Abstract
Alpha interferon (IFN-α) suppresses human immunodeficiency virus type 1 (HIV-1) replication in vitro by inducing cell-intrinsic retroviral restriction mechanisms. We investigated the effects of IFN-α/ribavirin (IFN-α/riba) treatment on 34 anti-HIV-1 restriction factors in vivo. Expression of several anti-HIV-1 restriction factors was significantly induced by IFN-α/riba in HIV/hepatitis C virus (HCV)-coinfected individuals. Fold induction of cumulative restriction factor expression in CD4+ T cells was significantly correlated with viral load reduction during IFN-α/riba treatment (r2 = 0.649; P < 0.016). Exogenous IFN-α induces supraphysiologic restriction factor expression associated with a pronounced decrease in HIV-1 viremia.
TEXT
Detailed analyses of the molecular and immunologic effects of the cytokine alpha interferon (IFN-α) may provide insights that contribute to the development of novel prophylactic, therapeutic, and curative strategies for HIV-1 infection. Induction of IFN-α expression is a critical first step in the defense against a range of viral infections (1, 2). The antiretroviral activity of IFN-α was demonstrated in vitro soon after the discovery of HIV-1 (3), and several studies have reported that exogenous IFN-α treatment potently suppresses HIV-1 in vivo (4–8). Moreover, IFN-α therapy was recently associated with significant reduction in the size of the HIV-1 latent reservoir (9), suggesting that interferon-associated pathways may be exploited to achieve HIV-1 eradication. The mechanisms underlying the in vivo anti-HIV-1 capacity of IFN-α remain to be thoroughly elucidated.
Cell-intrinsic immune mechanisms likely contribute to the beneficial effects of type I interferon (10). On this front, our laboratory recently reported that induction of the BST2/tetherin, APOBEC3G, and APOBEC3F cell-intrinsic immune defenses contributes to the IFN-α-mediated suppression of HIV-1 in vivo (6). A large number of additional host restriction factors with anti-HIV-1 activity in vitro have now been identified and characterized. In this study, we performed a comprehensive analysis of the effects of exogenous IFN-α treatment on all established anti-HIV-1 host restriction factors and HIV-1 viremia in vivo.
We examined restriction factor gene expression patterns in longitudinal samples from HIV/HCV-coinfected individuals undergoing pegylated IFN-α/ribavirin (IFN-α/riba) combination therapy and additionally characterized a separate population of non-interferon-treated control individuals. We designed and implemented a custom TLDA to measure the expression of 34 anti-HIV-1 restriction genes (11). We relied on the following two minimal criteria for inclusion in our CuRe TLDA: (i) peer-reviewed, published evidence of direct inhibition of HIV-1 replication in vitro and (ii) detectable expression in human peripheral blood mononuclear cells. All factors in the CuRe array meet the essential, minimal definition of a host restriction factor and function in a cell-autonomous manner to suppress HIV-1 replication.
Abbreviations: APOBEC, apolipoprotein B mRNA editing enzyme; ART, antiretroviral therapy; BST2, bone marrow stromal cell antigen 2; CT, cycle threshold; CuRe, cumulative restriction; SAMHD1, SAM domain and HD domain-containing protein 1; TRIM, tripartite motif; ISG, interferon-stimulated gene; CDKN1A, cyclin-dependent kinase inhibitor 1A; PAF1, RNA polymerase II-associated factor; EIF2AK2, eukaryotic translation initiation factor 2-alpha kinase 2; HCV, hepatitis C virus; HERC5, HECT domain and RLD 5; HIV-1, human immunodeficiency virus type 1; IFITM, interferon-induced transmembrane; IFN-α/riba, alpha interferon/ribavirin; LCMV, lymphocytic choriomeningitis virus; MOV10, Moloney leukemia virus 10, homolog; PBMC, peripheral blood mononuclear cell; SLFN11, Schlafen family member 11; TLDA, TaqMan low-density array; GAPDH, glyceraldehyde 3-phosphate dehydrogenase;18S, 18S ribosomal RNA; ACTB, beta-actin; PPIA, peptidylprolyl isomerase A; RPLP0, 60S acidic ribosomal protein, large, P0; UBC, ubiquitin C.
Subjects and specimen processing.
Longitudinal samples were collected from 15 HIV/HCV-coinfected individuals enrolled in the Swiss HIV Cohort Study ([SHCS]; www.shcs.ch) (12) who underwent IFN-α/riba treatment (see Table S1 in the supplemental material). All subjects had PBMC available before, during, and after IFN-α/riba treatment, were ART naïve, and had detectable HIV-1 RNA at baseline. Blood was collected prospectively from 12 HIV-1-infected (ART-naïve) viremic individuals and 12 HIV-1-uninfected individuals enrolled in the University of California San Francisco (UCSF) SCOPE cohort (see Table S2). PBMCs were isolated with Ficoll-Paque Plus. CD4+ T cells were enriched from fresh PBMCs using an EasySep Human CD4+ T cell enrichment kit (StemCell Technologies), according to the manufacturer's instructions. The research was approved by the relevant institutional review boards, and all human participants gave written informed consent.
Gene expression profiling.
Total RNA was extracted from PBMCs and CD4+ T cells directly after enrichment using TRIzol reagent (Invitrogen). RNA was transcribed into cDNA using a SuperScript VILO cDNA synthesis kit (Invitrogen). Quantitative real-time PCR utilized custom-made TLDA from Life Technologies, following the manufacturer's instructions. All assays and their respective target genes are listed in Table S3 in the supplemental material. Thermal cycling was performed using an ABI ViiA 7 real-time PCR System. Data were analyzed using ABI ViiA 7 software. A panel of six housekeeping genes (GAPDH, 18S, ACTB, PPIA, RPLP0, and UBC) was included in the TLDA plates. RPLP0 was identified as the most stably expressed housekeeping gene using the GeNorm algorithm (13). Therefore, raw CT numbers of amplified gene products were normalized to the housekeeping gene, RPLP0, to control for cDNA input amounts (see Fig. S1 in the supplemental material). Fold induction was determined using the comparative CT method (13). APOBEC3G, APOBEC3F, BST2/tetherin, and ISG15 relative copy numbers were recalculated from our previous work (6).
CuRe score calculation.
Missing (undetectable) values were imputed using the minimum expression value across samples for each gene. The expression value for the ith gene is notated as ei. A reference sample was selected based on having the maximum number of genes that were closest to the median gene expression profile. The reference expression value for the ith gene is notated as ri. The CuRe score for a sample is the cumulative fold difference in antiviral gene expression with respect to the value determined for the reference individual, expressed by the following formula:
Statistical analysis.
The paired Wilcoxon test, Mann-Whitney U test, and Pearson's r correlation coefficient were applied to data using GraphPad Prism v5.0c.
We initially examined the effects of IFN-α/riba treatment on plasma HIV-1 load. The 15 IFN-α/riba-treated individuals included in this study represent a subset of individuals studied in our previous work on IFN-α effects, chosen based on sample availability (6). Subject characteristics and IFN-α/riba treatment regimens are described in Table S1 in the supplemental material. IFN-α/riba treatment reduced plasma viral load by 0.91 (± 0.70) log10 copies/ml during treatment, and viremia typically returned to approximate pretreatment levels following therapy cessation (see Fig. S2 in the supplemental material). This effect is consistent with previous IFN-α/riba and IFN-α monotherapy studies (4, 5, 7, 8). Seven of 15 individuals were included in analyses of PBMC gene expression, and the remaining eight individuals were included in analyses of CD4+ T cell gene expression. A separate population of 24 IFN-α-untreated individuals (12 HIV-1 uninfected and 12 HIV-1 infected and ART naïve) enrolled in the SCOPE cohort was additionally characterized as a control group (see Table S2).
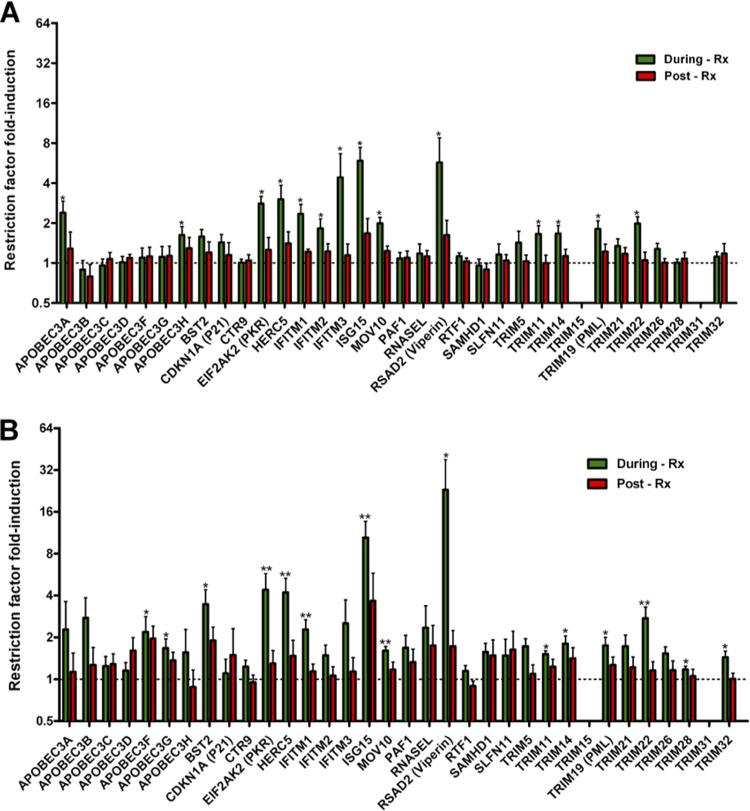
We next implemented the CuRe array to examine the effects of IFN-α/riba treatment on the expression of 34 anti-HIV-1 restriction factors (described in Table S3 in the supplemental material) in PBMCs and CD4+ T cells. Expression of 14 (APOBEC3A, APOBEC3H, IFITM1, IFITM2, IFITM3, ISG15, PKR, HERC5, MOV10, RSAD2 [viperin], TRIM11, TRIM14, TRIM19, and TRIM22) of 34 restriction genes was significantly elevated in unfractionated PBMCs during the IFN-α/riba treatment period with respect to pretreatment levels (Fig. 1A). Expression of an overlapping but distinct set of 15 restriction factors (APOBEC3F, APOBEC3G, BST2/tetherin, IFITM1, ISG15, PKR, HERC5, MOV10, RSAD2 [viperin], TRIM11, TRIM14, TRIM19, TRIM22, TRIM28, and TRIM32) was significantly induced by IFN-α/riba treatment in isolated CD4+ T cells (Fig. 1B). In both unfractionated PBMCs and CD4+ T cells, expression of induced genes returned to approximate baseline levels postcessation of IFN-α/riba treatment. Gene-by-gene fold induction levels (and statistics) observed in PBMCs and CD4+ T cells are presented in Table S4 in the supplemental material.
FIG 1.
IFN-α/riba induction of anti-HIV-1 restriction factors. (A) Fold induction in unfractionated PBMCs. (B) Fold induction in negatively selected CD4+ T cells. Values corresponding to fold induction during IFN-α/riba (green bars) and post-IFN-α/riba (red bars) treatment were normalized to the pretreatment expression level, as indicated by the dashed line. The mean and standard error are represented in each bar. Asterisks indicate statistically significant differences between the expression levels determined during and pretherapy based on a paired Wilcoxon test (* = P < 0.05; ** = P < 0.01).
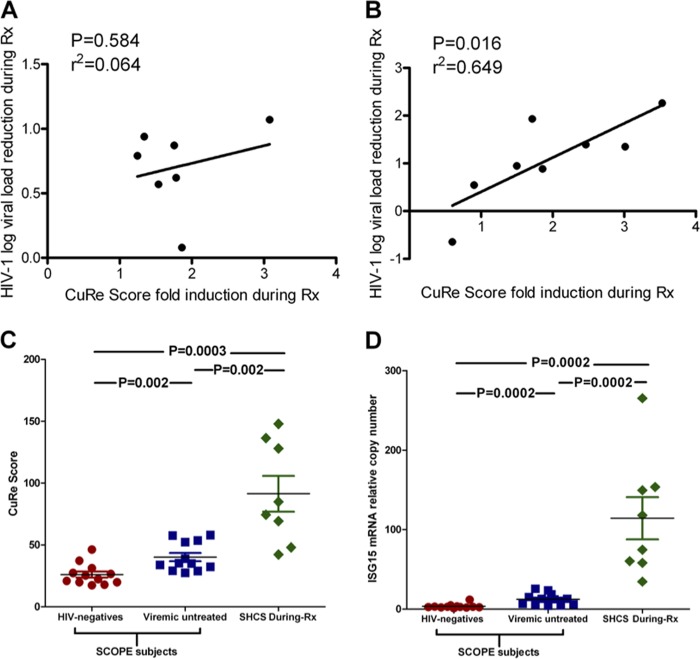
To infer the contribution of anti-HIV-1 restriction factors to the observed IFN-α/riba-mediated suppression of HIV-1, we examined correlations between the induction of restriction genes and reduction in HIV-1 viremia during the treatment period. We defined the CuRe score (explained above in greater detail) to represent overall, cumulative restriction factor gene expression across our 34 measured targets. IFN-α/riba-mediated viral load reduction did not exhibit any correlation with induction of the CuRe score in unfractionated PBMCs (r2 = 0.064, P = 0.584) (Fig. 2A) or induction of any of the 34 restriction factors in PBMCs considered on an individual gene-by-gene basis. IFN-α/riba-mediated viral load reduction exhibited a significant, pronounced correlation with induction of the CuRe score in CD4+ T cells (r2 = 0.649, P = 0.016) (Fig. 2B). Viral load reduction was significantly correlated with the induction of nine individual anti-HIV-1 restriction genes in CD4+ T cells (see Fig. S3 in the supplemental material), listed here hierarchically according to the strength of statistical association: TRIM22 (r2 = 0.64, P = 0.008), PKR (r2 = 0.533, P = 0.019), TRIM32 (r2 = 0.499, P = 0.025), TRIM11 (r2 = 0.534, P = 0.031), HERC5 (r2 = 0.435, P = 0.038), APOBEC3F (r2 = 0.42, P = 0.041), RSAD2 (viperin) (r2 = 0.41, P = 0.043), BST2/tetherin (r2 = 0.402, P = 0.046), and IFITM1 (r2 = 0.401, P = 0.046).
FIG 2.
IFN-α/riba induction of cumulative restriction (CuRe) score to supraphysiologic levels correlates with viral load reduction. (A) Relationship between HIV-1 load reduction during IFN-α/riba therapy and CuRe score fold induction in PBMCs; no significant correlation was observed. (B) Relationship between HIV-1 load reduction during IFN-α/riba therapy and CuRe score fold induction in CD4+ T cells. Significance was assessed using Pearson's r correlation coefficient. (C) Comparison between CuRe scores of SHCS subjects undergoing IFN-α/riba therapy and of HIV-1-negative and HIV-1-infected (ART-naïve) viremic individuals enrolled in the SCOPE cohort. (D) Comparison of levels of expression of ISG15 (a marker of interferon exposure) between SHCS subjects undergoing IFN-α/riba therapy and HIV-1-negative and HIV-1-infected (ART-naïve) viremic individuals enrolled in the SCOPE cohort. Mann-Whitney tests were employed to determine significance.
To investigate the hypothesis that exogenous IFN-α drives supraphysiologic expression of restriction factors, we compared CD4+ T cell anti-HIV-1 restriction factor expression in our SHCS interferon-treated population to CD4+ T cell restriction factor expression in a separate population of 12 HIV-1-infected (ART-naïve) viremic individuals and 12 HIV-1-uninfected individuals enrolled in the UCSF SCOPE cohort. There were no significant differences between SHCS subjects pre-IFN-α/riba treatment and SCOPE viremic subjects in terms of CuRe score (P = 0.464) or ISG15 expression (P = 0.203). However, there was a significant elevation in the CuRe score in SHCS subjects during IFN-α/riba treatment with respect to SCOPE viremic (P = 0.002) and uninfected (P = 0.0003) subjects, respectively (Fig. 2C). Similarly, ISG15 expression was significantly higher in SHCS subjects during IFN-α/riba treatment than in SCOPE viremic (P = 0.0002) and uninfected (P = 0.0002) subjects (Fig. 2D), supporting the hypothesis that exogenous IFN-α drives antiviral gene expression to supraphysiologic levels. Gene-by-gene analyses for each of the nine genes associated with IFN-α/riba-mediated suppression of HIV-1 viremia in our study are presented in Fig. S4 in the supplemental material.
Several recent reports have rejuvenated interest in deciphering the molecular pathways associated with the antiviral effects of IFN-α (6, 7, 9, 14–16). In particular, two recent reports on LCMV infection suggest that IFN-α is associated with both beneficial and detrimental disease outcomes and that the overall balance between the various, diverse effects of type I interferon ultimately determines the course of disease (14, 15). This is mirrored in studies of interferon within the context of HIV-1 infection, whereby IFN-α is known to induce several antiretroviral mechanisms that suppress viral replication but that may result in poor disease outcomes due to association with T cell activation and inflammation (17–19). Taken together, these observations suggest that additional work needs to be performed to dissect interferon-associated molecular pathways to identify critical antiretroviral mechanisms and to avoid possible proinflammatory consequences.
Endogenous IFN-α is often associated with rapid HIV-1 disease progression and high viral load (17, 18). We hypothesized that the inverse relationship between IFN-α and viral load observed within the context of exogenous IFN-α administration may result from the induction of antiviral genes to supraphysiologic levels not typically encountered in the absence of pharmacological manipulation. Our data strongly support this hypothesis and suggest that the induction of several restriction factors contributes to IFN-α suppression of HIV-1 in vivo. It is provocative that individual N in our study exhibited the greatest IFN-α/riba induction of the CuRe score and was the only individual to suppress viral load to undetectable levels for the entire duration of our study (see Fig. S2 in the supplemental material). Moreover, exogenous IFN-α induces these antiviral mechanisms without appreciably increasing CD4+ T cell activation, which promotes viral transactivation and replication (20). The lack of a relationship between restriction factor induction in PBMCs and viral load reduction suggests that specific consideration of HIV-1 target cells may be important when evaluating cell-intrinsic immune effects. In addition, a number of innate and adaptive immune mechanisms are likely triggered by IFN-α which may influence disease outcomes as well. The contribution of non-cell-intrinsic mechanisms to IFN-α anti-HIV-1 effects should be explored in greater detail.
These data support the concept that the induction of particular intrinsic immune mechanisms may constitute a promising antiretroviral strategy, complementing our previous in vitro work (21) and our translational studies of restriction mechanisms in HLA-B*57-positive individuals (22) and HIV-1 elite controllers (11).
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients who participate in the SHCS, UCSF SCOPE Cohort patients who participated in our study, the physicians and study nurses for excellent patient care, and the UCSF CFAR core laboratories for processing and storage of the samples.
This study was supported by grants from the National Institutes of Health (1K01DA024654 and R01 AI081668), the University of California, San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research (P30 AI027763), the DARE: Delaney AIDS Research Enterprise (DARE; U19AI096109), and the CFAR Network of Integrated Systems (R24 AI067039). Additional support was provided by Swiss HIV Cohort Study Project 594 and Swiss National Science Foundation grant 324730-130865. The Swiss HIV Cohort Study is supported by Swiss National Science Foundation grant 33CS30-134277 and the Swiss HIV Cohort Study Research Foundation.
The members of the Swiss HIV Cohort Study are V. Aubert, J. Barth, M. Battegay, E. Bernasconi, J. Böni, H. C. Bucher, C. Burton-Jeangros, A. Calmy, M. Cavassini, M. Egger, L. Elzi, J. Fehr, J. Fellay, H. Furrer (Chairman of the Clinical and Laboratory Committee), C. A. Fux, M. Gorgievski, H. Günthard (President of the SHCS), D. Haerry (deputy of “Positive Council”), B. Hasse, H. H. Hirsch, I. Hösli, C. Kahlert, L. Kaiser, O. Keiser, T. Klimkait, H. Kovari, R. Kouyos, B. Ledergerber, G. Martinetti, B. Martinez de Tejada, K. Metzner, N. Müller, D. Nadal, G. Pantaleo, A. Rauch (Chairman of the Scientific Board), S. Regenass, M. Rickenbach (Head of Data Center), C. Rudin (Chairman of the Mother & Child Substudy), P. Schmid, D. Schultze, F. Schöni-Affolter, J. Schüpbach, R. Speck, C. Staehelin, P. Tarr, A. Telenti, A. Trkola, P. Vernazza, R. Weber, and S. Yerly.
We declare that we have no conflicts of interest.
M.A.-M., T.L., J.C.G., and S.K.P. performed gene expression experiments and X.D. performed statistical analyses. A.R., B.L., S.G.D., and H.F.G. coordinated patient recruitment. M.A.-M., S.K.P., J.K.W., S.G.D., J.C.G., M.S.S., H.E.-D.A.G., and H.F.G. designed the studies. M.A.-M. and S.K.P. wrote the paper. We all read and approved the final manuscript.
Footnotes
Published ahead of print 23 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02687-13.
REFERENCES
- 1.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267. 10.1098/rspb.1957.0048 [DOI] [PubMed] [Google Scholar]
- 2.Stetson DB, Medzhitov R. 2006. Type I interferons in host defense. Immunity 25:373–381. 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 3.Poli G, Orenstein JM, Kinter A, Folks TM, Fauci AS. 1989. Interferon-a but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575–577. 10.1126/science.2470148 [DOI] [PubMed] [Google Scholar]
- 4.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, Lissen E, Gonzalez-Garcia J, Lazzarin A, Carosi G, Sasadeusz J, Katlama C, Montaner J, Sette H, Jr, Passe S, De Pamphilis J, Duff F, Schrenk UM, Dieterich DT. 2004. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N. Engl. J. Med. 351:438–450. 10.1056/NEJMoa040842 [DOI] [PubMed] [Google Scholar]
- 5.Aguilar Marucco D, Veronese L, de Requena DG, Bonora S, Calcagno A, Cavecchia I, Sinicco A, De Rosa FG, Cariti G, Di Perri G. 2007. Antiretroviral activity of pegylated interferon alfa-2a in patients co-infected with HIV/hepatitis C virus. J. Antimicrob. Chemother. 59:565–568. 10.1093/jac/dkl497 [DOI] [PubMed] [Google Scholar]
- 6.Pillai SK, Abdel-Mohsen M, Guatelli J, Skasko M, Monto A, Fujimoto K, Yukl S, Greene WC, Kovari H, Rauch A, Fellay J, Battegay M, Hirschel B, Witteck A, Bernasconi E, Ledergerber B, Gunthard HF, Wong JK. 2012. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proc. Natl. Acad. Sci. U. S. A. 109:3035–3040. 10.1073/pnas.1111573109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asmuth DM, Murphy RL, Rosenkranz SL, Lertora JJ, Kottilil S, Cramer Y, Chan ES, Schooley RT, Rinaldo CR, Thielman N, Li XD, Wahl SM, Shore J, Janik J, Lempicki RA, Simpson Y, Pollard RB. 2010. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J. Infect. Dis. 201:1686–1696. 10.1086/652420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbard JJ, Greenwell-Wild T, Barrett L, Yang J, Lempicki RA, Wahl SM, Asmuth DM, Murphy RL, Pollard RB, Kottilil S. 2012. Host gene expression changes correlating with anti-HIV-1 effects in human subjects after treatment with peginterferon Alfa-2a. J. Infect. Dis. 205:1443–1447. 10.1093/infdis/jis211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzoni L, Foulkes AS, Papasavvas E, Mexas AM, Lynn KM, Mounzer K, Tebas P, Jacobson JM, Frank I, Busch MP, Deeks SG, Carrington M, O'Doherty U, Kostman J, Montaner LJ. 2013. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J. Infect. Dis. 207:213–222. 10.1093/infdis/jis663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neil S, Bieniasz P. 2009. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29:569–580. 10.1089/jir.2009.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Mohsen M, Raposo RA, Deng X, Li M, Liegler T, Sinclair E, Salama MS, Ghanem HE, Hoh R, Wong JK, David M, Nixon DF, Deeks SG, Pillai SK. 2013. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology 10:106. 10.1186/1742-4690-10-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoeni-Affolter F, Ledergerber B, Rickenbach M, Rudin C, Gunthard HF, Telenti A, Furrer H, Yerly S, Francioli P. 2010. Cohort profile: the Swiss HIV Cohort study. Int. J. Epidemiol. 39:1179–1189. 10.1093/ije/dyp321 [DOI] [PubMed] [Google Scholar]
- 13.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340:207–211. 10.1126/science.1235214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks DG. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340:202–207. 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Arriaza J, Arnáez P, Gómez CE, Sorzano CO, Esteban M. 2013. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS One 8:e66894. 10.1371/journal.pone.0066894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. 2010. Interferon-alpha drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS 24:1415–1423. 10.1097/QAD.0b013e32833ac623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, Shianna KV, Ge D, Gunthard HF, Goldstein DB, Telenti A, Swiss HIVCS, Center for HIVAVI 2010. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6:e1000781. 10.1371/journal.ppat.1000781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy GA, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, Mudd J, Schacker T, Funderburg NT, Pilch-Cooper HA, Debernardo R, Rabin RL, Lederman MM, Harding CV. 2013. Interferon-alpha is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS One 8:e56527. 10.1371/journal.pone.0056527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manion M, Rodriguez B, Medvik K, Hardy G, Harding CV, Schooley RT, Pollard R, Asmuth D, Murphy R, Barker E, Brady KE, Landay A, Funderburg N, Sieg SF, Lederman MM. 2012. Interferon-alpha administration enhances CD8+ T cell activation in HIV infection. PLoS One 7:e30306. 10.1371/journal.pone.0030306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raposo RA, Abdel-Mohsen M, Bilska M, Montefiori D, Nixon DF, Pillai SK. 21 August 2013. Effects of cellular activation on anti-HIV-1 restriction factor expression profile in primary cells. J. Virol. 10.1128/JVI.02128-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raposo RA, Abdel-Mohsen M, Holditch SJ, Kuebler PJ, Cheng RG, Eriksson EM, Liao W, Pillai SK, Nixon DF. 2013. Increased expression of intrinsic antiviral genes in HLA-B*57-positive individuals. J. Leukoc. Biol. [Epub ahead of print.] 10.1189/jlb.0313150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.