Abstract
Sterile alpha motif and HD domain-containing protein 1 (SAMHD1) restricts human immunodeficiency virus type 1 (HIV-1) infection in myeloid cells but is inactivated by certain classes of simian immunodeficiency virus (SIV) Vpx proteins. Vpx proteins recruit the DCAF1-CRL4 E3 ubiquitin ligase to trigger species-specific SAMHD1 degradation. Determinants of SIV Vpx-mediated primate SAMHD1 degradation have been mapped to its C terminus. In this study, we have identified the N terminus of human SAMHD1 as a major species-specific determinant of Vpx-mediated suppression. The SIVmnd2 and SIVrcm Vpx proteins recognize the N terminus of rhesus, but not human, SAMHD1. We have also demonstrated that variation of two primate lineage-specific residues between human and rhesus SAMHD1 proteins determine resistance to SIVmnd2 and SIVrcm Vpx proteins. These residues (Cys15 and Ser52) are sequentially mutated to Phe in different lineages of Old World monkeys. Consequently, SIVmnd2 and SIVrcm Vpx proteins that could recognize Phe15- and Phe52-containing SAMHD1 could not inactivate human SAMHD1, which contains Cys15 and Ser52. In contrast, SIVmac Vpx, which targets the C terminus of SAMHD1 molecules, could inactivate various primate SAMHD1 molecules with divergent C-terminal sequences. Both C terminus-targeted SIVmac Vpx and N terminus-targeted SIVrcm Vpx require DCAF1 for the induction of SAMHD1 degradation. The ability of SIV Vpx to restrict SAMHD1 among different primate species is a manifestation of the SAMHD1 evolutionary pattern among those species.
INTRODUCTION
Human immunodeficiency virus type 2 (HIV-2) and selected simian immunodeficiency virus (SIV) lineages encode Vpx as a virion-associated protein packaged through specific interaction with Gag proteins (1–6). Vpx is required for efficient viral replication in macrophages (7–10) and dendritic cells (11, 12). The primary function of virion-associated Vpx is to promote the accumulation of viral DNA during reverse transcription (13–16). The Aicardi-Goutières syndrome-related gene product sterile alpha motif (SAM) and HD domain-containing protein 1 (SAMHD1) was recently identified as a potent inhibitor of HIV-1 in myeloid cells and resting CD4+ T cells (17–24) and as a regulator of endogenous retroelements (25).
SAMHD1 is a deoxynucleotide triphosphohydrolase and blocks HIV-1 reverse transcription by depleting the intracellular pool of deoxynucleoside triphosphates (23, 24, 26–29). Oligomer formation (26, 29), in particular, tetramer formation (30, 31), has been reported to be important for SAMHD1 deoxynucleotide triphosphohydrolase function and HIV-1 restriction. Recent studies suggested that the phosphorylation status of SAMHD1 (T592) is also important for its anti-HIV activity (32–34) but not its deoxynucleotide triphosphohydrolase activity (33, 34).
Vpx neutralizes the antiviral activity of SAMHD1 by promoting its proteasome-dependent degradation. Vpx binds DCAF1 by using conserved motifs in helix 1 and helix 3, which in turn recruit other components of the CRL4(DCAF1) E3 ubiquitin ligase (35–41) to facilitate SAMHD1 ubiquitination and subsequent degradation (18, 20, 22, 35, 36, 38, 39, 41). Nuclear targeting of SAMHD1 is required for Vpx-mediated SAMHD1 degradation (38, 39, 41).
SAMHD1 has been under positive selection during primate evolution (21, 42–44). Various SIV Vpx and Vpr proteins have been shown to inactivate primate SAMHD1 proteins. Species-specific recognition of SAMHD1 by Vpx and Vpr has been proposed as the driving force for SAMHD1 positive selection. Previous studies have indicated that Vpx loads SAMHD1 onto CRL4(DCAF1) E3 ubiquitin ligase, thereby facilitating its subsequent degradation through recognition of C-terminal sequences of SAMHD1 (35, 38, 39, 41). Consistent with this concept, SAMHD1 mutants with C-terminal truncations are resistant to Vpx-mediated degradation (38, 39, 41).
In the present study, we identified the N terminus of primate SAMHD1 as a major species-specific determinant of Vpx-mediated suppression. In contrast to SIVmac Vpx, the SIVrcm and SIVmnd2 Vpx proteins recognize the N terminus of rhesus (Rh) SAMHD1 molecules. We have also demonstrated that the SIVrcm and SIVmnd2 Vpx proteins fail to antagonize human (Hu) SAMHD1 because of the existence of two primate lineage-specific residues in the SAM domain and its upstream region. Thus, adaptation to SAMHD1 molecules of the Old World monkeys (OWMs) by the N terminus-targeted SIVrcm and SIVmnd2 Vpx proteins may have limited their ability to inactivate SAMHD1 from hominoids and New World monkeys (NWMs).
MATERIALS AND METHODS
Plasmid construction.
SIVmac239 Vpx-hemagglutinin (HA) in the pCG vector was a gift from J. Skowronski. Human pSAMHD1-HA was constructed in our laboratory as previously described (41, 45). Rhesus macaque (Macaca mulatta), Pongo abelii, Canis lupus familiaris, Mus musculus, and Bos taurus SAMHD1 expression vectors were obtained from the Changchun Boyi Biological Science and Technology Company. N-terminally HA-tagged SAMHD1 ΔN(110-626) and ΔC(1-611) constructs were generated by PCR and subcloned into the VR1012 vector. Hu-Rh SAMHD1 chimeric constructs were generated from pHu SAMHD1-HA and pRh SAMHD1-HA by using the internal PstI site (around 570 bp). Hu or Rh SAMHD1 point mutants were constructed from pHu SAMHD1-HA and pRh SAMHD1-HA by PCR-based site-directed mutagenesis. All constructs were confirmed to be correct by DNA sequencing. To generate an expression vector encoding the mCherry-SAMHD1 fusion protein, the SAMHD1-HA fragment was digested with SalI and XbaI and cloned into pmCherry-C1 to generate pmCherry-SAMHD1-HA. The expression vectors for SIVmnd2 Vpx, SIVrcm Vpx, and SIVsmm Vpx were obtained from the Changchun Boyi Biological Science and Technology Company.
Cell culture and antibodies.
HEK293T cells (NIH AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin. All cultured cell lines were maintained at 37°C in a humid atmosphere containing 5% CO2. The following antibodies were used: anti-HA monoclonal antibody (MAb; Covance MMS-101R), anti-Vprbp (DCAF1; Shanghai Genomics SG4220-28), anti-FLAG M2 antibody (Sigma F1804), anti-Myc MAb (Covance MMS-150R), and anti-actin MAb (Sigma A3853).
Transfection, coimmunoprecipitation, and immunoblotting.
DNA transfection was carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. HEK293T cells were harvested at 48 h after transfection, washed twice with cold phosphate-buffered saline (PBS), and lysed in lysis buffer. Cell extracts were then analyzed by SDS-PAGE and immunoblotting with the appropriate antibodies as previously described (41). For immunoprecipitation, HEK293T cells were harvested at 48 h after transfection, washed twice with cold PBS, and lysed in lysis buffer (150 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, complete protease inhibitor cocktail tablets [Roche]) at 4°C for 30 min and then centrifuged at 10,000 × g for 30 min. Precleared cell lysates were mixed with anti-HA antibody-conjugated agarose beads (Roche 190-119) or anti-c-Myc agarose affinity gel (Sigma A7470) and incubated at 4°C for 3 h or overnight. Samples were then washed eight times with washing buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.05% Tween 20). The beads were eluted with elution buffer (0.1 M glycine-HCl, pH 2.0). The eluted materials were then analyzed by SDS-PAGE and immunoblotting with the appropriate antibodies, and protein band intensities were quantified with Image J software (http://rsbweb.nih.gov/ij/index.html) as previously described (41).
RNA interference.
RNA interference against DCAF1 was carried out with a pool of four duplexed small interfering RNAs (siRNAs; Dharmacon) as previously described (46). HEK293T cells were transfected with the DCAF1 siRNA pool at a final concentration of 100 nM with Lipofectamine 2000 (Invitrogen). Nontargeting siRNA no. 2 (Dharmacon) was used as a negative control. Protein expression was monitored by immunoblotting 2 to 3 days after transfection.
Live-cell imaging.
Plasmids pEYFP-Nuc (0.25 μg; a gift from T. Inoue) and pmCherry-SAMHD1-HA (2 μg) were transfected into HEK293T cells with PEI Max (Polysciences) according to the manufacturer's protocol. For live-cell imaging, HEK293T cells were transfected in six-well coverslipped glass bottom cell culture dishes (InVitro Scientific) when the cells were ∼80% confluent and then visualized after 24 h with a Zeiss LSM510-Meta confocal imaging system equipped with four argon lasers (458-, 477-, 488-, and 514-nm lines), two HeNe lasers (542 and 633 nm), and one diode laser (405 nm). All images were acquired from a 63× objective, and image analysis and manipulation were performed with Zen 2009 software as previously described (47).
Phylogenetic analysis.
Alignment was achieved with the MEGA4 program (48) by the ClustalW method and optimized by hand with the BioEdit program, version 5.0.9. The lengths of the amino acid sequences used for analysis varied, depending on the purpose of the particular analysis, and are indicated in Results. All phylogenetic analyses were also performed with the MEGA4 program by the neighbor-joining method on the basis of the Kimura two-parameter model.
RESULTS
SIVrcm/mnd2 Vpx targets the N terminus of SAMHD1 for degradation.
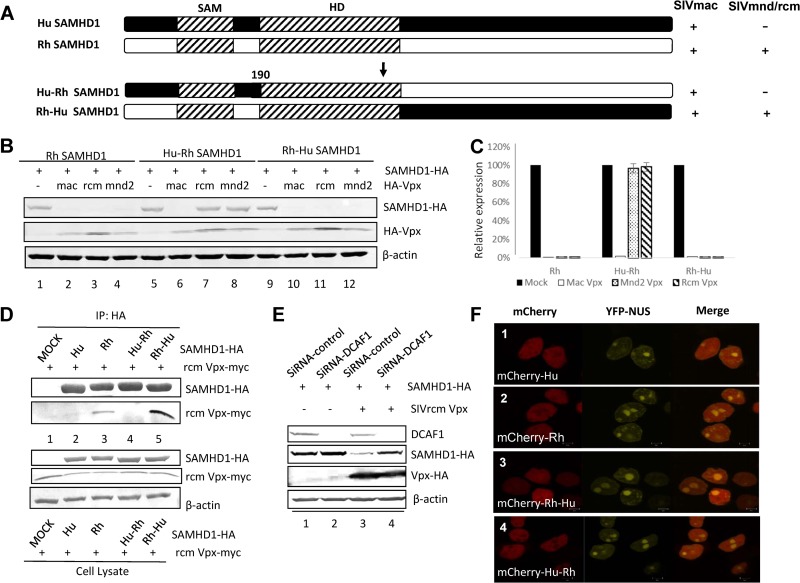
To study the functional interaction of Hu and Rh SAMHD1 constructs with various SIV Vpx proteins, Hu and Rh SAMHD1 constructs with N- and C-terminal deletions were generated (Fig. 1A). Unlike Rh SAMHD1, which is sensitive to degradation induced by Vpx proteins of diverse SIV strains (Fig. 1B and C), Hu SAMHD1 is resistant to SIVmnd2 and SIVrcm Vpx proteins (Fig. 1B and C). It has been reported that SIVmac Vpx targets the C terminus of Hu SAMHD1. C-terminal deletion of Hu and Rh SAMHD1 made these proteins resistant to SIVmac Vpx-induced degradation (Fig. 1D and E). On the other hand, C-terminally truncated Rh SAMHD1 could still be degraded in the presence of SIVmnd2 and SIVrcm Vpx (Fig. 1D and E). We have observed that SIVmac Vpx could induce the degradation of Hu SAMHD1 with the N-terminal region deleted (Fig. 1F and G), consistent with the idea that SIVmac Vpx targets the SAMHD1 C terminus. However, Rh SAMHD1 mutant protein with an N-terminal region deletion became more resistant to SIVrcm/mmd2 Vpx (Fig. 1F), although it was still sensitive to SIVmac Vpx-induced degradation (Fig. 1F, lane 6). Interestingly, the SIV Vpx proteins that could not induce the degradation of Hu SAMHD1 also lost the ability to induce the degradation of Rh SAMHD1 with the N-terminal region deleted. These results suggest that Vpx from SIVrcm and SIVmnd2 may recognize the SAMHD1 N-terminal region while Vpx proteins from SIVmac and SIVsmm target the SAMHD1 C-terminal region.
FIG 1.
The N-terminal region of Rh SAMHD1 is required for SIVrcm/mnd2 Vpx-mediated SAMHD1 degradation. (A) Sensitivities of various Hu and Rh SAMHD1 constructs (ΔN or ΔC) to Vpx-mediated degradation. Vpx proteins of both SIVmac and SIVmnd2/rcm were used. SAMHD1 proteins that are sensitive to the indicated Vpx-induced degradation are indicated by plus signs. Schematic representations of full-length Hu and Rh SAMHD1 and of Hu and Rh SAMHD1 with the N- or C-terminal region deleted (ΔN or ΔC). (B and C) Rh SAMHD1 proteins, but not Hu SAMHD1 proteins, are sensitive to degradation mediated by diverse SIV Vpx proteins. HA-tagged SAMHD1 constructs were coexpressed with the indicated expression vector for HA-tagged Vpx or the empty vector in HEK293T cells. Cell extracts were harvested 48 h later and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA and HA-Vpx; β-actin was used as the loading control. The data shown represent one of three independent experiments. Error bars represent the standard deviation of the mean of triplicate samples. (D and E) Rh SAMHD1 proteins with the C-terminal region deleted are sensitive to degradation mediated by Vpx of SIVmnd2 and SIVrcm. The data shown represent one of three independent experiments. Error bars represent the standard deviation of the mean of triplicate samples. (F and G) Rh SAMHD1 proteins with the N-terminal region deleted are resistant to degradation mediated by Vpx of SIVmnd2 and SIVrcm. The data shown represent one of three independent experiments. Error bars represent the standard deviation of the mean of triplicate samples.
To further evaluate this issue, we constructed chimeric Hu-Rh SAMHD1 expression vectors (Fig. 2A). Interestingly, Hu SAMHD1 with the N-terminal region from Rh SAMHD1 (Rh-Hu SAMHD1) became sensitive to SIVrcm/mnd2 Vpx-mediated degradation (Fig. 2B, lanes 11 and 12). In contrast, Rh SAMHD1 with the N-terminal region from Hu SAMHD1 (Hu-Rh SAMHD1) became resistant to SIVrcm/mnd2 Vpx-mediated degradation (Fig. 2B, lanes 7 and 8). In all assay systems, SIVmac Vpx induced SAMHD1 degradation (Fig. 2B). Thus, the N terminus controls the sensitivity of SAMHD1 to SIVrcm/mnd2 but not SIVmac Vpx proteins.
FIG 2.
The N-terminal region of Rh SAMHD1 increases the sensitivity of chimeric SAMHD1 proteins to SIVrcm/mnd2 Vpx-induced degradation. (A) Schematic representation of chimeric Hu-Rh SAMHD1 constructs. (B and C) Effects of Hu-Rh SAMHD1 chimeric proteins on Vpx-induced degradation. Expression vectors for HA-tagged Hu SAMHD1 protein and Hu-Rh SAMHD1 chimeric protein were coexpressed with HA-tagged Vpx in HEK293T cells. Cell extracts were harvested 48 h later and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA and HA-Vpx. β-Actin was used as the loading control. The results were confirmed in repeated experiments. (D) Effects of Hu-Rh SAMHD1 chimeric proteins on Vpx-SAMHD1 interaction. Expression vectors for HA-tagged Hu SAMHD1 protein and Hu-Rh SAMHD1 chimeric protein were coexpressed with Myc-tagged SIVrcm Vpx in HEK293T cells. Cell lysates were prepared 48 h after transfection and immunoprecipitated (IP) with an anti-HA affinity matrix (Roche). The interaction of SIVrcm Vpx with Hu-Rh SAMHD1 chimeric protein was detected by immunoblotting with anti-Myc or anti-HA antibody. (E) Effect of endogenous DCAF1 on SIVrcm Vpx-induced SAMHD1 degradation. Control siRNA or siRNA targeting DCAF1 was cotransfected with HA-SAMHD1 and HA-Vpx or the empty vector into HEK293T cells. Cell extracts were harvested 48 h later and analyzed by SDS-PAGE, followed by immunoblotting to detect endogenous DCAF1, SAMHD1-HA, and HA-Vpx; β-actin was used as the loading control. (F) Plasmids pEYFP-Nuc (Clontech) and pmCherry-SAMHD1 WT or the variants indicated were cotransfected into HEK293T cells with PEI Max (Polysciences). For live-cell imaging, HEK293T cells were transfected in six-well coverslipped glass bottom cell culture dishes and visualized after 24 h with a Zeiss LSM510-Meta confocal imaging system equipped with four argon lasers (458-, 477-, 488-, and 514-nm lines), two HeNe lasers (542 and 633 nm), and one diode laser (405 nm). All images were acquired from a 100× objective, and image analysis and manipulation were performed with Zen 2009 software. YFP, yellow fluorescent protein; NUS, nuclear targeting sequences.
It has been established that SIVsmm/mac Vpx recognizes the C terminus of SAMHD1 (21, 35, 41). Our data suggest that SIVrcm/mnd2 Vpx specifically targets the N-terminal region of Rh SAMHD1. Coimmunoprecipitation experiments confirmed that the N-terminal region of Rh SAMHD1 is important for Vpx interaction. SIVrcm Vpx interacted with Rh SAMHD1 (Fig. 2D, lane 3) but not Hu SAMHD1 (lane 2). Addition of the N-terminal region of Rh SAMHD1 to Hu SAMHD1 (Rh-Hu SAMHD1) restored this interaction (Fig. 2D, lane 5).
DCAF1 is required for SIVmac Vpx-mediated SAMHD1 degradation. We have also examined whether SIVrcm Vpx, which targets a different region of SAMHD1, also requires DCAF1 to function. Silencing of endogenous DCAF1 in HEK293T cells by specific siRNA impaired the ability of SIVrcm Vpx to deplete Rh SAMHD1 (Fig. 2E, lane 4) compared to that of cells containing DCAF1 (lane 3). Thus, both the N and C terminus-targeted Vpx proteins maintain a conserved strategy of hijacking DCAF1-containing E3 ubiquitin ligase for SAMHD1 inactivation.
Live-cell imaging results indicated that Hu, Rh, and Hu-Rh chimeric SAMHD1 proteins all maintained a nuclear accumulation ability (Fig. 2F), indicating that the divergent sensitivities of these various SAMHD1 variants to Vpx are not due to their altered intracellular distribution.
Primate lineage-specific amino acid variation in the N-terminal region of SAMHD1 proteins.
It is interesting that Hu SAMHD1 showed a species-specific resistance to the SIVrcm/SIVmnd2 lineage but not the SIVsmm lineage Vpx. SIVrcm/SIVmnd2 Vpx proteins have broad activity against diverse OWM SAMHD1 proteins (21, 43). However, hominoid and NWM SAMHD1 proteins are largely resistant to SIVrcm/SIVmnd2 Vpx proteins (21, 43).
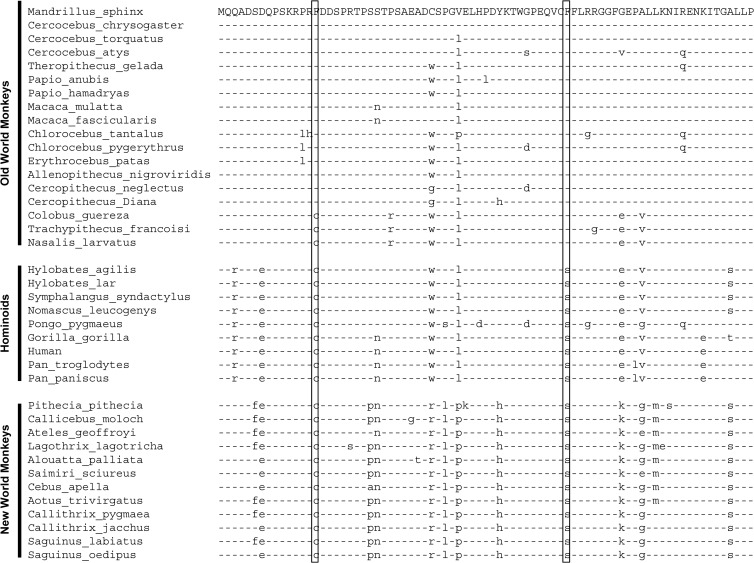
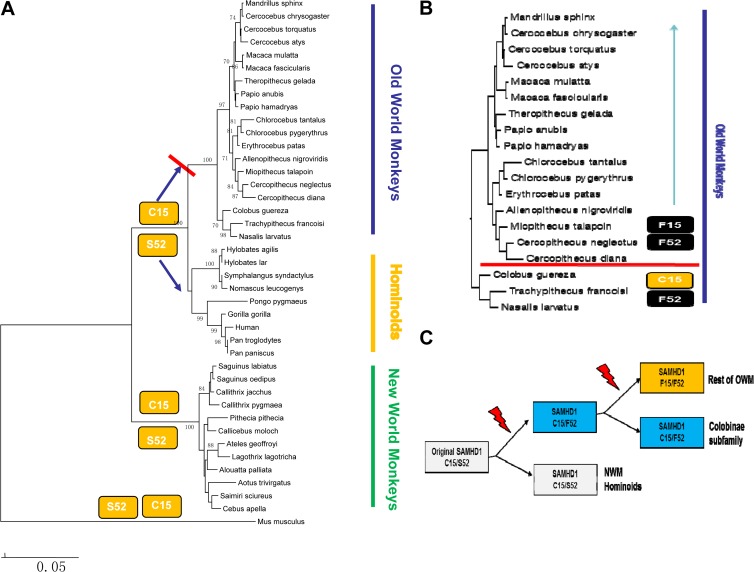
To investigate the species specificity of SIVrcm/SIVmnd2 Vpx proteins, the amino acid sequences of diverse OWM, hominoid, and NWM SAMHD1 proteins were compared (Fig. 3). Two particular amino acids (at positions 15 and 52) unique to OWM SAMHD1 proteins that are sensitive to SIVrcm/SIVmnd2 Vpx proteins were identified (Fig. 3). Comparing the protein sequences of the SAMHD1 proteins from NWMs, hominoids, and OWMs suggested that the ancestral primate SAMHD1 protein contained C15 and S52 (Fig. 4A). S52 was changed to F52 in the subfamily of Colobinae monkey SAMHD1 proteins (Fig. 4B). C15 and S52 were both changed to F15 and F52 in the remainder of the OWMs (Fig. 4B). It is likely that stepwise SAMHD1 mutations at positions 15 and 52 occurred during OWM evolution. S52F was mutated first in the ancestor of Colobinae monkeys and other OWMs (Fig. 4C). This mutation was preserved in the Colobinae monkeys. A separate selection led to the C15F mutation in the rest of the OWMs (Fig. 4C). C15 and S52 are preserved in hominoids and NWMs (Fig. 4C).
FIG 3.
Alignment of N-terminal regions and SAM domains of SAMHD1 proteins from various primate species, including OWMs, hominoids, and NWMs. positions 15 and 52, which displayed unique differences among SAMHD1 from OWMs, hominoids, and NWMs, are boxed.
FIG 4.
Unique patterns of C15F and S52F selection during primate SAMHD1 evolution. (A) The coding sequences of 41 primate SAMHD1 proteins and the Mus musculus SAMHD1 protein were retrieved from GenBank. Phylogenetic analysis was conducted by MEGA4 software by the neighbor-joining method with 1,000 replications on the basis of the Kimura two-parameter model. Bootstrap values of >70% are shown. (B) Phylogenetic analysis of SAMHD1 proteins from OWMs. C15 and F52 are unique to Colobinae monkeys, whereas F15 and F52 substitutions are prevalent among the members of the Cercopithecinae subfamily. (C) Possible evolution pathways for the emergence of F52 and F15 substitutions in SAMHD1 of OWMs.
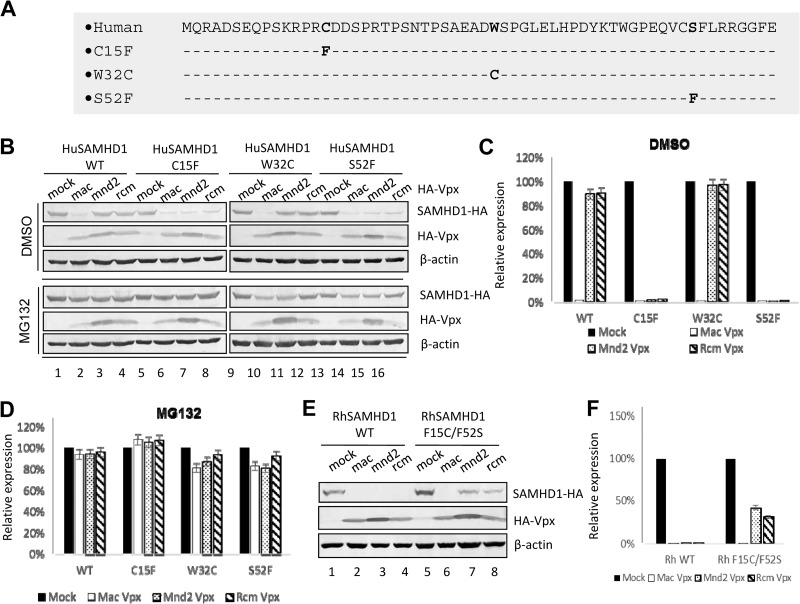
Residues C15 and S52 in the N-terminal region of Hu SAMHD1 are responsible for resistance to SIVrcm/mnd2 Vpx-induced degradation.
To evaluate the roles of C15 and S52 in the sensitivity of Hu SAMHD1 to SIVrcm/mnd2 Vpx, each individual amino acid was mutated (Fig. 5A). W32 has been suggested to be under positive selection in primate SAMHD1 (43). A W32C mutant was also constructed. HEK293T cells were cotransfected with different pSAMHD1-HA expression vectors to reflect the different mutant proteins that were created (wild-type [WT] and C15F, W32C, and S52F mutant proteins). Expression vectors for pSIVmac Vpx, SIVrcm Vpx, and SIVmnd2 Vpx and the empty vector were also generated. Transfected cells were harvested 48 later for SDS-PAGE and immunoblot analysis with the antibodies indicated (Fig. 5B). Unlike WT SAMHD1, which was resistant to SIVmnd2 and SIVrcm Vpx-mediated degradation (Fig. 5B, lanes 3 and 4), mutation of either residue (C15F or S52F) in SAMHD1 resulted in mutant SAMHD1 proteins sensitive to SIVmnd2 and SIVrcm Vpx-mediated degradation (Fig. 5B). On the other hand, W32C mutant SAMHD1 was still resistant to SIVmnd2 and SIVrcm Vpx-mediated degradation (Fig. 5B). Thus, we have identified two highly mutated sites in the N terminus of Hu SAMHD1 that are responsible for the phenotype of resistance to the degradation induced by SIVrcm/mnd2 Vpx (Fig. 5C). The SIVmnd2 and SIVrcm Vpx-mediated degradation of these SAMHD1 molecules could be suppressed by the proteasome inhibitor MG132 (Fig. 5B and D), indicating that a similar mechanism of SAMHD1 depletion is used by SIVmac Vpx and SIVmnd2/SIVrcm Vpx, although they target different regions of primate SAMHD1. We also performed the experiment with the reciprocal Rh SAMHD1 construct. We found that mutations of F15 and F52 of Rh SAMHD1 (F15C/F52S) reduced the ability of SIVmnd2 and SIVrcm Vpx to mediate the degradation of mutant Rh SAMHD1 proteins (Fig. 5E, lanes 7 and 8) compared to WT Rh SAMHD1 (lanes 3 and 4).
FIG 5.
Influence of F15 and F52 substitutions in Hu SAMHD1 on Vpx-mediated degradation. (A) Schematic representation of Hu SAMHD1 mutant constructs. (B to D) Effects of Hu SAMHD1 mutations on Vpx-induced degradation. Expression vectors for HA-tagged WT and C15F, W32C, and S52F mutant Hu SAMHD1 were coexpressed with expression vectors for the indicated HA-tagged Vpx in HEK293T cells. Cell extracts were harvested 48 h later and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA and HA-Vpx. β-Actin was used as the loading control. Expression vectors for HA-tagged Hu SAMHD1 mutant proteins were coexpressed with HA-tagged Vpx in HEK293T cells. DMSO, dimethyl sulfoxide. (E and F) Effects of Rh SAMHD1 mutations on degradation induced by diverse Vpx proteins. HA-tagged WT or F15C/F52S mutant Rh SAMHD1 was coexpressed with the indicated expression vector for HA-tagged Vpx or the empty vector in HEK293T cells. Cell extracts were harvested 48 h later and analyzed by SDS-PAGE, followed by immunoblotting to detect SAMHD1-HA and HA-Vpx; β-actin was used as the loading control. The data shown represent one of three independent experiments. Error bars represent the standard deviation of the mean of triplicate samples.
SIVmac Vpx could inactivate diverse OWM, hominoid, and NWM SAMHD1 proteins with divergent C-terminal sequences.
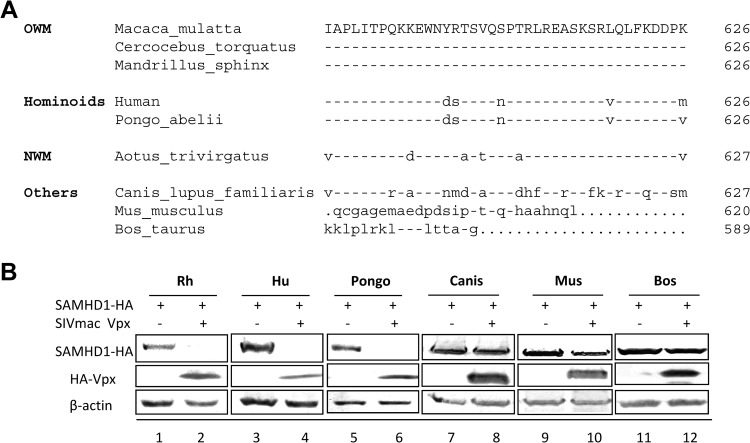
Amino acid variations in the N-terminal region of primate SAMHD1 molecules are involved in the species-specific recognition of SIVrcm/mnd2 Vpx. SIVmac Vpx targets the C-terminal region of the SAMHD1 molecule (35). To determine whether the C-terminal region of primate SAMHD1 molecules can also influence the species-specific recognition of SIVmac Vpx, the ability of SIVmac Vpx to mediate the degradation of SAMHD1 from an OWM (M. mulatta), hominoids (Homo sapiens and Pongo abelii), and a NWM (Aotus trivirgatus) was compared. These primate SAMHD1 molecules have significant variations in the C terminus (Fig. 6A). Degradation of these diverse primate SAMHD1 molecules induced by SIVmac Vpx was observed (Fig. 6B), suggesting that SIVmac Vpx recognizes a motif that is conserved in diverse primate SAMHD1 proteins. However, SIVmac Vpx did not induce the degradation of dog, cow, and mouse SAMHD1 molecules (Fig. 6B). The unique differences between the last 20 amino acids of dog and primate SAMHD1 molecules may provide some clues regarding the residues of primate SAMHD1 that are recognized by SIVmac Vpx.
FIG 6.
SIVmac Vpx could inactivate diverse primate but not nonprimate SAMHD1 proteins with divergent C-terminal sequences. (A) Alignment of the C-terminal regions of the indicated mammalian SAMHD1 protein sequences. (B) Degradation of coexpressed Rh, Hu, and Pongo SAMHD1 proteins was observed in SIVmac Vpx-transfected HEK293T cells. The Canis, Mus, and Bos SAMHD1 proteins were, however, resistant to SIVmac Vpx-induced degradation.
DISCUSSION
In this study, we discovered that the N-terminal region of Hu SAMHD1confers resistance to SIVrcm/mnd2 Vpx-induced degradation. Chimeric Rh-Hu SAMHD1 containing the SAM domain and the surrounding regions from Rh SAMHD1 became sensitive to SIVrcm/mnd2 Vpx-induced degradation. Furthermore, although full-length Rh SAMHD1 is sensitive to SIVrcm/mnd2 Vpx-induced degradation, Rh SAMHD1 with N-terminal but not C-terminal deletions became resistant to SIVrcm/mnd2 Vpx-induced degradation. On the other hand, both Hu and Rh SAMHD1 with N-terminal deletions are still sensitive to SIVmac Vpx-induced degradation. Thus, SIV Vpx proteins have evolved to target different regions of primate SAMHD1 molecules. While SIVmac Vpx targets the C terminus of SAMHD1, SIVrcm/mnd2 Vpx proteins target the N terminus of SAMHD1.
Examination of SAMHD1 proteins from NWMs, hominoids, and OWMs reveals primate lineage-specific variation in the N terminus of SAMHD1 molecules. In particular, C15 and S15 variation shows unique evolution patterns in OWMs. The ancestor primate SAMHD1 protein likely contained C15 and S52. S52 was changed to F52 in the SAMHD1 proteins of the subfamily Colobinae (Fig. 4). C15 and S52 were both changed to F15 and F52 in the Cercopithecinae SAMHD1 proteins (Fig. 4). Interestingly, SAMHD1 proteins from the natural host of SIVrcm and SIVmnd2 contain F15 and F52 and are sensitive to SIVrcm and SIVmnd2 Vpx-induced degradation. We have observed that these two specific amino acids in the SAM domain (S52) and the N-terminal region before the SAM domain (C15) are both required to control the species-specific resistance of Hu SAMHD1 to SIVrcm/mnd2 Vpx-induced degradation. It is possible that adaptation of these SIV viruses in their natural host resulted in the Vpx proteins of these viruses losing the ability to recognize Hu SAMHD1, which carries the evolutionarily more primitive C15 and S52 signature sequence.
Unlike C15 and S52, which are uniformly mutated in all or most OWMs, unique residues in the N-terminal region of SAMHD1 have been identified as being positively selected only in certain OWMs as a countermeasure against Vpx. For example, both positions D46 and Q69 in African green monkey (AGM) SAMHD1 are required to mediate resistance to SIVmnd2 Vpx-induced degradation. A single amino acid substitution, either D46G or Q69R, made mutant AGM SAMHD1 sensitive to SIVmnd2 Vpx. Hu SAMHD1 contains R69 but is still resistant to SIVmnd2 Vpx (Fig. 2B). In this case, the C15F or S52F mutation in Hu SAMHD1 made it again sensitive to SIVmnd2 Vpx (Fig. 5B). It appears that primate SAMHD1 molecules (such as AGM and Hu SAMHD1) can use different combinations of amino acid variations to confer resistance to a particular SIV Vpx (such as SIVmnd2 Vpx).
A single amino acid substitution at either position D46 or Q69 in AGM SAMHD1 could be modified to enhance the mutant protein's sensitivity to SIVmnd2 Vpx (43). Similarly, a position C15 or S52 change to F in Hu SAMHD1 can make the mutant protein sensitive to SIVmnd2 Vpx-induced degradation. These data suggest that SIVmnd2 Vpx is rather flexible in terms of substrate recognition. It is not clear whether other SIV Vpx proteins have evolved to have a similar property. Structural determination of SAMHD1 and Vpx complexes will likely be informative on this issue.
The force driving the C15F and S52F changes in the OWMs is not clear. The selection process is likely to have been gradual. Monkeys carrying C15 or F52 are represented by the Colobinae subfamily. Monkey of two of these genera (Trachypithecus and Nasalis) reside in Asia and are not known to carry any lentivirus. One possibility is that the original selection force was not a lentivirus but other pathogens such as DNA viruses or intracellular bacteria (Fig. 4). Another possibility is that the original selection force was a lentivirus carrying a Vpx protein that could inactivate C15- or S52-containing SAMHD1. As an adaptation by the host, the ancestor monkeys developed the S52-to-F52 mutation. A follow-up assault by SIV containing Vpx or other pathogens that could target C15- or F52-containing SAMHD1 molecules led to a subsequent C15-to-F15 change in the host SAMHD1 proteins (F15 and F52). F15 and F52 have more bulky side chains than C15 and S52, which may alter the interactions between SAMHD1 and its antagonists. Subsequent adaptation of contemporary SIV Vpx to F15- or F52-containing SAMHD1 molecules led to a loss of the ability to recognize the precursor C15- or S52-containing SAMHD1 molecules. It will be interesting to determine whether Vpx proteins that could recognize the precursor C15- or S52-containing SAMHD1 molecules still exist.
Although a Vpx protein that could degrade C15- or S52-containing Hu SAMHD1 has not been identified, SIVsmm-related lentiviruses (including the SIVmac and HIV-2 Vpx proteins) have developed a different strategy to inactivate Hu SAMHD1. These Vpx proteins target the C-terminal region of monkey SAMHD1. They have broad activity against SAMHD1 from OWMs and hominoids, as well as NWMs (21, 43). Thus, Vpx proteins of SIVmac and HIV-2 do not have the more restricted species specificity of the Vpx proteins of SIVmnd2 and SIVrcm, which target the N-terminal region of monkey SAMHD1 molecules. These results suggest that SIVmac Vpx targets a conserved motif in the C terminus of primate SAMHD1 proteins. However, SIVmac could not target dog SAMHD1 for degradation. Several residues uniquely different between dog and primate SAMHD1 molecules were observed. It will be interesting to determine whether these residues are the target of SIVmac Vpx.
We and others have found that SIVmac and SIVrcm Vpx (Fig. 2E)-mediated SAMHD1 degradation is DCAF1 dependent. Furthermore, SIVmac and SIVrcm Vpx-mediated SAMHD1 degradation is proteasome dependent (Fig. 5B). Thus, both the N and C terminus-targeted Vpx proteins maintain a conservative strategy of hijacking DCAF1-containing E3 ubiquitin ligase for SAMHD1 inactivation.
ACKNOWLEDGMENTS
We thank Jacek Skowronski for critical reagents, Xianjun Liu and Chunyan Dai for technical assistance, and Deborah McClellan for editorial assistance.
This work was supported in part by funding from the Chinese Ministry of Science and Technology (No 2012CB911100 and 2013ZX0001-005), the Chinese Ministry of Education (No IRT1016), and the Key Laboratory of Molecular Virology, Jilin Province, China (20102209). We also gratefully acknowledge scholarship support from the China Scholarship Council for Wei Wei.
Footnotes
Published ahead of print 30 October 2013
REFERENCES
- 1.Accola MA, Bukovsky AA, Jones MS, Gottlinger HG. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J. Virol. 73:9992–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson LE, Sowder RC, Copeland TD, Benveniste RE, Oroszlan S. 1988. Isolation and characterization of a novel protein (X-ORF product) from SIV and HIV-2. Science 241:199–201. 10.1126/science.3388031 [DOI] [PubMed] [Google Scholar]
- 3.Paxton W, Connor RI, Landau NR. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selig L, Pages JC, Tanchou V, Preveral S, Berlioz-Torrent C, Liu LX, Erdtmann L, Darlix J, Benarous R, Benichou S. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J. Virol. 73:592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Conway JA, Kim J, Kappes JC. 1994. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J. Virol. 68:6161–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu XF, Ito S, Essex M, Lee TH. 1988. A naturally immunogenic virion-associated protein specific for HIV-2 and SIV. Nature 335:262–265. 10.1038/335262a0 [DOI] [PubMed] [Google Scholar]
- 7.Guyader M, Emerman M, Montagnier L, Peden K. 1989. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J. 8:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kappes JC, Parkin JS, Conway JA, Kim J, Brouillette CG, Shaw GM, Hahn BH. 1993. Intracellular transport and virion incorporation of vpx requires interaction with other virus type-specific components. Virology 193:222–233. 10.1006/viro.1993.1118 [DOI] [PubMed] [Google Scholar]
- 9.Marcon L, Michaels F, Hattori N, Fargnoli K, Gallo RC, Franchini G. 1991. Dispensable role of the human immunodeficiency virus type 2 Vpx protein in viral replication. J. Virol. 65:3938–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu XF, Yu QC, Essex M, Lee TH. 1991. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65:5088–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. 10.1186/1742-4690-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangeot PE, Duperrier K, Negre D, Boson B, Rigal D, Cosset FL, Darlix JL. 2002. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther. 5:283–290. 10.1006/mthe.2002.0541 [DOI] [PubMed] [Google Scholar]
- 13.Fujita M, Otsuka M, Miyoshi M, Khamsri B, Nomaguchi M, Adachi A. 2008. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82:7752–7756. 10.1128/JVI.01003-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramberg T, Sunseri N, Landau NR. 2010. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J. Virol. 84:1387–1396. 10.1128/JVI.01437-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, Stevenson M. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4:e1000057. 10.1371/journal.ppat.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, Skowronski J. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4:e1000059. 10.1371/journal.ppat.1000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18:1682–1687. 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7:e1002425. 10.1371/journal.ppat.1002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 9:87. 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. 10.1016/j.chom.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228. 10.1038/nrg3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9:105. 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, Cheung LE, Wang G, Kazazian HH, Jr, Yu XF. 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 4:1108–1115. 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- 27.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287:21570–21574. 10.1074/jbc.C112.374843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell RD, Holland PJ, Hollis T, Perrino FW. 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286:43596–43600. 10.1074/jbc.C111.317628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White TE, Brandariz-Nuñez A, Carlos Valle-Casuso J, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. 2013. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436:81–90. 10.1016/j.virol.2012.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J, Kaur S, DeLucia M, Hao C, Mehrens J, Wang C, Golczak M, Palczewski K, Gronenborn AM, Ahn J, Skowronski J. 2013. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288:10406–10417. 10.1074/jbc.M112.443796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Chunfeng, G W, Zhao Ke, Qing Xiaohong, Zhang Yinjie, Peng Xin, Zhang Lei, Dong Yuhui, Zhang Wenyan, Li Peng, Wei Wei, Gong Yong, Yu Xiao-Fang. 2013. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4:2722. [DOI] [PubMed] [Google Scholar]
- 32.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3:1036–1043. 10.1016/j.celrep.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 33.Welbourn S, Dutta SM, Semmes OJ, Strebel K. 2013. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87:11516–11524. 10.1128/JVI.01642-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White TE, Brandariz-Nuñez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. 10.1016/j.chom.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. 2012. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287:12550–12558. 10.1074/jbc.M112.340711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayinde D, Casartelli N, Schwartz O. 2012. Restricting HIV the SAMHD1 way: through nucleotide starvation. Nat. Rev. Microbiol. 10:675–680. 10.1038/nrmicro2862 [DOI] [PubMed] [Google Scholar]
- 37.Berger G, Turpin J, Cordeil S, Tartour K, Nguyen XN, Mahieux R, Cimarelli A. 2012. Functional analysis of the relationship between Vpx and the restriction factor SAMHD1. J. Biol. Chem. 287:41210–41217. 10.1074/jbc.M112.403816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandariz-Nuñez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR. 2012. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86:12552–12560. 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laguette N, Benkirane M. 2012. How SAMHD1 changes our view of viral restriction. Trends Immunol. 33:26–33. 10.1016/j.it.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei W, Guo H, Han X, Liu X, Zhou X, Zhang W, Yu XF. 2012. A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell Microbiol. 14:1745–1756. 10.1111/j.1462-5822.2012.01835.x [DOI] [PubMed] [Google Scholar]
- 42.Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. 2013. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 9:e1003496. 10.1371/journal.ppat.1003496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. 2012. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11:194–204. 10.1371/journal.ppat.1003496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, de Silva S, Wang JH, Wu L. 2012. Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS One 7:e37477. 10.1371/journal.pone.0037477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo H, Wei W, Wei Z, Liu X, Evans SL, Yang W, Wang H, Guo Y, Zhao K, Zhou JY, Yu XF. 2013. Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation. PLoS One 8(7):e66201. 10.1371/journal.pone.0066201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. 10.1016/j.chom.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 47.Guo H, Wei W, Wei Z, Liu X, Evans SL, Yang W, Wang H, Guo Y, Zhao K, Zhou JY, Yu XF. 2013. Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation. PLoS One 8:e66201. 10.1371/journal.pone.0066201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]