Abstract
Dengue virus (DENV), a flavivirus of global importance, is transmitted to humans by mosquitoes. In this study, we developed in vitro and in vivo models of saliva-mediated enhancement of DENV infectivity. Serine protease activity in Aedes aegypti saliva augmented virus infectivity in vitro by proteolyzing extracellular matrix proteins, thereby increasing viral attachment to heparan sulfate proteoglycans and inducing cell migration. A serine protease inhibitor reduced saliva-mediated enhancement of DENV in vitro and in vivo, marked by a 100-fold reduction in DENV load in murine lymph nodes. A saliva-mediated infectivity enhancement screen of fractionated salivary gland extracts identified serine protease CLIPA3 as a putative cofactor, and short interfering RNA knockdown of CLIPA3 in mosquitoes demonstrated its role in influencing DENV infectivity. Molecules in mosquito saliva that facilitate viral infectivity in the vertebrate host provide novel targets that may aid in the prevention of disease.
INTRODUCTION
Flaviviruses cause substantial morbidity and mortality throughout the world. Dengue virus (DENV), transmitted by Aedes aegypti mosquitoes, causes the most significant public health impact (1). When A. aegypti probe during a blood meal, saliva is injected primarily into host extravascular dermal spaces (2, 3). To efficiently feed, A. aegypti must overcome host barriers, including the physiological responses elicited by hemostatic and inflammatory systems (4–9). Mosquito saliva contains a potent mixture of secreted molecules that can affect vascular constriction, blood coagulation, platelet aggregation, inflammation, immunity, and angiogenesis (9–12). Interestingly, mosquito saliva enhances West Nile virus pathogenesis in a mouse model, possibly by modulating TH1/TH2 responses, suggesting that certain salivary proteins are important for enhancing flavivirus infectivity during transmission from vector to host (2, 4, 13–15).
The vast majority of DENV research uses laboratory techniques, such as needle inoculation, that may either compromise or miss key elements of natural infectivity, such as the cointroduction of mosquito saliva proteins into the inoculation site (16). Because of this, the effect of mosquito saliva on DENV transmission and pathogenesis is largely unknown. Previous research on how A. aegypti saliva affects DENV infectivity of human myeloid dendritic cells (DCs) showed a reduction in infectivity in vitro (17). In contrast, saliva treatment showed a modest increase in DENV infectivity in human keratinocytes in vitro (18). Interestingly, the human immune response to specific mosquito saliva proteins has been correlated to disease outcome (19), which is supported by work with humanized mice showing that previous exposure to mosquito saliva enhanced pathogenesis (20). In this study, we developed in vitro and in vivo models to test the effect of mosquito saliva on DENV infection and then used these models to identify a class of molecules that participate in the enhancement of virus infectivity upon entry into the mammalian host. Identification of molecules that mediate infectivity enhancement will allow for the production of vector-based vaccines and therapeutics that will target arthropod saliva components and interfere with viral transmission.
MATERIALS AND METHODS
Ethics statement.
Animal care and treatment complied with National Institutes of Health policy, were in accordance with institutional guidelines, and were approved by the Yale University and University of Pittsburgh Institutional Animal Care and Use Committee.
Ab and reagents.
Antibodies (Ab) used were anti-DENV type 2 3H5.1 from Millipore, anti-DENV NS1 (MA1-71254) from Pierce, and ab6328, ab34710, ab11575, ab3099, and ab2891 from Abcam. Protease inhibitors used were antipain 2HCl at 50 μg/ml, bestatin at 40 μg/ml, chymostatin at 60 μg/ml, E-64 at 10 μg/ml, leupeptin at 5 μg/ml, pepstatin at 0.7 μg/ml, phosphoramidon at 330 μg/ml, Pefabloc SC at 1 mg/ml, EDTA at 0.5 mM, and aprotinin at 2 μg/ml from Roche. Batimastat was used at 200 nM (Santa Cruz Biotechnology). A protease inhibitor cocktail was used at a 1:250 dilution (P8340; Sigma). Heparan sulfate, chondroitin sulfate A, and chondroitin sulfate B were used at concentrations of 100 μg/ml (Sigma).
Mosquito rearing and saliva material collection.
Aedes aegypti and Culex tarsalis were provided by staff at the Connecticut Agricultural Experiment Station. Mosquitoes were maintained in a sugar solution at 27°C and 80% humidity according to standard rearing procedures. Salivary glands were dissected as described previously (2). Salivary gland extracts were prepared by placing 100 salivary glands in 100 μl sterile phosphate-buffered saline (PBS), freeze-thawing by placing on dry ice three times, and then removing insoluble debris by centrifugation at 5,000 × g for 10 min (2). One μl is equal to 1 salivary gland extract equivalent (SGE). Midgut, ovary, head, and carcass extracts were prepared similarly. Mosquito saliva was collected by the immersion oil technique (21). Saliva was extracted from immersion oil with PBS. All salivary material was stored at −80°C.
Cell culture and virus production.
All mammalian cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum and antibiotics at 37°C with 5% CO2. Insect cells were maintained in DMEM containing 10% fetal bovine serum, tryptose phosphate, and antibiotics at 30°C. All viruses were passaged in C6/36 cells. Mouse-adapted dengue virus type 1 (DENV1) Hawaii strain was obtained from the ATCC (VR1254). Mouse-adapted dengue virus type 2 (DENV2) TH-36 strain was obtained from the ATCC (VR345). DENV2 New Guinea C strain was obtained from the CAES. Dengue virus type 3 (DENV3) was obtained from the CAES. Dengue virus type 4 (DENV4) strain H241 was obtained from the ATCC (VR1490). DENV2 PLO46 and E124/128 were obtained from Sujan Shresta as virus stocks from infectious clones as previously described (22). Approximately 1 × 105 genome equivalents (GE) of each DENV2 PLO46 and E124/128 were used for in vitro infections of mouse embryonic fibroblasts (MEFs). West Nile virus strain NY99 was obtained from the CAES. This strain was passaged only three times before use. This strain was originally isolated from an infected Culex pipiens mosquito in Connecticut. Virus titers were determined by a modified focus-forming immunoperoxidase assay using Vero cells as described previously (23). Briefly, undiluted and 10-fold-diluted virus stocks or infectious supernatants were inoculated onto confluent Vero cell monolayers in a 6-well format in a total volume of 2 ml of the mammalian cell culture medium described above. After 1 h of adsorption at 37°C, the inoculum was removed and the cell monolayers were overlaid with 1% SeaPlaque agarose (Lonza) in 90% low-glucose DMEM containing 1% penicillin-streptomycin and 10% fetal bovine serum (FBS) warmed to 37°C. Cells were incubated for 4 to 5 days at 37°C and 5% CO2. Agarose plugs were then moved by centrifugation. Cells were washed with PBS and then fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were washed in PBS three times and then incubated with blocking buffer (1× PBS, 0.1% Tween, 1% bovine serum albumin [BSA]) for 1 h at room temperature. Anti-DENV NS1 (MA1-71254) from Pierce was then added to blocking buffer at 1:200 and incubated for 1 h at room temperature. Unbound antibody was removed by washing cells with PBS three times, and then primary antibody was stained with goat anti-mouse IgG (horseradish peroxidase [HRP] polymer; ab2891) by adding one drop to 300 μl PBS and incubating for 30 min at room temperature. Unbound antibody was removed by washing cells in PBS three times, and then DENV NS1 positive signal was detected using an AEC peroxidase substrate kit (SK-4200) from Vector Laboratories. NS1-positive foci were counted manually using a light microscope. At 18 h postinfection (hpi), NS1 immunoperoxidase assays using MEF cells were performed, the protocol described above was scaled down to a 48-well format, and no agarose plug was necessary.
In vitro SGE-mediated infectivity enhancement assays.
MEFs were seeded at 25,000 cells/well in 48-well plates (uncoated or poly-d-lysine-coated polystyrene) and grown to approximately 70% confluence overnight. Medium was aspirated, and cells were washed with PBS. Mosquito tissue extracts were diluted in a total volume of 100 μl PBS and inoculated into cells at room temperature for 10 min. Saliva material was removed, and then approximately 0.1 to 1 multiplicities of infection (MOI) of DENV or 100,000 GE of DENV2 PLO46 and E124/128 was inoculated into cells in a total volume of 500 μl for 1 h at 37°C. Unbound virus was then removed, and fresh medium was added. Infections progressed for up to 18 h. For the three-dimensional (3D) SGE-mediated infectivity enhancement assay, 106 MEFs were suspended in 1.0 ml rat-tail type-1 collagen, and plugs were allowed to solidify in Transwell dishes. Two ml of medium was added to the top of each collagen plug. In a total volume of 25 μl, 103 FFU of DENV2 were coinjected into the synthetic dermis with PBS or 2 SGE. At 18 hpi, collagen was solubilized in RLT buffer and dounce homogenized. Total RNA was extracted using RNeasy kits (Qiagen).
For analysis of total DENV vRNA, total mRNA was harvested using RNeasy kits (Qiagen). Amplification of both the viral target and reference gene target was performed using a duplex format in 0.2-ml, 96-well PCR plates (Bio-Rad) with a total reaction volume of 25 μl. Reverse transcription and quantitative PCR (RT-qPCR) were performed in the same closed tube with 250 ng of total RNA per reaction using the Quantitect RT-PCR kit (Qiagen).
All primers were used at a final concentration of 4 μM and were synthesized by the Keck Facility at Yale University. Primer sequences are available upon request. Primers were developed using Gene Link Software (OligoAnalyzer 1.2 and OligoExplorer 1.2). All RT-qPCRs were performed using an iQ5 machine (Bio-Rad). Cycling conditions were 50°C for 30 min (reverse transcription) and 95°C for 15 min, followed by 42 cycles of 94°C for 15 s and 54.5°C for 1 min. Relative quantities of viral target cDNA were determined using REST software.
Immunofluorescence.
MEFs were maintained and treated with saliva material, followed by infection with DENV as described above. Cells were either fixed directly or removed to isolate underlying extracellular matrix (ECM) by multiple washes with 100 mM EDTA in PBS for 20 min at room temperature. Both cells and isolated ECM were fixed by treatment with 4% paraformaldehyde in PBS for 20 min at room temperature. Cells or ECM were then washed with PBS and blocked with 1% BSA in PBS for 20 min. BSA was removed, and then cells or ECM were incubated with a 1:200 dilution of anti-DENV envelope antibody for 20 min at room temperature (MAb 3H5-1; Millipore). Cells or ECM were washed three times with PBS and incubated with a 1:1,000 dilution of anti-mouse secondary Ab conjugated to tetramethyl rhodamine isothiocyanate (TRITC) for 20 min at room temperature. Cells or ECM were washed three times with PBS, and cells were incubated alone with a 1:2,000 dilution of 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature. Cells were washed three times with PBS and visualized using a Zeiss Axiovert 200 M fluorescence microscope.
Western blot analysis.
Cell lysates were boiled for 5 min in reducing SDS-PAGE buffer and run on a 4 to 12% SDS-PAGE gel for 1.5 h at 15 mA per gel. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% milk in 1% PBS-Tween (PBST) for 1 h at room temperature and then incubated with the appropriate primary antibody overnight. The membrane was then washed extensively with PBST and stained with the appropriate horseradish peroxidase secondary antibody for 30 min at room temperature. After extensive washing with PBST, immunoblots were incubated with ECL substrate (Amersham, NJ) for 1 min at room temperature. Signal was detected using Kodak film.
HPLC fractionation and LC-MS/MS.
One hundred salivary glands were dissected from female A. aegypti organisms and placed in 100 μl PBS. The sample was freeze-thawed three times at −80°C, and insoluble debris was pelleted by centrifugation at 5,000 × g for 10 min. The supernatant was reserved. SGE was fractionated by high-performance liquid chromatography (HPLC) on a nonporous reverse-phase column with a TFA buffer system into 80 100-μl fractions. Ten μl of each fraction was diluted into 90 μl PBS and used as SGE treatments for in vitro SGE-mediated infectivity enhancement assays as described above. The remaining 90 μl of fractionated SGE from enhancing fractions was submitted for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Proteins were digested with trypsin and analyzed using LC-MS/MS on a Thermo Scientific LTQ-Orbitrap XL mass spectrometer using Waters nanoACQUITY ultra-high-pressure liquid chromatographs (UPLC) for peptide separation. MS/MS spectra were searched in-house using the Mascot algorithm for uninterpreted MS/MS spectra after using the Mascot Distiller program to generate Mascot-compatible files. An A. aegypti database was used for searching. The Keck Biotechnology Resource at Yale University performed both HPLC and LC-MS/MS.
siRNA knockdown in Aedes aegypti salivary glands.
For short interfering RNA (siRNA) knockdown experiments, 1-week-old adult female A. aegypti organisms were kept on ice and then transferred to a cold tray to receive an intrathoracic microinjection of ∼500 ng/200 nl PBS CLIPA3 siRNA or control siRNA. The following CLIPA3 and control siRNAs were synthesized by Thermo Scientific: CLIPA3 forward, 5′ CCG CAA AUU UAC AGA ACG AUU 3′; reverse, 5′ UCG UUC UGU AAA UUU GCG GUU 3′; control forward, 5′ CCU GAU AUU UCG AAG AUG AUU 3′; reverse, 5′ UCA UCU UCG AAA UAU CAG GUU 3′. Following a 7-day recovery period, salivary glands were dissected and placed in PBS for SGE production and in vitro SGE-mediated infectivity enhancement assays or in RLT buffer for RNA extraction and RT-qPCR. RNA was extracted using Qiashredders and an RNeasy kit (Qiagen). Primers used for CLIPA3 RT-qPCR were the following: forward, 5′GGA TTT CCC GAC GAC AAA C 3′; reverse, 5′ TCC AGT GAT AAT GCC TGT GC 3′. Primers for actin as the reference gene were the following: forward, 5′ GAA CAC CCA GTC CTG CTG ACA 3′; reverse, 5′ TGC GTC ATC TTC TCA CGG TTA G 3′. Amplification of both the viral target and reference gene target was performed using a duplex format in 0.2-ml, 96-well PCR plates (Bio-Rad) with a total reaction volume of 25 μl. Reverse transcription and quantitative PCR were performed in the same closed tube with ∼5 ng of total RNA per reaction using the Quantitect RT-PCR kit (Qiagen) and the cycling parameters listed above.
Dengue and West Nile virus mouse models.
For the DENV mouse model, four groups of 10 15-week-old, age-matched, mixed-sex interferon I receptor-deficient (IFNAR−/−) C57BL/6 (AB6) mice were each inoculated in a single rear footpad with 20 μl containing 107 GE of either DENV2 and 1% dimethylsulfoxide (DMSO), 107 GE DENV2 with 1 mM Pefabloc SC in DMSO, 107 GE DENV2 with 1.0 SGE and 1% DMSO, or 107 GE with 1 mM Pefabloc SC in DMSO and 1.0 SGE. Twenty-four hpi, mice were anesthetized and blood samples were processed for total RNA using Qiashredders and an RNeasy kit (Qiagen). Mice were then euthanized, and footpads, lymph nodes, and spleens were dissected and processed for total RNA using Qiashredders and an RNeasy kit (Qiagen). Nanograms of DENV vRNA were normalized to total ng RNA by RT-qPCR.
For the WNV mouse model, 6-week-old female C57BL/6 mice were inoculated with WNV into the back of each footpad at a dose of 500 PFU per footpad. Mice were inoculated with either WNV plus 1% DMSO, WNV plus 1 mM Pefabloc SC in DMSO, WNV plus 1.0 Culex tarsalis SGE and 1% DMSO, or WNV plus 1.0 Culex tarsalis SGE and 1 mM Pefabloc SC in DMSO. Twenty-four hpi, mice were anesthetized and blood samples were processed for total RNA using an RNeasy kit (Qiagen). Mice were then euthanized and footpads were dissected and processed for total RNA using an RNeasy fibrous tissue kit (Qiagen). Relative WNV vRNA in the blood and in footpads was assessed by RT-qPCR, as mentioned above, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference gene.
Amplification of targets was performed using a duplex format in 0.2-ml, 96-well PCR plates (Bio-Rad) with a total reaction volume of 25 μl. Reverse transcription and quantitative PCR were performed in the same closed tube with 5 to 100 ng of total RNA per reaction using the Quantitect RT-PCR kit (Qiagen) and the cycling parameters listed above.
RESULTS
Mosquito saliva enhances dengue virus infectivity in fibroblasts at an early stage of infection.
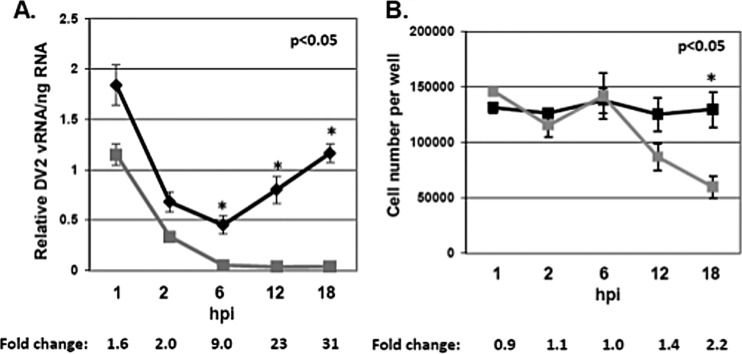
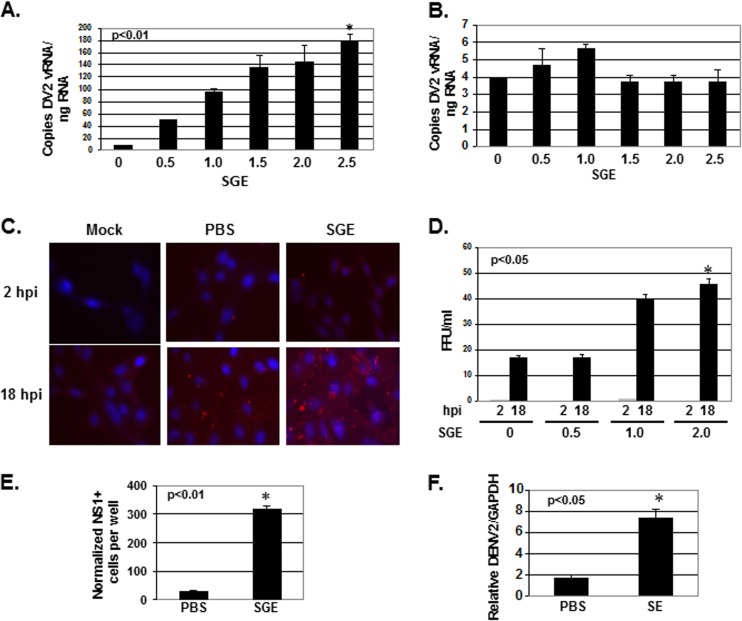
To test the hypothesis that mosquito saliva can enhance DENV infectivity, multiple cell lines were treated with a range of salivary gland equivalents (SGEs) from female A. aegypti organisms for 10 min at room temperature. After treatment, SGE was removed from cells, and approximately 0.1 to 1.0 MOI of dengue virus type 2 New Guinea strain (DENV2) was inoculated into cells for 1 h at 37°C. After 1 h, unbound virus was removed and fresh medium was added. Cells were allowed to incubate for up to 18 h at 37°C to allow a single complete cycle of virus infection. SGE treatment increased the amount of DENV2 vRNA per cell in NIH 3T3 MEFs, wild-type 129 MEFs, IFNAR−/− 129 MEFs, and primary IFNAR−/− C57BL/6 MEFs (Table 1). To determine the time point during the virus life cycle when enhancement of vRNA in each cell occurred, total RNA was isolated from cells at 1, 2, 6, 12, and 18 h postinfection (hpi) and the relative amount of DENV2 vRNA was normalized to total RNA (Fig. 1A). Cell number was also determined at these time points (Fig. 1B). Although a 2-fold increase in vRNA per cell was observed at 1 and 2 hpi, this was not statistically significant. Interestingly, a steady increase in vRNA per cell was observed starting a 6 hpi, leading to a 31-fold increase in vRNA per cell by 18 hpi (Fig. 1A). This increase corresponded to the cell migration phenotype observed by light microscopy and an eventual loss in cell number, yet the loss in cell number did not account for the entire fold increase in vRNA per cell (Table 1 and Fig. 1B). The largest increase in DENV2 vRNA took place in NIH 3T3 MEF cells, and it was a dose-dependent response (Fig. 2A). No increase was present using RAW 264.7 mouse macrophage (Fig. 2B). SGE-mediated enhancement of infectivity in IFNAR−/− 129 and primary IFNAR−/− C57BL/6 MEFs suggested that increased vRNA per cell did not result from SGE-mediated inhibition of the interferon pathway (Table 1).
TABLE 1.
Characteristics of cell lines used and their responses to salivary gland extract treatment regarding DENV2 infectivity enhancement and cell migration
| Cell line | Species, tissue, cell type | Enhancement | Cell migration |
|---|---|---|---|
| NIH 3T3 | Mouse, embryo, fibroblast | Yes | Yes |
| 129 IFNAR+/+ | Mouse, embryo, fibroblast | Yes | Yes |
| 129 IFNAR−/− | Mouse, embryo, fibroblast | Yes | Yes |
| C57BL/6 IFNAR−/− | Mouse, embryo, fibroblast | Yes | Yes |
| RAW 264.7 | Mouse, blood, macrophage | No | No |
FIG 1.
Mosquito saliva temporally enhances dengue virus vRNA in mouse embryonic fibroblasts. (A) NIH 3T3 MEFs were either left untreated (gray squares) or were treated (black diamonds) with SGE for 10 min, inoculated with DENV2 at an MOI of 0.1 for 1 h, and then incubated for 1, 2, 6, 12, and 18 h. Total RNA was extracted, and relative DENV2 vRNA values were normalized to 250 ng of input RNA by RT-qPCR. (B) NIH 3T3 MEFs were untreated (black squares) or treated (gray squares) with SGE for 10 min, inoculated with DENV2 at an MOI of 0.1 for 1 h, and then incubated for 1, 2, 6, 12, and 18 h. Cells were detached from wells by trypsin treatment, and cells were counted by light microscopy. Fold change between SGE-treated and untreated time points are indicated for both panels. Data represent averages from three independent experiments, including standard errors of the means. Student's t tests were performed for individual time points to assess statistical significance.
FIG 2.
Mosquito saliva enhances dengue virus infectivity in mouse embryonic fibroblasts. NIH 3T3 MEFs (A) and RAW 264.7 macrophage cells (B) were treated with SGE for 10 min, inoculated with DENV2 at an MOI of 0.1 for 1 h, and then incubated for 18 h. Total RNA was extracted, and the copy numbers of DENV2 vRNA were normalized to 250 ng of input RNA by RT-qPCR. (C) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 2.5 SGE for 10 min, inoculated with DENV2 at an MOI of 0.1 for 1 h, and then incubated for 18 h. A mock-infected control was included. Unbound virus was washed from cells, and cells were fixed with 4% paraformaldehyde and stained with anti-DENV2 envelope antibody (red) and DAPI (blue). (D) NIH 3T3 MEFs were treated with 0, 0.5, 1.0, and 2.0 SGE for 10 min, inoculated with DENV2 at an MOI of 0.1 for 1 h, and then incubated for 2 and 18 h. Focus-forming immunoperoxidase assays were performed on Vero cells using NIH 3T3 MEF supernatants in a 6-well format. (E) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 1.0 SGE for 10 min and then inoculated with DENV2 at an MOI of 0.1 for 1 h. Cells were stained for DENV2 NS1 using an immunoperoxidase assay, and NS1-positive cells were counted 18 hpi. These data were not normalized to the total amount of cells on the plate. (F) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 2.0 saliva equivalents (SE) for 10 min and then inoculated with DENV2 at an MOI of 0.1 for 1 h. Total RNA was extracted, and expression of DENV2 vRNA and GAPDH was assessed by RT-qPCR 18 hpi. DENV2 vRNA was normalized to GAPDH. Data represent the averages from at least six independent experiments, including standard errors of the means. Student's t tests were performed on samples treated with the highest concentration of SGE to assess statistical significance.
Increased DENV2 vRNA in total RNA resulted from more cells being infected, more virus binding to the cell culture plate, increased replication within cells, or a reduction in total RNA. When cells were treated with 2.5 SGE, RT-qPCR data of DENV2 vRNA showed a cycle threshold (CT) value of 24.9 (±0.1), and untreated samples showed a CT of 27.2 (±0.2). This suggested that at least a 4-fold amount of infectivity enhancement was not due to a reduction in total RNA. Accordingly, immunofluorescence analysis with antibody against DENV2 envelope protein found significantly more DENV2 antigen in cells that were pretreated with SGE at 18 hpi (Fig. 2C). Further, SGE-treated cells secreted significantly more infectious virus into the supernatant 18 hpi than nontreated cells, and no infectious virus was detected in the supernatant at 2 hpi (Fig. 1D). Importantly, more infectious virus was detected in SGE-treated cell supernatants despite a loss in cell number. Data shown in Fig. 2D were not normalized to cell number.
To determine if more cells were infected when pretreated with SGE and to control for the low productivity of cells infected for 18 h, we stained for nonstructural protein 1 (NS1)-positive cells 18 hpi and found a statistically significant increase in the number of cells infected during SGE treatment (Fig. 2E). These data were significant with and without normalization to cell number (data not shown). To verify that the enhancing activity in SGE was not an artifact of mosquito proteins found in salivary gland tissue but in saliva, A. aegypti saliva was collected by the oil immersion technique (21). Saliva collected in this manner induced cell migration (data not shown) and enhanced DENV2 vRNA similar to SGE treatment (Fig. 2F).
To test if SGE-mediated enhancement of infectivity was a unique trait specific for tissue culture-adapted DENV2, we performed in vitro SGE-mediated infectivity enhancement assays using mouse-adapted DENV1 and DENV2, low-passage-number tissue culture-adapted DENV3 and DENV4 isolates, and tissue culture-adapted DENV2 with high (DENV2 PLO46) and low (DENV2 E124/E128) affinities for heparan sulfate proteoglycans (22). Interestingly, SGE treatment significantly enhanced the infectivity of all DENV isolates tested except for DENV2 E124/E128, suggesting that the mechanism of SGE-mediated infectivity enhancement in vitro included attachment to heparan sulfate proteoglycans (Table 2).
TABLE 2.
Relative RT-qPCR data from SGE treatments of mouse embryonic fibroblasts followed by infection with multiple dengue virus serotypes and isolatesa
| Isolate | Result by SGE level |
P valuee | |
|---|---|---|---|
| 0 | 1.0 | ||
| DENV1 Hawaiib (m) | 1.1 (± 0.0) | 4.5 (±0.3) | <0.05 |
| DENV2 TH-36 (m) | 0.9 (± 0.0) | 10.6 (±1.6) | <0.05 |
| DENV3 CAESc (tc) | 1.7 (± 0.2) | 14.2 (±0.4) | <0.05 |
| DENV4 H241 (tc) | 1.0 (± 0.0) | 51.3 (±3.5) | <0.05 |
| DENV2 NGC (tc) | 1.0 (± 0.1) | 12.9 (±0.3 | <0.05 |
| DENV2 PLO46d (tc) | 1.2 (± 0.3) | 30.0 (±3.9) | <0.05 |
| DENV2 E124/E128 (tc) | 1.7 (± 0.4) | 1.9 (±0.3) | NS |
Data represent averages from at least three independent in vitro SGE-mediated enhancement assays (see Materials and Methods). Untreated samples were set to 1.0. Student's t tests were performed to assess statistical significance.
Mouse-adapted DENV types 1 and 2 were obtained from the ATCC as detailed in Materials and Methods.
Tissue culture-adapted DENV types 2 to 4 were obtained from the ATCC and the Connecticut Agricultural Experiment Station as detailed in Materials and Methods.
Stocks from infectious clones of tissue culture-adapted DENV type 2 PLO46 and E124/128 were obtained from Sujan Shresta.
Data represent averages from three or more independent experiments and include standard errors of the means. Student's t tests were performed to assess statistical significance. NS, not significant.
Because low affinity to heparan sulfate increases in vivo pathogenesis of dengue and a number of other viruses (22), and because high-affinity interactions with heparan sulfate can evolve during tissue culture passaging, we aligned tissue culture-adapted DENV2 NGC, DENV2 PLO46, and DENV2 E124/E128 envelope protein with 21 envelope proteins previously sequenced directly from the serum of patients with secondary dengue fever, dengue hemorrhagic fever, and dengue septic shock 1 to 5 days postinfection (dpi) and analyzed residues previously proposed to interact with heparan sulfate (22, 24, 25). The resolving of the crystal structure of the dengue virus envelope, in addition to mutational studies, has identified multiple basic residues that may interact with heparan sulfate (22, 25). All residues predicted to interact with heparan sulfate were conserved between DENV2 PLO46 and the 21 envelope proteins sequenced directly from patient serum. DENV2 NGC had an additional basic residue at E126K. This substitution was not present in DENV2 PLO46. DENV2 PLO46 lost an acidic residue at D203N. However, neither of these amino acids has been implicated in heparan sulfate binding, and none of the 21 envelope proteins sequenced directly from patient serum contained the charge changes in K124 or K128 seen in the DENV2 E124/E128 mutant. Further analysis of clones sequenced directly from patient serum mosquito salivary glands, in addition to the production of wild-type infectious clones, is required to determine the heparan sulfate binding status of physiologically relevant virus.
Mosquito saliva increases virus attachment and induces cell migration by the proteolysis of extracellular matrix proteins.
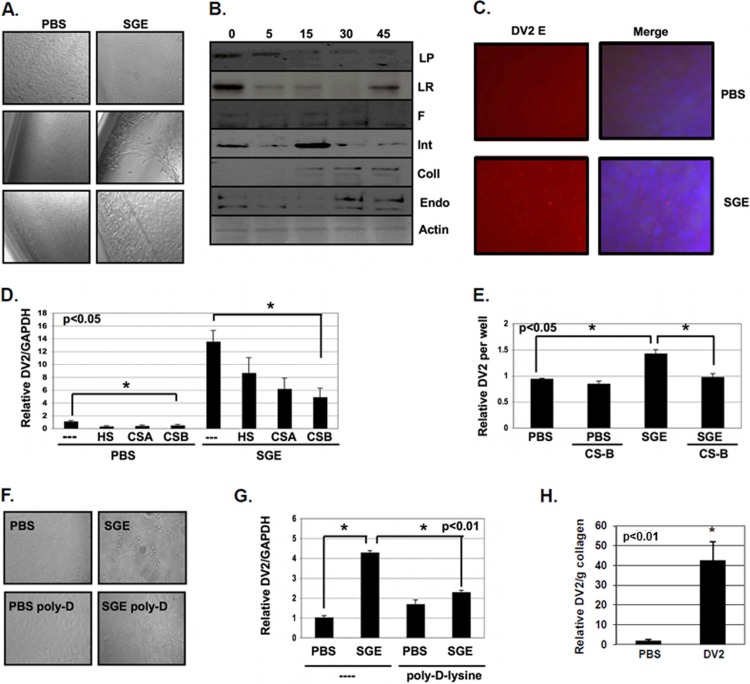
By light microscopy, NIH 3T3 MEFs begin to migrate away from the edges of the cell culture plate by 2 h after SGE treatment (Fig. 3A). Western blot analysis showed that many extracellular matrix (ECM)-related proteins were affected by SGE treatment (Fig. 3B). After 5 min of SGE treatment, reductions in levels of laminin protein, laminin receptor, fibronectin, and integrin alphaVbeta3 were observed (Fig. 3B). SGE treatment also induced the uPARAP/Endo180 collagen-recycling pathway, with uPARAP/Endo180 and collagen type I expression being enhanced after 30 and 45 min of SGE treatment (Fig. 3B).
FIG 3.
SGE enhances virus attachment to exposed glycosaminoglycans and induces cell migration. (A) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 1.0 SGE for 10 min and then allowed to incubate for 4 h at 37°C before observation by phase-contrast light microscopy. (B) NIH 3T3 MEFs were treated with 1.0 SGE for 0, 5, 15, 30, and 45 min. Equal volumes of protein lysates were loaded onto SDS-PAGE gels, protein was transferred to PVDF membranes, and the expression of multiple proteins was analyzed by Western blotting. Laminin protein (LP), laminin receptor (LR), fibronectin (F), integrin (Int), collagen type I (Coll), endo180 (Endo), and actin were used. (C) NIH 3T3 MEFs were treated with buffer alone (PBS) or with 2.5 SGE for 10 min and inoculated with DENV2 at an MOI of 1 for 1 h. Unbound virus was washed from cells, and cells were fixed with paraformaldehyde and stained for DENV2 envelope (red) and DAPI (blue). (D) DENV2 was untreated (−) or pretreated with 100 μg/ml heparan sulfate (HS), chondroitin sulfate A (CS-A), and chondroitin sulfate B (CS-B) for 30 min at room temperature. NIH 3T3 MEFs were then treated with buffer alone (PBS) or with 1.0 SGE for 10 min and then inoculated with glycosaminoglycan-treated or untreated DENV2 at an MOI of 1 for 1 h at 37°C. Unbound virus was removed, and then cells were allowed to incubate for 18 h. Total RNA was extracted, and DENV2 vRNA and GAPDH expression was analyzed by RT-qPCR. DENV2 vRNA was normalized to GAPDH. (E) ECM material was isolated on cell culture plates by washing off monolayers of NIH 3T3 MEF cells with 10 mM EDTA solution in PBS for 10 min at 37°C. Wells were either treated with buffer alone (PBS) or treated with 2.0 SGE for 10 min at 37°C. Wells were then inoculated with untreated or CS-B-treated DENV2 for 1 h at 37°C. Unbound virus was removed, total RNA was extracted, and DENV2 vRNA expression was analyzed by RT-qPCR and normalized per well. (F and G) MEFs were seeded onto either untreated (−) or poly-d-lysine-treated (poly-d-lysine) polystyrene cell culture plates and treated with buffer alone (PBS) or with 1.0 SGE and then inoculated with DENV2 at an MOI of 1 for 1 h. (F) Cell migration was observed by light microscopy. (G) Total RNA was extracted, and the expression of DENV2 vRNA and GAPDH was assessed by RT-qPCR 18 hpi. DENV2 vRNA was normalized to GAPDH. (H) Total RNA was extracted from MEFs suspended in collagen matrices, and the expression of DENV2 vRNA was assessed by RT-qPCR 18 hpi and normalized to grams of collagen. Data represent the averages from more than three independent experiments. Student's t tests were performed to analyze statistical significance.
Immunofluorescent analysis of DENV2 envelope antigen showed that more viral antigen accumulated at the site of cell stripping than in the rest of the cell culture plate (Fig. 3C). Accordingly, a 2-fold enhancement of DENV2 vRNA per cell occurred within 2 h of infection (Fig. 1A). Pretreatment of virus with the glycosaminoglycans (GAGs) heparan sulfate and chondroitin sulfates A and B reduced infectivity of DENV2. This occurred with and without SGE treatment, suggesting that DENV2 utilizes GAGs for enhanced attachment during SGE treatment (Fig. 3D). We tested this hypothesis by inoculating chondroitin sulfate B-treated DENV2 to isolated ECM material that was pretreated with SGE. Chondroitin sulfate B blocked enhanced attachment to the ECM (Fig. 3E).
Although SGE enhanced attachment 1.5- to 2-fold, this did not account for the total increase in infectivity. To determine if induction of cell migration was also important for SGE-mediated infectivity enhancement, NIH 3T3 MEFs were plated on untreated and poly-d-lysine-treated polystyrene cell culture plates. We chose poly-d-lysine as an alternative to treatment with actin or microtubule destabilizing agents, assuming that poly-d-lysine immobilization would have fewer pleiotropic effects. However, it is unclear how poly-d-lysine interacts with specific saliva molecules. Interestingly, poly-d-lysine completely inhibited cell migration and SGE-mediated infectivity enhancement. These data suggest that a combination of enhanced attachment and interaction with migrating cells leads to SGE-mediated infectivity enhancement in vitro (Fig. 3F and G). Enhanced attachment to heparan sulfate proteoglycans is critical in a two-dimensional in vitro environment, preventing removal of attached virus during a washing step. This step would not be required in a three-dimensional in vivo environment where there is no step to remove unbound virus.
The physiological relevance of cell migration on a 2D polystyrene cell culture plate is questionable. Therefore, we tested if SGE-mediated enhancement of infectivity was an artifact of 2D cell culture conditions. To do this, we simulated the 3D environment of human skin, whereby mosquito saliva and virus are deposited during transmission by suspending MEFs in collagen plugs. Briefly, MEFs were embedded in a rat tail type I collagen matrix to form a synthetic dermis, and then 103 FFU of DENV2 was coinjected into the synthetic dermis with PBS or 2 SGE. Collagen plugs were dounce homogenized in RLT buffer, and total RNA was extracted 18 hpi. RT-qPCR analysis showed a significant increase in DENV2 vRNA per cell when virus was coinjected with salivary gland material (Fig. 3H). No decrease in mouse-specific GAPDH housekeeping gene expression was detected in these experiments, suggesting that there was no cell loss when suspended in the collagen matrix, yet DENV2 vRNA per gram of collagen was still enhanced. Specifically, the raw RT-qPCR values for the DENV2 target were 26.6 (±0.2) and 22.2 (±0.4) for untreated and SGE-treated samples, respectively. The raw RT-qPCR values for the GAPDH housekeeping gene were 17.6 (±0.1) and 17.9 (±0.2) for untreated and SGE-treated samples, respectively.
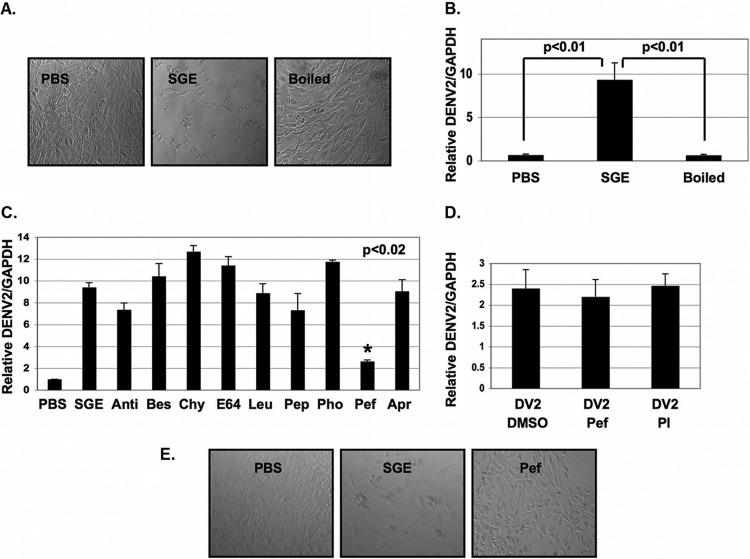
In vitro mosquito saliva-mediated enhancement of dengue virus infectivity is due to serine protease activity.
Boiling SGE material completely inhibited SGE-mediated infectivity enhancement and cell migration, suggesting that a protein was responsible (Fig. 4A and B). To determine if a protease influenced infectivity enhancement, we screened multiple protease inhibitors for their ability to inhibit SGE activity. The serine protease inhibitor Pefabloc SC was able to significantly inhibit SGE activity (Fig. 4C and D). Pefabloc SC had no effect on DENV2 infectivity (Fig. 4D). Inhibition of SGE-mediated infectivity enhancement also correlated with inhibition of cell migration (Fig. 4E).
FIG 4.
SGE-mediated enhancement of dengue virus infectivity is inhibited by Pefabloc SC in vitro. (A and B) NIH 3T3 MEFs were treated with buffer (PBS), pretreated with 1.0 SGE, or pretreated with 1.0 boiled SGE (Boiled), followed by inoculation with DENV2 at an MOI of 0.1 for 1 h. (A) Cells were observed by light microscopy 18 hpi (B) and then total RNA was extracted, and the expression of DENV2 vRNA and GAPDH was assessed by RT-qPCR. DENV2 vRNA was normalized to GAPDH. (C) SGEs were prepared and individually treated with the protease inhibitors antipain-2HCl (Anti), bestatin (Bes), chymostatin (Chy), E-64 (E64), leupeptin (Leu), pepstatin (Pep), phosphoramidon (Pho), pefabloc SC (Pef), and aprotinin (Apr). MEFs were either treated with buffer alone (PBS) or treated with 1.0 SGE with and without protease inhibitors for 10 min and inoculated with DENV2 at an MOI of 0.1 for 1 h. Unbound virus was removed, and cells were allowed to incubate for 18 h. Total RNA was extracted, and the expression of DENV2 vRNA and GAPDH was assessed. DENV2 vRNA was normalized to GAPDH by RT-qPCR. (D) MEFs were treated with Pef or a protease inhibitor cocktail containing Pef (PI) for 10 min and then inoculated with DENV2 at an MOI of 0.1 for 1 h. Unbound virus was removed, and cells were allowed to incubate for 18 h. Total RNA was extracted, and the expression of DENV2 vRNA and GAPDH was assessed by RT-qPCR. DENV2 vRNA was normalized to GAPDH. (E) Prior to extracting total RNA as described for panel C, cells were observed by phase-contrast light microscopy. Data represent averages from at least three independent experiments. Student's t tests were performed to assess statistical significance when appropriate.
Saliva-mediated flavivirus infectivity enhancement is due to serine protease activity in vivo.
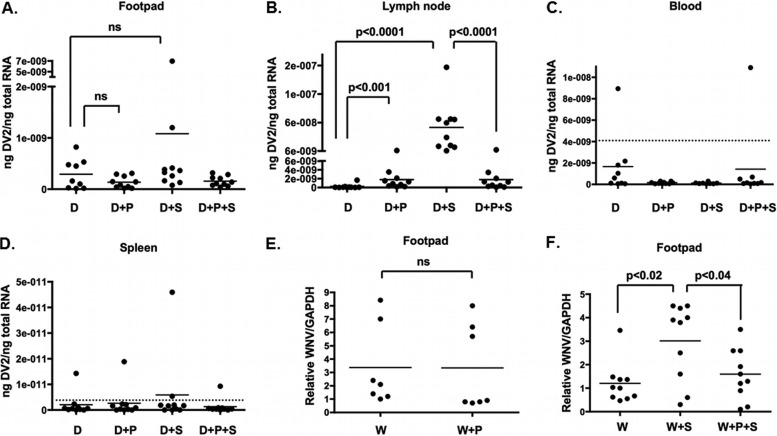
To determine if a serine protease was also required for SGE-mediated infectivity enhancement in vivo, we determined if Pefabloc SC was able to block SGE-mediated infectivity enhancement in a dengue virus mouse model. Four groups of 10 15-week-old, age-matched, mixed-sex IFNAR−/− C56/B6 (AB6) mice were each inoculated into a single rear footpad with 20 μl containing either 107 genome equivalents (GE) of either DENV2 and 1% DMSO, 107 GE DENV2 with 1 mM Pefabloc SC in DMSO, 107 GE DENV2 with 1.0 SGE and 1% DMSO, or 107 GE with 1 mM Pefabloc SC in DMSO and 1.0 SGE. RNA was harvested from footpads, blood, draining lymph nodes, and spleen 24 hpi. Although there was a higher trend of DENV2 vRNA in footpads when SGE was present, there was no significant difference between groups (Fig. 5A). In contrast, SGE enhanced DENV2 vRNA 100-fold in lymph nodes, and this enhancement was completely inhibited by Pefabloc SC treatment (Fig. 5B). Only a few samples detected DENV2 vRNA above background levels in the blood and spleen (Fig. 5C and D).
FIG 5.
Pefabloc SC inhibits SGE-mediated enhancement of dengue and West Nile viruses in vivo. Four groups of 10 IFNAR−/− C57BL/6 mice were inoculated into a single rear footpad with 20 μl containing 107 genome equivalents (GE) of DENV2 and 1% DMSO (D), 107 GE DENV2 with 1 mM Pefabloc SC in DMSO (D+P), 107 GE DENV2 with 1.0 SGE and 1% DMSO (D+S), or 107 GE with 1 mM Pefabloc SC in DMSO and 1.0 SGE (D+P+S). Twenty-four hpi, footpads (A), lymph nodes (B), blood (C), and spleens (D) were harvested into RLT buffer, and RNA was extracted for RT-qPCR analysis. DENV vRNA was normalized to total RNA per reaction. (E) C57BL/6 mice were inoculated in a single rear footpad (7 per group) with 500 PFU WNV and 1% DMSO (W) or with 1 mM Pefabloc SC in DMSO (W+P). Relative viral load was assessed in blood by RT-qPCR 24 hpi. (F) Three groups of 10 C57BL/6 mice were inoculated in rear footpads with 500 PFU WNV and 1% DMSO (W), with 1.0 C. tarsalis SGE and 1% DMSO (W+S), or with both 1.0 C. tarsalis SGE and 1 mM Pefabloc SC in DMSO (W+P+S). Relative viral load was assessed in footpads by RT-qPCR 24 hpi. Each symbol represents one mouse. The dotted line represents the background threshold. Mann-Whitney analysis was performed to assess statistical significance between groups. Data represent one experiment.
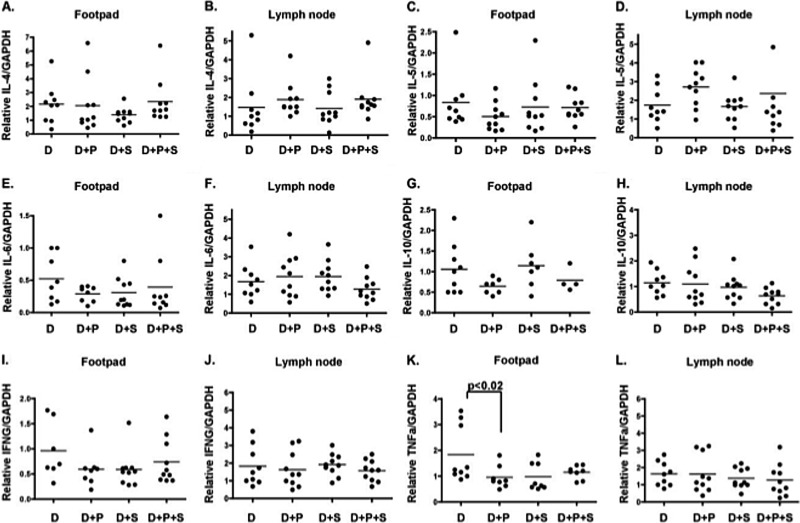
To determine if polarization toward TH2 cytokines was correlated with SGE-mediated infectivity enhancement, mRNA expression of TH2 cytokines interleukin-4 (IL-4), IL-5, IL-6, and IL-10 and TH1 cytokines IFN-γ and tumor necrosis factor alpha (TNF-α) were examined in footpads and lymph nodes 24 hpi in the four groups of mice listed above. No significant change in mRNA expression was detected in TH2 or TH1 cytokines (Fig. 6A to L).
FIG 6.
RT-qPCR analysis of TH2/TH1 cytokine expression in footpads and lymph nodes from IFNAR−/− C57BL/6 mice infected with DENV2 and treated with SGE and Pefabloc SC. Relative expression of IL-4 (A and B), IL-5 (C and D), IL-6 (E and F), IL-10 (G and H), IFN-γ (I and J), and TNF-α (K and L) was examined in footpads and draining lymph nodes 24 hpi in mice inoculated into a single rear footpad with 20 μl containing 107 genome equivalents (GE) of either DENV2 and 1% DMSO (D), 107 GE DENV2 with 1 mM Pefabloc SC in DMSO (D+P), 107 GE DENV2 with 1.0 SGE (D+S), or 107 GE with 1 mM Pefabloc SC in DMSO and 1.0 SGE (D+S+P). Relative cytokine levels were normalized to GAPDH. Each dot represents a single mouse. Mann-Whitney tests were performed between groups to assess statistical significance. Data represent one experiment.
West Nile virus (WNV) is an important flavivirus that has emerged in North America, and it is primarily transmitted by Culex spp. To determine if our studies were applicable to other flaviviruses and mosquitoes, we assessed the ability of Pefabloc SC to block Culex tarsalis SGE-mediated infectivity enhancement in a WNV mouse model. As a control, we tested if Pefabloc SC alone had any effect on WNV vRNA in footpads 24 hpi. Pefabloc SC had no effect on WNV vRNA levels in the footpad (Fig. 5E). However, Pefabloc SC did significantly reduce SGE-mediated infectivity enhancement of WNV in footpads, suggesting that a salivary serine protease is important for SGE-mediated infectivity enhancement of WNV in vivo (Fig. 5F).
Identification of CLIPA3 as a serine protease important for saliva-mediated infectivity enhancement.
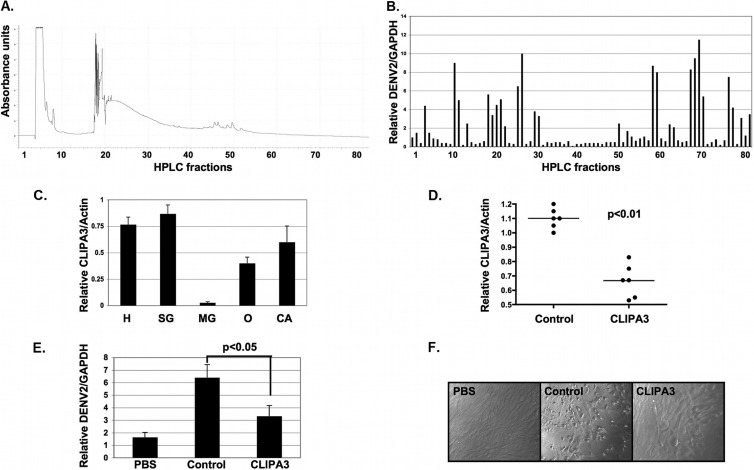
To identify the serine protease responsible for SGE-mediated enhancement of dengue virus infectivity, we fractionated 100 salivary gland equivalents on a reverse-phase HPLC column into 80 fractions (Fig. 7A). We then screened each fraction for the ability to induce NIH 3T3 MEF cell migration and enhance DENV2 infectivity in vitro (Fig. 7B). Multiple clusters were identified that both enhanced and inhibited DENV2 infectivity. All clusters that enhanced infectivity also induced cell migration. No other clusters induced cell migration (data not shown). We classified clusters 10-11, 18-22, 25-26, 57-58, 66-69, and 75-76 as enhancing fractions 1 to 6 and performed LC-MS/MS on the remaining volume of one fraction from each of these clusters. Score values were low. However, 7 proteins were identified in multiple fractions. One protein had a signal peptide and was annotated as a serine protease (i.e., CLIPA3 and AAEL005718) (Table 3). BLAST analysis showed that CLIPA3 had homology to noncatalytic stubble-like proteins. CLIPA3 is noncatalytic due to a glycine substitution at the catalytic serine residue.
FIG 7.
Identification of CLIPA3 as a component involved in SGE-mediated infectivity enhancement. (A) HPLC trace showing protein concentrations in 80 100-μl fractions after reverse-phase HPLC fractionation of 100 A. aegypti SGE. (B) Ten μl of each HPLC fraction was diluted in 90 μl PBS and used as SGE for in vitro SGE-mediated infectivity enhancement assays. Eighteen hpi, RNA was harvested and RT-qPCR was performed. Relative levels of DENV2 were normalized to GAPDH. (C) RT-qPCR assay measuring relative expression of CLIPA3 in female A. aegypti head (H), salivary gland (SG), midgut (MG), ovary (O), and carcass (CA) tissues. Relative levels of CLIPA3 were normalized to actin. (D) RT-qPCR assay measuring relative expression of CLIPA3 in female A. aegypti salivary glands 7 days after microinjection with 500 ng control or CLIPA3 siRNA. Relative levels of CLIPA3 were normalized to actin. Each dot represents 3 pooled salivary glands. (E) In vitro SGE-mediated infectivity enhancement assay using buffer alone (PBS), 1.0 SGE from control siRNA-treated mosquitoes (Control), or 1.0 SGE from CLIPA3 siRNA-treated mosquitoes. Relative levels of DENV2 were normalized to GAPDH. Data represent averages from at least 6 independent experiments. (F) Light microscopy images of NIH 3T3 MEFs shown in panel E 6 h post-SGE treatment. Student's t tests were performed to assess statistical significance when appropriate.
TABLE 3.
LC-MS/MS hits from enhancing HPLC fractions 1 to 6
| Identifier | Comment | Fraction(s) | Score | Signal peptide predictiona |
|---|---|---|---|---|
| AAEL012658 | Rgs-gaip interaction protein | 4 | 25 | Yes |
| UGP | UDP-glucose pyrophosphorylase | 6 | 25 | No |
| AAEL012336 | Putative uncharacterized protein | 1, 2, 3, 5 | 24–29 | No |
| AAEL003131 | Putative uncharacterized protein | 1, 3 | 26–29 | No |
| AAEL005718 | Serine protease | 1, 2, 3, 4, 5 | 23–27 | Yes |
| AAEL008557 | Putative uncharacterized protein | 1 | 22 | No |
| AAEL000217 | Serine/threonine protein kinase | 1, 2, 3, 4, 5, 6 | 21–25 | No |
| AAEL013989 | Protein translocation complex beta | 2 | 98 | No |
| AAEL008188 | 60S ribosomal protein L6 | 2 | 74 | No |
| SGP1A2 | Putative secreted protein | 2 | 70 | Yes/No |
| AAA62350 | Actin | 2 | 69 | No |
| AAEL011194 | Fatty acid synthase | 2 | 36 | No |
| AAEL001593 | Glycerol-3-phosphate dehydrogenase | 2 | 33 | No |
| AAEL010796 | Transmembrane protein 1 | 2, 3, 6 | 26–28 | No |
| RpS14 | 40S ribosomal protein S14 | 2 | 26 | No |
| AAEL007898 | Calmin | 2, 3, 6 | 20–25 | No |
| AAEL009720 | Putative uncharacterized protein | 2 | 25 | No |
| AAEL003324 | Acidic ribosomal protein P1 | 2 | 23 | No |
| AAEL006016 | Putative uncharacterized protein | 2 | 22 | No |
| AAEL001233 | Putative uncharacterized protein | 2 | 21 | No |
| AAEL002560 | Putative uncharacterized protein | 3, 6 | 28 | No |
| AAEL008528 | Protein tyrosine phosphatase n11 | 3 | 25 | No |
| AAEL001516 | Vesicle-associated protein | 5 | 28 | No |
| AAEL001760 | Putative uncharacterized protein | 6 | 31 | No |
| AAEL004039 | Blooms syndrome DNA helicase | 3 | 24 | No |
| AAEL012430 | AMP-dependent ligase | 6 | 22 | No |
| AAEL010205 | Putative uncharacterized protein | 6 | 21 | No |
In silico predictions of signal peptides were performed using the SignalP 3.0 server from the Center for Biological Sequence Analysis at the Technical University of Denmark DTU.
To determine if CLIPA3 was expressed in the salivary glands, we performed RT-qPCR against CLIPA3 and assessed the relative levels of CLIPA3 expression in the head, salivary glands, midgut, ovary, and carcass. CLIPA3 expression was highest in the head and salivary gland with minimal expression in the midgut (Fig. 7C). Supporting the expression data, analysis of midgut lysates showed no cell migration or DENV enhancing activities (data not shown).
We then targeted CLIPA3 in an in vivo knockdown experiment by intrathoracically microinjecting 500 ng of control and CLIPA3 siRNA. Salivary glands were dissected 7 days after microinjection and analyzed for CLIPA3 expression by RT-qPCR (Fig. 7D). A significant knockdown in mRNA expression was observed. We then used SGE from control and CLIPA3 siRNA samples for SGE enhancement assays in vitro. SGE from mosquitoes with CLIPA3 siRNA enhanced DENV2 infectivity to a lesser degree and failed to induce cell migration of NIH 3T3 MEFs (Fig. 7E and F).
DISCUSSION
To elucidate factors in A. aegypti saliva involved in DENV transmission, we developed in vitro and in vivo models of saliva-mediated infectivity enhancement. We noticed significant enhancement of DENV2 infectivity in mouse embryonic fibroblasts (MEFs) at 18 hpi using multiple measures of infectivity. This occurred in 2D and 3D in vitro environments. Importantly, immobilizing cells on poly-d-lysine plates completely inhibited infectivity enhancement, suggesting that SGE-mediated induction of cell migration played a role in infectivity enhancement. Infectivity of a DENV2 mutant with low affinity for heparan sulfate proteoglycans (DENV2 E124/128) was also not enhanced by SGE treatment, suggesting that engagement of heparan sulfate proteoglycans was critical for SGE-mediated infectivity enhancement in vitro (22). We hypothesize that enhanced heparan sulfate binding is only beneficial in a two-dimensional in vitro environment, preventing removal of attached virus during a washing step. This step would not be required in a three-dimensional in vivo environment. We also note that enhancement occurred in MEFs isolated from IFNAR−/− 129 and IFNAR−/− C57BL/6 mice and in the lymph nodes of IFNAR−/− C57BL/6 mice, suggesting that modulation of the IFN response was not responsible for the phenotype. SGE-mediated infectivity enhancement was also not correlated with TH2 polarization in vivo.
Adaptation to heparan sulfate proteoglycans during cell culture passaging has been reported for a number of viruses, and low affinity to heparan sulfate can increase in vivo pathogenesis of DENV and a number of other viruses (22, 26, 27). An alignment with tissue culture-adapted DENV2 NGC, DENV2 PLO46, and DENV2 E124/E128 envelope protein with 21 envelope proteins sequenced directly from the serum of patients showed that residues proposed to interact with heparan sulfate were conserved (22, 24, 25). This analysis suggested that low- and high-affinity heparan sulfate binding viruses exist in nature. Further, the form of virus selected in the mosquito and deposited into the host skin is largely unknown. A recent cryoelectron microscopy analysis of DENV virions at temperatures expected in the mosquito and the host suggested that the conformation of epitopes on envelope protein is different (28). Definitive determination of the HS binding phenotype of DENV at different points in the replication/transmission cycle will require additional sequence obtained directly from patient serum and mosquito salivary glands and recapitulation of the natural sequences in a DENV cDNA clone.
Pefabloc SC significantly inhibited SGE-mediated infectivity enhancement in vivo using both DENV and WNV mouse models. In the DENV mouse model, enhancement was only observed in the draining lymph nodes at 24 hpi. In the WNV mouse model, enhancement was observed in footpads and in blood (data not shown) at 24 hpi. The tissue differences observed between DENV and WNV SGE-mediated enhancement may be due to technical issues or reflect biological differences between these viruses or the genetic backgrounds of the mouse models. We hypothesize that mosquito saliva serine proteases liquefy dermal tissue by proteolyzing ECM components. Proteolysis increases cellular mobility and inflammatory signals within the dermis and enhances interaction between immobilized virions and permissive cell types, such as Langerhans cells or macrophage (29). Irritant and allergen-dependent modulation of the dermal environment has previously been shown to induce migration of Langerhans cells to draining lymph nodes (29–32). This is correlated with a fibroblast- and IL-10-dependent switch of Langerhans cells to a macrophage-like phenotype and may require the breakdown of integrin-mediated interactions with extracellular matrix components (32, 33). In this context, a mosquito serine protease may induce Langerhans cell migration to draining lymph nodes by disrupting interactions between Langerhans cells and the extracellular matrix.
CLIPA3 was identified as an important component of SGE-mediated infectivity enhancement by screening biochemical fractions of SGE for DENV-enhancing activity, followed by siRNA knockdown of CLIPA3 in adult mosquitoes. Interestingly, RNA Seq analysis detected expression of CLIPA3 in salivary glands 14 days postinfection with DENV2 but not in uninfected salivary glands (34). CLIPA3 expression was not detected in salivary glands using less sensitive transcriptomic approaches without blood meal administration (12). We hypothesize that CLIPA3 has a low basal expression level in salivary glands and is induced upon blood feeding or DENV infection.
CLIPA3 is a member of the CLIP-domain serine protease family, a family exclusively found in invertebrates. This family is important in a wide range of physiological events, such as ECM degradation, food digestion, coagulation, and immunity (35). The function of the CLIP domain is unknown; however, it may form protein-protein interactions required for regulating proteolytic cascades (35). CLIPA3 has homology to noncatalytic stubble-like proteins. Previous studies of Drosophila identified a critical role for stubble in regulating epithelial morphogenesis during development. Specifically, stubble is a protease that detaches imaginal discs from the ECM by coordinating the proteolytic modification of ECM components (36, 37). Although similar to stubble, CLIPA3 is noncatalytic due to a glycine substitution at the catalytic serine residue. Since Pefabloc SC irreversibly inhibits active serine proteases by forming covalent sulfonyl derivatives of the active-site serine residue, we hypothesize that CLIPA3 is a cofactor rather than the sole effector involved in SGE-mediated infectivity enhancement. Multiple reports have identified noncatalytic CLIP-domain-containing serine proteases as both activating and inhibiting cofactors (38, 39). Future investigations into the role of CLIPA3 in SGE-mediated infectivity enhancement will require determining if CLIPA3 is secreted into saliva and if activity requires the presence of other proteins or specific processing events.
This study demonstrates that DENV coopts the proteolytic activity of A. aegypti saliva to enhance infectivity in the vertebrate host. This phenomenon may be generalizable to additional flavivirus and/or mosquitoes, as saliva from C. tarsalis mosquitoes has the same influence on West Nile virus infectivity. It is theoretically possible that the inhibition of this specific mosquito activity, perhaps by administering a specific blocking antibody to the host, could modulate viral infectivity, perhaps in conjunction with a traditional pathogen-based vaccine. The discovery of mosquito saliva proteins that are critical for pathogen transmission will enable us to better understand how arthropod-borne pathogens establish early infection in the mammalian host and facilitate the production of vector protein-based vaccines and therapeutics.
ACKNOWLEDGMENTS
We thank Ruth Montgomery for informative discussions and the Fikrig laboratory for general support. We also thank Sujan Shresta for sharing DENV2 PLO46 and E124/E128 virus stocks and Nicolas Garcia for technical help (22).
We acknowledge financial support through an NIH 5T32AI07019-35 training grant administered by David Schatz and the Yale Interdisciplinary Immunology Training Program.
Footnotes
Published ahead of print 16 October 2013
REFERENCES
- 1.Gubler DJ, Clark GG. 1995. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg. Infect. Dis. 1:55–57. 10.3201/eid0102.952004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Styer LM, Lim PY, Louie KL, Albright RG, Kramer LD, Bernard KA. 2011. Mosquito saliva causes enhancement of West Nile virus infection in mice. J. Virol. 85:1517–1527. 10.1128/JVI.01112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Styer LM, Kent KA, Albright RG, Bennett CJ, Kramer LD, Bernard KA. 2007. Mosquitoes inoculate high doses of West Nile virus as they probe and feed on live hosts. PLoS Pathog. 3:1262–1270. 10.1371/journal.ppat.0030132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider BS, Higgs S. 2008. The enhancement of arbovirus transmission and disease by mosquito saliva is associated with modulation of the host immune response. Trans. R. Soc. Trop. Med. Hyg. 102:400–408. 10.1016/j.trstmh.2008.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geering K. 1975. Haemolytic activity in the blood clot of Aedes aegypti. Acta Trop. 32:145–151 [PubMed] [Google Scholar]
- 6.Stark KR, James AA. 1995. A factor Xa-directed anticoagulant from the salivary glands of the yellow fever mosquito Aedes aegypti. Exp. Parasitol. 81:321–331 [DOI] [PubMed] [Google Scholar]
- 7.Champagne DE, Ribeiro JM. 1994. Sialokinin I and II: vasodilatory tachykinins from the yellow fever mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 91:138–142. 10.1073/pnas.91.1.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasserman HA, Singh S, Champagne DE. 2004. Saliva of the Yellow Fever mosquito, Aedes aegypti, modulates murine lymphocyte function. Parasite Immunol. 26:295–306. 10.1111/j.0141-9838.2004.00712.x [DOI] [PubMed] [Google Scholar]
- 9.Calvo E, Mans BJ, Andersen JF, Ribeiro JM. 2006. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 281:1935–1942 [DOI] [PubMed] [Google Scholar]
- 10.Calvo E, Sanchez-Vargas I, Favreau AJ, Barbian KD, Pham VM, Olson KE, Ribeiro JM. 2010. An insight into the sialotranscriptome of the West Nile mosquito vector, Culex tarsalis. BMC Genomics 11:51. 10.1186/1471-2164-11-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thangamani S, Wikel SK. 2009. Differential expression of Aedes aegypti salivary transcriptome upon blood feeding. Parasit. Vectors 2:34. 10.1186/1756-3305-2-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. 2007. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics 8:6. 10.1186/1471-2164-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider BS, McGee CE, Jordan JM, Stevenson HL, Soong L, Higgs S. 2007. Prior exposure to uninfected mosquitoes enhances mortality in naturally-transmitted West Nile virus infection. PLoS One 2:e1171. 10.1371/journal.pone.0001171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider BS, Soong L, Girard YA, Campbell G, Mason P, Higgs S. 2006. Potentiation of West Nile encephalitis by mosquito feeding. Viral Immunol. 19:74–82. 10.1089/vim.2006.19.74 [DOI] [PubMed] [Google Scholar]
- 15.Colpitts TM, Conway MJ, Montgomery RR, Fikrig E. 2012. West Nile virus: biology, transmission, and human infection. Clin. Microbiol. Rev. 25:635–648. 10.1128/CMR.00045-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thangamani S, Higgs S, Ziegler S, Vanlandingham D, Tesh R, Wikel S. 2010. Host immune response to mosquito-transmitted chikungunya virus differs from that elicited by needle inoculated virus. PLoS One 5:e12137. 10.1371/journal.pone.0012137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ader DB, Celluzzi C, Bisbing J, Gilmore L, Gunther V, Peachman KK, Rao M, Barvir D, Sun W, Palmer DR. 2004. Modulation of dengue virus infection of dendritic cells by Aedes aegypti saliva. Viral Immunol. 17:252–265. 10.1089/0882824041310496 [DOI] [PubMed] [Google Scholar]
- 18.Surasombatpattana P, Ekchariyawat P, Hamel R, Patramool S, Thongrungkiat S, Denizot M, Delaunay P, Thomas F, Luplertlop N, Yssel H, Misse D. 2013. Aedes aegypti saliva contains a prominent 34-kDa protein that strongly enhances dengue virus replication in human keratinocytes. J. Investig. Dermatol. [Epub ahead of print.] 10.1038/jid.2013.251 [DOI] [PubMed] [Google Scholar]
- 19.Machain-Williams C, Mammen MP, Jr, Zeidner NS, Beaty BJ, Prenni JE, Nisalak A, Blair CD. 2012. Association of human immune response to Aedes aegypti salivary proteins with dengue disease severity. Parasite Immunol. 34:15–22. 10.1111/j.1365-3024.2011.01339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox J, Mota J, Sukupolvi-Petty S, Diamond MS, Rico-Hesse R. 2012. Mosquito bite delivery of dengue virus enhances immunogenicity and pathogenesis in humanized mice. J. Virol. 86:7637–7649. 10.1128/JVI.00534-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson SL, Richards SL, Smartt CT. 2010. A simple method for determining arbovirus transmission in mosquitoes. J. Am. Mosq. Control Assoc. 26:108–111. 10.2987/09-5935.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S. 2008. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J. Virol. 82:8411–8421. 10.1128/JVI.00611-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pletnev AG, Bray M, Hanley KA, Speicher J, Elkins R. 2001. Tick-borne Langat/mosquito-borne dengue flavivirus chimera, a candidate live attenuated vaccine for protection against disease caused by members of the tick-borne encephalitis virus complex: evaluation in rhesus monkeys and in mosquitoes. J. Virol. 75:8259–8267. 10.1128/JVI.75.17.8259-8267.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. 2012. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J. Virol. 86:8546–8558. 10.1128/JVI.00736-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modis Y, Ogata S, Clements D, Harrison SC. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231. 10.1128/JVI.79.2.1223-1231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 75:5627–5637. 10.1128/JVI.75.12.5627-5637.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 2013:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816–820. 10.1038/77553 [DOI] [PubMed] [Google Scholar]
- 30.Jacobs JJ, Lehe CL, Hasegawa H, Elliott GR, Das PK. 2006. Skin irritants and contact sensitizers induce Langerhans cell migration and maturation at irritant concentration. Exp. Dermatol. 15:432–440. 10.1111/j.0906-6705.2006.00420.x [DOI] [PubMed] [Google Scholar]
- 31.Lehe CL, Jacobs JJ, Hua CM, Courtellemont P, Elliott GR, Das PK. 2006. Subtoxic concentrations of allergenic haptens induce LC migration and maturation in a human organotypic skin explant culture model: a novel method for identifying potential contact allergens. Exp. Dermatol. 15:421–431. 10.1111/j.0906-6705.2006.00415.x [DOI] [PubMed] [Google Scholar]
- 32.Ouwehand K, Oosterhoff D, Breetveld M, Scheper RJ, de Gruijl TD, Gibbs S. 2011. Irritant-induced migration of Langerhans cells coincides with an IL-10-dependent switch to a macrophage-like phenotype. J. Investig. Dermatol. 131:418–425. 10.1038/jid.2010.336 [DOI] [PubMed] [Google Scholar]
- 33.Price AA, Cumberbatch M, Kimber I, Ager A. 1997. Alpha 6 integrins are required for Langerhans cell migration from the epidermis. J. Exp. Med. 186:1725–1735. 10.1084/jem.186.10.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. 2012. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One 7:e50512. 10.1371/journal.pone.0050512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang IH, Nam HJ, Lee WJ. 2008. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 41:102–107. 10.5483/BMBRep.2008.41.2.102 [DOI] [PubMed] [Google Scholar]
- 36.Hammonds AS, Fristrom JW. 2006. Mutational analysis of Stubble-stubbloid gene structure and function in Drosophila leg and bristle morphogenesis. Genetics 172:1577–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Appel LF, Prout M, Abu-Shumays R, Hammonds A, Garbe JC, Fristrom D, Fristrom J. 1993. The Drosophila Stubble-stubbloid gene encodes an apparent transmembrane serine protease required for epithelial morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 90:4937–4941. 10.1073/pnas.90.11.4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SY, Kwon TH, Hyun JH, Choi JS, Kawabata SI, Iwanaga S, Lee BL. 1998. In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur. J. Biochem. 254:50–57. 10.1046/j.1432-1327.1998.2540050.x [DOI] [PubMed] [Google Scholar]
- 39.Shin SW, Zou Z, Raikhel AS. 2011. A new factor in the Aedes aegypti immune response: CLSP2 modulates melanization. EMBO Rep. 12:938–943. 10.1038/embor.2011.130 [DOI] [PMC free article] [PubMed] [Google Scholar]