Abstract
Envelope glycoprotein (Env) reactivity (ER) describes the propensity of human immunodeficiency virus type 1 (HIV-1) Env to change conformation from the metastable unliganded state in response to the binding of ligands (antibodies and soluble CD4 [sCD4]) or incubation in the cold. To investigate Env properties that favor in vivo persistence, we inoculated rhesus macaques with three closely related CCR5-tropic simian-human immunodeficiency viruses (SHIVs) that differ in ER to cold (ERcold) and ER to sCD4 (ERsCD4); these SHIVs were neutralized by antibodies equivalently and thus were similar in ERantibody. All three SHIVs achieved high levels of acute viremia in the monkeys without alteration of their Env sequences, indicating that neither ERcold nor ERsCD4 significantly influences the establishment of infection. Between 14 and 100 days following infection, viruses with high ERcold and ERsCD4 were counterselected. Remarkably, the virus variant with low ERcold and low ERsCD4 did not elicit a neutralizing antibody response against the infecting virus, despite the generation of high levels of anti-Env antibodies in the infected monkeys. All viruses that achieved persistent viremia escaped from any autologous neutralizing antibodies and exhibited low ERcold and low ERsCD4. One set of gp120 changes determined the decrease in ERcold and ERsCD4, and a different set of gp120 changes determined resistance to autologous neutralizing antibodies. Each set of changes contributed to a reduction in Env-mediated entry. During infection of monkeys, any Env replication fitness costs associated with decreases in ERcold and ERsCD4 may be offset by minimizing the elicitation of autologous neutralizing antibodies.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is the cause of AIDS, which results from the depletion of CD4-positive T lymphocytes (1, 2). Infection of HIV-1 target cells is mediated by the trimeric viral envelope glycoprotein (Env) spike. The Env spike is composed of three protomers consisting of two subunits (gp120 and gp41) that are generated by cleavage of a gp160 precursor glycoprotein (3–5). HIV-1 Env mediates entry into CD4+ T lymphocytes by first engaging the receptor CD4, which initiates a conformational change in gp120 that allows the binding to chemokine receptors, typically CCR5 or CXCR4 (6–16). The gp120 interactions with CD4 and CCR5/CXCR4 also trigger conformational changes in gp41 that promote membrane fusion and virus entry (5, 17). This two-receptor system allows HIV-1 to bury vulnerable Env elements until engagement with the target cell, thus providing a steric mechanism to escape immune recognition by potentially neutralizing antibodies (18).
Neutralizing antibodies are elicited during natural HIV-1 infection. Most early-arising neutralizing antibodies are strain restricted and drive the evolution of Env changes that allow virus escape (19). More broadly neutralizing antibodies are elicited in only a minority of HIV-1-infected people, usually arising several years after the initiation of infection (20–23). Although passive administration of broadly neutralizing antibodies can prevent immunodeficiency virus infections in animal models (24–28), the contribution of neutralizing antibodies to the control of viremia in an established infection and their impact on disease progression are unknown. HIV-1 variants clearly escape early autologous neutralizing antibodies, but the impact of the accumulating escape-associated Env changes on viral fitness is still uncertain. Evidence has been presented that donor neutralization escape phenotypes revert in recipients upon transmission, suggesting that potential fitness costs associated with autologous escape are selected against in the absence of neutralization pressure in the new hosts (29–32). Likewise, virus escape mutants that arose in HIV-1-infected individuals passively treated with broadly neutralizing antibodies reverted after antibody levels diminished (33). On the other hand, another study found little evidence that replication rates of neutralization-resistant viruses consistently differ from those of early or transmitted/founder viruses from the same individual (34). Thus, the contribution of the neutralizing antibody response to the control of an already established HIV-1 infection remains unclear.
Chimeric simian-human immunodeficiency viruses (SHIVs) have been used to investigate the role of HIV-1-specific proteins in virus evolution, interaction with the host immune system, and pathogenesis (35–41). Although simian immunodeficiency virus SIVmac infection of rhesus macaques exhibits many parallels with HIV-1 infection of humans, the HIV-1 and SIVmac Envs differ in the degree of dependence on the CD4 receptor for entry, in the use of alternative coreceptors (besides CCR5), and in the composition of most neutralizing antibody epitopes (42). Therefore, SHIVs with both CCR5-using (R5), CXCR4-using (X4), and dual-tropic (R5X4) HIV-1 Envs have been created and studied (40, 43–48). The vast majority of transmitted/founder and early isolates of HIV-1 are R5 and infect the activated subset of CD4-positive T lymphocytes; in some HIV-1 subtypes, R5X4 or X4 variants that can infect naive T lymphocytes arise later in the course of infection (49–53). Some R5X4 and X4 SHIVs are efficient pathogens, causing rapid CD4-positive T-lymphocyte depletion and AIDS-like illness in nearly all infected monkeys (43, 54, 55); however, the rapidity with which the host immune system is compromised in these animals often results in the lack of generation of neutralizing antibodies (36, 39, 56). Therefore, infection of monkeys with R5 SHIVs is thought to mimic early HIV-1 infection more closely with respect to target cells, the pattern and timing of antiviral immune responses, and immunopathogenesis. However, R5 SHIVs are inconsistent with respect to establishing persistent infections that lead to immunodeficiency and AIDS-like illness. An understanding of the HIV-1 Env properties that determine in vivo consistency would expedite the development of R5 SHIVs with diverse transmitted/founder HIV-1 Envs for vaccine and other prevention studies.
HIV-1 Env has evolved several features (variable surface loops, carbohydrates, and structural lability) that minimize the generation and impact of potentially neutralizing antibodies. Both the trimer apex, where the gp120 V1/V2 variable regions are thought to reside, and the gp120 outer domain can be targeted by neutralizing antibodies with various degrees of breadth, and both regions are shielded by glycans (57, 58). The Env trimer apex “opens” upon CD4 engagement, consistent with the V1/V2 stem changing conformation to form the gp120 bridging sheet that, along with the V3 loop, engages the coreceptor (59, 60). The propensity (“intrinsic Env reactivity” [ER]) of HIV-1 Env variants to change their conformation from the unliganded state globally influences viral sensitivity to inactivation by neutralizing antibodies, soluble CD4 (sCD4), CD4-mimetic compounds, and prolonged exposure to cold (61–63). ER is inversely related to the magnitude of the activation energy barrier between the unliganded state of Env and lower-energy downstream states.
Although HIV-1 Env properties related to ER have been studied in tissue culture assays (61–63), their contribution to the establishment and persistence of infections in vivo is not understood. Here, we study the infection of rhesus macaques inoculated with three closely related R5 SHIVs. Two of the R5 SHIVs (SHIV-KA and SHIV-KY) were created by grafting the tropism-determining gp120 V3 loops from the R5 HIV-1ADA and HIV-1YU2 strains, respectively, into the dual-tropic, pathogenic SHIV-KB9 (44). The third R5 SHIV, SHIV-KY62, was derived from a rhesus macaque that had been inoculated 62 weeks earlier with SHIV-KY. These three SHIVs were found to be equally sensitive to neutralization by antibodies. However, the infectivity of SHIV-KA and SHIV-KY was inhibited more effectively by sCD4 binding and incubation in the cold than that of SHIV-KY62. These results corroborate previous observations (64, 65) suggesting that some HIV-1 Env variants exhibit global sensitivity to cold, sCD4, and antibodies, whereas other Env variants exhibit specific sensitivity to one or two of these agents. Here, we use ER to refer to global Env reactivity to multiple agents and introduce the terms ERcold, ERsCD4, and ERantibody to indicate Env reactivity to each of these specific agents. A high level of global ER implies that a virus variant exhibits high levels of ERcold, ERsCD4, and ERantibody. Thus, relative to SHIV-KY62, SHIV-KY and SHIV-KA exhibit increased ERcold and ERsCD4 but are comparable in ERantibody and ER. This provides an opportunity to investigate the specific contribution of ERcold and ERsCD4 to the efficiency of SHIV infection in monkeys. We examined the evolution in monkeys of Env phenotypes such as in vitro fitness and sensitivity to neutralizing antibodies, sCD4, and cold. The env genetic determinants of these properties were mapped. The results provide insights into the properties of Env that allow the establishment of persistent infection in monkeys and reveal differences between these in vivo fitness requirements and Env properties that dictate replication efficiency in tissue culture systems.
MATERIALS AND METHODS
Cells and virus.
Rhesus monkey peripheral blood mononuclear cells (PBMC) were isolated by Ficoll enrichment from 10 ml of blood-EDTA. Cells were stimulated for 72 h with 1 to 2 μg/ml of phytohemagglutinin (PHA) (Sigma-Aldrich) and cultured in the presence of 10% interleukin-2 (IL-2) (Hemagen Diagnostics) in RPMI plus 10% fetal bovine serum (FBS). CCR5+ CEMx174 5.25 M7 cells were a kind gift from Nathaniel Landau and were cultured in RPMI plus 10% FBS. CD4+ CCR5+ and CD4+ CXCR4+ Cf2Th cells were cultured with Dulbecco's modified Eagle's medium (DMEM) plus 10% FBS in the presence of 400 μg/ml G418 and 150 μg/ml hygromycin (66). Ghost cell lines expressing CD4 and several chemokine receptors were cultured in DMEM plus 10% FBS supplemented with 500 μg/ml G418, 100 μg/ml hygromycin, and 1 μg/ml puromycin (from Vineet N. KewalRamani and Dan R. Littman, NIH AIDS Research and Reference Reagent Program) (67). 293T, COS-1, and HOS cells were cultured in DMEM plus 10% FBS. Penicillin-streptomycin was added to all culture media. Cells were grown at 37°C with 5% CO2.
SHIV-KA and SHIV-KY were constructed by introducing the env sequences encoding the gp120 V3 loop from HIV-1ADA and HIV-1YU2, respectively, into the env sequence in the 3′ KB9 proviral clone (44). Viruses were generated by ligation of 5′ and 3′ proviral halves, as previously described for SHIV-KB9 (68). Briefly, 10 μg of 5′ KB9 and either 3′ KA or 3′ KY was digested with SphI (New England BioLabs) for 3 h before ligation with 2,000 U of high-concentration T4 DNA ligase (New England BioLabs) for 5 min at room temperature. Ligation products were immediately transfected by using Lipofectamine 2000 (Invitrogen) into HEK293T cells. Viral supernatants were harvested after 24 h, passed through a 0.45-μm filter, and transferred to CEMx174 5.25 M7 cell cultures for 24 h. Virus was removed from cultures after 24 h, and the medium was replaced with fresh RPMI plus 10% FBS. Viral infection was monitored by observing green fluorescent protein (GFP) expression in the cells under a fluorescence microscope and by assaying reverse transcriptase (RT) activity in cell supernatants for several days. Peak RT activity was typically detected 10 to 14 days after infection, and virus was harvested at this time. Cells were removed by centrifugation and 0.45-μm filtration of the virus-containing supernatant.
Single-round reporter viruses were produced by transfection of HEK293T cells seeded onto 10-cm2 plates with 2 μg of a packaging plasmid (pCMVΔP1ΔenvpA), 6 μg of an HIV-1 long terminal repeat (LTR)-driven luciferase reporter plasmid (pHIvec2.luc), and 2 μg of an Env expression plasmid (pSVIIIenv), using Lipofectamine 2000. Viral supernatants were harvested after 48 h, passed through a 0.45-μm filter, frozen in a dry ice-ethanol bath, and stored at −80°C. Virus stocks were normalized for RT activity prior to use.
Animals.
PBMC from Mamu A01-negative rhesus macaques were prescreened for susceptibility to infection by SHIV-KA, SHIV-KY, and SHIV-KY62. Approximately 1 million Ficoll-enriched PBMC were infected with 50,000 RT units of virus following PHA stimulation. Virus was removed after 24 h, and RT activity in the culture supernatants was monitored for 17 days. The monkeys whose PBMC supported the highest level of in vitro replication were chosen for inoculation. Two animals were intravenously inoculated with approximately 5,000,000 RT units of each virus. Blood samples were drawn from each animal twice weekly for the first 2 weeks, weekly for weeks 2 to 4, biweekly for weeks 4 to 10, and monthly after week 10.
Antibodies and soluble CD4.
The IgG1 b12 and b6 antibodies, which recognize the CD4-binding site of gp120; the PG9 antibody, which recognizes a quaternary structure-dependent epitope; and PGT128, which recognizes a carbohydrate-dependent gp120 epitope, were kind gifts from Dennis Burton (57, 58, 69, 70). The F105 and VRC01 antibodies, which also recognize the gp120 CD4-binding site, were kindly provided by Marshall Posner and John Mascola, respectively (71, 72). James Robinson provided the 17b antibody, which recognizes a CD4-induced gp120 epitope, and the 39F antibody, which binds to the gp120 V3 region (73). The 2G12 antibody, which targets a carbohydrate-dependent gp120 epitope, and the 4E10 antibody, which recognizes the membrane-proximal external region (MPER) of gp41, were provided by Hermann Katinger (74, 75). The anti-CD4 antibody OKT4 was purchased from eBioscience.
sCD4 was purified from transfected HEK293F supernatants, as previously described (76), and quantified by Bradford analysis. The anti-HIV-1 activity of each sCD4 preparation was normalized to that of earlier stocks by comparative sCD4 sensitivity assays.
Plasmids and site-directed mutagenesis.
All envelope glycoprotein expression vectors were derived from pSVIIIenv-KB9, which contains the entire KB9 env and rev genes flanked by HIV-1HXB2 LTR elements (68). The Asp718-BamHI fragments of some single-genome amplicons of SHIV-KA and SHIV-KY from days 70 to 98 after inoculation of monkeys (see below) were subcloned into pSVIIIenv-KB9. Other amplicons from days 154 to 434 after infection exhibited changes in regions outside the Asp718-BamHI fragment and required complete env sequence cloning by introducing SphI and BclI sites at the 5′ and 3′ extremities, respectively, of env. Digested fragments were ligated with 2,000 U of high-concentration T4 DNA ligase (New England BioLabs) for 5 min at room temperature.
Mutations specifying single-amino-acid changes were introduced into the pSVIIIenv-KY or relevant plasmids according to the QuikChange II XL site-directed mutagenesis protocol (Stratagene). All plasmids were transformed into Top10 cells (Invitrogen), cultured at 37°C, and completely sequenced.
Viremia measurements.
The numbers of viral RNA (vRNA) copies per ml of plasma were quantified by using an ultrasensitive branched DNA detection assay with a detection limit of 125 RNA copies per ml (Siemens). For monkeys 331-08 and 401-08, viral loads were determined by using a Qiagen QIAsymphony Virus/Bacteria Midi kit and QIAgility Applied Biosystems StepOne quantitative real-time PCR (CHAVI Viral Core Laboratory, Duke Human Vaccine Institute, Durham, NC); viral loads in these two monkeys were always well above the detection limit of the assay (250 to 500 copies/ml).
Single-genome analysis.
Amplification of single env segments was modified slightly from a previously described method (77). Briefly, viral RNA was isolated from infected plasma by using a QiaAmp kit (Qiagen) and eluted with AVE buffer diluted 1:5 with nuclease-free water. cDNA was generated from vRNA templates by using 0.24 μM reverse primer 1 (SHIV U3/R [CAGAGCGACTGAATACAGAGCG]) and Superscript III (Invitrogen), as recommended by the polymerase manufacturer. Limiting dilutions were applied to cDNA templates for the first round of amplification to provide an average of a single DNA copy per reaction in a 96-well plate. Each 20-μl PCR mixture included 0.2 mM deoxynucleoside triphosphate (dNTP), 2 mM MgSO4, and 0.5 U of High Fidelity Platinum Taq polymerase (Invitrogen). Primers used for the first round of amplification were 0.25 μM primer 2F (GTAGCATTAGTAGTAGCAAT) and SHIV U3/R, which anneal to positions 6203 and 9673 of the SHIV-KB9 genome, respectively. Templates were amplified through 35 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min. Second-round PCR products were amplified from 2 μl of first-round PCR products using 0.5 μM primers 4F (GACAGGTTAATTGATAGACTA) and 1R (TGGGTTTCTCCATGGAGT), which anneal to positions 6293 and 9000, respectively, through 45 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 3 min. A positive ratio of 1:3 or less decreases the likelihood of multiple template amplification, thereby eliminating founder bias and recombination via primer-template switching during PCR (77). Each amplicon was purified by using the Qiagen PCR purification kit before sequencing.
Single-round infectivity assays.
Cf2Th or GHOST cells expressing CD4 and the coreceptor were plated at a density of 6,000 cells/well in a 96-well flat-bottom luminometer plate (PerkinElmer) 24 h prior to infection with HIV-luc pseudovirus. Viruses were prepared at 50,000 or 150,000 RT units/ml and serially diluted 1:2 with media to deliver 1,250 to 10,000 or 3,750 to 30,000 RT units/well in a total volume of 200 μl/well. Each infection was performed twice in triplicate. Cells were lysed with passive lysis buffer (Promega) after 48 h, and luciferase activity was measured in a luminometer (Berthold), as previously described (61, 64).
Assays to measure virus inhibition by autologous plasma, monoclonal antibodies, sCD4, and cold.
CD4+ CCR5+ Cf2Th cells were preplated for infection, as described above. The HIV-luc pseudovirus, normalized by RT, and heat-inactivated plasma, monoclonal antibody, or soluble CD4 were preincubated for 1 h at 37°C prior to infection. For autologous neutralization assays, 5,000 RT units/well of virus was preincubated with a 1:50 dilution of heat-inactivated plasma. Neutralization by a single dose of broadly acting monoclonal antibodies was performed with 10 μg/ml of 2G12, b12, VRC01, 4E10, 17b, or 39F and 5 μg/ml of PGT128. Virus was exposed to 0 to 20 μg/ml of sCD4 prior to infection for 1 h at 37°C. Virus was tested for cold sensitivity by incubation on ice for either 0, 24, 48, and 96 h or 0, 2, 4, 8, 16, and 24 h before infection. All infected cells were incubated at 37°C for 48 h and assayed for luciferase activity, as described above.
Env expression, processing, and recognition by autologous and heterologous plasma antibodies.
Env variants were expressed in HEK293T cells by transfection of pSVIIIenv and pTat at a ratio of 2:1 using Lipofectamine 2000 (Invitrogen). After 24 h, cells were radiolabeled with 100 μCi/ml of 35S protein-labeling mix (PerkinElmer) following depletion with cysteine- and methionine-free DMEM supplemented with 10% dialyzed FBS (Gibco Invitrogen). Cell-associated Env was harvested by NP-40 lysis of cells 24 h after labeling. Env was immunoprecipitated from 100 μl of cleared lysates with 3 μl of a mixture of polyclonal sera from HIV-1-infected individuals or inactivated plasma from the infected monkeys along with protein A-Sepharose beads (Roche) and washed three times with NP-40 buffer. Steady-state levels of expressed Env were determined by resolving gp160 and gp120 on a 10% Bis-Tris denaturing polyacrylamide gel (Invitrogen) run with morpholinepropanesulfonic acid (MOPS) buffer. Gels were visualized by PhosphorImager scanning (GE Healthcare Life Sciences).
Assays measuring sCD4 binding to the Env trimer.
Assays to measure soluble CD4 binding to trimeric Env complexes on cell surfaces were performed as previously described (61, 64). Briefly, plasmid pSVIIIenv was cotransfected with the HIV-1 Tat-expressing plasmid pTat into COS-1 or HOS cells. After 3 days, cells were washed with blocking buffer before incubation with sCD4 or the 2G12 anti-Env antibody (for normalization of Env expression) for 45 min at room temperature and then washed again with blocking buffer. Cells were incubated with an anti-human IgG or anti-CD4 (OKT4) antibody conjugated with horseradish peroxidase (HRP) for 45 min at room temperature and washed several times with buffer. HRP activity was measured in a luminometer after addition of Western Lightning oxidizing and luminol reagents (PerkinElmer Life Sciences).
The sCD4 reactivity of Env (ERsCD4) was calculated by using the formula , where IC50sCD4 is the 50% inhibitory concentration of sCD4.
RESULTS
Tropism and replicative abilities of the SHIVs.
The SHIVs used in this study were derived from the dual-tropic, pathogenic SHIV-KB9 (44). The tropism-determining gp120 V3 regions from the R5 primary viruses HIV-1ADA and HIV-1YU2 replace the SHIV-KB9 gp120 V3 region in SHIV-KA and SHIV-KY, respectively. The env gene of SHIV-KY62 was derived from a virus isolated 62 weeks after the infection of a monkey with SHIV-KY. The Envs of SHIV-KA, SHIV-KY, and SHIV-KY62 were tested for the ability to infect CD4+ GHOST cell lines expressing different chemokine receptors, including CXCR4, Bob/Gpr15, Bonzo/Strl33, CCR1, CCR2b, CCR3, CCR4, CCR5, CCR8, and CX3CR1. Only the CD4+ CCR5+ cells were infected by viruses bearing the SHIV-KA, SHIV-KY, and SHIV-KY62 Envs (data not shown). Thus, the substitution of the SHIV-KB9 gp120 V3 region with the V3 region of the R5 HIV-1 isolates resulted in SHIVs that utilize CCR5 but not CXCR4 as a coreceptor.
The ability of the SHIV Envs to mediate virus infection was tested. In single-round infections, recombinant HIV-1 with the KY Envs infected cells more efficiently than viruses with the KA Envs, and these were more efficient than viruses with the KY62 Envs (Fig. 1A). Multiple-round replications of SHIV-KA, SHIV-KY, and SHIV-KY62 in CD4+ CCR5+ CEMx174 5.25 M7 cells and in rhesus macaque PBMC were compared. In these cells, SHIV-KY replicated equivalently to or slightly better than SHIV-KA; both SHIV-KY and SHIV-KA replicated more efficiently than SHIV-KY62 (Fig. 1B and data not shown). Thus, in multiple assays, the Envs from passaged SHIV-KY62 supported virus replication less efficiently than the Envs of either SHIV-KY or SHIV-KA.
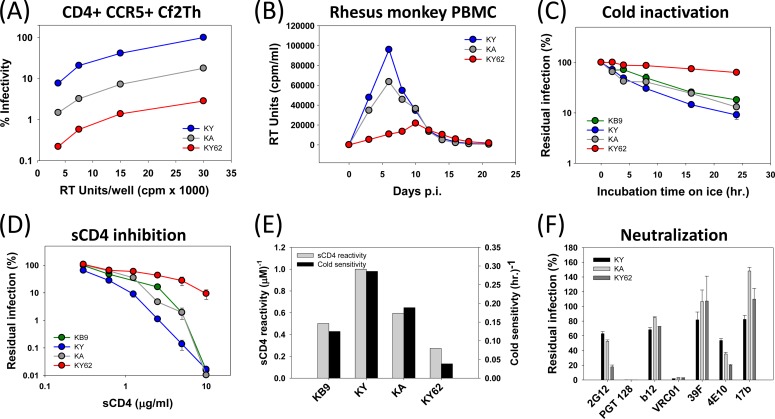
FIG 1.
Infectivity and ER-related properties of SHIV Envs. All virus preparations were normalized by reverse transcriptase (RT) activity and used to test infectivity and sensitivity to inhibition. (A) Infectivity of KY, KA, and KY62 Env-pseudotyped reporter viruses was tested in CD4+ CCR5+ Cf2Th cells. The Cf2Th CD4+ CCR5+ cells were lysed 48 h after infection and assayed for luciferase activity. All values are expressed as percent infection at a given virus level compared to the value observed for the maximum input of the KY virus. (B) The infectivity of full-length SHIVs was tested in PBMC pooled from three rhesus macaques. Cell supernatants were collected at the times indicated and assayed for RT activity. (C to F) Cold inactivation (C and E), sCD4 inhibition (D and E), and neutralization sensitivity (F) experiments were performed by using reporter viruses pseudotyped with the KB9, KY, KA, or KY62 Envs. (C) Single-round reporter viruses were incubated on ice for 0 to 24 h before infection. (D and F) Viruses were preincubated at 37°C with 0 to 20 μg/ml of sCD4 (D) or with 10 μg/ml of 2G12, b12, 39F, 17b, 4E10, or VRC01 or 5 μg/ml of PGT128 (F) 1 h before infection. All infections were performed in triplicate with Cf2Th CD4+ CCR5+ target cells, using RT-normalized levels of input virus. Cells were lysed after 48 h and evaluated for luciferase activity. Error bars represent standard errors of the means from two separate experiments. (E) sCD4 reactivity and cold sensitivity of the indicated SHIV Envs are shown. sCD4 reactivity was calculated as described in Materials and Methods and normalized to the ERsCD4 of viruses pseudotyped with KY Env, which was set at a value of 1. Cold sensitivity was calculated as the reciprocal of the time (IT50) on ice required to inactivate 50% of the viral infectivity.
Properties related to ER of SHIVs.
Global changes in intrinsic Env reactivity (ER) of HIV-1 Env influence HIV-1 sensitivity to cold, soluble CD4, and antibodies (61). Thus, a high-ER Env will, by definition, exhibit high levels of ERcold, ERsCD4, and ERantibody. Specific changes in ERcold, ERsCD4, and ERantibody are also possible for individual HIV-1 Env variants that are not globally reactive (64, 65). To evaluate properties of the parental SHIV-KB9, SHIV-KA, SHIV-KY, and SHIV-KY62 related to ER, recombinant viruses pseudotyped with the Envs were tested for infectivity following incubation on ice and following incubation with sCD4 and neutralizing antibodies. The susceptibility to inactivation in the cold (ERcold) exhibited the rank order KY > KA > KB9 ≫ KY62 (Fig. 1C).
Env reactivity to sCD4 (ERsCD4) reflects the inhibition of HIV-1 infectivity by sCD4 relative to the binding of sCD4 to the trimeric Env complex (61). Recombinant viruses with the KY and KA Envs as well as parental KB9 Env were more sensitive to sCD4 than were viruses with KY62 Env (Fig. 1D). When sCD4 binding to the Env complex was measured, the sCD4 reactivity of each Env could be calculated and exhibited the rank order KY > KA > KB9 > KY62 (Fig. 1E). As has been previously observed (61), cold sensitivity and sCD4 reactivity correlated (Fig. 1E).
Increases in ER result in globally increased HIV-1 sensitivity to neutralizing antibodies (64). Viruses pseudotyped with the KB9, KA, KY, and KY62 Envs were neutralized comparably by a number of monoclonal antibodies directed against gp120 or gp41 (Fig. 1F). Viruses with KY62 Env were slightly more sensitive than the other viruses to the glycan-directed anti-gp120 antibody 2G12 and to the MPER-directed anti-gp41 antibody 4E10. All four viruses were resistant to the weakly neutralizing anti-CD4BS antibody F105 or b6 (70, 71) and to the quaternary structure-dependent antibody PG9/PG16 (due to a lack of a critical glycosylation site at asparagine 160 [data not shown]) (58, 78). Thus, these viruses exhibited comparable sensitivity to neutralizing monoclonal antibodies, suggesting that the KB9, KA, KY, and KY62 Envs have similar levels of global intrinsic reactivity (ER) and ERantibody.
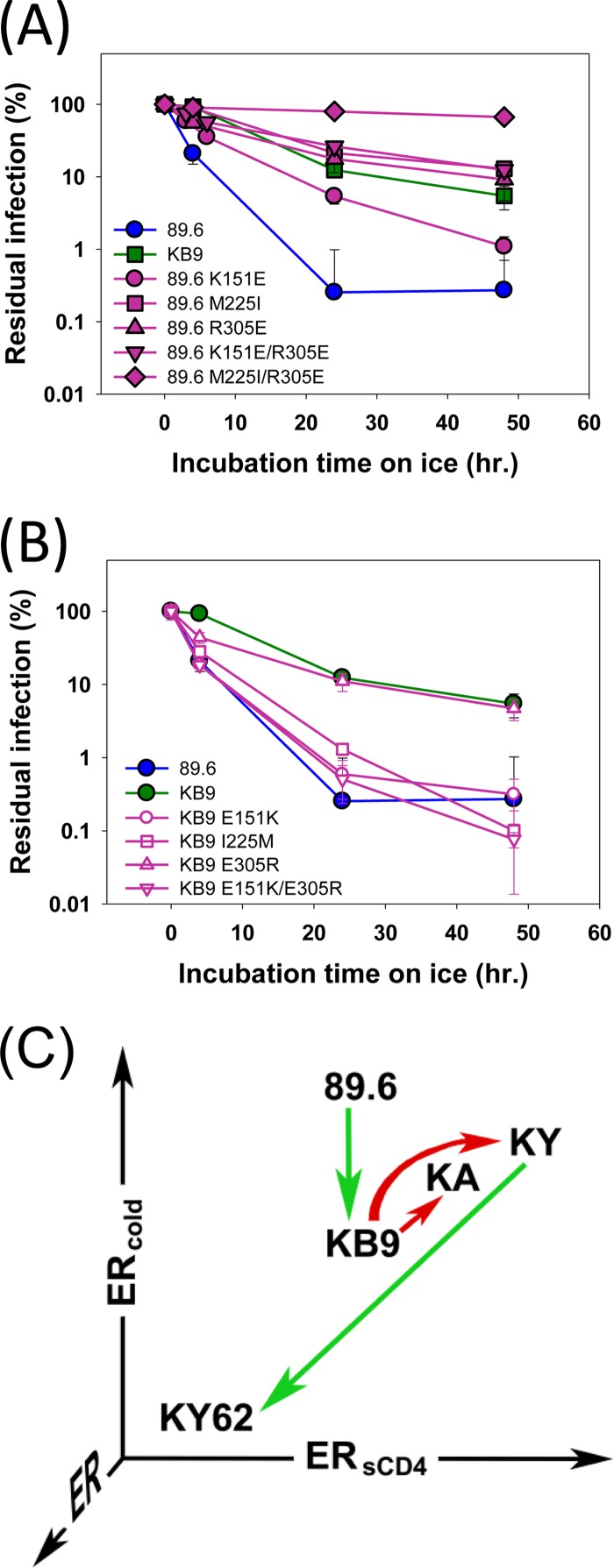
The parental virus for these studies, SHIV-KB9, is highly pathogenic and was derived by serial passage of the less virulent SHIV-89.6 in rhesus macaques (44). The Env ectodomains of SHIV-KB9 and SHIV-89.6 differ by only 12 amino acid residues. We compared 89.6 and KB9 Env properties related to ER. Sensitivity to sCD4 and CD4-binding affinity, and therefore ERsCD4, are similar for KB9 and 89.6 (64). Viruses with the KB9 and 89.6 Envs exhibited comparable sensitivity to most neutralizing antibodies, with the exception of IgG1 b12 and AG1121 (38). Thus, the KB9 and 89.6 Envs exhibit similar global ER and ERantibody. However, the cold sensitivities of viruses with these Envs differed significantly, with the average half-life on ice being >10-fold higher for KB9 than for 89.6 (Fig. 2A and B). The Env residues responsible for this difference in ERcold mapped to residues 151 (V1), 225 (C2), and 305 (V3) (Fig. 2A and B). Of note, residues 225 and 305 also contribute to the increased virulence and syncytium-forming ability of KB9 relative to those of 89.6 (79). As was observed for SHIV-KY and SHIV-KY62, passage of SHIV-89.6 in monkeys produced a virus, SHIV-KB9, with a significant reduction in ERcold.
FIG 2.
Env determinants of differences in cold sensitivity between SHIV-89.6 and SHIV-KB9. (A and B) Residues in 89.6 Env that differ from those in KB9 Env were altered, alone or in combination, to those found in KB9 Env and vice versa. Recombinant luciferase-expressing HIV-1 pseudotyped with the designated Envs was incubated on ice for the indicated times, and infectivity was determined following incubation with Cf2Th CD4+ CCR5+ cells. Experiments were performed as indicated in the Fig. 1 legend. (C) The approximate ERcold and ERsCD4 values of the SHIV Envs used in this study are plotted. The red arrows indicate phenotypic changes that resulted from modifications of Env introduced in the laboratory. The green arrows indicate phenotypic changes that arose during passage of the virus in monkeys. Note that the ERantibody of all the studied Envs is low.
The Env properties of the SHIVs are summarized in Fig. 2C. The KY and KA Envs exhibited higher ERsCD4 and ERcold than did parental KB9 Env. Chimerism in the V3 loop of SHIV-KY and SHIV-KA presumably contributed to the observed increases in ERsCD4 and ERcold. Infection of a monkey with SHIV-KY resulted in a virus, SHIV-KY62, with decreases in both ERsCD4 and ERcold. Likewise, after monkey passage, SHIV-89.6 with a high ERcold gave rise to SHIV-KB9, with a significantly reduced ERcold. The ERsCD4 of 89.6 is low and did not change upon in vivo passage in monkeys. Thus, high levels of ERcold and ERsCD4 are apparently counterselected in vivo. This implies that low ERcold and ERsCD4 are advantageous in the host, even when global neutralization sensitivity and ER are similar (as is the case for all five SHIVs in this study).
Infection of rhesus monkeys with SHIV variants.
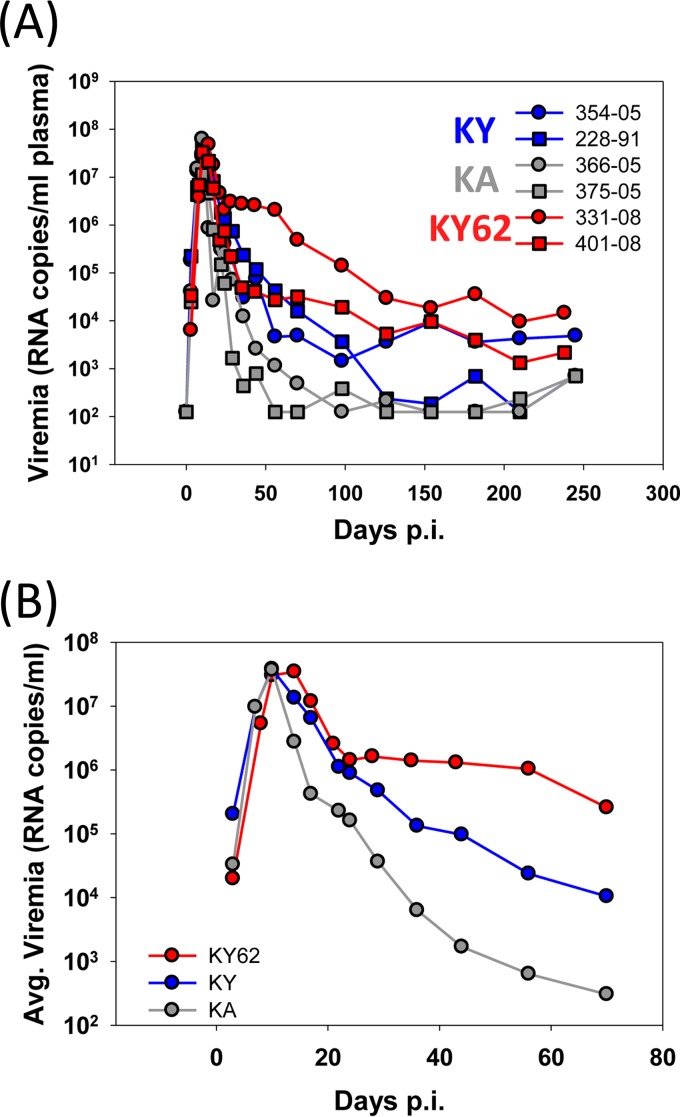
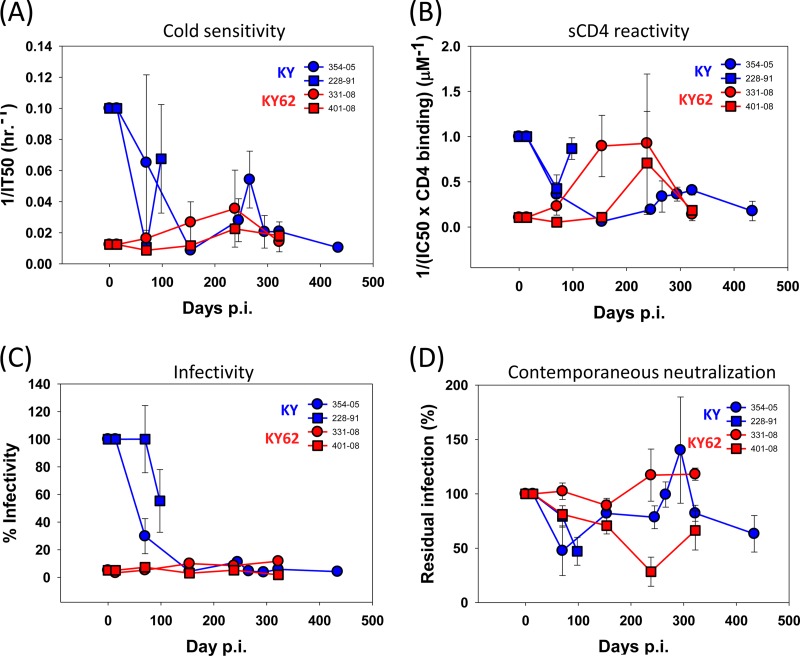
To test the hypothesis that low ERcold and ERsCD4 values are advantageous in vivo and to investigate the coevolution of the virus and host immune responses, SHIV-KY62 was inoculated intravenously into each of two rhesus macaques. We retrospectively studied the infection of four monkeys, two inoculated intravenously with the same dose of SHIV-KY and two inoculated with SHIV-KA. All six monkeys exhibited high levels (1 × 107 to 5 × 107 RNA copies/ml plasma) of viremia at day 10 postinoculation (p.i.) (Fig. 3A and B). Thus, no significant differences in the abilities of these SHIVs to replicate during the earliest, ramp-up phase of infection were apparent (Fig. 3A and B). Subsequently, the level of viremia decreased in all the animals, with the rate of decrease being dependent upon the particular virus. Plasma viral RNA levels diminished in both SHIV-KA-infected monkeys to near or below the limit of detection (125 RNA copies/ml plasma) by 100 days p.i. (Fig. 3A). Of the two animals inoculated with SHIV-KY, one monkey (monkey 354-05) maintained a level of viremia of between 103 and 104 RNA copies/ml plasma for at least 440 days p.i. (Fig. 3A and data not shown). Viral loads in the other SHIV-KY-infected monkey (monkey 228-91) decreased to between 100 and 1,000 RNA copies/ml plasma by 150 days p.i. The highest levels of chronic viremia (between 103 and 105 RNA copies/ml plasma) at between 100 and 200 days p.i. were seen for the two monkeys inoculated with SHIV-KY62. Thus, differences in ERcold and ERsCD4 among the SHIV variants did not apparently affect the ability of the virus to establish an acute infection. However, differences among the KA, KY, and KY62 Envs resulted in different rates of decline of viremia after the peak. The area under the curve (AUC) of viremia versus time was calculated for days 14 to 98 p.i. The average AUC was lowest for the SHIV-KA-infected monkeys and highest for the SHIV-KY62-infected animals (Table 1). Thus, SHIV-KY62, although containing an Env with lower fitness in tissue culture and lower ERcold and ERsCD4 than the KY and KA Envs, successfully persisted in the infected monkeys.
FIG 3.
Viremia in SHIV-infected monkeys. (A) Plasma virus levels in rhesus macaques intravenously inoculated with SHIV-KA, SHIV-KY, and SHIV-KY62. (B) Average viremia levels in SHIV-infected monkeys.
TABLE 1.
Viremia and autologous neutralization in infected monkeys
| Monkey | SHIV strain | Postpeak viremia (AUC) (106 RNA copies/ml)a | Avg postpeak viremia (AUC) (106 RNA copies/ml)b | Chronic viremia (RNA copies/ml)c | Autologous neutralization onset (days)d |
|---|---|---|---|---|---|
| 366-05 | KA | 6.43 | 8.7 | 6,020 | 28 |
| 375-05 | KA | 10.96 | 8,624 | 32 | |
| 354-05 | KY | 49.24 | 58.42 | 544,698 | 42 |
| 228-91 | KY | 67.6 | 82,404 | 59 | |
| 331-08 | KY62 | 262 | 161.2 | 4,433,000 | ≫70 |
| 401-08 | KY62 | 60.41 | 816,718 | ≫70 |
Area under the viral load-time curve (AUC) from days 14 to 98 p.i. Viral loads were measured in RNA copies/ml of plasma.
Average AUC for each virus group.
AUC from days 98 to 210 p.i.
Days p.i. when autologous neutralization potency reached 99% inhibition of input virus at a plasma dilution of 1:50.
Host antibody response in SHIV-infected monkeys.
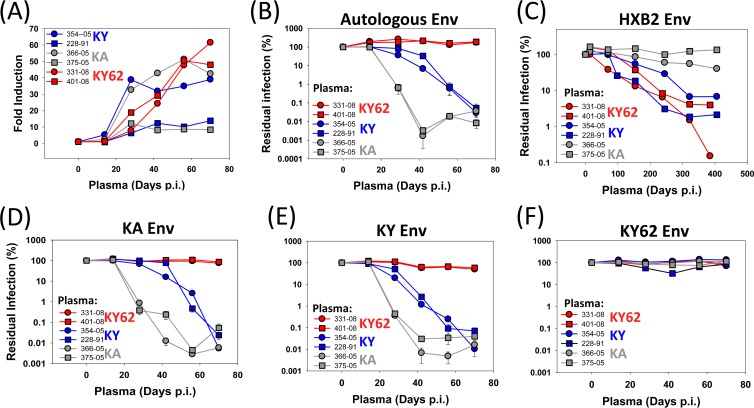
Early antibody responses to the infecting SHIVs in the monkeys were examined. All of the SHIV-infected monkeys generated detectable antibodies against Env by 28 days p.i. (Fig. 4A). By 70 days p.i., the anti-Env antibody titer was slightly higher in SHIV-KY62-infected monkeys than in monkeys infected with SHIV-KA or SHIV-KY (Fig. 4A).
FIG 4.
Generation of anti-Env antibodies in SHIV-infected monkeys. (A) Levels of antibodies in the plasma of SHIV-infected monkeys that recognize Env of the inoculated SHIV. (B to F) Reporter viruses pseudotyped with the indicated Env were preincubated for 1 h at 37°C with a single dilution of autologous or heterologous plasma (1:50) from the indicated times postinfection. After 1 h at 37°C, the virus-plasma mixtures were incubated with CD4+ CCR5+ Cf2Th cells. Luciferase activity was measured in the target cells. Residual infection is shown as the percentage of reporter activity observed with day 0 (uninfected) plasma. Error bars represent the standard errors of the means.
To evaluate the initial neutralizing antibody response in the SHIV-infected monkeys, single-round infection assays were performed in the presence of plasma from the infected animals. Recombinant HIV-1 carrying the KA, KY, or KY62 Envs was incubated with autologous and heterologous plasma from several time points during the first 70 days of infection and then incubated with CD4+ CCR5+ Cf2Th cells. Antibodies that neutralized viruses with the KA and KY Envs were generated by day 28 p.i. in both SHIV-KA-infected monkeys and further increased in potency by days 42 to 70 p.i. (Fig. 4B, D, and E). Neutralizing activity against viruses with KY and KA Envs appeared in the plasma of the SHIV-KY-infected monkeys by day 42 p.i. but was less potent than that seen in the SHIV-KA-infected animals. The plasma from the SHIV-KA- and SHIV-KY-infected monkeys did not neutralize viruses with the KY62 Envs (Fig. 4F). Remarkably, despite the generation of antibodies against Env in SHIV-KY62-infected monkeys, no neutralizing activity against viruses with KA, KY, or KY62 Envs was detected in the plasma of these monkeys (Fig. 4B and F). Thus, differences in Env apparently influence the generation of and/or susceptibility to the early neutralizing antibody response in the infected monkeys. Moreover, the potency of the early neutralizing antibody response against the infecting virus correlated inversely with the average level of chronic viremia at between 98 and 210 days p.i. (Spearman = 0.948; P < 0.05) (Table 1). These results are consistent with a contribution of neutralizing antibodies to the control of viremia at between 40 and 200 days p.i. in this model system.
Antibodies that neutralize heterologous strains of HIV-1 typically arise later than autologous neutralizing antibodies in the course of SHIV infection of monkeys or HIV-1 infection of humans (21, 37, 57, 80, 81). To examine whether any heterologous neutralizing antibodies were generated during infection with the SHIV-KA, SHIV-KY, and SHIV-KY62 variants, the plasma of the infected monkeys was tested for the ability to neutralize HIV-1 vectors pseudotyped with HXB2 Env. HXB2 Env is derived from a laboratory-adapted HIV-1 strain, so viruses with HXB2 Env are generally sensitive to neutralization (82). In this sensitive assay, antibodies neutralizing the viruses with HXB2 Env were detected in the plasma of monkeys infected with SHIV-KY and SHIV-KY62 by days 70 to 406 p.i. (Fig. 4C). In contrast, the plasma from monkeys infected with SHIV-KA, which did not achieve a detectable level of chronic viremia, did not efficiently neutralize viruses with HXB2 Env. We also examined the Env-directed antibody response in infected monkeys from the late chronic stage (day 322 p.i.) by immunoprecipitating HXB2 Env with various concentrations of plasma; a trend was observed between the titers of anti-Env antibodies at this stage and the level of chronic viremia, although this association did not reach statistical significance (Spearman rS = 0.83; P = 0.058). However, the level of cross-neutralization of viruses with HXB2 Env seen at this time point strongly correlated with the level of postacute viremia (Spearman rS = 0.94; P = 0.017); this suggests a possible relationship between the broadening of the neutralization response to the HXB2 virus and the level of early infection. Higher levels of anti-Env antibodies in the plasma, however, did not correlate with more potent HXB2 cross-neutralization (Spearman rS = 0.54; P = 0.30).
We also examined the ability of the monkey plasma from 70 to 406 days p.i. to inhibit infection mediated by Envs from heterologous primary HIV-1 isolates (YU2, ADA, and JRFL), which are generally more resistant to neutralization than the HXB2 isolate. None of the sera efficiently neutralized these viruses (data not shown). Thus, the HXB2-neutralizing activity observed in the plasma of SHIV-KY- and SHIV-KY62-infected monkeys at 70 to 406 days p.i. was not sufficiently broad or potent to inhibit the entry of neutralization-resistant primary viruses.
Timing of in vivo phenotypic changes related to ER.
The hypothesis that low values of ERsCD4 and ERcold are advantageous in vivo is supported by our observation that SHIVs with high ERsCD4 and/or ERcold were apparently counterselected in the infected monkeys and replaced by SHIV variants with lower ERsCD4 and ERcold. To investigate this selection further, we examined the ERsCD4 and ERcold of viruses containing Env variants PCR amplified from SHIV-KY- and SHIV-KY62-infected monkeys at multiple time points of infection. As the Env sequences of the viruses at the time of peak viremia (14 days p.i.) did not differ from those of the inoculated SHIVs (see below), ERsCD4 and ERcold did not change during the first 2 weeks of infection. Notably, by 70 days p.i., the circulating SHIV-KY viruses in monkeys 228-91 and 354-05 had Envs that were cold and sCD4 resistant, relative to KY Env in the inoculated SHIV (Fig. 5A and B). In the monkey in which infection persisted (monkey 354-05), the Envs from viruses at later time points remained relatively resistant to cold and sCD4. Transient increases in ERsCD4 and ERcold were observed in Envs from monkey 354-05 at around 250 to 300 days p.i.; however, by day 434 p.i., the Envs from the circulating viruses returned to a more resistant state. In contrast, in monkey 228-91, which controlled SHIV-KY infection, the Envs from circulating viruses became more sensitive to cold and sCD4 at 98 days p.i. compared with the day 70 Envs (Fig. 5A and B). These increases in ERcold and ERsCD4 coincided with a decrease of viremia in this monkey to undetectable levels. Envs from the monkeys infected with SHIV-KY62, a virus with low ERcold and low ERsCD4, exhibited slight increases in ERcold and more substantial increases in ERsCD4 between days 150 and 250 p.i. However, both ERcold and ERsCD4 returned to low levels by 322 days p.i.
FIG 5.
ERcold, ERsCD4, infectivity, and autologous neutralization of envelope glycoprotein isolates. Env variants derived from SHIV-KY- or -KY62-infected monkeys at the indicated times after inoculation were tested for infectivity and sensitivity to inhibition by incubation of pseudotyped reporter viruses on ice or with inhibitors (sCD4 and contemporaneous plasma). The viruses were then added to Cf2Th CD4+ CCR5+ cells, and luciferase activity was measured 48 h later. (A) Cold sensitivity is shown as the reciprocal of the average time on ice required for a 50% reduction in virus infectivity (IT50). (B) Average soluble CD4 reactivity was calculated as described in Materials and Methods. (C) Average infectivity of variants was calculated from infectivity of maximum virus input levels relative to KY virus infectivity observed with the same RT-normalized virus input. Each experiment was performed with four virus dilutions to ensure that subsaturating infection concentrations were compared. (D) Average autologous neutralization of variants was performed by measuring inhibition of infectivity in the presence of 2% contemporaneous plasma compared to preinfection plasma from the same monkey. All experiments were performed as described in the Fig. 1 legend.
Our results indicate that SHIVs with low or high ERcold and ERsCD4 can efficiently establish infection. However, between days 14 and 100 p.i., as the monkeys seroconverted, SHIVs with high ERcold and high ERsCD4 were counterselected. Transient increases in ERcold and ERsCD4 were apparently tolerated, but long-term-persistent SHIVs generally exhibited low values of these parameters.
Viral fitness and neutralization resistance.
The ability of recombinant HIV-1 pseudotyped with the Envs derived from SHIV-infected monkeys to infect target cells was examined. Compared with single-round viruses pseudotyped with the KY Envs, viruses pseudotyped with Envs derived from SHIV-KY-infected monkeys beyond 70 days p.i. were less infectious for CD4+ CCR5+ Cf2Th cells (Fig. 5C). Viruses pseudotyped with the parental SHIV-KY62 Envs, as well as Envs derived from the SHIV-KY62-infected monkeys (monkeys 331-08 and 401-08), were also significantly impaired for the ability to infect CD4+ CCR5+ Cf2Th cells (Fig. 5C). The proteolytic maturation and subunit association of the Envs were comparable (data not shown), indicating that the basis for the poor replication of the Envs that evolved in the monkeys did not involve a global disruption of the Env trimer. Thus, shortly after the peak of acute viremia, the SHIV-KY Envs exhibited decreases in ERcold, ERsCD4, and the ability to support virus entry into cultured CD4+ CCR5+ cells.
To examine whether the observed evolution of Env sequences in the SHIV-KY-infected monkeys might have resulted from pressure exerted by neutralizing antibodies, recombinant HIV-1 viruses pseudotyped with the Envs obtained at different times of infection were tested for sensitivity to neutralization by plasma from infected monkeys. Compared with viruses bearing the parental KY Envs (Fig. 4D), viruses with Envs from day 70 p.i. were resistant to neutralization by contemporaneous plasma from the SHIV-KY-infected monkeys (Fig. 5D). The ability of plasma from the SHIV-KY- and SHIV-KY62-infected monkeys to neutralize contemporaneous viruses was limited (Fig. 5D). These results suggest that SHIVs that establish chronic infections escape from neutralizing antibodies present in the plasma.
As the viruses in SHIV-KY-infected monkey 354-05 evolved resistance to autologous neutralizing antibodies, the viruses from days 70 to 245 p.i. became more sensitive to neutralization by the b12 CD4-binding-site antibody (data not shown). This increased sensitivity was specific, as the sensitivity of the viruses to the gp41-directed 4E10 antibody did not change during the course of infection (data not shown). Following the appearance of antibodies in the plasma of monkey 354-05 that neutralized viruses with the heterologous HXB2 Envs (beginning at days 245 to 434 p.i.), the SHIV-KY viruses in this animal reacquired resistance to the b12 antibody. These observations suggest that antibodies that drive this viral resistance may arise in this monkey only after 250 days p.i.
Evolution of SHIV Env sequences in infected monkeys.
The HIV-1 env genes described above were derived by a single-genome-amplification (SGA) procedure from SHIV-infected monkeys, allowing an assessment of the diversification of the viruses over time. As mentioned above, env sequences analyzed near the time of peak viremia (day 10 p.i.) exhibited no nonsynonymous changes compared with the env sequences of the parental SHIVs (data not shown). This result implies that SHIV-KA, SHIV-KY, and SHIV-KY62 were all efficient in establishing infection in the inoculated monkeys and that no Env adaptation was required to achieve high levels of viremia.
The env sequences of the SHIVs circulating in plasma during later phases of infection (70 to 434 days p.i.) were analyzed by SGA. The env sequences were amplified from the plasma of both SHIV-KY62-infected monkeys at multiple time points. Although env sequences were obtained from the plasma of both SHIV-KY-infected macaques at 70 days p.i. and from one of the monkeys (monkey 228-91) at 98 days p.i., env sequences could not be detected in monkey 228-91 at any subsequent time. In contrast, in the SHIV-KY-infected monkey (monkey 354-05) with higher levels of chronic viremia, env sequences were amplified from plasma on days 154 to 434 p.i. Between 8 and 40 amplicons were sequenced at each time point. No env sequences could be detected in the plasma of the SHIV-KA-infected monkeys on day 70 p.i., and no sequence divergence from the inoculate was found in amplicons from day 24 or 36 p.i., the last time point of amplifiable levels of vRNA in monkey 375-05 or 366-05, respectively. Thus, as might be expected, the success rate of SGA reflects the abundance of viruses in plasma.
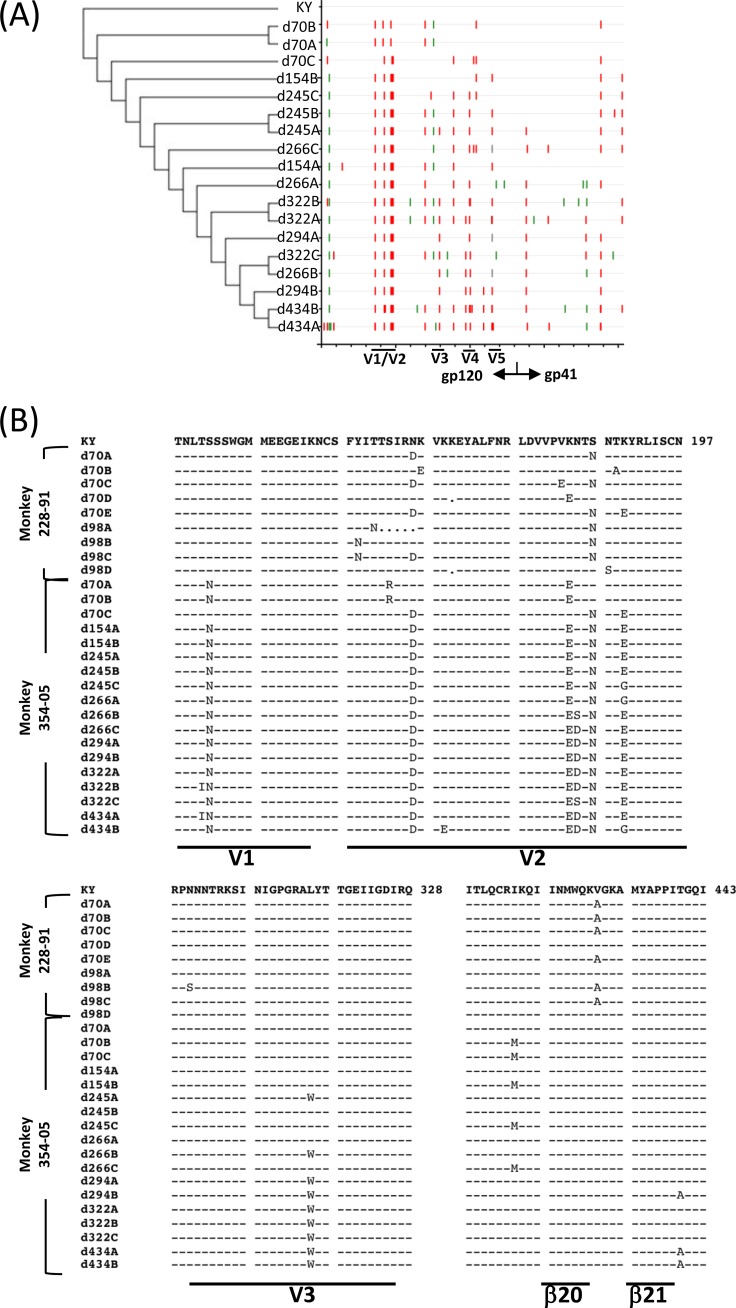
Amplicons from SHIV-KY-infected monkey 354-05 on days 28, 42, and 56 p.i. demonstrated minimal but diverse changes, particularly in the gp120 V2 and β20-β21 regions (data not shown). This phylogenetic “rake” of diverse viruses is “purified” by days 70 and 154 p.i., so that only three and two clones, respectively, were identified at each time point (Fig. 6A). This pattern is similar to the Env amplicon assortment from monkey 228-91 on day 70 p.i., which demonstrates charge changes and glycosylation rearrangements in V2 (Fig. 6B).
FIG 6.
Phylogeny and alignment of SHIV-KY and -KY62 Env sequences. (A) Cladogram (left) and Highlighter analysis (right) of env sequences from animal 354-05 showing synonymous (green) and nonsynonymous (red) mutations. Relative positions of gp120 variable regions are shown, and the gp120/gp41 cleavage point is delineated. (B and C) Alignment of Env sequences spanning the V1/V2, V3, and C4 gp120 regions from SHIV-KY-infected animals 228-91 (days 70 and 98) and 354-05 (days 70 to 434) (B) or SHIV-KY62-infected animals 331-08 and 401-08 (days 70 to 322) (C). Phylogenic tree construction was calculated with PhyML 3.0 (ATCG) (102). Alignments were generated by using Highlighter and SeqPublish (Los Alamos National Laboratory). All amino acid positions are numbered according to the HXB2 HIV-1 prototype.
From days 70 to 434 p.i., specific Env clonal lineages emerged in the circulating SHIV-KY viruses; a partial summary of the analyzed sequences is presented in Fig. 6B. The SHIV-KY viruses in both monkey 228-91 and monkey 354-05 exhibited similar changes in the gp120 V2 variable region involving an acidic substitution (N167D), a shift of a glycan from Asn186 to Asn187 (S187N), and additional basic-to-acidic substitutions near the altered glycosylation site (K185E and K190E). These changes contribute to a general increase in the acidity of the V2 region as infection proceeds, a trend previously noted for monkeys infected with SHIV-89.6P (83). The SHIV-KY Envs in monkey 354-05 also gained an N-linked site of glycosylation in the V1 variable region (S142N). Thus, the SHIV-KY Envs in both monkeys exhibited alterations of an N-linked glycosylation site and the appearance of acidic residues in the gp120 V2 variable region. In addition, a late-arising gp120 V3 change (L317W) became fixed in the SHIV-KY viruses in monkey 354-05 by day 294 p.i. In both SHIV-KY-infected animals, sporadic changes in β19 to β21 (I420M, V430A, and T440A) were also observed. In monkey 354-05, env diversity increased after 154 days p.i. by both point mutations and recombination.
Single genomes from SHIV-KY62-infected monkeys were amplified on days 10, 70, 154, 238, and 322 p.i. The animal with the highest sustained viremia, monkey 331-08, contained Envs with the most sequence diversity and the highest degree of change from parental SHIV-KY62; this may reflect the requirement to escape the stronger neutralizing antibody response generated in this animal than that in monkey 401-08 (data not shown). As was observed for SHIV-KY infection, most early substitutions in KY62 Env involved alterations in the gp120 V2 and β21 regions (Fig. 6C). Both SHIV-KY62-infected monkeys exhibited β21 A433T changes early in infection, with the K432R change emerging later in monkey 331-08. Some common acidic-to-basic changes in the V2 region were observed for both SHIV-KY62-infected animals. Viruses from monkey 331-08 also acquired changes in the gp120 V3 region not associated with known tropism determinants, suggesting that these changes arise in response to other selective pressures. Other noteworthy changes include a deletion in V1 and a few hydrophobic residue changes in the V2 stem as well as the addition of a potential N-linked glycosylation site at asparagine 160. The glycan at this position is important for the integrity of the quaternary structure-dependent neutralizing antibodies PG9 and PG16 (58).
Env determinants of resistance to autologous neutralizing antibodies, soluble CD4, and cold.
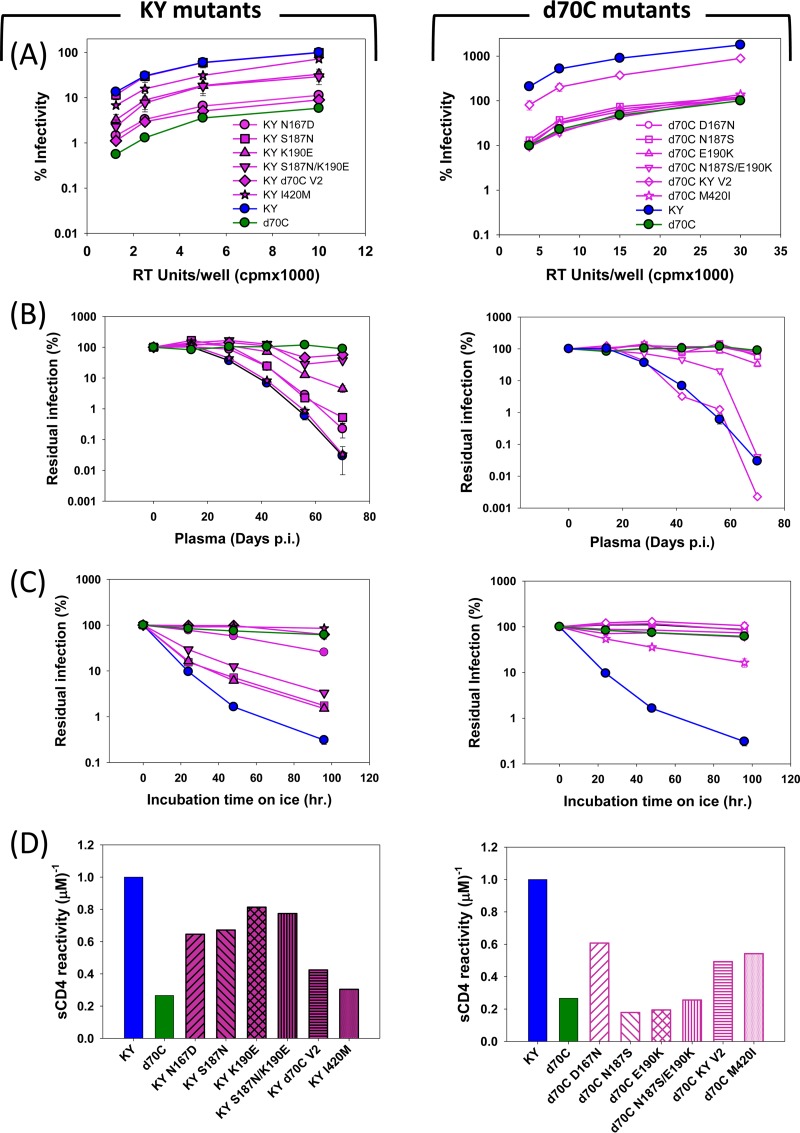
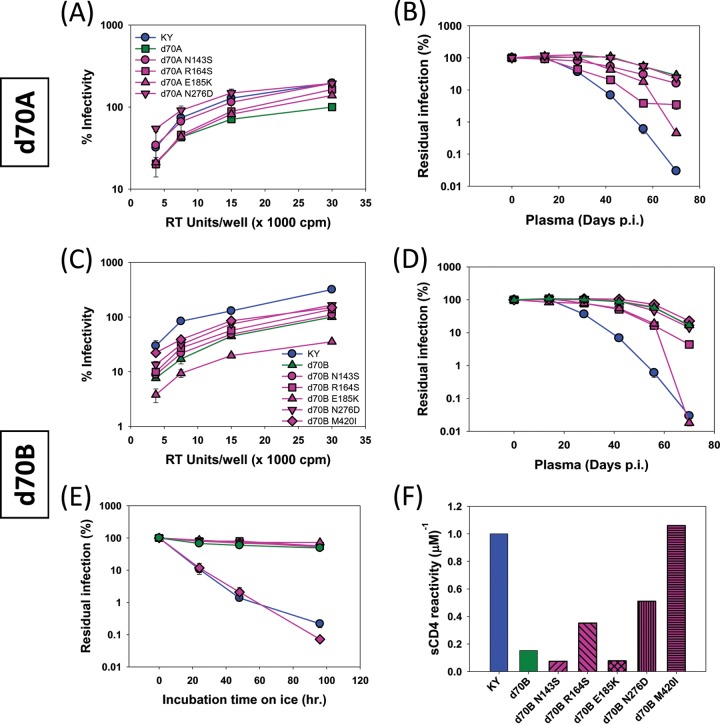
As mentioned above, Env sequences in SHIV-KY-infected monkey 354-05 exhibited greater sequence diversity between days 28 and 56 than at subsequent time points (days 70 and 154), suggesting the existence of a genetic bottleneck at this stage of the infection. This is consistent with the changes in fitness and resistance to autologous neutralizing antibodies, sCD4, and cold associated with the Envs derived from this animal. Of the three sequences recovered from monkey 354-05 on day 70, isolate C showed the highest phylogenetic relatedness to later isolates, including SHIV-KY62 (Fig. 6A). To investigate the Env determinants of the observed viral phenotypes, we studied the day 70 Env sequences because the relevant phenotypes were already established by this time, yet relatively few Env changes had occurred compared with the Env from the inoculated SHIV-KY. The amino acid residues in d70C Env that had changed during the course of infection were reverted to those found in KY Env, either individually or as a group. Conversely, the d70C Env changes were introduced into the KY Envs. The infectivity and sensitivity to autologous neutralizing antibodies, sCD4, and cold of the Env variants are shown in Fig. 7 and Tables 2 and 3. Collectively, the changes in the gp120 V2 variable region were necessary and sufficient to explain the resistance to autologous neutralizing antibodies of viruses with the d70C Envs (Fig. 7A and B). The shift in the V2 N-linked glycosylation site and nearby acidic substitution (S187N/K190E) were the major determinants of resistance to neutralizing antibodies. Other V2 region changes, S164R and K185E, were also found to contribute to the resistance to autologous neutralizing antibodies of the d70A and d70B Envs relative to that of the KY Env (Fig. 8B and D). Thus, gp120 V2 region changes determine resistance to autologous neutralizing antibodies in the SHIV-KY-infected monkey.
FIG 7.
Env mutant infectivity, autologous neutralization, ERcold, and ERsCD4. Viruses pseudotyped with KY (left) or d70C (right) Env recombinants were evaluated for infectivity and inhibition by plasma antibodies, cold, and sCD4 relative to the values observed for viruses pseudotyped with wild-type KY or d70C Envs, respectively. (A) Relative infectivity of the viruses pseudotyped with the indicated Env variants. (B) Neutralization of the recombinant viruses by monkey 354-05 plasma from days 0 to 70 p.i. (C and D) Cold sensitivity (C) and sCD4 reactivity (D) of the recombinants are shown. A summary of the KY and d70C Env phenotypes can be found in Tables 2 and 3, respectively. All experiments were performed as described in the Fig. 1 legend. The error bars indicate the standard errors of the means.
TABLE 2.
Infectivity, autologous neutralization, and sensitivity to cold and sCD4 inactivation of viruses pseudotyped with KY Env variantse
| Mutant | Location of gp120 change | Infectivity (%)a | Cold IT50 (h)b | sCD4 reactivityc | Neutralization (%)d |
|---|---|---|---|---|---|
| KY | Wild type | 100 | 7.1 | 1.0 | 99.9 |
| KY N167D | V2 region | 12 | 55 | 0.65 | 99.8 |
| KY S187N | V2 region | 99 | 9.4 | 0.67 | 99.5 |
| KY K190E | V2 region | 33 | 9.5 | 0.82 | 99.5 |
| KY S187N/K190E | V2 region | 29 | 14 | 0.77 | 62.8 |
| KY d70C V2 | V2 region | 11 | >96 | 0.42 | 42.6 |
| KY I420M | β19 | 79 | >96 | 0.30 | 99.9 |
Infectivity levels represent the percentage of mutant reporter virus readouts at maximum virus input compared with equivalent RT levels of reporter virus pseudotyped with KY Env.
Cold IT50 is calculated as the half-life of virus on ice, in hours.
sCD4 reactivity (ERsCD4) was calculated as described in Materials and Methods and is reported relative to the ERsCD4 of the KY Env, which is set at a value of 1.
Percent neutralization of virus with autologous contemporaneous plasma from monkey 354-05 at a 1:50 dilution.
Values in boldface type indicate phenotypes differing significantly from the wild type.
TABLE 3.
Infectivity, autologous neutralization, and sensitivity to cold and sCD4 inactivation of viruses pseudotyped with KY d70C Env variantse
| Mutant | Location of gp120 change | Infectivity (%)a | Cold IT50 (h)b | sCD4 reactivityc | Neutralization (%)d |
|---|---|---|---|---|---|
| d70C | Wild type | 100 | >96 | 0.27 | 11 |
| d70C D167N | V2 region | 117 | >96 | 0.61 | 33 |
| d70C N187S | V2 region | 121 | >96 | 0.18 | 42 |
| d70C E190K | V2 region | 110 | >96 | 0.19 | 67 |
| d70C N187S/E190K | V2 region | 119 | 80 | 0.26 | 99.9 |
| d70C KY V2 | V2 region | 890 | >96 | 0.49 | 99.9 |
| d70C G355R | LE | 100 | >96 | 0.15 | 41 |
| d70C T413I | V4 region | 78 | >96 | 0.23 | 10 |
| d70C M420I | β19 | 137 | 25 | 0.54 | 21 |
Infectivity levels represent the percentage of mutant reporter virus readout at maximum virus input compared with equivalent RT levels of reporter virus pseudotyped with d70C Env.
Cold IT50 is calculated as the half-life of virus on ice, in hours.
sCD4 reactivity (ERsCD4) was calculated as described in Materials and Methods and is reported relative to the ERsCD4 of KY Env, which is set at a value of 1.
Percent neutralization of virus with autologous contemporaneous plasma from monkey 354-05 at a 1:50 dilution.
Values in boldface type indicate phenotypes differing significantly from the wild type.
FIG 8.
Early mutant infectivity, autologous neutralization, ERcold, and ERsCD4. (A and C) Viruses pseudotyped with d70A or d70B Env mutants were evaluated for infectivity, relative to the values observed for wild-type d70A or d70B Envs, respectively. (B and D) Sensitivity of the pseudotyped viruses to neutralization by autologous plasma from days 0 to 70 is shown. (E and F) d70B mutants were also tested for cold sensitivity (E) and sCD4 reactivity (F). All experiments were performed as described in the Fig. 1 legend. Standard errors of the means are indicated by error bars.
Determinants of resistance to cold and sCD4 in the d70C Envs, relative to the Envs of the inoculated SHIV-KY, were investigated. The I420M change in the β19-β20 turn of gp120 made a major contribution to these phenotypes (Fig. 7C and D). Likewise, the I420M change was the major determinant of the relative cold and sCD4 resistance of the d70B Envs (Fig. 8E and F). Changes in residue 420 also made small contributions to the decrease in Env entry efficiency (Fig. 7A). Independently of the I420M change, the V2 region contributed to the resistance of the d70C Envs to cold and sCD4 relative to the KY Envs (Fig. 7C and D). The N167D change in V2 alone made a significant contribution to these phenotypes. Thus, changes in gp120 residue 420 made major contributions to resistance to cold and sCD4, and in some contexts, V2 changes could independently determine these phenotypes. Of note, the V2 changes that determined lower ERcold and ERsCD4 (the N167D change) and neutralizing antibody resistance (the S187N/K190E changes) differed.
The decreased infectivity of viruses with the d70C Envs relative to that of viruses with the KY Envs was investigated (Fig. 7A). Together, the changes in the gp120 V2 region were necessary and sufficient to explain the decreased infectivity of the Envs from day 70 p.i. The V2 change associated with decreased ERcold and ERsCD4 (N167D) and the V2 changes associated with resistance to neutralizing antibodies (S187N/K190E and K190E) individually resulted in decreased infectivity when introduced into the KY Env.
In summary, between peak viremia and day 70 p.i., the SHIV-KY Envs in monkey 354-05 exhibited a number of changes. The S187N/K190E changes in the gp120 V2 region, which involved a glycosylation shift and acidic substitution, conferred escape from autologous neutralizing antibodies. Another change in V2, N167D, which does not involve a change in glycosylation, decreased ERcold and ERsCD4. Changes in gp120 residue 420 in the β19-β20 turn independently decreased ERcold and ERsCD4. Changes associated with neutralization resistance and with reductions in ERcold and ERsCD4, particularly the V2 changes, contributed to the decreased ability of Env to mediate infection in tissue-cultured target cells. This fitness cost was apparently offset by the benefits of escaping neutralization by antibodies and other in vivo advantages associated with low ERcold and ERsCD4.
Late determinants of Env reactivity to cold and sCD4.
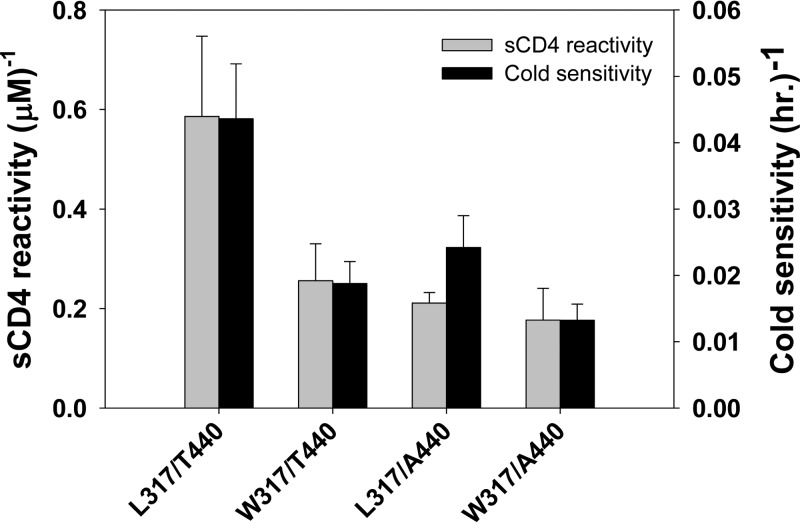
Between days 245 and 322 p.i., transient increases in ERcold and ERsCD4 were observed in the Envs of circulating SHIV-KY viruses in persistently infected monkey 354-05 (see above). These phenotypic changes correlated temporally with the emergence of Envs in the circulating viruses with two gp120 changes that became fixed in the circulating SHIVs: L317W near the tip of the V3 loop and T440A in the LF loop. These changes were introduced into d266A Envs that had neither change; leucine 317 and alanine 440 in d266B Envs were altered to tryptophan and threonine residues, respectively. Conversely, the tryptophan at residue 317 and alanine at residue 440 were reverted to leucine and threonine residues, respectively, in the d434A and d434B Envs. The most cold- and sCD4-resistant Envs in this mutant panel contained tryptophan 317 and alanine 440 (Table 4 and Fig. 9). Envs with leucine 317 and threonine 440 were most sensitive to cold and sCD4. Envs with only one of the resistance-associated residues exhibited intermediate phenotypes. Thus, the observed L317W and T440A changes contributed to the reemergence of SHIV-KY variants with low ERcold and ERsCD4 in monkey 354-05 beyond 322 days p.i.
TABLE 4.
Infectivity and sensitivity to cold and sCD4 inactivation of viruses pseudotyped with d266 and d434 Env variants from monkey 354-05
| Mutant | Residue at position: |
Infectivity (%)a | Cold IT50 (h)b | sCD4 reactivityc | |
|---|---|---|---|---|---|
| 317 | 440 | ||||
| d266A | L | T | 100 | 15 | 0.89 |
| d266A L317W | W | T | 102 | 43 | 0.15 |
| d266A T440A | L | A | 63 | 26 | 0.16 |
| d266A L317W/T440A | W | A | 255 | 50 | 0.11 |
| d266B | W | T | 100 | 76 | 0.46 |
| d266B W317L | L | T | 52 | 35 | 0.26 |
| d266B T440A | W | A | 52 | 84 | 0.14 |
| d266B W317L/T440A | L | A | 72 | 54 | 0.21 |
| d434A | W | A | 100 | >96 | 0.09 |
| d434A W317L | L | A | 72 | 54 | 0.21 |
| d434A A440T | W | T | 86 | 77 | 0.14 |
| d434A W317L/A440T | L | T | 72 | 28 | 0.36 |
| d434B | W | A | 100 | 79 | 0.37 |
| d434B W317L | L | A | 61 | 47 | 0.26 |
| d434B A440T | W | T | 90 | 39 | 0.27 |
| d434B W317L/A440T | L | T | 41 | 23 | 0.84 |
Infectivity levels represent the percentage of mutant reporter virus readout at maximum virus input compared with the infectivity levels of the reporter viruses pseudotyped with the corresponding wild-type Env.
Cold IT50 represents the half-life of virus on ice, in hours.
sCD4 reactivity (ERsCD4) was calculated as described in Materials and Methods and is reported relative to the ERsCD4 of KY Env, which is set at a value of 1.
FIG 9.
Changes in ERcold and ERsCD4 in late virus variants with L317W and/or T440A Env changes. The W317 and/or A440 residue was introduced into d266 Env or was changed to L317 and/or T440 in d434 Env from monkey 354-05. Cold inactivation and sCD4 reactivity of the pseudotyped viruses were measured as described in the Fig. 1 legend. A summary of the phenotypes, including relative infectivity, cold sensitivity, and sCD4 reactivity, for each late variant is described in Table 4.
DISCUSSION
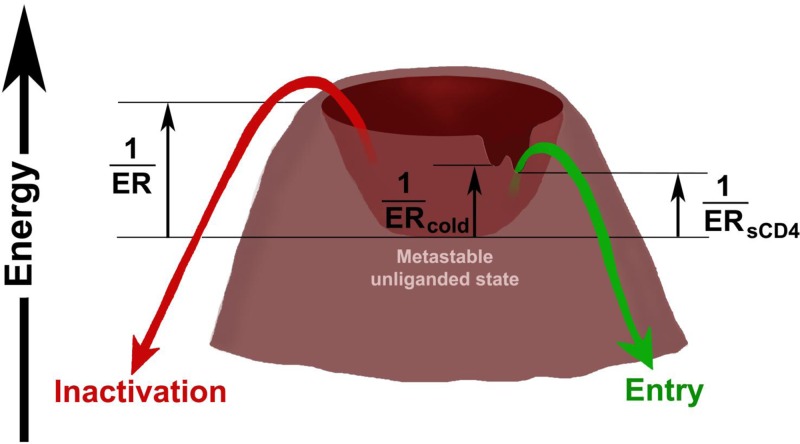
The infection of rhesus macaques with SHIVs represents an opportunity to study the properties of the HIV-1 Envs that contribute to the burst of acute viremia, establishment of persistent infection, evasion of the host immune response, and pathogenesis. Intrinsic envelope reactivity (ER) is a property of the HIV-1 Envs that reflects their propensity to negotiate transitions from the metastable unliganded state to other conformations (64). ER is inversely related to the activation barriers that separate the metastable, unliganded state of Env from lower-energy conformations (Fig. 10). ER can influence the consequences of the binding of ligands (e.g., antibodies) to the HIV-1 Env trimer and thereby exert an impact on virus sensitivity to neutralization (61). HIV-1 Envs with high ER are invariably sensitive to bound sCD4 and to cold, two properties that often correlate (64). By modulating the free energy of the metastable, unliganded state (Fig. 10, asterisk), Env reactivity to the binding of different ligands and to cold exposure could be globally altered. Not all cold-sensitive Envs are intrinsically reactive, however, and recent studies suggest that the global intrinsic envelope reactivity (ER), sensitivity to cold (ERcold), and reactivity to bound sCD4 (ERsCD4) can be independently regulated (64, 65). The viral phenotypes characterized in this study support this hypothesis and suggest that the heights of the activation barriers for virus inactivation by cold, sCD4, and antibodies can be individually modulated (Fig. 10). Viruses with high global ER or high ERantibody are expected to be counterselected by neutralizing antibodies in the host. However, as HIV-1 does not generally encounter either low temperature or sCD4 in human hosts, the importance of cold resistance and sCD4 reactivity for HIV-1 replication and persistence in vivo is unknown. Here, we inoculated rhesus monkeys with three R5 SHIV variants that exhibit similar sensitivities to neutralizing antibodies (and thus are similar in global ER and ERantibody) but differ in ERcold and ERsCD4. SHIV-KA and SHIV-KY differ only in the sequences of the gp120 V3 region, and SHIV-KY62 contains an Env that evolved in an infected monkey inoculated with SHIV-KY. Viruses with the KA, KY, and KY62 Envs were neutralized comparably by a panel of monoclonal antibodies directed against the HIV-1 Envs. However, the KA and KY Envs exhibited higher ERcold and ERsCD4 values than those of the KY62 Envs. In tissue-cultured cells, SHIV-KA and SHIV-KY replicated more efficiently than SHIV-KY62. Upon intravenous inoculation into monkeys, however, all three SHIVs were indistinguishable in their ability to replicate and achieve a high level of acute viremia. Thus, the basic capacities of SHIV-KA, SHIV-KY, and SHIV-KY62 to replicate in monkeys prior to the generation of an efficient antiviral response appear to be similar. This implies that the differences in the ERcold and ERsCD4 among the viruses exert negligible effects on the ability to initiate and establish an infection in monkeys. This is consistent with the observation that although transmitted/founder HIV-1 strains are all relatively resistant to neutralizing antibodies (i.e., they are “tier 2” viruses, with ER values in the low range), they exhibit a wide range of ERcold and ERsCD4 levels (64).
FIG 10.
Energy landscape of HIV-1 Env variants. The metastable, unliganded state of the HIV-1 Env glycoproteins exists in a local energy well. The heights of the activation barriers surrounding this local energy well are inversely related to the ER, ERantibody, ERsCD4, and ERcold (64). Changes in the energy of the metastable unliganded state (asterisk) result in global changes in ER, which in turn result in changes in ERantibody, ERsCD4, and ERcold. Note that “notches” in the hills surrounding the energy well allow ERantibody, ERsCD4, and ERcold to be independently regulated from each other and from ER. ERcold and ERsCD4 often correlate in HIV-1 Env variants (64). The activation energy associated with the binding of target cell CD4 can influence the level of CD4 required on the cell for HIV-1 entry. At the time of seroconversion in SHIV-infected monkeys, HIV-1 Envs with more shallow notches are apparently selected, lowering ERcold, ERsCD4, and entry efficiency. This is predicted to result in an Env that remains longer in the metastable, unliganded state.
Soon after the peak of viremia, host humoral immune responses emerged, and the courses of SHIV-KA, -KY, and -KY62 infection differed. By 14 to 28 days p.i., antibodies directed against Env were generated in the monkeys infected by SHIV-KA, SHIV-KY, and SHIV-KY62. By 28 to 42 days p.i., neutralizing antibodies directed against the inoculated virus were detected in the plasma of SHIV-KA- and SHIV-KY-infected monkeys. The generation of antibodies that neutralize the inoculated SHIV was dependent on the Envs of the inoculated virus. SHIV-KY62 did not elicit autologous neutralizing antibodies, despite the efficient elicitation of anti-Env antibodies in the infected monkeys and the comparable neutralizing antibody sensitivities of the KY62 and KA or KY Envs. Presumably, Env elements commonly targeted by strain-specific neutralizing antibodies during early infection are less accessible and/or immunogenic in the KY62 Envs. Indeed, the autologous neutralizing antibodies elicited in SHIV-KA- and SHIV-KY-infected monkeys did not neutralize viruses with the KY62 Envs. Envs with low ERcold and ERsCD4 are predicted to have higher activation barriers that decrease the spontaneous sampling of the CD4-bound or other low-energy conformations (Fig. 10) (64) and therefore may be less prone to elicit autologous neutralizing antibodies. Envs with low ERcold and ERsCD4 may also be less susceptible to any neutralizing antibodies generated. Thus, the low ERcold and ERsCD4 of the KY62 Envs may be relevant to the minimal elicitation of autologous neutralizing antibody responses by decreasing the number of Env conformations presented to the host immune system and by diminishing the impact of any elicited antibodies that can bind the Env trimer.
The generation of strain-specific neutralizing antibodies in HIV-1-infected humans is typically followed by the emergence of viruses that escape from inhibition by these antibodies (34, 84–90). The impact of neutralizing antibodies on HIV-1 fitness, viremia, and clinical course is uncertain (29, 30, 91–95). In the SHIV-infected monkeys, the decline in circulating virus after the peak of viremia was correlated directly with the potency of the neutralizing antibody response generated against the inoculated virus Envs. Furthermore, the emergence of SHIV variants resistant to the contemporaneous plasma neutralizing antibodies may contribute to the establishment of a persistent viremic state. SHIV-KA Envs did not exhibit any changes during the acute phase of infection and thus remained vulnerable to the potent neutralizing antibodies generated in the plasma of the infected monkeys. Perhaps, as a result, SHIV-KA loads in both infected macaques dropped to nearly undetectable levels by day 50 p.i. and remained there for the duration of the study. The levels of neutralizing antibodies directed against the inoculated SHIV-KY Envs were intermediate between those in SHIV-KA-infected monkeys and those in SHIV-KY62-infected animals. Although the SHIV-KY variants that emerged by 70 days p.i. in both infected monkeys were relatively resistant to the autologous neutralizing antibodies, this resistance decreased in the Envs of the viruses in monkey 228-91, which subsequently controlled viremia. In contrast, in the other SHIV-KY-infected monkey (monkey 354-05) that developed persistent viremia, viruses resistant to autologous neutralizing antibodies arose and predominated throughout infection. Finally, SHIV-KY62-infected monkeys did not develop neutralizing antibodies against the inoculated virus until much later, even though these animals efficiently generated antibodies directed against Env early in infection and subsequently made antibodies that cross-neutralized the heterologous HXB2 virus. The SHIV-KY62 viruses established a persistent viremia in these monkeys. Apparently, minimizing the elicitation of host neutralizing antibodies and/or evolving resistance to any such antibodies that are generated facilitates the persistence of these SHIVs in rhesus macaques.
Whereas the in vivo fitness benefit of resistance to neutralizing antibodies is intuitively obvious, the advantages of low ERcold or low ERsCD4 are less apparent. Our observations support the proposition that an Env with low ERcold and ERsCD4 is conducive to the ability of a SHIV to persist in monkeys. First, viruses bearing Envs with lower ERcold and ERsCD4 were selected within the first 70 days of SHIV-KY infection of monkeys, concurrently with the generation of neutralizing antibodies. The sCD4 reactivities and cold sensitivities of the SHIV-KY Envs that were selected in vivo generally covaried and were determined by identical Env amino acid residues, supporting the close relationship of these two properties (64). Second, the Env changes that led to lowered ERcold and ERsCD4 were distinct from those that resulted in resistance to autologous neutralizing antibodies. Thus, although low ERcold and ERsCD4 on the one hand and neutralization resistance on the other hand both evolved concurrently with the generation of a neutralizing antibody response in the infected monkeys, they represent distinct and separable properties of Env. This is consistent with the observation that HIV-1 isolates with similar values of neutralization sensitivity can exhibit a wide range of ERcold and ERsCD4 and vice versa (64, 65). Finally, the SHIV-KY progeny that successfully persisted in monkeys (including SHIV-KY62) generally maintained ERcold and ERsCD4 levels lower than those of parental SHIV-KY. The temporal relationship between the appearance of autologous neutralizing antibodies and decreases in the ERcold and ERsCD4 of the viral Envs is consistent with the hypothesis that low ERcold and low ERsCD4 help the virus to evade the humoral immune response.
Of note, the initial effectiveness of the neutralizing antibody response against the inoculated virus was inversely correlated with the emergence of cross-neutralizing antibodies against the laboratory-adapted HXB2 virus. Animals inoculated with SHIV-KA developed a robust autologous neutralizing antibody response that coincided with early viral clearance, but these antibodies did not cross-neutralize the HXB2 virus. SHIV-KY Env, with higher ERcold and ERsCD4, induced neutralizing antibodies against the inoculated virus, but these arose more slowly and were of lower titers than those in SHIV-KA-infected monkeys. Apparently, this allowed the rapid selection of neutralization-resistant and low-ERcold and low-ERsCD4 variants in SHIV-KY-infected animals, which may have contributed to longer persistence and the subsequent elicitation of HXB2-neutralizing antibodies. Autologous neutralizing antibodies were not detectable in SHIV-KY62-inoculated animals until very late in the infection, but these animals produced more antibodies that cross-neutralized the HXB2 virus. Envs with low ERcold and ERsCD4 are predicted to have higher activation barriers that decrease the spontaneous sampling of the CD4-bound or other low-energy conformations (64) (Fig. 10). Therefore, these low-ERcold and low-ERsCD4 Envs may present the unliganded Env trimer to the host immune system, rather than other Env conformers, predisposing to the generation of neutralizing antibodies with some breadth. This situation is reminiscent of the observation that Env immunogens from transmitted/founder HIV-1, which generally exhibit low ER values, elicit antibodies that can neutralize divergent tier 1 HIV-1 isolates (96).
Changes in the V2 variable region of gp120 allowed viral escape from the strain-restricted neutralizing antibodies generated in SHIV-KY-infected monkeys. Alterations in a potential N-linked glycosylation site at residue 187 and adjacent residue 190 explained most of the V2 phenotype with respect to neutralization escape. Changes in the V2 region also contributed to lowered ERcold and ERsCD4, but in this case, the most important residue was asparagine/aspartic acid 167. Low ERcold and low ERsCD4 could also be achieved by changes in methionine/isoleucine 420 in the gp120 β19 strand. Thus, two redundant pathways to achieve low values of ERcold and ERsCD4 apparently were available to the KY Envs. An understanding of how these changes affect Env neutralization sensitivity, ERcold, and ERsCD4 will require structures of these elements in the context of the Env trimer.
The Env changes associated with neutralizing antibody escape and those associated with lowered ERcold and ERsCD4 resulted in a decrease in the ability of Env to mediate virus entry into tissue-cultured cells. The appearance of Env variants with decreases in neutralization sensitivity, ERcold, and ERsCD4 in SHIV-KY-infected monkeys is consistent with the positive contribution of these Env properties to virus persistence in vivo. Also, consistent with this proposed in vivo advantage of low ERcold/ERsCD4, SHIV-KY62 exhibited decreased elicitation/susceptibility to autologous neutralizing antibodies. Thus, SHIV-KY62 very efficiently established persistent infections in monkeys, despite the associated decrease in the ability of KY62 Env to mediate infections in PBMC and other cell types in tissue culture. The advantages of neutralizing antibody resistance and decreased ERcold and ERsCD4 to the establishment of persistent infection apparently offset any associated decreases in the ability of the evolved Envs to mediate virus infection.
Unexpectedly, concomitant with the evolution of SHIV-KY variants that escaped autologous neutralizing antibodies, the viral Envs became more sensitive to neutralization by b12, an antibody directed against the CD4-binding site of gp120. Comparison of the sequences of Envs from monkey 228-91 that were sensitive or resistant to b12 neutralization indicates that the Env determinant is in the V2 variable region. Changes in V2 residues (Asp182 and Asn186) were previously shown to influence HIV-1 susceptibility to b12 neutralization (97–99). Also of note, SHIV-89.6 acquired resistance to b12 neutralization during the passage in monkeys that gave rise to SHIV-KB9 (38). Determinants of b12 resistance in SHIV-KB9 Env were narrowed to the V2 changes E185K and N187S, both of which reverted in d70 variants from SHIV-KY-inoculated animals 354-05 and 228-91. Thus, alterations in the V2 region likely contributed to changes in b12 sensitivity early in the course of SHIV-KY infection, but later determinants of b12 resistance remain to be characterized. As the V2 structures are distant from the b12 epitope, the mechanism whereby these V2 changes affect sensitivity to b12 inhibition also requires further studies. Of interest, when SHIV-KY-infected monkey 354-05 generated antibodies that could neutralize the heterologous HXB2 virus, the SHIV-KY variants in this animal became more resistant to b12 neutralization. This observation raises the possibility that during the chronic stage of infection (days 250 to 430 p.i.), weakly neutralizing antibodies directed against the CD4-binding site of gp120 in the plasma of monkey 354-05 may contribute to the counterselection of SHIV-KY variants susceptible to these antibodies.
Later in the course of SHIV-KY infection of monkey 354-05, transient increases in ERcold and ERsCD4 appeared. These increases were partial and did not return either ERcold or ERsCD4 to preinfection values. A subsequent lowering of ERcold and ERsCD4 was observed after day 300 p.i., with changes in the V3 tip (L317W) and LF loop (T440A) mediating this restoration. The late fluctuations in ERcold and ERsCD4 of the SHIV-KY Envs are reminiscent of those described for a chronic R5 SHIV infection just before conversion of the virus to an X4 variant and rapid disease progression (100, 101). Such transient changes in ERcold and ERsCD4 may arise as HIV-1 Envs adapt to altered environmental circumstances (e.g., the generation of new neutralizing antibodies or infection of different cell types) and then need to reequilibrate to optimize fitness.
An understanding of the importance of low envelope reactivity to SHIV persistence and pathogenicity should assist efforts to develop this animal model of HIV-1 infection further.
ACKNOWLEDGMENTS
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
This work was supported by the National Institutes of Health (AI 24755 and the Center for HIV/AIDS Vaccine Immunology [AI 67854]), by the International AIDS Vaccine Initiative, and by the late William F. McCarty-Cooper. K.M. and H.H. were supported by an NRSA postdoctoral training program in AIDS research (NIH T32 AI007386).
Footnotes
Published ahead of print 16 October 2013
REFERENCES
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871. 10.1126/science.6189183 [DOI] [PubMed] [Google Scholar]
- 2.Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp PM, Shaw GM, Hahn BH. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID networks for HIV isolation and characterization. J. Virol. 70:1651–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593–595. 10.1126/science.2984774 [DOI] [PubMed] [Google Scholar]
- 4.Veronese FD, DeVico AL, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG. 1985. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science 229:1402–1405. 10.1126/science.2994223 [DOI] [PubMed] [Google Scholar]
- 5.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884 [DOI] [PubMed] [Google Scholar]
- 6.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. 10.1126/science.272.5270.1955 [DOI] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. 10.1016/S0092-8674(00)81313-6 [DOI] [PubMed] [Google Scholar]
- 8.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. 10.1038/312763a0 [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. 10.1038/381661a0 [DOI] [PubMed] [Google Scholar]
- 10.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. 10.1038/381667a0 [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. 10.1126/science.272.5263.872 [DOI] [PubMed] [Google Scholar]
- 12.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. 10.1038/312767a0 [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDougal JS, Maddon PJ, Dalgleish AG, Clapham PR, Littman DR, Godfrey M, Maddon DE, Chess L, Weiss RA, Axel R. 1986. The T4 glycoprotein is a cell-surface receptor for the AIDS virus. Cold Spring Harb. Symp. Quant. Biol. 51(Part 2):703–711 [DOI] [PubMed] [Google Scholar]
- 15.Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. 1991. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 65:5007–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810. 10.1146/annurev.biochem.70.1.777 [DOI] [PubMed] [Google Scholar]
- 18.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565. 10.1128/JVI.77.19.10557-10565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519. 10.1073/pnas.0504658102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636. 10.1128/JVI.01482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. 10.1038/nature12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769. 10.1128/JVI.02036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348. 10.1128/JVI.00110-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951–954. 10.1038/nm.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. 10.1371/journal.ppat.1000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, Forthal DN, Koff WC, Poignard P, Watkins DI, Burton DR. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 84:1302–1313. 10.1128/JVI.01272-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. U. S. A. 109:18921–18925. 10.1073/pnas.1214785109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, Ketas T, Marx PA, Klasse PJ, Burton DR, Moore JP. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343–346. 10.1038/nm833 [DOI] [PubMed] [Google Scholar]
- 29.Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528–6531. 10.1128/JVI.79.10.6528-6531.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. 10.1126/science.1093137 [DOI] [PubMed] [Google Scholar]
- 31.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790. 10.1128/JVI.01730-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sagar M, Wu X, Lee S, Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586–9598. 10.1128/JVI.00141-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, Manrique A, Huber M, Rehr M, Oxenius A, Weber R, Stiegler G, Vcelar B, Katinger H, Aceto L, Gunthard HF. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615–622. 10.1038/nm1244 [DOI] [PubMed] [Google Scholar]
- 34.Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941. 10.1128/JVI.00757-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng-Mayer C, Brown A, Harouse J, Luciw PA, Mayer AJ. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford JM, Earl PL, Moss B, Reimann KA, Wyand MS, Manson KH, Bilska M, Zhou JT, Pauza CD, Parren PW, Burton DR, Sodroski JG, Letvin NL, Montefiori DC. 1999. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J. Virol. 73:10199–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Etemad-Moghadam B, Karlsson GB, Halloran M, Sun Y, Schenten D, Fernandes M, Letvin NL, Sodroski J. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72:8437–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etemad-Moghadam B, Sun Y, Nicholson EK, Karlsson GB, Schenten D, Sodroski J. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 73:8873–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montefiori DC, Reimann KA, Wyand MS, Manson K, Lewis MG, Collman RG, Sodroski JG, Bolognesi DP, Letvin NL. 1998. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J. Virol. 72:3427–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura Y, Shingai M, Willey R, Sadjadpour R, Lee WR, Brown CR, Brenchley JM, Buckler-White A, Petros R, Eckhaus M, Hoffman V, Igarashi T, Martin MA. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J. Virol. 84:4769–4781. 10.1128/JVI.02279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Si Z, Cayabyab M, Sodroski J. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208–4218. 10.1128/JVI.75.9.4208-4218.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato S, Johnson W. 2007. Antibody-mediated neutralization and simian immunodeficiency virus models of HIV/AIDS. Curr. HIV Res. 5:594–607. 10.2174/157016207782418515 [DOI] [PubMed] [Google Scholar]
- 43.Harouse JM, Tan RC, Gettie A, Dailey P, Marx PA, Luciw PA, Cheng-Mayer C. 1998. Mucosal transmission of pathogenic CXCR4-utilizing SHIVSF33A variants in rhesus macaques. Virology 248:95–107. 10.1006/viro.1998.9236 [DOI] [PubMed] [Google Scholar]
- 44.Karlsson GB, Halloran M, Li J, Park IW, Gomila R, Reimann KA, Axthelm MK, Iliff SA, Letvin NL, Sodroski J. 1997. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 71:4218–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li JT, Halloran M, Lord CI, Watson A, Ranchalis J, Fung M, Letvin NL, Sodroski JG. 1995. Persistent infection of macaques with simian-human immunodeficiency viruses. J. Virol. 69:7061–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pal R, Taylor B, Foulke JS, Woodward R, Merges M, Praschunus R, Gibson A, Reitz M. 2003. Characterization of a simian human immunodeficiency virus encoding the envelope gene from the CCR5-tropic HIV-1 Ba-L. J. Acquir. Immune Defic. Syndr. 33:300–307. 10.1097/00126334-200307010-00003 [DOI] [PubMed] [Google Scholar]
- 47.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J. Virol. 80:8729–8738. 10.1128/JVI.00558-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan RC, Harouse JM, Gettie A, Cheng-Mayer C. 1999. In vivo adaptation of SHIV(SF162): chimeric virus expressing a NSI, CCR5-specific envelope protein. J. Med. Primatol. 28:164–168. 10.1111/j.1600-0684.1999.tb00265.x [DOI] [PubMed] [Google Scholar]
- 49.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621–628. 10.1084/jem.185.4.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlsson A, Parsmyr K, Aperia K, Sandstrom E, Fenyo EM, Albert J. 1994. MT-2 cell tropism of human immunodeficiency virus type 1 isolates as a marker for response to treatment and development of drug resistance. J. Infect. Dis. 170:1367–1375. 10.1093/infdis/170.6.1367 [DOI] [PubMed] [Google Scholar]
- 51.Schuitemaker H, Kootstra NA, de Goede RE, de Wolf F, Miedema F, Tersmette M. 1991. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J. Virol. 65:356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuitemaker H, Kootstra NA, Groenink M, De Goede RE, Miedema F, Tersmette M. 1992. Differential tropism of clinical HIV-1 isolates for primary monocytes and promonocytic cell lines. AIDS Res. Hum. Retroviruses 8:1679–1682. 10.1089/aid.1992.8.1679 [DOI] [PubMed] [Google Scholar]
- 53.Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman DR, Fenyo EM, Albert J. 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181–188. 10.1006/viro.1997.8980 [DOI] [PubMed] [Google Scholar]
- 54.Joag SV, Li Z, Foresman L, Stephens EB, Zhao LJ, Adany I, Pinson DM, McClure HM, Narayan O. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimura Y, Igarashi T, Donau OK, Buckler-White A, Buckler C, Lafont BA, Goeken RM, Goldstein S, Hirsch VM, Martin MA. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. U. S. A. 101:12324–12329. 10.1073/pnas.0404620101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. 10.1126/science.1145373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. 10.1371/journal.ppat.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, III, Sodroski J. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J. Virol. 83:4476–4488. 10.1128/JVI.02110-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB, III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J. Virol. 83:8364–8378. 10.1128/JVI.00594-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. 10.1371/journal.ppat.1002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medjahed H, Pacheco B, Desormeaux A, Sodroski J, Finzi A. 2013. The HIV-1 gp120 major variable regions modulate cold inactivation. J. Virol. 87:4103–4111. 10.1128/JVI.03124-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LaBonte JA, Madani N, Sodroski J. 2003. Cytolysis by CCR5-using human immunodeficiency virus type 1 envelope glycoproteins is dependent on membrane fusion and can be inhibited by high levels of CD4 expression. J. Virol. 77:6645–6659. 10.1128/JVI.77.12.6645-6659.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karlsson GB, Halloran M, Schenten D, Lee J, Racz P, Tenner-Racz K, Manola J, Gelman R, Etemad-Moghadam B, Desjardins E, Wyatt R, Gerard NP, Marcon L, Margolin D, Fanton J, Axthelm MK, Letvin NL, Sodroski J. 1998. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J. Exp. Med. 188:1159–1171. 10.1084/jem.188.6.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. 10.1126/science.7973652 [DOI] [PubMed] [Google Scholar]
- 70.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, III, Burton DR. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325–4332 [PubMed] [Google Scholar]
- 72.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892–10905. 10.1128/JVI.75.22.10892-10905.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 78:12975–12986. 10.1128/JVI.78.23.12975-12986.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289. 10.1084/jem.20090378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521. 10.1128/JVI.00552-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 75:5646–5655. 10.1128/JVI.75.12.5646-5655.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251. 10.1371/journal.ppat.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shingai M, Donau OK, Schmidt SD, Gautam R, Plishka RJ, Buckler-White A, Sadjadpour R, Lee WR, Labranche CC, Montefiori DC, Mascola JR, Nishimura Y, Martin MA. 2012. Most rhesus macaques infected with the CCR5-tropic SHIVAD8 generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 109:19769–19774. 10.1073/pnas.1217443109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452. 10.1128/JVI.02108-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. 2006. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80:999–1014. 10.1128/JVI.80.2.999-1014.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo EM. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107–112. 10.1097/00002030-199002000-00002 [DOI] [PubMed] [Google Scholar]
- 85.Arendrup M, Nielsen C, Hansen JE, Pedersen C, Mathiesen L, Nielsen JO. 1992. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 5:303–307 [PubMed] [Google Scholar]
- 86.Moore PL, Gray ES, Morris L. 2009. Specificity of the autologous neutralizing antibody response. Curr. Opin. HIV AIDS 4:358–363. 10.1097/COH.0b013e32832ea7e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, Morris L. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. 10.1371/journal.ppat.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149. 10.1073/pnas.0630530100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. 10.1371/journal.ppat.1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- 91.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 8:e1002721. 10.1371/journal.ppat.1002721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bunnik EM, Lobbrecht MS, van Nuenen AC, Schuitemaker H. 2010. Escape from autologous humoral immunity of HIV-1 is not associated with a decrease in replicative capacity. Virology 397:224–230. 10.1016/j.virol.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 93.Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, Mulenga J, Allen S, Hunter E, Hahn BH, Shaw GM, Blackwell JL, Derdeyn CA. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211–5218. 10.1128/JVI.00201-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, Huang W, Quinn TC. 2009. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J. Infect. Dis. 199:580–589. 10.1086/596557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song H, Pavlicek JW, Cai F, Bhattacharya T, Li H, Iyer SS, Bar KJ, Decker JM, Goonetilleke N, Liu MK, Berg A, Hora B, Drinker MS, Eudailey J, Pickeral J, Moody MA, Ferrari G, McMichael A, Perelson AS, Shaw GM, Hahn BH, Haynes BF, Gao F. 2012. Impact of immune escape mutations on HIV-1 fitness in the context of the cognate transmitted/founder genome. Retrovirology 9:89. 10.1186/1742-4690-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, Lu X, Sutherland LL, Scearce RM, Bowman C, Parks R, Chen H, Blinn JH, Lapedes A, Watson S, Xia SM, Foulger A, Hahn BH, Shaw GM, Swanstrom R, Montefiori DC, Gao F, Haynes BF, Korber B. 2013. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J. Virol. 87:4185–4201. 10.1128/JVI.02297-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mo H, Stamatatos L, Ip JE, Barbas CF, Parren PW, Burton DR, Moore JP, Ho DD. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J. Virol. 71:6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Utachee P, Nakamura S, Isarangkura-Na-Ayuthaya P, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. 2010. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J. Virol. 84:4311–4320. 10.1128/JVI.02619-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watkins JD, Diaz-Rodriguez J, Siddappa NB, Corti D, Ruprecht RM. 2011. Efficiency of neutralizing antibodies targeting the CD4-binding site: influence of conformational masking by the V2 loop in R5-tropic clade C simian-human immunodeficiency virus. J. Virol. 85:12811–12814. 10.1128/JVI.05994-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhuang K, Finzi A, Tasca S, Shakirzyanova M, Knight H, Westmoreland S, Sodroski J, Cheng-Mayer C. 2011. Adoption of an “open” envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques. PLoS One 6:e21350. 10.1371/journal.pone.0021350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhuang K, Finzi A, Toma J, Frantzell A, Huang W, Sodroski J, Cheng-Mayer C. 2012. Identification of interdependent variables that influence coreceptor switch in R5 SHIVSF162P3N-infected macaques. Retrovirology 9:106. 10.1186/1742-4690-9-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]