vpx-mediated degradation of SAMHD1.
The CRL4(DCAF1) E3 ligase has been shown to be essential for DNA replication, cell cycle regulation, and embryonic development (10–12). Its ubiquitination capacity was found to be usurped by different proteins of diverse viruses (13, 14). So far, there is little direct proof for the importance of neddylation to cellular and virally hijacked CRL4(DCAF1). Therefore, we examined the ability of MLN4924 to inhibit the neddylation of the Vpx-CRL4(DCAF1) E3 complex (Fig. 1A).
ln monocytes, lentiviral Vpx hijacks CRL4(DCAF1) E3 and mediates the proteasome-dependent degradation of SAMHD1, thereby permitting viral replication (9, 15–19). However, the cellular regulation of this SAMHD1 degradation is still unclear, and even the evidence for the neddylation of Cullin4 is not straightforward. To evaluate the role of neddylation in the function of the Vpx-CRL4(DCAF1) E3 ligase complex, we tested the effects of MLN4924 on Vpx-mediated SAMHD1 degradation. HEK293T cells were transfected with pSAMHD1-HA and/or pHA-Vpxmac239. Twenty-four hours later, 300 nM MLN4924 was added to the cell culture. After another 24 h, the cells were harvested for immunoblotting with an antihemagglutinin (anti-HA) antibody to detect HA-tagged SAMHD1. The results showed that MLN4924 strongly impaired the ability of Vpx to induce SAMHD1 degradation; it functioned as well as the specific proteasome inhibitor MG132 (Fig. 1B). Further analysis showed that MLN4924 restored over 60% of the expression of SAMHD1 in the presence of Vpx (Fig. 1C).
Vpr and Vpx recruit the Cullin4A-containing E3 ubiquitin ligase to promote G2 arrest and myeloid cell infection (13, 17, 18, 20–25). Vpr but not Vpx has been observed to enhance the neddylation of Cullin4A (13, 18, 22). To evaluate the effect of MLN4924 on the Cullin4A nedd8 modification in the presence of Vpx, we exposed HEK293T cells transiently transfected with HA-Vpx or Myc-Cullin4A to MLN4924. Immunoblotting revealed a drastic reduction in neddylated Cullin4A after MLN4924 treatment, compared to dimethyl sulfoxide (DMSO)-treated cells (Fig. 1D), suggesting that MLN4924 inhibits Vpx-mediated degradation of SAMHD1 by interfering with the neddylation of Cullin4A. Treatment with MLN4924 did not affect the proliferation of HEK293T cells (Fig. 1E), indicating that it did not cause significant cytotoxicity under our experimental conditions. These data indicated that the pharmacological NAE inhibitor MLN4924, which is already being used in human clinical trials, dramatically reduced Vpx-downregulated SAMHD1 by directly regulating Vpx-CRL4(DCAF1) E3 function.
MLN4924 inhibits Vpx-dependent SIVmac239 infection in macrophages.
By overcoming the restriction imposed by SAMHD1, the virion-packaged Vpx protein plays a critical role in the early phase of the viral replication cycle, enhancing the lentiviral infection of macrophages (5, 6, 25–28). We therefore investigated the role of MLN4924 in Vpx-dependent viral infection in phorbol 12-myristate 13-acetate (PMA)-treated macrophage-like THP-1 cells. The THP-1 cells were cultured in 100 nM PMA for 48 h to promote differentiation and then treated with 300 nM MLN4924 or DMSO for 12 h. The treated cells were then infected with the same viral titer of the G protein of vesicular stomatitis virus (VSV-G)-pseudotyped SIVmac-GFP or SIVmacΔVpx-GFP virus-like particles (VLPs). Compared to control samples, MLN4924-treated cells showed significantly reduced viral infection with SIVmac-GFP; the ratio of green fluorescent protein (GFP)-positive cells decreased by more than 80% (Fig. 2A). As expected, SIVmacΔVpx-GFP infection was inhibited in these cells (Fig. 2A). SIVmac-GFP virus infection of THP-1 cells resulted in SAMHD1 degradation (Fig. 2B, lane 3), which was blocked by MLN4924 treatment (Fig. 2B, lane 4).
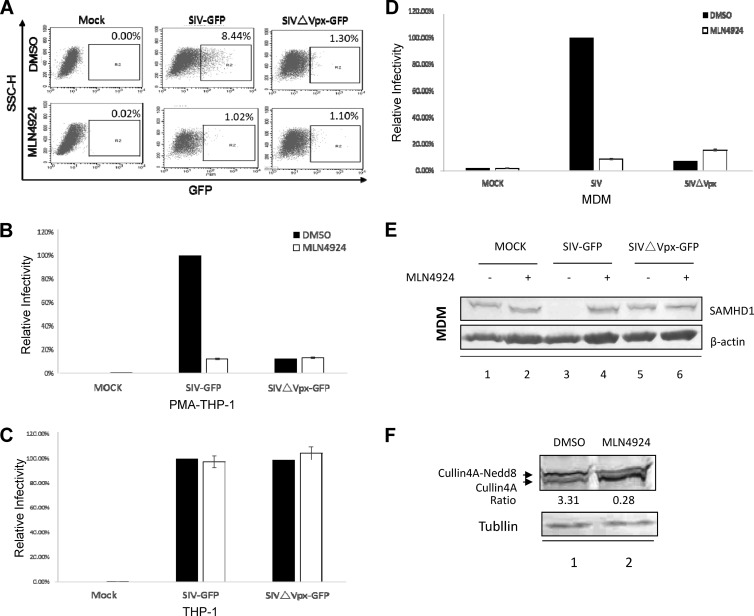
FIG 2.
Inhibition of SIV infection in macrophages by MLN4924. (A) MLN4924 is a highly effective inhibitor of Vpx-dependent SIVmac infection in PMA-treated THP-1 cells. Human monocytic THP-1 cells were stimulated with PMA (100 ng/ml for 48 h) and then treated with 300 nM MLN4924 or DMSO for 12 h. The two groups of treated cells were then infected with an equivalent amount of SIVmac239-GFP or SIVmacΔVpxGFP VLPs. The percentage of GFP-positive cells was determined by flow cytometry. Representative flow cytometry data are shown. (B) The rate of SIVmac239-GFP virus infection in the absence of MLN4924 was set to 100%. The bar charts are representative of three independent experiments, and the error bars indicate the standard deviations of three replicates within one experiment. (C) MLN4924 treatment had little effect on SIVmac infection in undifferentiated THP-1 cells. (D) MLN4924 is a highly effective inhibitor of SIVmac infection in primary macrophages. Primary macrophages were isolated and infected with SIVmac or SIVmacΔVpx viruses as previously described (26). Virus infection was determined by monitoring GFP-positive cells 2 days after infection. (E) Effect of MLN4924 treatment on SAMHD1 expression during SIVmac infection of primary macrophages. Cell extracts were harvested 2 days after infection and analyzed by SDS-PAGE, followed by immunoblotting with anti-SAMHD1 antibody to detect SAMHD1. β-Actin was used as the loading control. (F) Effect of MLN4924 treatment on Cullin4A expression. Endogenous Cullin4A from SIVmac virus-infected U937/SAMHD1 cells treated with MLN4924 or DMSO was detected with an anti-Cullin4A antibody from Abcam. Tubulin was used as the loading control.
It has been reported that undifferentiated THP-1 cells cannot confer resistance to SIVmacΔVpx infection. We observed that infection with SIVmacΔVpx-GFP was similar to that with SIVmac-GFP in undifferentiated THP-1 cells (Fig. 2C). Similarly, MLN4924 did not manifest any antiviral activity in undifferentiated THP-1 cells (Fig. 2C). On the other hand, MLN4924 treatment strongly inhibited the infection of primary macrophages with SIVmac-GFP (Fig. 2D). These primary macrophages were refractory to SIVmacΔVpx-GFP infection (Fig. 2D). Thus, we demonstrated that a first-in-class cancer drug targeting the nedd8 modification pathway efficiently blocked Vpx-dependent SIV replication in macrophages.
SIVmac-GFP virus infection resulted in the depletion of SAMHD1 in primary macrophages (Fig. 2E, lane 3), and this effect was blocked by MLN4924 treatment (lane 4). The impaired depletion of SAMHD1 from MLN4924 -treated macrophages during SIVmac-GFP virus infection was also associated with a reduction in Cullin4A nedd8 modification (Fig. 2F, lane 2) compared to untreated cells (lane 1).
MLN4924-mediated SIV inhibition requires SAMHD1.
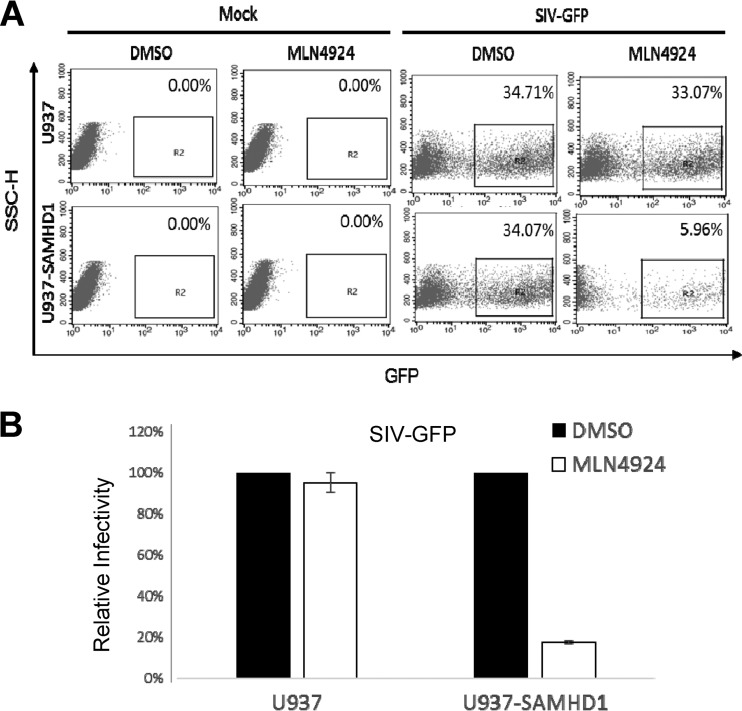
We next examined whether SAMHD1 was required for MLN4924-mediated SIV inhibition. U937 cells do not normally express detectable SAMHD1. We therefore established SAMHD1-expressing U937 cells (U937/SAMHD1) by using lentiviral vectors expressing SAMHD1. In control U937 cells, MLN4924 treatment had no significant effect on SIVmac-GFP virus infection (Fig. 3A). However, in SAMHD1-positive U937 cells, MLN4924 significantly inhibited infection with the SIVmac-GFP virus (Fig. 3A). In repeated experiments, MLN4924 only inhibited SIVmac-GFP virus infection of U937/SAMHD1 cells, and not U937 cells (Fig. 3B). SIVmac-GFP virus infection resulted in the depletion of SAMHD1 in U937/SAMHD1 cells (data not shown), and this effect was blocked by MLN4924 treatment (data not shown).
FIG 3.
SAMHD1 is required for MLN4924-mediated inhibition of SIVmac infection. (A) SAMHD1 was transduced into U937 cells by using lentiviral vectors, as previously described (35) to generate U937/SAMHD1 cells. U937 or U937/SAMHD1 cells were stimulated with PMA (100 ng/ml for 48 h) and then treated with 300 nM MLN4924 or DMSO for 12 h. The two groups of treated cells were infected with equivalent amounts of SIVmac239-GFP VLPs. The percentage of GFP-positive cells was determined by flow cytometry 2 days after infection. (B) The rate of SIVmac239-GFP virus infection in the absence of MLN4924 was set to 100%. The bar charts are representative of three independent experiments, and the error bars indicate the standard deviations of three replicates within one experiment.
MLN4924 is a broad inhibitor of Vpx/Vpr-mediated SAMHD1 degradation.
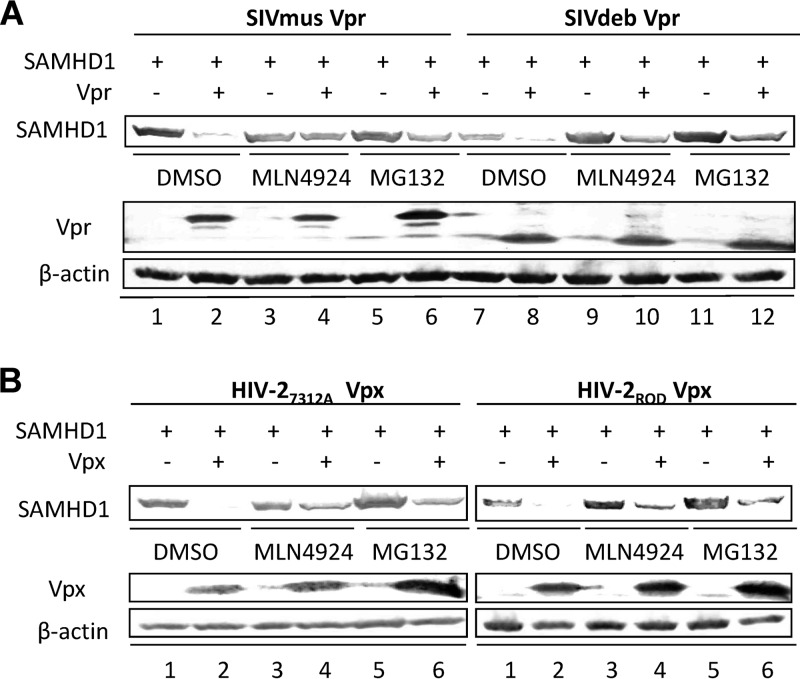
Recent studies of the coevolution of HIV accessory proteins and host resistance factors have revealed a complex balance between the ability of these interactions to protect the host and their ability to promote viral adaptation (29, 30). Unlike HIV-1 Vpr, the Vpr proteins of the ancient lentiviral strains SIVmus and SIVdeb, which predate SIV Vpx, have been shown to be able to induce SAMHD1 degradation, suggesting an evolutionary trajectory for the viral proteins Vpr and Vpx from closely related lentiviruses. To determine whether the neddylation-regulated CRL4(DCAF1) complex is required for the function of these ancient Vprs, we examined the SAMHD1 degradation efficiency of SIVmus and SIVdeb Vprs, with or without treatment with MLN4924. MLN4924 distinctly disrupted Vpr-induced degradation of SAMHD1 by both SIVmus and SIVdeb (Fig. 4A), at a level equivalent to that observed with control samples treated with MG132. These results indicated that the ancestral Vpr also utilizes the same modified ubiquitin-proteasome system to overcome the SAMHD1 protein. We also found that MLN4924 maintained a strong ability to inhibit the SAMHD1 degradation induced by HIV-2ROD and HIV-27312A Vpx (Fig. 4B). These results indicated that the activation of CRL4(DCAF1) E3 ligase by neddylation is pivotal for the function of an extended lineage of Vpx/Vpr proteins, which in all cases are susceptible to the ability of MLN4924 to interrupt upstream ubiquitin cascades.
FIG 4.
MLN4924 inhibits ancient SIV Vpr- and HIV-2 Vpx-mediated degradation of SAMHD1. (A and B) HEK293T cells were cotransfected with a SAMHD1-HA expression vector and Vpx or Vpr or their respective control vector. At 24 h after transfection, the cells were DMSO treated or treated with MG132/MLN4924 for 24 h. Expression levels of SAMHD1-HA and Vpx-HA were assessed by immunoblotting with an anti-HA antibody. β-Actin was used as the loading control.
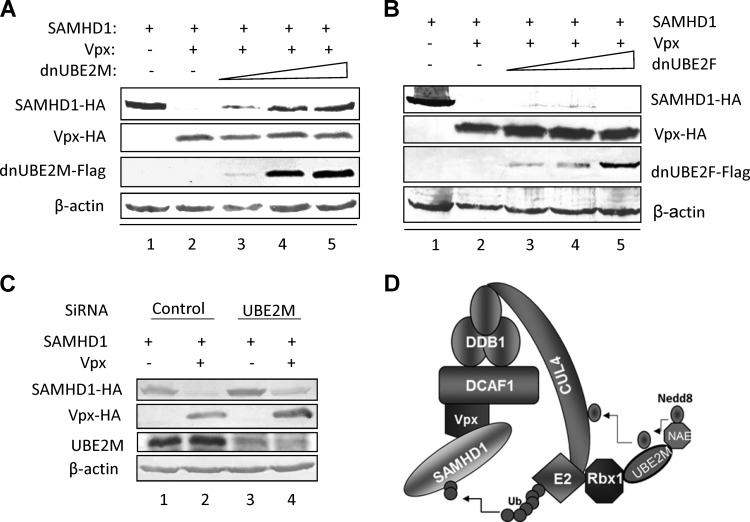
A dominant negative mutant of UBE2M disrupts Vpx function.
We next evaluated the role of neddylation in Vpx's ability to counteract the SAMHD1 protein. There are two well-known pathways for the neddylation system (31): Rbx1-UBE2M and Rbx2-UBE2F. Interestingly, we found that the expression of the UBE2M (Ubc12) dominant negative mutant (C111S) in HEK293T cells impaired Vpx-induced SAMHD1 degradation in a dose-dependent manner (Fig. 5A). However, the expression of the UBE2F dominant negative mutant (C116S) did not affect the function of Vpx (Fig. 5B). Silencing UBE2M with specific small interfering RNA targeted to the UBE2M coding regions also inhibited Vpx-mediated SAMHD1 degradation (Fig. 5C). We concluded that the Rbx1/UBE2M-dependent neddylation system is essential for the activation of the Vpx-CRL4(DCAF1) E3 ligase complex (Fig. 5D).
FIG 5.
UBE2M is essential for Vpx-mediated SAMHD1 degradation. (A) A UBE2M mutant inhibited Vpx-mediated SAMHD1 degradation. In HEK293T cells, SIVmac239 Vpx failed to induce SAMHD1 degradation in the presence of increasing amounts of UBE2M(C111S). Cell lysates were harvested and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA. β-Actin was used as the loading control. (B) The UBE2F mutant did not inhibit Vpx-mediated SAMHD1 degradation. (C) Silencing of UBE2M inhibited the Vpx-mediated degradation of SAMHD1. (D) Schematic diagram of Nedd8-activated Vpx-CRL4 E3 ligase to induce SAMHD1 degradation.
Activation of CRL E3 ligases by neddylation has been shown to change the conformation of the CRL complex and thus facilitate the movement of substrates closer to E2 for engagement by the ubiquitin cascade (32). During the first step in neddylation, NAE exhibits a high selectivity for its cognate Nedd8 and mediates a cascade that includes Nedd8 maturation and activation (32). In the present study, we established that the Nedd8-NAE-targeting cancer drug MLN4924 blocks Vpx-dependent viral replication in a macrophage cell line, indicating that host neddylation activity is also critical for the viral protein Vpx's hijacking of the CRL4 ubiquitin system. In addition, we demonstrated that a dominant negative mutant of UBE2M, but not of UBE2F, also disrupted Vpx function, further supporting the contention that the Rbx1-UBE2M Nedd8 modification pathway is critical for Vpx function and that cellular regulation of CRL4 function is also important for the Vpx-bound CRL4(DCAF1) E3 ligase (31). We also concluded that, although Vpx/Vpr has coevolved with host factors (both inhibitors and partners), the core strategy of maintaining the interaction with the cellular CRL4 E3 ligase to enhance early viral replication has been conserved.
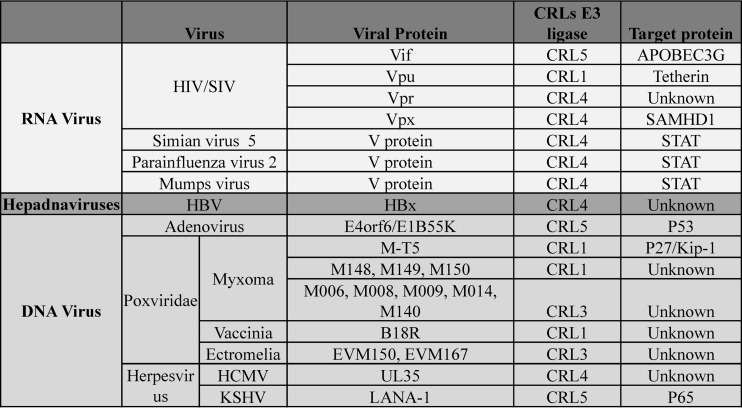
A key function of viral accessory proteins is to inactivate host restriction factors by binding with Cullin family protein-associated E3 ligases to induce degradation of the restriction factors. Here, we have demonstrated that MLN4924 affects the function of the Vpx-CRL4 E3 complex and Vpx-dependent viral replication in a macrophage cell line, suggesting the feasibility of using MLN4924 for antiviral therapy. Coincident with our findings, Stanley et al. recently reported that MLN4924 restores APOBEC3G accumulation by preventing Vif-mediated degradation (33). It was equally interesting to determine whether this first-in-class cancer drug could disrupt other viral infections, since the Cullin-Ring E3 ubiquitin ligases are required for the integrated infection of diverse viruses, including hepatitis B virus X protein (HBx) and the V protein of simian virus 5 (SV5-V), which use CRL4(DDB1) to avoid immune activation and enhance viral infection (34) (Fig. 6). Thus, the neddylation of cellular CRL ubiquitin ligases that is required for HIV and other viral replication may constitute a promising target for new antiviral drug discovery. MLN4924 is a potential candidate for further development as an antiviral inhibitor, if its cytotoxic effect can be further reduced. Whether or not that can be achieved, it is a useful drug, as demonstrated here, for identifying potential targets for therapeutic intervention.
FIG 6.
Table of viral proteins that usurp Nedd8-activated CRL E3 ligase to modulate host factors.
ACKNOWLEDGMENTS
We thank Jacek Skowronski for critical reagents, Chunyan Dai for technical assistance, and Deborah McClellan for editorial assistance.
This work was supported in part by funding from the Chinese Ministry of Science and Technology (2012CB911100 and 2013ZX0001-005), the Chinese Ministry of Education (IRT1016), and the Key Laboratory of Molecular Virology, Jilin Province (20102209). We also gratefully acknowledge scholarship support from the China Scholarship Council to Wei Wei.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 2.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736. 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- 3.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- 4.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. 10.1038/nature06553 [DOI] [PubMed] [Google Scholar]
- 5.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228. 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18:1682–1687. 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR. 2012. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86:12552–12560. 10.1128/JVI.01657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCall CM, Miliani de Marval PL, Chastain PD, II, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. 2008. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 28:5621–5633. 10.1128/MCB.00232-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, Chung CH, Seo SB, Yoon JB, Ko E, Noh DY, Kim KI, Kim KK, Baek SH. 2012. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 48:572–586. 10.1016/j.molcel.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 12.Kassmeier MD, Mondal K, Palmer VL, Raval P, Kumar S, Perry GA, Anderson DK, Ciborowski P, Jackson S, Xiong Y, Swanson PC. 2012. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 31:945–958. 10.1038/emboj.2011.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrofelbauer B, Hakata Y, Landau NR. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U. S. A. 104:4130–4135. 10.1073/pnas.0610167104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, Frappier L. 2012. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. J. Virol. 86:806–820. 10.1128/JVI.05442-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. 2012. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287:12550–12558. 10.1074/jbc.M112.340711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W, Guo H, Han X, Liu X, Zhou X, Zhang W, Yu XF. 2012. A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell. Microbiol. 14:1745–1756. 10.1111/j.1462-5822.2012.01835.x [DOI] [PubMed] [Google Scholar]
- 17.Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, Stevenson M. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4(5):e1000057. 10.1371/journal.ppat.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, Skowronski J. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4(5):e1000059. 10.1371/journal.ppat.1000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan L, Ehrlich E, Yu XF. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81:10822–10830. 10.1128/JVI.01380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188. 10.4161/cc.6.2.3732 [DOI] [PubMed] [Google Scholar]
- 22.Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:11778–11783. 10.1073/pnas.0702102104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen EA. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3(7):e85. 10.1371/journal.ppat.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Feng Y, Narayan O, Zhao LJ. 2001. Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 263:131–140. 10.1016/S0378-1119(00)00583-7 [DOI] [PubMed] [Google Scholar]
- 25.Guo H, Wei W, Wei Z, Liu X, Evans SL, Yang W, Wang H, Guo Y, Zhao K, Zhou JY, Yu XF. 2013. Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation. PLoS One 8(7):e66201. 10.1371/journal.pone.0066201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu XF, Yu QC, Essex M, Lee TH. 1991. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65:5088–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu XF, Ito S, Essex M, Lee TH. 1988. A naturally immunogenic virion-associated protein specific for HIV-2 and SIV. Nature 335:262–265. 10.1038/335262a0 [DOI] [PubMed] [Google Scholar]
- 28.Henderson LE, Sowder RC, Copeland TD, Benveniste RE, Oroszlan S. 1988. Isolation and characterization of a novel protein (X-ORF product) from SIV and HIV-2. Science 241:199–201. 10.1126/science.3388031 [DOI] [PubMed] [Google Scholar]
- 29.Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. 2012. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11:194–204. 10.1016/j.chom.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. 10.1016/j.chom.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada H, Yeh ET, Kamitani T. 2000. A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. J. Biol. Chem. 275:17008–17015. 10.1074/jbc.275.22.17008 [DOI] [PubMed] [Google Scholar]
- 32.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134:995–1006. 10.1016/j.cell.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jager S, Mason-Herr J, Hayashi F, Yokoyama S, Krogan NJ, Harris RS, Peterlin BM, Gross JD. 2012. Inhibition of a NEDD8 cascade restores restriction of HIV by APOBEC3G. PLoS Pathog. 8(12):e1003085. 10.1371/journal.ppat.1003085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. 2010. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 17:105–111. 10.1038/nsmb.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. 10.1038/nature10718 [DOI] [PubMed] [Google Scholar]