Abstract
Dengue virus (DENV), a mosquito-borne virus, is responsible for millions of cases of infections worldwide. There are four DENV serotypes (DENV1 to -4). After a primary DENV infection, the antibodies elicited confer lifetime protection against that DENV serotype. However, in a secondary infection with another serotype, the preexisting antibodies may cause antibody-dependent enhancement (ADE) of infection of macrophage cells, leading to the development of the more severe form of disease, dengue hemorrhagic fever. Thus, a safe vaccine should stimulate protection against all dengue serotypes simultaneously. To facilitate the development of a vaccine, good knowledge of different DENV serotype structures is crucial. Structures of DENV1 and DENV2 had been solved previously. Here we present a near-atomic resolution cryo-electron microscopy (cryo-EM) structure of mature DENV4. Comparison of the DENV4 structure with similar-resolution cryo-EM structures of DENV1 and DENV2 showed differences in surface charge distribution, which may explain their differences in binding to cellular receptors, such as heparin. Also, observed variations in amino acid residues involved in interactions between envelope and membrane proteins on the virus surface correlate with their ability to undergo structural changes at higher temperatures.
INTRODUCTION
Dengue virus (DENV) (1) may be responsible for up to 400 million infections worldwide annually (2). It causes febrile illness accompanied by rashes and joint and muscle pain (1). A fraction of the patients develop the more severe forms of the disease: dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), which are fatal if left untreated (1). To date, only symptomatic treatments are available, although there are several vaccine candidates currently undergoing clinical trials.
DENV is a member of the family Flaviviridae. There are four cocirculating serotypes, DENV1 to -4 (3, 4). Usually, an infection with DENV leads to a long-term immunity against that serotype but short-term to no immunity against other serotypes. It has also been shown (5) that an infection with a serotype different from that of the previous infection could potentially lead to the development of DHF/DSS. This may occur in part due to antibody-dependent enhancement (ADE) of infection of macrophage cells, facilitated by cross-reactive, nonneutralizing antibodies or by subneutralizing concentrations of antibodies binding to virus (5, 6).
Numerous attempts have been made to reduce infection via the eradication of its mosquito vector, Aedes aegypti. These efforts started in the 1970s and had been shown to be successful for a period of time in Singapore and Cuba (7). However, such efforts were proven to be unsustainable (8) by the explosion of dengue fever cases in recent years (4). A long-term solution to fully eradicate the disease can be achieved by making a safe and effective vaccine, which can simultaneously stimulate protection against all four serotypes. This would eliminate the possibility of an individual vaccinated against one DENV serotype becoming potentially primed to develop the more severe form of the disease when exposed to another serotype.
DENV is a positive-sense RNA virus with a particle diameter of about 500 Å. The glycoprotein shell consists of envelope (E) and membrane (M) proteins embedded in a host-derived bilipid membrane. Inside the virus particle is the 10.5-kb single-stranded RNA genome complexed with capsid proteins. The surface of DENV particles had been previously shown to be highly dynamic. It can undergo major structural changes at various stages of the infection cycle (9). Previous studies of interactions of DENV2 with antibodies demonstrated that the usually inaccessible parts of its shell proteins could be exposed when the incubation temperature is raised to 37°C (10, 11). Also, cryo-electron microscopy (cryo-EM) reconstruction of DENV2 particles that were incubated at 37°C showed that the particles had expanded and acquired a bumpy surface (12, 13).
During infection, the virus first attaches to cell surface receptors, such as DC-SIGN (14), heparan sulfate (15, 16), and others. After clathrin-mediated endocytosis, the low pH in the endosome induces the E proteins on the virus surface to undergo structural rearrangement from dimers to trimers (9). This leads to fusion of the virus particle with the endosomal membrane, releasing its RNA genome into the cell cytosol. The RNA genome is translated into a single polyprotein, which is cleaved by a series of host and viral proteases into three structural proteins (capsid, premembrane [prM], and E) and seven nonstructural proteins. Virus particles are assembled in the endoplasmic reticulum. The surface of the newly synthesized immature virus particles contains trimeric spikes made from heterodimers of E and prM proteins, with the pr peptide of the prM positioned at the tips of the spikes (17). The virus matures when transported through the acidic compartments of the trans-Golgi network. In this process, the virus undergoes dramatic structural change from the originally spiky-looking surface to a smooth-looking one. This structural rearrangement involves the reorganization of the E-prM trimers into E-prM dimers, which exposes the furin protease cleavage site on the prM. After furin cleavage, the pr molecules stay associated with the virus particles. The virus becomes fully infectious after release from the cell, when the pr molecules dissociate from the virus in the neutral-pH extracellular environment (18).
To develop a safe dengue vaccine and effective therapeutic, it will be an advantage if the high-resolution structures of all DENV serotypes are available. So far, cryo-EM structures of the mature DENV1 (19) and DENV2 (20) are solved to 4.5-Å and 3.6-Å resolutions, respectively. Here we present the structure of mature DENV4 at a 4.1-Å resolution. While the overall organization of the shell proteins of DENV4 is very similar to that of the previously reported DENV serotypes, the surface characteristics are markedly different. Also, the number of contacts between the shell proteins varies between serotypes, which may explain their differences in ability to undergo structural changes at increased temperature.
MATERIALS AND METHODS
Virus sample preparation.
DENV is classified as a biosafety level 2 (BSL2) agent in Singapore; therefore, all procedures involving virus sample were carried out in BSL2-certified facilities. The method has been described previously (21). Briefly, C6/36 cells were infected with DENV4 strain SG/06K2270DK1/2005 at a multiplicity of infection of 0.1 and incubated at 29°C. The virus supernatant was harvested after 4 days and centrifuged at 6,000 rpm for 30 min. Virus was precipitated with 8% (wt/vol) polyethylene glycol 8000 (PEG8000) (Sigma-Aldrich) at 4°C overnight. The virus was collected by centrifugation, and the pellet was resuspended in NTE buffer (12 mM Tris, pH 8.0, 120 mM NaCl, and 1 mM EDTA). The virus was further purified through a 24% (wt/vol) sucrose cushion and then a linear 10 to 30% (wt/vol) potassium tartrate-glycerol gradient. The virus band was collected, buffer exchanged to NTE buffer, and concentrated in an Amicon Ultra centrifugal filter (100-kDa molecular weight cutoff [MWCO]; Millipore).
Cryo-electron microscopy and image data processing.
Lacey carbon copper grids with a layer of thin carbon were used. A 2.5-μl sample was pipetted onto the grid and then blotted with filter paper for 2 s before flash freezing in liquid ethane using an FEI Vitrobot Mark IV instrument. Image acquisitions were done on an FEI Titan Krios electron microscope operating at 300 kV at liquid nitrogen temperature with a nominal magnification of 75,000 and an electron dose of 18e−/Å2. The images were collected with a 4,000 (4K)-by-4K-resolution FEI Falcon direct electron detector. After calibration, the image pixel size was determined to be 1.08 Å. In total, 718 images were collected. Virus particles were selected semiautomatically, by using the e2boxer tool (Swarm mode) in the EMAN2 (22) software package, and 23,564 particles were picked. The astigmatic defocus parameters for each micrograph were estimated using the program CTFFIND3 (23).
Image reconstruction and protein model building.
Image reconstruction was done in an iterative manner: the orientation assignment for all images was carried out using a multipath simulated annealing protocol (24) followed by three-dimensional (3D) reconstruction using the make3d software program from EMAN (25), modified to handle astigmatic contrast transfer function (CTF). An 8-Å resolution structure of mature DENV1 was used as a starting model. After 20 iterations of the orientation search and reconstruction, the result converged. In the end, 16,602 particle images were selected from the data set to produce the final map. The resolution was determined by plotting the Fourier shell correlation coefficient between reconstructions generated from two half-data sets with a cutoff value of 0.5.
The cryo-EM structure of mature DENV2 asymmetric unit (PDB code 3J27) was used as a basis to manually build the model of DENV4 using the software program Coot (26). The structure was then refined using the CNS (27, 28) software package, using the simulated annealing protocol with icosahedral symmetry constraints.
Accession numbers.
The cryo-EM map of mature DENV4 has been deposited in the Electron Microscopy Data Bank (EMD-2485). The coordinates of the component proteins were deposited in the Protein Data Bank (PDB code 4CBF).
RESULTS AND DISCUSSION
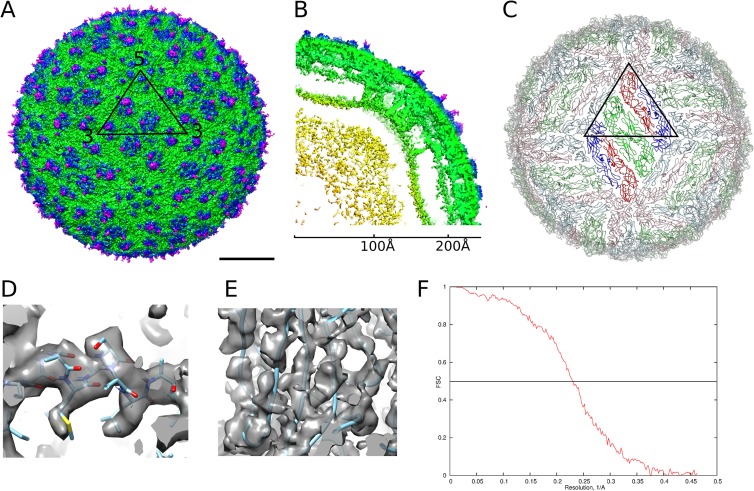
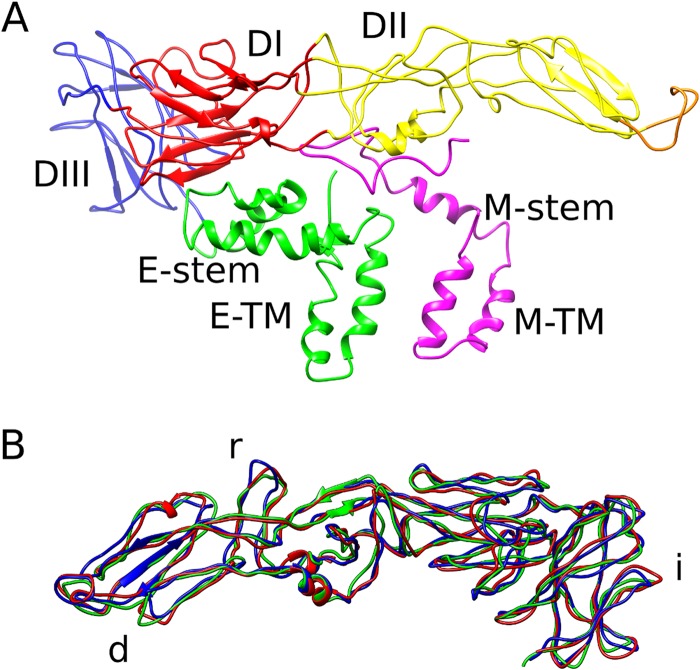
The DENV4 structure (Fig. 1A to C) shows a 480-Å-diameter icosahedral virion with three copies each of the E and M proteins in an asymmetric unit. Three parallel-lying dimers assemble into a raft, and the rafts are arranged in a characteristic herringbone pattern (Fig. 1C). The E proteins form the outer shell and are anchored in the membrane together with the M proteins. The densities corresponding to the polypeptide chains of the component proteins are well resolved (Fig. 1D and E) in the 4.1-Å-resolution map (Fig. 1F). The E protein consists of five domains, of which domains I, II, and III form the mostly β-structural ectodomain, which is similar to the DENV4 E protein ectodomain crystal structure (29) but twisted to follow the virus shell curvature, and the α-helical stem and transmembrane regions form the membrane anchor (Fig. 2A). The M protein consists of a loop at the N terminus followed by membrane-associated α-helical stem and transmembrane regions (Fig. 2A). The N-terminal loop makes extensive contacts with the underside of the E ectodomain. As in other dengue virus structures, the capsid protein is not visible. The three individual E protein molecules in an icosahedral asymmetric unit are not completely identical, with the main structural variation in the loops at the edges of the molecules involved in contacts with the neighboring asymmetric units near the 3- and 5-fold icosahedral vertices (Fig. 2B).
FIG 1.
Cryo-EM structure of DENV4 solved to 4.1-Å resolution. (A) Surface of the DENV4 cryo-EM map. The black triangle shows the icosahedral asymmetric unit. Scale bar, 100 Å. (B) A quarter of a slice through the center, showing radial density distribution. The map is colored radially in panels A and B: up to 160 Å, yellow; 161 Å to 220 Å, green; 221 Å to 230 Å, blue; from 230 Å, magenta. (C) A view of the herringbone arrangement of the E proteins. The three individual E proteins in each asymmetric unit are colored in red, green, and blue. The E proteins in the raft in the center, which consist of two asymmetric units, are in brighter colors. (D and E) Views of the electron density map fitted with structures of the helical E stem region (D), showing densities corresponding to bulky side chains, and β-sheets (E), showing that the densities of β-strands are well separated. (F) A plot of Fourier shell correlation.
FIG 2.
Structure of the envelope and M proteins on DENV4. (A) Structural organization of the E and M protein heterodimer. Following are the colors of the E protein domains: DI, red; DII, yellow (with the fusion loop in orange); DIII, blue; stem and transmembrane regions, green. The M protein is colored magenta. (B) Superposition of the three individual E proteins in an asymmetric unit. The transmembrane regions are removed for clarity. The most variable parts of the E protein are the loops involved in intradimeric contacts (marked “d”), interraft contacts (marked “r”), and icosahedral 3- and 5-vertex formation (marked “i”).
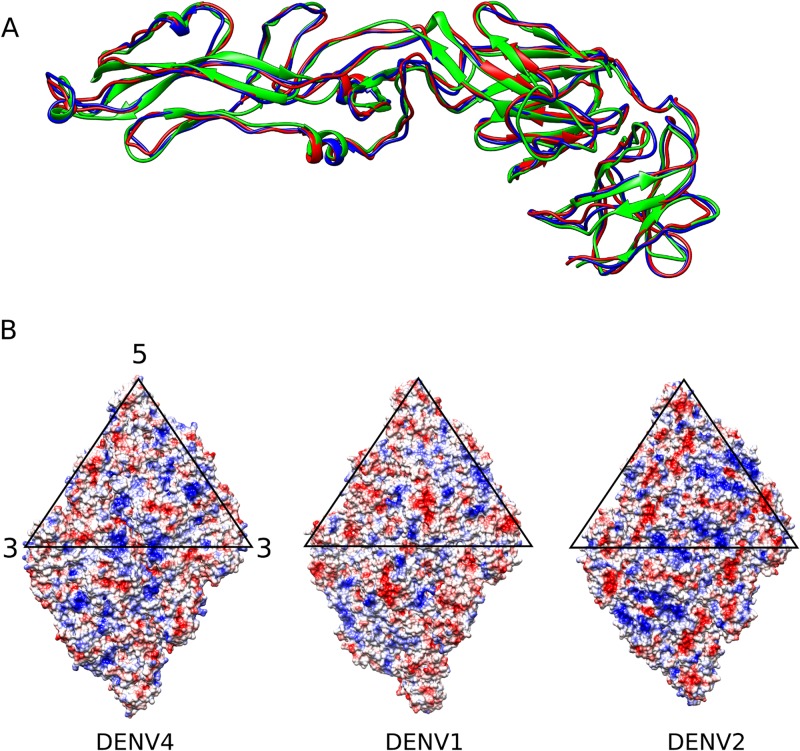
Superposition of the E proteins of DENV4 strain SG/06K2270DK1/2005 with DENV2 strain New Guinea C (30) and DENV1 strain PVP159 (19) does not show significant structural differences (Fig. 3A). The root mean square deviation (RMSD) value between the C-α chains of DENV4 and DENV2 is 1.5 Å, and that between DENV4 and DENV1 is 1.2 Å. The E protein sequence of DENV4 shares about 60% identity to that of both DENV2 and DENV1. Analysis of the surface electrostatic potential showed that the surface charge distribution of DENV4 is different from that of the others (Fig. 3B). Heparan sulfate, an ancillary DENV receptor, consists of repeating disaccharide units, imparting to it an overall negative charge. It has been shown to bind strongly to DENV2 and weakly to DENV1 and DENV4 (31). The surface potential on DENV2 showed larger continuous patches of positive charges compared to those for DENV1 and DENV4, and this may facilitate enhanced binding to heparan sulfate. It has been shown previously that passaging virus in certain cell lines (FRhL, SW13, and BHK21) may increase its affinity for heparan sulfate (32, 33). If this is the case, then the comparison between DENV2 and DENV4 with different passage histories might not be valid. However, Añez et al. (32) has also shown that the mutations did not occur when virus was passaged in C6/36 cells. The virus used in this study had been passaged only in C6/36 cells and therefore may not have acquired such changes.
FIG 3.
Comparing DENV4 with DENV2 and DENV1. (A) Superposition of the E protein structures from the three serotypes: DENV1 (red), DENV2 (green), and DENV4 (blue). There is very little structural variation, with an RMSD of less than 2 Å. (B) Surface charge distribution on a raft for DENV4 (left), DENV1 (center), and DENV2 (right). Red indicates negative charge; blue indicates positive charge. Despite high protein sequence similarity (more than 60% identity between DENV4 and DENV1 or DENV2), the charge distributions are different.
The DENV surface is stabilized by a combination of E protein intradimeric, interdimeric, and interraft interactions (Fig. 1C). The residues involved in these contacts are more conserved than those on the surface-exposed areas of the E proteins. The virus may have evolved in this way so that its surface can easily change without affecting its structural integrity in order to evade the immune system.
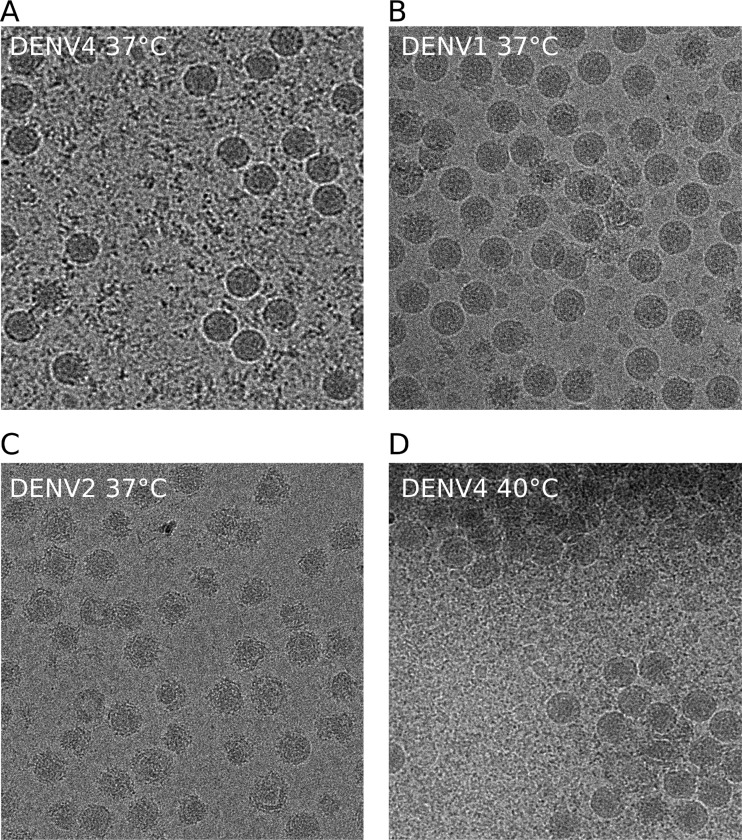
Particles of DENV2 had been observed to expand with incubation at 37°C (12, 13). To test whether this occurs in other serotypes, we incubated DENV1 strain PVP159 and DENV4 strain SG/06K2270DK1/2005 for 30 min at 37°C. The virus particles of DENV4 (Fig. 4A) and DENV1 (Fig. 4B), in contrast to those of DENV2 strain NGC (Fig. 4C), seemed to maintain their size and smooth appearance, indicating that both of these serotypes are more structurally stable at 37°C. Although DENV1 and DENV4 do not expand as does DENV2, we cannot eliminate the possibility that there may be small local domain movements. A further increase in temperature to 40°C caused DENV4 to aggregate (Fig. 4D), with the virus particle size unchanged and the surface still visibly not as bumpy as that in DENV2 (Fig. 4C).
FIG 4.
Abilities of the different DENV serotypes to undergo structural changes at elevated temperatures. Cryo-EM images of DENV4 strain SG/06K2270DK1/2005 (A), DENV1 strain PVP159 (B), and DENV2 strain NGC (C) after incubation at 37°C for 30 min are shown. The sizes of the DENV1 and DENV4 particles remained the same, and their surfaces appeared to be smooth, whereas DENV2 had expanded particles with a rough appearance. (D) DENV4 incubated at 40°C for 30 min. DENV4 particles aggregated together; however, the surfaces are not as bumpy as those of DENV2 at 37°C.
The previously reported structure of the DENV2 at 37°C (12) showed expanded structure where the E intradimer contacts were mostly maintained but the E interdimeric and interraft interactions were disrupted. We compared the structures of the DENV serotypes in terms of the numbers of possible residues that could be involved in hydrogen bonds and electrostatic interactions within E protein dimers, between E protein dimers, and between rafts (Table 1). We assumed that the distance of 8 Å or less between the C-α atoms of two residues indicates possible interaction. The results showed that DENV2 and DENV4 are similar to each other in terms of the number of contacts compared to DENV1. Overall, DENV1 shows stronger intradimeric but weaker interdimeric and interraft contacts than DENV2 and DENV4. DENV4 has the highest number of residues that could be involved in interraft contacts and strong interdimeric interactions, consistent with its observed structural stability at higher temperatures (Fig. 4D).
TABLE 1.
Comparison of numbers of possible electrostatic interactions and hydrogen bonds in E intradimeric, E interdimeric, E interraft, and E-M interface between different DENV serotypesa
| Interaction | No. of residues possibly involved in electrostatic interactions/H bonds |
||
|---|---|---|---|
| DENV1 | DENV2 | DENV4 | |
| E intradimeric | 38/40 | 30/36 | 28/38 |
| E interdimeric | 36/14 | 42/26 | 40/20 |
| E interraft | 74/28 | 74/44 | 86/24 |
| E-M protein | 36/26 | 22/26 | 32/30 |
Since most side chain densities are not resolved, it is assumed that the distance of 8 Å or less between the C-α atoms of two residues indicates possible interaction. Here we count the number of strongly charged residues (Asp, Glu, Lys, and Arg) and polar residues (Asn, Gln, and His) since they may form electrostatic interactions or hydrogen bonds, respectively, with nearby main chain or residues with suitable side chains. Numbers given are of interacting residues per raft.
Although the residues involved in the E and M protein interactions are the most conserved, with 88% identity between the three serotypes, the nonconserved residues are mostly charged. Analysis of the contacts showed that DENV2 had significantly fewer charged residues at its E-M interface than either DENV1 or DENV4. In the DENV2 expanded structure, the E protein layer moved to a higher radius, indicating that the E protein would have to dissociate from the M protein anchored in the membrane. The smaller number of contacts in this interface of DENV2 is consistent with its observed lower thermal structural stability of the virus surface. The strength of the E-M protein interactions may thus be indicative of the ability of virus to undergo expansion at human physiological temperature. This is also supported by the observation that DENV1, with fewer interdimeric and interraft contacts than DENV2, is structurally more stable at 37°C (Fig. 4B and C).
In conclusion, comparison of our near-atomic-resolution structure of DENV4 to the other DENV serotype structures at similar resolutions allow us to observe differences in their virus shell features. In addition, it also enables us to locate regions that are likely to be important in conferring structural stability. This structure can be used as a model for in silico studies of virus interaction with receptors, antibodies, and drugs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an NRF fellowship (R-913-301-015-281), a MOE tier 3 grant (R-913-301-146-112), and a Duke-NUS block grant (R913-200-039-304) awarded to S.-M.L.
The DENV4 strain was provided by E. E. Ooi. We are grateful to M. Wirawan for her help with the manuscript.
Footnotes
Published ahead of print 23 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02641-13.
REFERENCES
- 1.Simmons CP, Farrar JJ, Chau NVV, Wills B. 2012. Dengue. N. Engl. J. Med. 366:1423–1432. 10.1056/NEJMra1110265 [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen R, Vasilakis N. 2011. Dengue—quo tu et quo vadis? Viruses 3:1562–1608. 10.3390/v3091562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubler DJ. 2011. Dengue, urbanization and globalization: the unholy trinity of the 21st century. Trop. Med. Health 39:3–11. 10.2149/tmh.2010-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flipse J, Wilschut J, Smit JM. 2013. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic 14:25–35. 10.1111/tra.12012 [DOI] [PubMed] [Google Scholar]
- 6.Williams KL, Sukupolvi-Petty S, Beltramello M, Johnson S, Sallusto F, Lanzavecchia A, Diamond MS, Harris E. 2013. Therapeutic efficacy of antibodies lacking FcgammaR against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies. PLoS Pathog. 9:e1003157.. 10.1371/journal.ppat.1003157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubler DJ. 2011. Prevention and control of Aedes aegypti-borne diseases: lesson learned from past successes and failures. Asia Pac. J. Mol. Biol. Biotechnol. 19:111–114 [Google Scholar]
- 8.Ooi E-E, Goh K-T, Gubler DJ. 2006. Dengue prevention and 35 years of vector control in Singapore. Emerg. Infect. Dis. 12:887–893. 10.3201/10.3201/eid1206.051210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22. 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- 10.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317. 10.1038/nsmb.1382 [DOI] [PubMed] [Google Scholar]
- 11.Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. 2011. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7:e1002111. 10.1371/journal.ppat.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes in dengue virus when exposed to a temperature of 37°C. J. Virol. 87:7585–7592. 10.1128/JVI.00757-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 110:6795–6799. 10.1073/pnas.1304300110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124:485–493. 10.1016/j.cell.2005.11.042 [DOI] [PubMed] [Google Scholar]
- 15.Dalrymple N, Mackow ER. 2011. Productive dengue virus infection of human endothelial cells is directed by heparan sulfate-containing proteoglycan receptors. J. Virol. 85:9478–9485. 10.1128/JVI.05008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roehrig JT, Butrapet S, Liss NM, Bennett SL, Luy BE, Childers T, Boroughs KL, Stovall JL, Calvert AE, Blair CD, Huang CY. 2013. Mutation of the dengue virus type 2 envelope protein heparan sulfate binding sites or the domain III lateral ridge blocks replication in Vero cells prior to membrane fusion. Virology 441:114–125. 10.1016/j.virol.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 17.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834. 10.1126/science.1153263 [DOI] [PubMed] [Google Scholar]
- 18.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837. 10.1126/science.1153264 [DOI] [PubMed] [Google Scholar]
- 19.Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. 2013. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87:7700–7707. 10.1128/JVI.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 20:105–110. 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725. 10.1016/S0092-8674(02)00660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46. 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 23.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347. 10.1016/S1047-8477(03)00069-8 [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Jiang W, Jakana J, Chiu W. 2007. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J. Struct. Biol. 160:11–27. 10.1016/j.jsb.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT. 2007. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2:2728–2733. 10.1038/nprot.2007.406 [DOI] [PubMed] [Google Scholar]
- 29.Cockburn JJ, Navarro Sanchez ME, Goncalvez AP, Zaitseva E, Stura EA, Kikuti CM, Duquerroy S, Dussart P, Chernomordik LV, Lai CJ, Rey FA. 2012. Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J. 31:767–779. 10.1038/emboj.2011.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. 2013. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 20:105–110. 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YL, Lei HY, Lin YS, Yeh TM, Chen SH, Liu HS. 2002. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antiviral Res. 56:93–96. 10.1016/S0166-3542(02)00095-5 [DOI] [PubMed] [Google Scholar]
- 32.Añez G, Men R, Eckels KH, Lai CJ. 2009. Passage of dengue virus type 4 vaccine candidates in fetal rhesus lung cells selects heparin-sensitive variants that result in loss of infectivity and immunogenicity in rhesus macaques. J. Virol. 83:10384–10394. 10.1128/JVI.01083-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee E, Wright PJ, Davidson A, Lobigs M. 2006. Virulence attenuation of dengue virus due to augmented glycosaminoglycan-binding affinity and restriction in extraneural dissemination. J. Gen. Virol. 87:2791–2801. 10.1099/vir.0.82164-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.