Abstract
ha72 of Helicoverpa armigera nucleopolyhedrovirus (a homologue of ac78) was identified as a conserved late baculovirus gene and characterized. HA72 localizes in the intranuclear ring zone. By generating mutants, we showed that HA72 is essential for budded virus (BD) production and occlusion-derived virus (ODV) embedding. HA72 also interacted with P33, a baculoviral sulfhydryl oxidase. A point mutation of amino acid 22 from lysine to glutamic acid curtailed BV production and precluded ODV occlusion as well as interaction with P33.
TEXT
Baculoviruses are pathogens that infect insects from the orders Lepidoptera, Diptera, and Hymenoptera (1). To date, 60 baculovirus genomes have been fully sequenced, and all of them contain the 37 core genes (2) that play critical roles in baculovirus infection. ac78 is a newly identified baculovirus core gene (2). A previous study showed that deletion of the homologue of ac78 (bm64) in Bombyx mori nucleopolyhedrovirus (BmNPV) prevented the production of infectious budded virus (BV) (3). Its homologue in Helicoverpa armigera NPV (HA72) was shown to be a component of the occlusion-derived virus (ODV) envelope (4). However, its role in the viral life cycle is still unclear, and the present study was undertaken to provide initial biochemical, microscopic, and genetic characterizations of HA72.
HA72 contains an IPLKL motif and a putative fumarate reductase flavoprotein C-terminal motif.
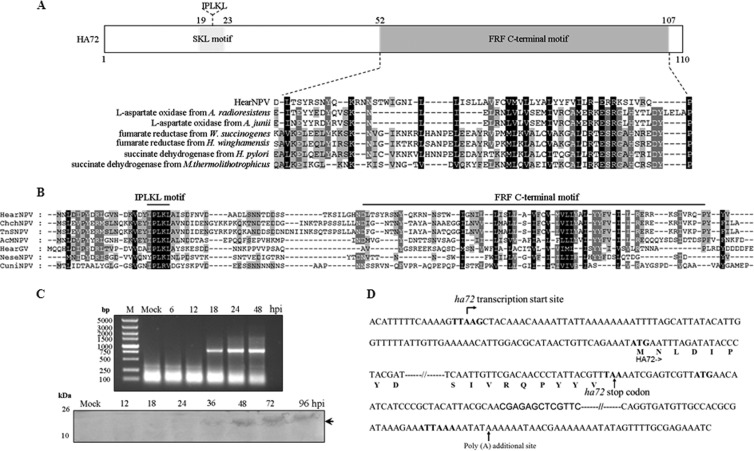
Bioinformatics analysis showed that HA72 has two recognized motifs (Fig. 1A): an IPLKL motif at the N terminus, required for a peroxisome-targeting signal in plants (5), and a putative fumarate reductase flavoprotein (FRF) C-terminal motif that has been implicated in electron transfer in the oxidation-reduction process (6, 7). The IPLKL motif is highly conserved in all baculoviral HA72 homologues, while the FRF-C is conserved at a low amino acid identity in all of the baculoviruses (Fig. 1A and B).
FIG 1.
Characterization of HA72. (A) Map of HA72, showing the location of the IPLKL motif and a putative FRF C-terminal motif. An alignment of a portion of HA72 with representative sequences containing FRF C-terminal motifs is shown. GenBank protein accession numbers: WP_005405373.1, l-aspartate oxidase from Acinetobacter radioresistens SK82; WP_004953071.1, l-aspartate oxidase from Acinetobacter junii NIPH 182; NP_907042.1, fumarate reductase from Wolinella succinogenes DSM 1740; WP_006801824.1, fumarate reductase from Helicobacter winghamensis ATCC BAA-430; WP_001941531.1, succinate dehydrogenase from Helicobacter pylori GAM231Ai; WP_018153189.1, succinate dehydrogenase from Methanothermococcus thermolithotrophicus. (B) Multiple sequence alignments of HA72 with selected baculoviral homologues. GenBank protein accession numbers: NP_075141.1, HearNPV; YP_249681.1, Chrysodeixis chalcites NPV (ChchNPV); YP_308961.1, Trichoplusia ni single NPV (TnSNPV); NP_054108.1, AcMNPV; YP_001649107.1, HearGV; YP_025153.1, Neodiprion sertifer NPV (NeseNPV); NP_203338.1, Culex nigripalpus NPV (CuniNPV). Alignments were constructed using the Lasergene program MegAlign and displayed with the GeneDoc software. (C) Temporal analysis of ha72 transcription and expression. HzAM1 cells were infected with vHaBac-egfp-ph, harvested at the indicated times, and subjected to RT-PCR (upper panel) or Western blot analysis (lower panel). A polyclonal antibody against full-length HA72 that had been raised in rabbits was used in the Western blotting. (D) Illustration of the 5′- and 3′-RACE results for ha72. The sequence of ha72 with its flanking regions is shown. The late promoter (TTAAG), the start and stop codons, and the poly(A) signals are shown in boldface. The arrow indicates the location of the transcription initiation site for ha72 mRNA or the additional poly(A) site.
Time course analyses of transcription and expression suggest that ha72 is a late gene.
Reverse transcription-PCR (RT-PCR) and Western blotting were performed to assess the temporal transcription and expression of ha72. The data summarized in Fig. 1C show that transcription was first detected at 18 hours postinfection (hpi). A rabbit polyconal antibody against the full length of HA72 was used for Western blotting, and a specific immunoreactive band was initially detected at 36 hpi, indicating that ha72 is a late gene. The gene-specific primer ha72up-R (Table 1) was used for 5′-random amplification of cDNA ends (5′-RACE) and, consistent with the above data, sequencing of the 5′-RACE products showed that expression of the ha72 transcript was initiated from the first A of a late gene transcriptional start site motif, TTAAG at 87 nucleotides upstream of the predicted start codon. For the 3′-RACE analysis, an oligo(dT)15 three-site adaptor primer was used for generating cDNA, and subsequent PCR amplification was performed by using a three-site adaptor primer and an ha72-specific forward primer (Table 1). The 3′-RACE analysis showed that ha72 uses a polyadenylation signal motif, ATTAAA at 480 nucleotides downstream of its stop codon (Fig. 1D). In Autographica californica multiple NPV (AcMNPV), ac78 expression is initiated at 12 hpi from the late transcriptional motif TTAAG, and its mRNA is polyadenylated (8).
TABLE 1.
Primers used in the study
| Primers | Sequencea | Restriction site(s) |
|---|---|---|
| Oligo(dT)15 three-site adaptor primer | 5′-CTGATCTAGAGGTACCGGATCCTTTTTTTTTTTTTTT-3′ | |
| Three-site adaptor primer | 5′-CTGATCTAGAGGTACCGGATCC-3′ | |
| ha72-specific forward primer | 5′- ATGAATTTAGATATACCCTACGA-3′ | |
| ha72up-F | 5′-CCCGGTACCCGATAGAATTAAATTCTTTGTTCA-3′ | KpnI site |
| ha72up-R | 5′-CCCCTCGAGGTGGACTATATTCCGCTAAAAT-3′ | XhoI site |
| ha72down-F | 5′-GGGTCTAGACCGAGACGATCGTAGGGTA-3′ | XbaI site |
| ha72down-R | 5′-GGGTCTAGAGATATCTGTGTCCAGTCGCAAATACC-3′ | XbaI and EcoRI sites |
| ha72R-F | 5′-CGCCTCGAGAGCGACGTTCCTCAACGCT-3′ | XhoI site |
| ha72R-R | 5′-CGCGCTAGCGTCCGTGGTCAATTCGAAAAT-3′ | NheI site |
| ha72K22E-R | 5′-CTACGTTAAAATCGGATATAGCCAATTCTAGCGGAATATAG-3′ | |
| ha72K22E-F | 5′-CTATATTCCGCTAGAATTGGCTATATCCGATTTTAACGTAG-3′ | |
| ha72K22R-F | 5′-CTATATTCCGCTAAGATTGGCTATATCCGATTTTAACGTAG-3′ | |
| ha72K22R-R | 5′-CTACGTTAAAATCGGATATAGCCAATCTTAGCGGAATATAG-3′ | |
| ha39-F | 5′-GAAATGCGAATCAGACAGATTACTCG-3′ | |
| ha39-R | 5′-CGCAACCTAACATTTGAGAACACAC-3′ |
Sequences of restriction sites are underlined.
HA72 is essential for BV production, and lysine at position 22 plays an important role in its function.
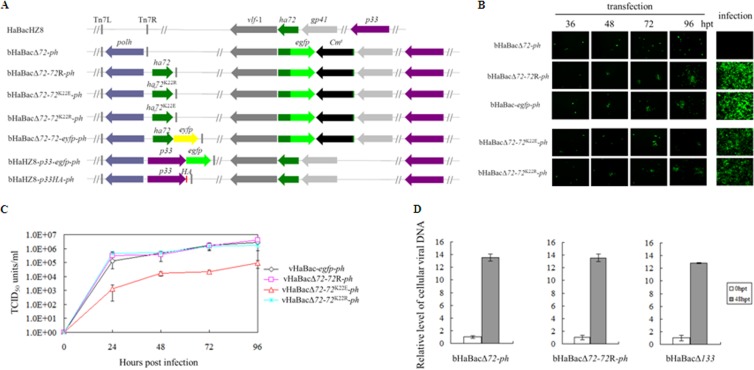
To generate an ha72-inactivated recombinant bacmid (bHaBacΔ72) as a tool to study the function of HA72, a portion (bp 33 to 45) of the coding sequence was replaced with egfp under the hsp70 promoter and also a Cmr cassette by utilizing homologous recombination. Briefly, an upstream fragment was amplified using the primer pair ha72up-F and ha72up-R. A downstream fragment was amplified with primers ha72down-F and ha72down-R. The amplicons were cloned in the pKS-egfp-Cmr vector flanking an egfp-Cmr cassette. The linear fragment, containing egfp, Cmr, and the ha72 flanking sequences, was amplified with ha72up-F and ha72down-R and subsequently electroporated into Escherichia coli BW25113 harboring bHaBacHZ8 and pKD46. Polyhedrin was reintroduced into the original ph locus (bHaBacΔ72-ph) to monitor morphogenesis of the occlusion bodies (OBs). A rescue bacmid, bHaBacΔ72-72R-ph, was also generated by using the Bac-to-Bac system to contain the ha72 open reading frame (ORF) and ph under the control of their native promoters. The ha72 coding sequence was amplified with ha72R-F and ha72R-R. In order to verify that the IPLKL motif is important for HA72 function, mutants with replacement of its lysine (K, at position 22) with positively charged arginine (R) or negatively charged glutamic acid (E) were constructed and designated bHaBacΔ72-72K22E-ph and bHaBacΔ72-72K22R-ph, respectively (Fig. 2A). An ha72 site-directed mutant was generated by overlap extension PCR. For the K22E mutant, the first round of PCR entailed two parallel PCRs with the following primer pairs: ha72R-F/ha72K22E-R and ha72K22E-F/ha72R-R. The second round of amplification was performed with ha72R-F and ha72R-R primers, using the annealed PCR products from the first round as the template. Similarly, the K22R mutant was generated with primers ha72K22R-F and ha72K22R-R. All the primers used in this studied are outlined in Table 1. Schematic diagrams for the generated constructs are shown in Fig. 2A.
FIG 2.
HA72 is essential for BV production but not DNA replication, and the K22E mutation curtails BV production. (A) Schematic diagrams of recombinant bacmids. The ha72 ORF was partially replaced with an egfp and Cm resistance gene cassette from pKS-egfp-Cmr, which contains an egfp gene driven by an hsp70 promoter and a Cmr gene under the control of the T7 promoter, to generate ha72-inactivated bacmid bHaBacΔ72. The recombinant bacmids bHaBacΔ72-ph, bHaBacΔ72-72R-ph, bHaBacΔ72-72K22E-ph, bHaBacΔ72-72K22R-ph, and bHaBacΔ72-72-eyfp-ph were generated by Tn7-based transposition of the indicated genes into the ph locus of bHaBacΔ72. bHaBacΔ72-p33-egfp-ph and bHaBacHZ8-p33HA-ph were generated by Tn7-based transposition of the indicated genes into bHaBacHZ8. (B) Analysis of virus replication and BV production in HzAM1 cells. Fluorescence microscopy images were taken at various time posttransfection or at 120 hpi. (C) One-step growth curve analysis of recombinant viruses. HzAM1 cells were infected with vHaBac-egfp-ph, vHaBacΔ72-72R-ph, vHaBacΔ72-72K22E-ph, or vHaBacΔ72-72K22R-ph at a multiplicity of infection of 5. Supernatants were harvested at the designated time points postinfection and quantified for production of infectious BV by the endpoint dilution method. Data points represent average titers from triplicate infections. Error bars represent standard errors. (D) Real-time PCR analysis of viral DNA replication. HzAM1 cells were transfected with 3 μg bHaBacΔ72-ph, bHaBacΔ72-72R-ph, or control bacmid, bHaBacΔ133 (null for the f protein gene) through three experiments with three replicates per experiment. At the indicated time points postinfection, cells were collected to extract total DNA. Prior to quantitative PCR, DNA was digested with DpnI to avoid interference from E. coli-derived viral DNA. Quantitative PCRs were carried out in triplicates, utilizing 1 μl of DpnI-treated DNA. Error bars represent the standard errors.
Cells transfected with bHaBacΔ72-ph showed only single fluorescent cells, while neighboring green fluorescent cells were observed for cells transfected with the control bacmids (bHaBac-egfp-ph and bHaBacΔ72-72R-ph) (Fig. 2B). When the supernatants from the transfection mixtures were used to infect a second batch of cells, no infection was observed with HaBacΔ72-ph, while control bacmids showed successful infection, suggesting that disruption of ha72 leads to a defect in the production of infectious BVs (Fig. 2B).
One-step growth curve assays were conducted to detect further effects of the recombinants on the production of progeny virus. Comparable viral growth kinetics were observed with vHaBacΔ72-72R-ph, vHaBac-egfp-ph, and vHaBacΔ72-72K22R-ph, but statistical analysis showed that vHaBacΔ72-72K22E-ph had significantly decreased BV titers from 24 hpi to 96 hpi compared to the rescue virus, vHaBacΔ72-72R-ph (Fig. 2C). The results indicated that K22 is needed for optimal production of infectious BV.
Inactivation of HA72 does not affect viral DNA replication.
Quantitative PCR was used to analyze the effect of ha72 on viral DNA replication by using the paired primers ha39-F and ha39-R. Heliothis zea AM1 (HzAM1) cells were transfected with bHaBacΔ72-ph, bHaBacΔ72-72R-ph, or the control bacmid, bHaBacΔ133, which does not contain the membrane fusion protein F (9). At 0 h posttransfection (hpt) and 48 hpt, intracellular viral DNA was extracted and analyzed. The data outlined in Fig. 2D show comparable levels of DNA synthesis for all constructs, suggesting that ha72 is not essential for DNA replication.
HA72 localizes in the intranuclear ring zone region during infection.
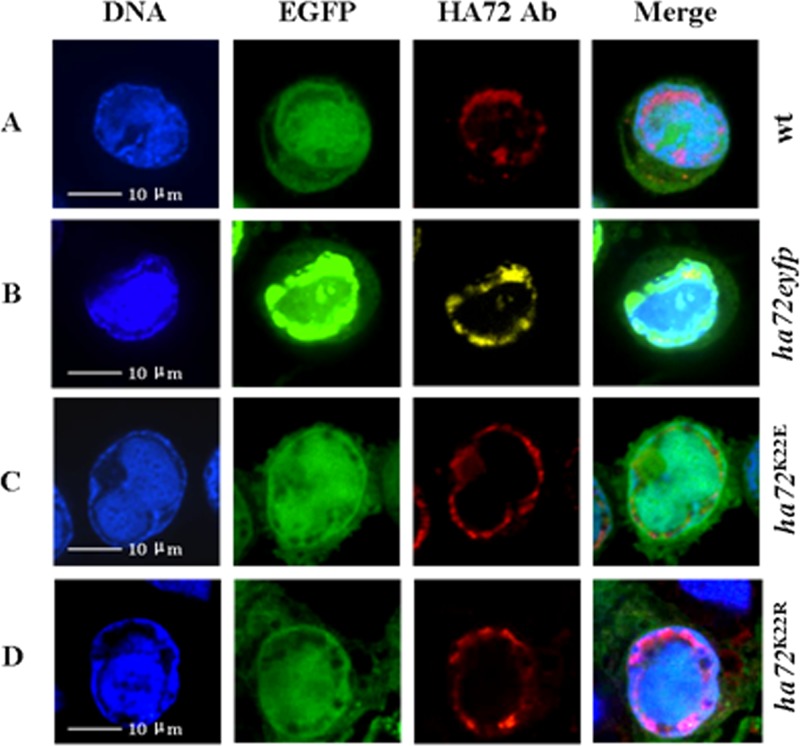
Confocal microscopy was carried out on HzAM1 cells infected with vHaBac-egfp-ph or vHaBacΔ72-72-eyfp-ph (Fig. 3A and B). Fluorescence was captured at 48 hpi by using a laser scanning confocal microscope (PerkinElmer UltraView VOX) with a laser wavelength of 488 nm for DNA (blue) stained with Hoechst 33258 (Beyotime), 561 nm for enhanced green fluorescent protein (EGFP), 605 nm for rhodamine-conjugated secondary antibody (red; Chemicon), and 527 nm for enhanced yellow fluorescent protein (EYFP; yellow). HA72 was mainly localized to the ring zone region (Fig. 3A and B). A small amount of fluorescence was also observed in the center of the nucleus (Fig. 3A and B). By mutating K to R or E, we did not observe changes in the intranuclear localization of HA72 (Fig. 3C and D).
FIG 3.
Subcellular localization of HA72 in HzAM1 cells. Cells were infected with vHaBac-egfp-ph, vHaBacΔ72-72-eyfp-ph, vHaBacΔ72-72K22E-ph, or vHaBacΔ72-72K22R-ph at a multiplicity of infection of 5 and prepared for confocal imaging at 48 hpi. For immunofluoresence assays, polyclonal antiserum against HA72 was used as the primary antibody and stained with anti-rabbit IgG (A, B, and D) or observed via autofluorescence of HA72-EYFP chimera (B). DNA was stained with Hoechst stain. The panels on the right are merged images. Bars, 10 μm.
Deletion of ha72 prevents ODV occlusion.
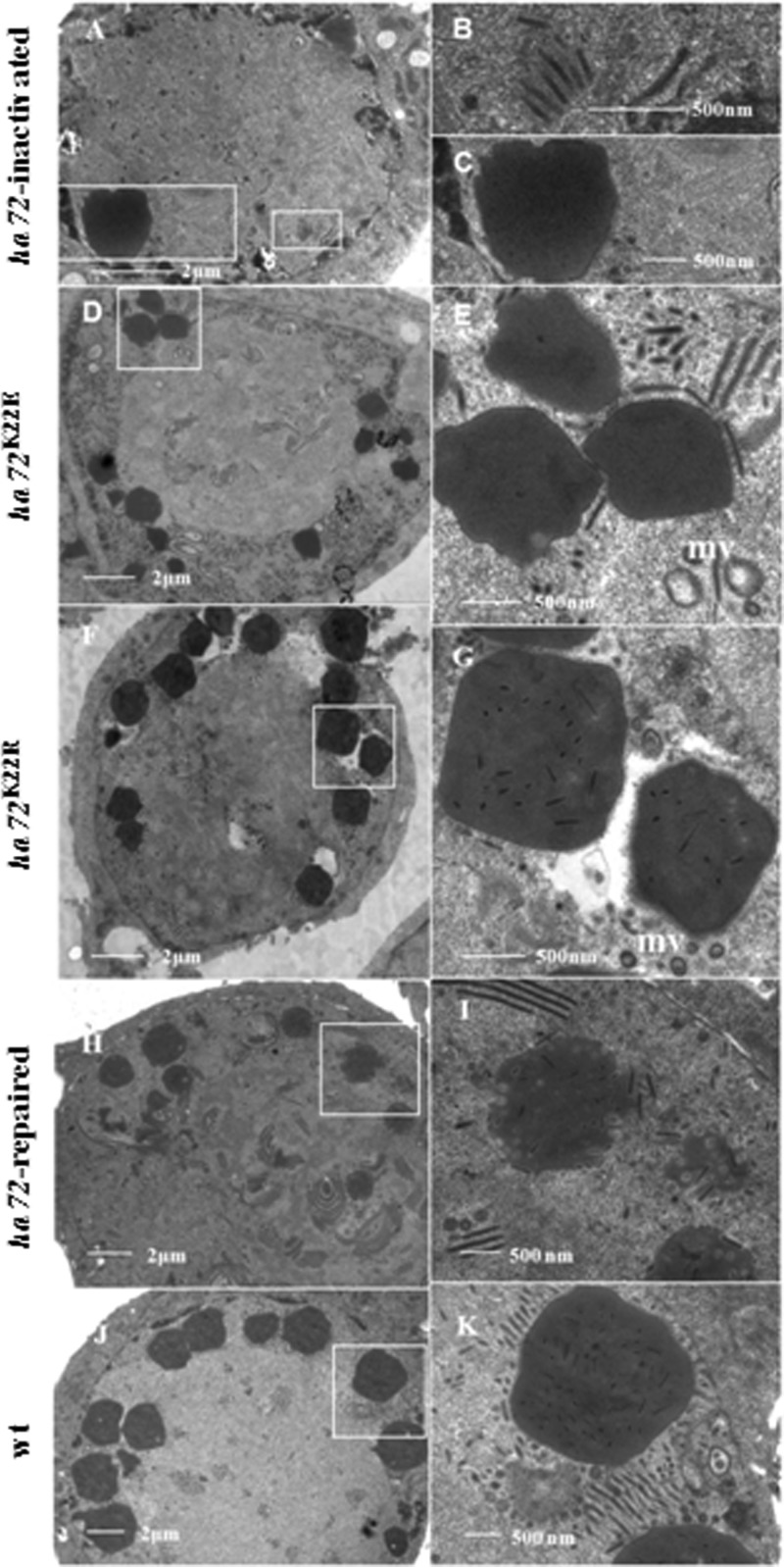
Cells were transfected with bHaBacΔ72-ph, harvested at 96 hpt, and prepared for electron microscopy to observe the effects of ha72 disruption on any morphological alterations. Typical symptoms of infection, such as nuclear hypertrophy and the presence of an electron-dense virogenic stroma (VS), were observed. Nucleocapsids with a normal appearance were also observed; however, they were not embedded into OBs (Fig. 4A, B, and C). Therefore, inactivation of HA72 prevented ODV occlusion.
FIG 4.
TEM analysis. HzAM1 cells were transfected with bHaBacΔ72-ph (A to C) or infected with vHaBacΔ72-72K22E-ph (D and E), vHaBacΔ72-72K22R-ph (F and G), vHaBacΔ72-72R-ph (H and I), or vHaBac-egfp-ph (J and K). Samples were collected at 96 hpt or 96 hpi for TEM.
The K22 mutation interferes with ODV occlusion.
Transmission electron microscopy (TEM) was also used to monitor the morphology of mutants generated from mutations in the IPLKL motif. In the K22E mutant, ODVs with a normal appearance were observed, but occlusion within OBs did not seem to take place (Fig. 4D and E). In contrast, in the K22R mutant, OBs with normal ODV occlusion were observed (Fig. 4F and G), similar to the control viruses vHaBacΔ72-72R-ph (Fig. 4H and I) and vHaBac-egfp-ph (Fig. 4J and K).
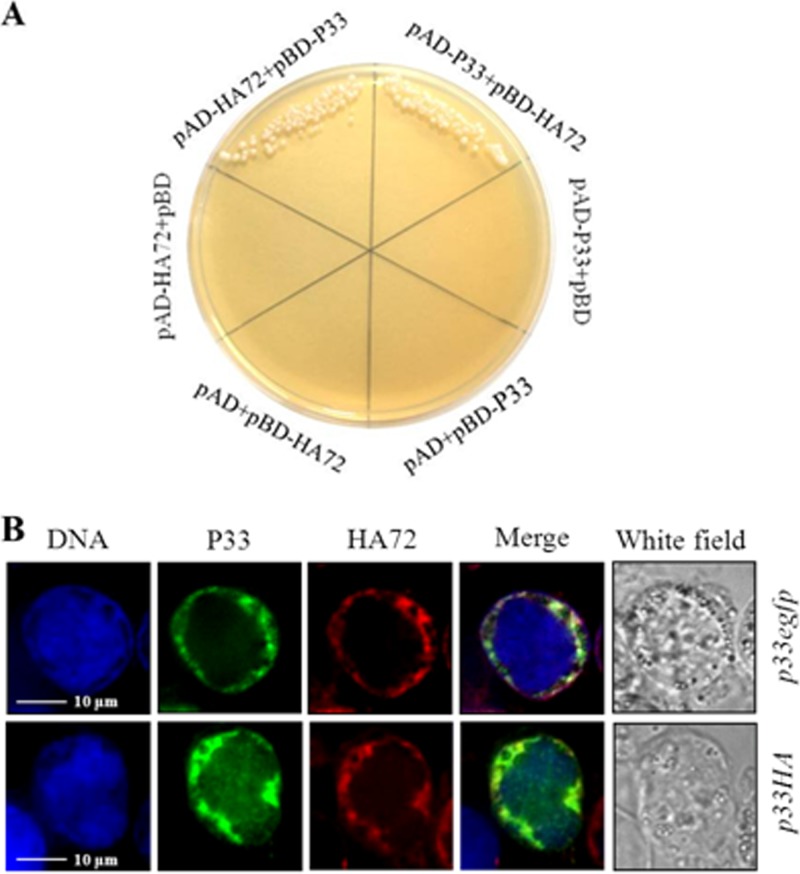
HA72 interacts with P33 in vitro and colocalizes with P33 in infected cells.
In order to investigate how HA72 participates in the process of ODV occlusion, a yeast two-hybrid (Y2H) assay was employed to screen interactions of HA72 with Helicoverpa armigera NPV (HearNPV) ODV structural proteins as described previously (10). We decided to test interactions with viral structural proteins, because it had been shown already that HA72 is part of the virion (4). The results showed that HA72 interacted with P33 (HA80) when it was expressed in the vectors containing either a binding domain or an activation domain (Fig. 5A). The K22E mutation abolished this interaction, while K22R did not affect the interaction (data not shown). During Y2H experiments, the interactions of HA72 with 38K or P49 were also detected but resulted in weak yeast growth (data not shown).
FIG 5.
Interaction and colocalization of HA72 and P33. (A) Results from the yeast two-hybrid assay revealed an interaction of HA72 and P33. The AD fusion plasmid and the BD fusion plasmid were used to cotransform AH109 yeast competent cells. The names of the cotransforming plasmids are indicated. (B) Immunofluorescent staining or autofluorescence images, showing colocalization of HA72 and P33. HzAM1 cells were infected with vHaBacHZ8-p33-egfp-ph or vHaBacHZ8-p33HA-ph, and at 48 hpi HA72 was stained with rhodamine-conjugated antibody. Bars, 10 μm.
Recombinant bacmids bHaBacHZ8-p33-egfp-ph and HaBacHZ8-p33HA-ph were constructed to study the colocalization of HA72 and P33 (Fig. 2A). Confocal microscopy of cells infected with vHaBacHZ8-p33-egfp-ph or vHaHZ8-p33HA-ph showed that P33 is located at the rim of nuclei at 48 hpi (Fig. 5B). For the hemagglutinin (HA)-tagged P33, the chimera was primarily probed with HA-tagged (6E2) mouse monoclonal antibody (Cell Signaling) and subsequently signaled with fluorescein isothiocyanate-conjugated Affinipure goat anti-mouse IgG (H+L; Proteintech). When anti-HA72 antibody was used as the probe, the colocalization of HA72 and P33 to the ring zone region was observed (Fig. 5B). The colocalization result supported the possibility of an interaction between HA72 and P33.
In summary, this study characterized the functions of a baculovirus core gene, ha72. We revealed that K22 is important for the function of HA72, as the K22E mutation curtailed BV production and precluded ODV occlusion. As K22 is located at the conserved IPLKL motif, it will be interesting to see if the motif is important for protein function. Furthermore, protein-protein interactions showed that HA72 interacted with P33, a baculoviral sulfhydryl oxidase that plays an important role in BV production and multiple-enveloped ODV formation (11, 12). As HA72 contains a putative FRF C-terminal motif, we suggest that HA72 and P33 work together in a redox process during baculovirus infection. At this stage, we are expending much effort to obtain enough soluble expressed HA72 to study its enzymatic activity, and we hope to demonstrate its possible role in the proposed redox process in the future. During the process of our manuscript submission, a similar result was published by Tao et al. regarding AC78 (13). Our data corroborate their data indicating that HA72/AC78 is a core baculoviral gene essential for the production of infectious BV and ODV occlusion but is not involved in viral DNA replication.
ACKNOWLEDGMENTS
This research was supported in part by the National Science Foundation of China (31130058 to Z.H.), the Ministry of Science and Technology of China (2009CB118903 and 2008DFB30220 to Z.H.), and a Chinese Academy of Sciences visiting professorship for senior international scientists (2012T1S0019 to B.A.).
We thank Xiulian Sun for statistical analysis and Xijia Liu for making the polyclonal antibody against HA72.
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.Miller LK. (ed). 1997. The baculoviruses. Plenum, New York, NY [Google Scholar]
- 2.Garavaglia MJ, Miele SAB, Iserte JA, Belaich MN, Ghiringhelli PD. 2012. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 86:12069–12079. 10.1128/JVI.01873-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono C, Kamagata T, Taka H, Sahara K, Asano S-I, Bando H. 2012. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res. 165:197–206. 10.1016/j.virusres.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 4.Hou D, Zhang L, Deng F, Fang W, Wang R, Liu X, Guo L, Rayner S, Chen X, Wang H. 2013. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J. Virol. 87:829–839. 10.1128/JVI.02329-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychaudhuri A, Tipton PA. 2002. Cloning and expression of the gene for soybean hydroxyisourate hydrolase. Localization and implications for function and mechanism. Plant Physiol. 130:2061–2068. 10.1104/pp.011049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritz G, Roth A, Schiffer A, Büchert T, Bourenkov G, Bartunik HD, Huber H, Stetter KO, Kroneck PM, Ermler U. 2002. Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-Å resolution. Proc. Natl. Acad. Sci. U. S. A. 99:1836–1841. 10.1073/pnas.042664399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattevi A, Tedeschi G, Bacchella L, Coda A, Negri A, Ronchi S. 1999. Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure 7:745–756. 10.1016/S0969-2126(99)80099-9 [DOI] [PubMed] [Google Scholar]
- 8.Chen Y-R, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 87:6391–6405. 10.1128/JVI.00194-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Tan Y, Yin F, Deng F, Vlak JM, Hu Z, Wang H. 2008. The F protein of Helicoverpa armigera single nucleopolyhedrovirus can be substituted functionally with its homologue from Spodoptera exigua multiple nucleopolyhedrovirus. J. Gen. Virol. 89:791–798. 10.1099/vir.0.83466-0 [DOI] [PubMed] [Google Scholar]
- 10.Peng K, Wu M, Deng F, Song J, Dong C, Wang H, Hu Z. 2010. Identification of protein-protein interactions of the occlusion-derived virus-associated proteins of Helicoverpa armigera nucleopolyhedrovirus. J. Gen. Virol. 91:659–670. 10.1099/vir.0.017103-0 [DOI] [PubMed] [Google Scholar]
- 11.Wu W, Passarelli AL. 2010. Autographa californica multiple nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply enveloped occlusion-derived virus formation. J. Virol. 84:12351–12361. 10.1128/JVI.01598-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long C, Rohrmann G, Merrill G. 2009. The conserved baculovirus protein p33 (Ac92) is a flavin adenine dinucleotide-linked sulfhydryl oxidase. Virology 388:231–235. 10.1016/j.virol.2009.04.006 [DOI] [PubMed] [Google Scholar]
- 13.Tao XY, Choi JY, Kim WJ, Lee JH, Liu Q, Kim SE, An SB, Lee SH, Woo SD, Jin BR. 2013. The Autographa californica multiple nucleopolyhedrovirus ORF78 is essential for BV production and general occlusion body formation. J. Virol. 87:8441–8450. 10.1128/JVI.01290-13 [DOI] [PMC free article] [PubMed] [Google Scholar]