Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes over 90 genes and 25 microRNAs (miRNAs). The KSHV life cycle is tightly regulated to ensure persistent infection in the host. In particular, miRNAs, which primarily exert their effects by binding to the 3′ untranslated regions (3′UTRs) of target transcripts, have recently emerged as key regulators of KSHV life cycle. Although studies with RNA cross-linking immunoprecipitation approach have identified numerous targets of KSHV miRNAs, few of these targets are of viral origin because most KSHV 3′UTRs have not been characterized. Thus, the extents of viral genes targeted by KSHV miRNAs remain elusive. Here, we report the mapping of the 3′UTRs of 74 KSHV genes and the effects of KSHV miRNAs on the control of these 3′UTR-mediated gene expressions. This analysis reveals new bicistronic and polycistronic transcripts of KSHV genes. Due to the 5′-distal open reading frames (ORFs), KSHV bicistronic or polycistronic transcripts have significantly longer 3′UTRs than do KSHV monocistronic transcripts. Furthermore, screening of the 3′UTR reporters has identified 28 potential new targets of KSHV miRNAs, of which 11 (39%) are bicistronic or polycistronic transcripts. Reporter mutagenesis demonstrates that miR-K3 specifically targets ORF31-33 transcripts at the lytic locus via two binding sites in the ORF33 coding region, whereas miR-K10a-3p and miR-K10b-3p and their variants target ORF71-73 transcripts at the latent locus through distinct binding sites in both 5′-distal ORFs and intergenic regions. Our results indicate that KSHV miRNAs frequently target the 5′-distal coding regions of bicistronic or polycistronic transcripts and highlight the unique features of KSHV miRNAs in regulating gene expression and life cycle.
INTRODUCTION
In eukaryotes, a transcript usually encodes a single polypeptide, whereas a polycistronic transcript that can encode several polypeptides is uncommon (1, 2). Viruses, on the other hand, often encode bicistronic and polycistronic transcripts to increase coding efficiency (3). Such gene structure is essential for viruses because of their relatively small genome sizes. In bicistronic and polycistronic transcripts, it is usually the 5′-proximal open reading frames (ORFs) that are translated into proteins by cap-dependent mechanism (1, 2). Although the 5′-distal ORFs have the potentials for encoding proteins, a mechanism other than the cap-dependent initiation for protein translation is required (1, 2). Therefore, the function of the extended 3′ untranslated regions (3′UTRs) containing the 5′-distal ORFs in the bicistronic and polycistronic transcripts is often unclear.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus etiologically associated with Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease, malignancies commonly found in immunocompromised patients (4). Several studies have shown that KSHV encodes a number of bicistronic and polycistronic transcripts (5); however, other than the 5′-proximal ORFs, only two 5′-distal ORFs have been shown to be translated into proteins by mechanisms of initiation at an internal ribosomal entry site and termination-reinitiation, respectively (6, 7). For other KSHV bicistronic and polycistronic transcripts, whether there are alternative functions besides encoding the 5′-proximal proteins remain unclear.
Like other herpesviruses, KSHV has two replication phases: latent and lytic (4). Only a few KSHV genes are expressed during latency, while most of viral genes are expressed in lytic replication. After acute infection in an immunocompetent host, KSHV establishes latency to evade immunosurveillance. In KSHV-associated tumors, most KSHV-infected cells are tightly maintained in latency, but a few of them also undergo spontaneous lytic replication (4). Latency is essential for KSHV-induced tumorigenesis, while lytic replication is required for virus spread and promotion of tumor development (8). Therefore, the fine balance of latency and lytic replication is critical for successful KSHV persistent infection and the development of KSHV-induced malignancies and hence is tightly regulated.
MicroRNAs (miRNAs) are ∼22 nucleotides long, single-stranded noncoding RNAs (9). miRNAs regulate protein expression by translation repression, direct cleavage of mRNAs or both based on an imperfect complementarity between miRNAs and the target mRNA transcripts (10, 11). KSHV encodes 12 pre-miRNAs generating 25 mature miRNAs (12–15). All viral miRNAs are expressed during both viral latency and lytic replication, indicating their important roles in viral life cycle and KSHV-induced malignancies. Indeed, recent studies have shown that KSHV miRNAs regulate viral life cycle in addition to cell cycle, apoptosis, inflammation, angiogenesis, and immune evasion by targeting cellular genes (16–39). Several KSHV miRNAs directly target lytic genes, including ORF50, ORF56, ORF57, and ORF-K2 (19, 34, 40, 41). However, whether any other KSHV genes and transcripts are targeted by the viral miRNAs remains unclear.
miRNAs primarily target the 3′UTRs regions (42). Thus far, the 3′UTR sequences have only been mapped for ∼30% of KSHV gene transcripts. To identify additional viral targets of KSHV miRNAs, we performed 3′RACE (3′ rapid amplification of cDNA ends) to map the 3′UTRs of 74 KSHV genes representing more than 80% of KSHV genes. Of 83 3′UTRs, 34 (41%) are from viral genes with bicistronic and polycistronic transcripts, many of which are identified for the first time. Screening of the 3′UTR reporters has identified 28 3′UTRs targeted by KSHV miRNAs, 11 (39%) of which belong to viral genes with bicistronic and polycistronic transcripts with targeting sites downstream of their 5′-proximal genes. Among them, transcripts from the ORF30-33 and ORF71-73 loci have been confirmed. The miRNA targeting sites, including those in the coding regions of 5′-distal genes, have been further identified. These results indicate the complexity of regulation of viral genes and that the 3′UTR regions downstream of the 5′-proximal genes in bicistronic and polycistronic transcripts can serve as miRNA regulatory elements.
MATERIALS AND METHODS
Cell culture.
PEL cell lines BCBL-1, BCP-1, and BC-3 were maintained in RPMI 1640 medium containing 15% fetal bovine serum (FBS) and 100 μg of gentamicin/ml. Human embryonic kidney 293T cells were grown in Dulbecco modified Eagle medium supplemented with 10% FBS and 100 μg of gentamicin/ml. Recombinant KSHV BAC36-infected 293T cells (293T-BAC36) were cultured as described for 293T cells but with 50 μg of hygromycin/ml (43). To induce viral lytic replication, cells were treated with 20 ng of tetradecanoyl phorbol acetate (TPA)/ml for 48 h.
Construction of KSHV 3′UTR reporters.
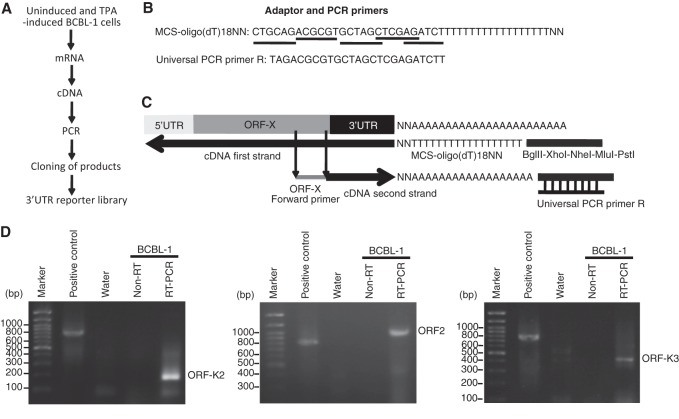
The strategy and procedures for mapping the 3′UTRs and cloning the 3′UTR reporters are shown in Fig. 1. Total RNA was isolated from equal numbers of uninduced BCBL-1 cells and TPA-induced BCBL-1 cells using TRIzol reagent according to the manufacturer's instructions (Life Technologies, Grand Island, NY). After DNase I treatment, cDNA synthesis was carried out with 10 μg of total RNA using the SuperScript III first-strand synthesis system (Life Technologies). A modified oligo(dT) 18NN primer with a tail of multiple cloning sites (MCS) at the 5′ end was used as an adaptor for reverse transcription (RT) to obtain the full-length 3′UTR sequences (Table 1). The 3′UTR sequence from each viral ORF was amplified by PCR using a universal primer complementary to the MCS tail of the RT primer and a specific primer anchored to the downstream sequence of the viral ORF (Fig. 1B and C). The resulting DNA bands were recovered, digested with restriction enzymes, and cloned into a modified pGL-control reporter vector as previously described (26). For a positive control, we used a primer from the human prohibitin gene (PHB) to amplify the 3′UTR sequence from the total RNA of HL60 cells. Samples without RT (non-RT) and water were used as negative controls. Examples of 3′RACE products for ORF-K2, ORF2, and ORF-K3 are shown in Fig. 1D. We were not able to amplify the 3′UTRs of several ORFs despite the use of different primers. For those that were amplified and cloned, we sequenced 10 clones for each PCR product. All cloned 3′UTRs were then aligned with the KSHV genome (GenBank accession no. U75698).
FIG 1.
Schematic illustration of the strategy and procedures used for mapping KSHV 3′UTRs. (A) Procedures used for mapping and cloning KSHV 3′UTRs. (B) Adaptor and PCR primers used for cloning KSHV 3′UTRs. The restriction enzyme sites in the adaptor are underlined. (C) Alignment of adaptor and PCR primers with a hypothetical gene transcript. (D) Examples of 3′RACE products ORF-K2, ORF2, and ORF-K3 resolved on agarose gels. A primer from PHB was used to amplify the 3′UTR sequence from the total RNA of HL60 cells and served as a positive control. Samples without reverse transcription (RT), indicated by “non-RT,” and water were used as negative controls.
TABLE 1.
Primers used in RT and PCR cloning of 3′UTRs
| Gene/application | Primera | Sequence (5′–3′) |
|---|---|---|
| MCS-oligo(dT)18NN | RT | CTGCAGACGCGTGCTAGCTCGAGATCTTTTTTTTTTTTTTTTTTNN |
| Universal R | PCR-R | TAGACGCGYGCTAGCTCGAGATCTT |
| ORF-K1 | PCR-F | TCTTGGTACCCTACACGATTTGTGCACGGAAGAC |
| ORF4 | PCR-F | GCGGGTACCAAACAACACAGTTGCCATCCACTAAT |
| ORF6 | PCR-F | ATTAGGTACCCCCAGAAGAGCCTTGCCTTATCCA |
| ORF9 | PCR-F | TCTTCTCGAGCTTACCGGGGTGGATATAGCAAG |
| ORF10 | PCR-F | TTTTGGTACCGTGGCGGTGGACCTGTACTTCGAC |
| ORF11 | PCR-F | TCTTGGTACCGTTTACGCAGACTGACTCGCT |
| ORF-K2 | PCR-F | AAAGTTACCGCGGTGCGTGTTTTGGACTCTATCCC |
| ORF2 | PCR-F | AGAGGTACCGACGTGTTTCTCTCGCATGATAGCTT |
| ORF-K3 | PCR-F | TCTCGGTACCGCGGGTTGAAGTGTTTCCATATAG |
| ORF70 | PCR-F | AGAGGTACCTACTGCCCGCATCCTACCATTCGTAT |
| ORF-K4 | PCR-F | AAAGGTACCACACCAAGGGCATCCTGCTCGTCGCT |
| ORF-K5 | PCR-F | AATGGTACCGTCGCCTCTGGAGACAAAGAACGTGA |
| ORF-K6 | PCR-F | TTTGGTACCCAGCGGCTGCCTGCCATAGCTTAGA |
| ORF-K7 | PCR-F | TTTGGTACCTACAGTCACCCCTTGCGGGTTATTG |
| ORF16 | PCR-F | TCTGTTACCCACTATATTGGCAGCGGTCGCGATGA |
| ORF17 | PCR-F | TTTGGTACCCAAGATGTTCTGCGAGGAGCTCCTGA |
| ORF18 | PCR-F | TTTGGTACCGCTGGCCTACCTTGGTGCGTTTAACA |
| ORF19 | PCR-F | TTTACCGGTCGGCGATGACTACGATAGAGCGTACT |
| ORF21 | PCR-F | TCTACCGGTGGCTATTAAACCCAGGGCCATCAACT |
| ORF22 | PCR-F | TCTACCGGTTGGCCTGCAGTCTCTCATGTATGTCA |
| ORF23 | PCR-F | TCTACCGGTGAGGTGCTCGGATTTCTTGCCAAGTA |
| ORF24 | PCR-F | TCTACCGGTTTGTCCGTCACGCCATTCGCAAACTA |
| ORF25 | PCR-F | TTTACCGGTTTGAAGAGGTGGCGCCGATGAAGAGA |
| ORF26 | PCR-F | TTTACCGGTAATCTTGGCCCCAGACTGCACGTGTA |
| ORF27 | PCR-F | TCTACCGGTAATCTATCCACCCCTCCCTTGATTCT |
| ORF29b | PCR-F | TCTGAGCTCGTGTGCTGATGATTCCGATGATTCA |
| ORF30 | PCR-F | TTTGAGCTCCTCGGTCTCGCTGACTCTCTATGTC |
| ORF31 | PCR-F | TCTGGTACCTGCACGTTATCGTCTCCATCTATTC |
| ORF32 | PCR-F | TCTGGTACCGTGTTACCTGGGGGTTTTGCTATTA |
| ORF33 | PCR-F | TCTACCGGTGTCTTCTGTTCGATCCCATTGTCCA |
| ORF34 | PCR-F | TTTACCGGTGTCCAACTACGGGCGACTATCTAAT |
| ORF35 | PCR-F | TTTACCGGTTTCGGAGATCAAGGACACAATCGTC |
| ORF36 | PCR-F | TTTACCGGTCTGGCGTTTCAGAAGCAGTGCTATT |
| ORF37 | PCR-F | TTTACCGGTTGCTAATCGTAACCCCCGTCTACTT |
| ORF38 | PCR-F | TTTACCGGTTGCTAATCGTAACCCCCGTCTACTT |
| ORF39 | PCR-F | TTTACCGGTGCGCCAAAGTGAAGGACATATCGAC |
| ORF40 | PCR-F | TTTACCGGTCCCACGCCTGTCACTGACATTAATA |
| ORF41 | PCR-F | AAAACCGGTGCCCTAAACATGCCTCCCGACACTTC |
| ORF42 | PCR-F | TTTACCGGTACAAACCTGCGCGGACCCAGTAAAC |
| ORF43 | PCR-F | TTTACCGGTGACTGACTGGAGTGCCAATGAGTAC |
| ORF44 | PCR-F | TCTACCGGTACCGCAATATCAGAGCCAGTCTAGT |
| ORF45 | PCR-F | TCTACCGGTCCCTGTGTGGAAACGGTGCATATAA |
| ORF46 | PCR-F | TCTGAGCTCCCAGGGCTGTAATCACTTTAACCTA |
| ORF47 | PCR-F | TCTGAGCTCCGAAGTTTGACGGCCTATACTGTAG |
| ORF48 | PCR-F | TCTGAGCTCGTGTGCTGATGATTCCGATGATTCA |
| ORF49 | PCR-F | TTTGAGCTCTAGGTTCCGCGGCTTTGGTCAAGTA |
| ORF50 | PCR-F | AAAGGTACCACGAGGTACAGGAGTCCGGCACACT |
| ORF-K8 | PCR-F | ACAGGTACCCAAGAGGACCACACATTTCGCAACA |
| ORF52 | PCR-F | TCTGAGCTCCCTGAAGGGACTTTCGCTTAGAATC |
| ORF53 | PCR-F | TCTGAGCTCACTACCCCAGGATTCTATGACGTTG |
| ORF54 | PCR-F | TTTACCGGTGGTAGCCGCATATGCCAGATTGTGT |
| ORF55 | PCR-F | AAAACCGGTGGCGTCAGACGACTCGGTAATATGG |
| ORF56 | PCR-F | TCTGAGCTCCTTCATTCCATTAGGGTGAGCGAAGT |
| ORF57 | PCR-F | TTTGAGCTCGCTGCTCTTGGCCTTTGTCCTAACTA |
| ORF-K9 | PCR-F | TCTGGTACCCCGACGAGGATATTTAACCCGAATAC |
| ORF-K10 | PCR-F | TTTGGTACCAGCAGCTGTTCAACACCGCGCGATAC |
| ORF-K10.5 | PCR-F | TTTGAGCTCGACGCTTGCCAGGTGAAGAGTACGAG |
| ORF58 | PCR-F | TTTGGTACCGCAATGGGGGTCGAGATTCAGCTAAT |
| ORF59 | PCR-F | TTTGGTACCCAGGAGGCAGCCGGAATTGGAGTCTC |
| ORF60 | PCR-F | TCTGGTACCTTGAGCGAGACAATTCTGATTACACC |
| ORF61 | PCR-F | TCTGGTACCAGAGTCAGGAATGCGAGCTATCTTAG |
| ORF62 | PCR-F | TCTGAGCTCTGCCTGTACTGGTGTTTGCGAACTTT |
| ORF64 | PCR-F | AAAGGTACCCGTGGTGCCCATGCCCCATCCAATTA |
| ORF65 | PCR-F | AAAGGTACCCACTGGGGGTCTCGGGAAGCAGTATA |
| ORF66 | PCR-F | ATTGGTACCGCCCTTGCCATTCGAGACCTAGATCA |
| ORF67 | PCR-F | ATCGGTACCTGAAGTCGCTGCTGGAGACAGTGTAC |
| ORF69 | PCR-F | AAAGAGCTCCGCACTTGTCAACGCCCTATAACCTC |
| ORF-K12 | PCR-F | TTTGAGCTCGTTTGTGGCAGTTCATGTCCCGGATG |
| ORF71 | PCR-F | TTTGAGCTCCTCTTGCGCGACCTGCTTCACTTAGA |
| ORF72 | PCR-F | GGTGGTACCGATCTGCGCATTCTGGACAGCTATTA |
| ORF73 | PCR-F | ATAGGTACCCCAGACCAGTCGCCCATAACTTATTG |
| ORF-K14 | PCR-F | TAAGAGCTCCGTGAGGCTGCCGCGTGGGGATAATA |
| ORF74 | PCR-F | GGCGAGCTCTTGTGTTTTGCTTCCCTTACCACGTA |
| ORF75 | PCR-F | AAAGAGCTCAGACCCTGCCAGCCAGACACTATCCC |
| PHB | PCR-F | ATCTTTGACTGCCGTTCTCGACC |
RT, reverse transcription; PCR-F, forward PCR primer; PCR-R, reverse PCR primer.
Plasmids and site-directed mutagenesis.
KSHV miRNA expression plasmids, miRNA “sensor reporter” plasmids, and the p3×FLAG plasmid were previously described (26, 44). To express the 3×FLAG fusion proteins with gene-specific 3′UTRs, ORF30, ORF31, ORF32, ORF33, and ORF73 coding regions with their full-length 3′UTR sequences were amplified from their respective cDNAs and cloned into the p3×FLAG vector (Table 2).
TABLE 2.
Primers used for cloning of expression plasmids and mutagenesis of the 3′UTR plasmids
| Primer | Sequence (5′–3′)a |
|---|---|
| ORF31-33-Mutant1-F | CTCGAATTCAGAGACGTCAACCCTTTTGTTTGGCTC |
| ORF31-33-Mutant1-R | GAGCCAAACAAAAGGGTTGACGTCTCTGAATTCGAG |
| ORF31-33-Mutant2-F | CCACGCACGTCGAAGACGTCCTAACAGGAGTGCTC |
| ORF31-33-Mutant2-R | GAGCACTCCTGTTAGGACGTCTTCGACGTGCGTGG |
| ORF71-73-Mutant1-F | TATCTGATTTAATAAACTGATACAAGTTTTGTAAGAA |
| ORF71-73-Mutant1-R | TTCTTACAAAACTTGTATCAGTTTATTAAATCAGATA |
| ORF71-73-Mutant2-F | CCATCACACTCCCAACACTATCGCCATACACCATAGA |
| ORF71-73-Mutant2-R | TCTATGGTGTATGGCGATAGTGTTGGGAGTGTGATGG |
| ORF71-73-Mutant3-F | GCTGGGGGGCTCCCAAGTGGTGGACTTTTGGCACCAC |
| ORF71-73-Mutant3-R | GTGGTGCCAAAAGTCCACCACTTGGGAGCCCCCCAGC |
| ORF71-73-Mutant4-F | TGGCACCACGAGGTCAAGTGGCTGATTACAAAAGCC |
| ORF71-73-Mutant4-R | GGCTTTTGTAATCAGCCACTTGACCTCGTGGTGCCA |
| 3Flag-ORF30-F | AAAGGGCCCTGGGTGAGCCAGTGGATCCTGGACA |
| 3Flag-ORF31-F | AAAGGGCCCTGTCACAAAACAGAAAGACTCTGCCT |
| 3Flag-ORF32-F | AATGGGCCCTGGATGCGCATGCTATCAACGAAAGA |
| 3Flag-ORF33-F | ATAGGGCCCTGGCTAGCCGGAGGCGCAAACTTC |
| 3Flag-ORF30-33-Universal-R | GCCAAGCTTTTAAGCAGTACTTCGGTTTATTTGA |
| 3Flag-ORF73a-F | TATGCTAGCGGCGCCCCCGGGAATGCGCCTGAGGT |
| 3Flag-ORF73a-R | AGTACGCTAGCCGTTGTCCGTGTGTTCA |
| 3Flag-ORF73b-F | TGAACACACGGACAACGGCTAGCGTACT |
| 3Flag-ORF73b-F | CCCCTCGAGTTCTTACAAAACTTGTTAGTGTTTATTAAATCAGA |
| 3Flag-Luc-F | GCGGCTAGCGGAAGACGCCAAAAACATAAAGAAAG |
| 3Flag-Luc-F | GCGGAATTCCGACTCTAGAATTACACGGCGATCT |
| ORF30-ApaI-F | AAAGGGCCCTGGGTGAGCCAGTGGATCCTGGACA |
| ORF33-HindIII-R | GCCAAGCTTTTAAGCAGTACTTCGGTTTATTTGA |
| ORF31-ApaI-F | AAAGGGCCCTGTCACAAAACAGAAAGACTCTGCCT |
| ORF32-ApaI-F | AATGGGCCCTGGATGCGCATGCTATCAACGAAAGA |
| ORF33-ApaI-F | ATAGGGCCCTGGCTAGCCGGAGGCGCAAACTTC |
Mutated nucleotides and restriction sites are indicated by underlining.
Mutagenesis of the predicted putative miRNA binding sites in reporter plasmids and viral gene expression plasmids was performed by site-directed mutagenesis as previously described (26). Briefly, two complementary 36- to 40-nucleotide primers with mutated nucleotides in the center were used for 18 cycles of PCR amplification (Table 2). The extension time of each cycle varied with the sizes of the amplification fragments, usually at 2 kb/min. After removal of the template DNA by DpnI digestion, PCR products were cloned and confirmed by sequencing.
KSHV miRNA sponge plasmids were constructed by using a lentiviral vector pNLSIN kindly provided by Bryan R. Cullen at Duke University Medical Center. Six copies of miR-K10a-3p, together with six copies of miR-K10b-3p sequences, were inserted into a modified pEGFP-C1 shuttle vector (Clontech Laboratories, Mountain View, CA) in which the stop codon TAA was added after the enhanced green fluorescent protein (EGFP) coding sequence. The fragment covering cytomegalovirus promoter, sponge sequence and a poly(A) sequence was PCR amplified and cloned into the SnaBI and XhoI sites of the pNLSIN vector.
miRNA mimics and suppressors.
miRNA mimics and suppressors were as previously described (38). The sequences are listed in Table 3.
TABLE 3.
Sequences of miRNA mimics and suppressors
| Mimic or suppressor | Sequence (5′-3′)a |
|---|---|
| Mimics | |
| miR-K3-5p | UCACAUUCUGAGGACGGCAGCGA |
| miR-K10a | UAGUGUUGUCCCCCCGAGUGGC |
| miR-K10b | UGGUGUUGUCCCCCCGAGUGGC |
| miR-K10a-3p_+1_5 | UUAGUGUUGUCCCCCCGAGUGGC |
| miR-K10b-3p_+1_5 | UUGGUGUUGUCCCCCCGAGUGGC |
| Suppressors | Sequence |
| Scrambled control | CATTAAT+G+T+C+G+G+A+C+AACTCAAT |
| Anti-miR-K10a | GCCACTC+G+G+G+G+G+G+A+CAACACTA |
A “+” before a letter indicates an LNA-modified base.
3′UTR reporter luciferase assay.
To examine the effect of a miRNA on a 3′UTR reporter, 293T cells grown to ca. 60 to 70% confluence in each well of 24-well plate were cotransfected with 15 ng of a 3′UTR reporter plasmid, a reporter vector control or a “sensor reporter,” 50 ng of pSV-β-galactosidase plasmid, and 400 ng of a miRNA expression plasmid or miRNA expression vector control using F2 transfection reagent (Targeting System, El Cajon, CA) or Lipofectamine 2000 transfection reagent (Life Technologies). The “sensor reporters” were used to monitor the expression of the miRNAs (45). To examine the effect of a miRNA mimic on a reporter, cells were first transfected with the miRNA mimic or a scrambled control for 16 h and then with the 3′UTR reporter plasmid or a reporter vector control. Cells were lysed directly with 150 μl of lysis buffer at 48 h posttransfection. The luciferase and β-galactosidase activities were determined as previously described (26). The experiments were repeated at least three times, with the results presented as averages and standard deviation (STD) by setting the values of miRNA expression vector control as “1”. In the initial screening experiments, three rounds of independent screening were performed, each with three repeats. The results of the three rounds of screenings were combined and analyzed.
Manipulation of miRNA functions.
To examine the inhibition of a viral protein expression by a miRNA, 293T cells grown to 70% confluence were cotransfected with the indicated viral gene expression plasmid, together with the miRNA expression plasmid or a vector control using Lipofectamine 2000 (Life Technologies). Alternatively, 293T cells grown to 50% confluence were first transfected with a miRNA mimic or a scrambled mimic control purchased from Ambion (Life Technologies) for 16 h and then the indicated plasmid. For 293T-BAC36 cells, only miRNA mimics or the scrambled mimic control were transfected. To inhibit the function of a KSHV miRNA in PEL cells, a specific LNA-based miRNA suppressor or a scrambled suppressor control purchased from Exiqon (Vedbaek, Denmark) was transfected into cells by using siPORT NeoFX transfection reagent (Life Technologies) for 48 h, and the expression of the targeted protein was examined by Western blotting.
Western blotting.
293T cells plated in 12-well plate were cotransfected with 5 to 100 ng of a 3×FLAG tag-fused viral gene expression plasmid, 0.8 μg of a miRNA expression plasmid or a miRNA expression control vector, and 5 ng of 3×FLAG-luciferase as an internal control for each well. miRNA mimics or the scrambled control were transfected as described above. At 48 h after the last transfection, cells were lysed in radioimmunoprecipitation assay buffer with 1% of a cocktail of protease inhibitors (Sigma, St. Louis, MO). Protein samples separated by using a 10% NuPAGE gel or a 4 to 12% Bis-Tris gel were transferred onto a nitrocelluose membrane. The membrane was blocked with 5% nonfat milk in 1× Tris-buffered saline at room temperature for 1 h, incubated with a primary antibody at a 1:1,000 dilution overnight at 4°C, and then incubated with a horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Specific bands were revealed with chemiluminescence substrates and recorded with an IS2000MM imaging scanner (Eastman Kodak Company, Rochester, NY). Mouse monoclonal antibody to FLAG was purchased from Sigma. Rat monoclonal antibody to LANA was purchased from ABI (New York, NY). Rabbit peptide antibodies to vCyclin and vFLIP were raised with peptides TKALVDPKTGSLC and HLDPRFLERHLAGTC, respectively.
Lentivirus production.
293T cells grown to 70 to 80% confluence in a 100-mm dish were cotransfected with 18 μg of a lentiviral sponge plasmid or a control vector, together with 1 μg of pcTat, 1 μg of pcRev, and 0.5 μg of pHITG plasmids. Culture medium was changed at 2 h posttransfection. Culture medium with viruses was collected at 48, 72, and 96 h posttransfection, cleared off cell debris by centrifugation at 3,000 × g for 10 min, filtered with a 0.45-μm-pore-size filter, and concentrated by ultracentrifugation. Virus titer was determined by counting of GFP-positive cells after infection of 293T cells with 10×-diluted viruses.
RESULTS
Determination of the 3′UTR sequences and polyadenylation [poly(A)] signals (PAS) sites of KSHV transcripts.
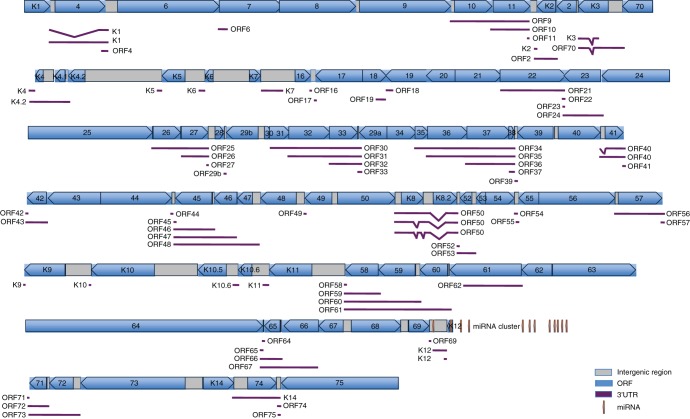
KSHV genome encodes close to 90 genes (8). Only ∼30% of the viral 3′UTRs have been determined. To identify the 3′UTR sequences of the remaining KSHV genes, we performed 3′RACE and cloned the 3′UTR sequences as luciferase reporter constructs. The strategy, procedures, and primers used for the cloning process are illustrated in Fig. 1A to C. Examples of the amplified 3′UTR products for ORF-K2, ORF2, and ORF-K3 are shown in Fig. 1D. For each PCR product, we sequenced up to 10 clones. A total of 83 independent constructs covering 74 KSHV genes were obtained, sequenced, and mapped to the KSHV genome. Among them, the 3′UTRs and their poly(A) sites for 40 KSHV genes were newly identified. Several viral genes had more than one 3′UTR. The structures and positions of the 3′UTRs are summarized in Table 4, and their maps are shown in Fig. 2. The details of the individual 3′UTRs are presented Fig. S1 to Fig. S17 in the supplemental material.
TABLE 4.
Mapping of 3′UTRs of KSHV genesa
| Gene | Pol | 3′UTR cloning | ORF position | PAS sequence | PAS position | TTS from this study | TTS from other studies | Source or reference |
|---|---|---|---|---|---|---|---|---|
| ORF-K1 | + | Y | 105–974 | AAUACA | 2953–2958 | 2970 | 2972 | 64 |
| ORF4 | + | Y | 1142–2794 | AAUACA | 2953–2958 | 2972 | 64 | |
| ORF6 | + | Y | 3210–6611 | AUUAAA | 7008–7013 | 7035 | This study | |
| ORF7 | + | N | 6628–8715 | Not mapped | ||||
| ORF8 | + | N | 8699–11236 | Not mapped | ||||
| ORF9 | + | Y | 11363–14401 | AAUAAA | 17055–17060 | 17079 | This study | |
| ORF10 | + | Y | 14519–15775 | AAUAAA | 17055–17060 | 17070 | This study | |
| ORF11 | + | Y | 15790–17013 | AAUAAA | 17055–17060 | 17070 | 17080 | 69 |
| ORF-K2 | – | Y | 17875–17261 | AAUAAA | 17205–17200 | 17184 | 17182 | 70 |
| ORF2 | – | Y | 18553–17921 | AAUAAA | 17205–17200 | 17181 | This study | |
| ORF-K3 | – | Y | 19609–18608 | AUUAAA | 18612–18607 | 18593 | 18595, 18577 | 71 |
| ORF70 | – | Y | 21104–20091 | AUUAAA | 18612–18607 | 18594 | This study | |
| ORF-K4 | – | Y | 21832–21548 | AAUAAA | 21348–21343 | 21324 | This study | |
| ORF-K4.1 | – | N | 22529–22185 | AAUAAA | 21348–21343 | 21324 | 52, 72, 73 | |
| ORF-K4.2 | – | N | 23077–22745 | AAUAAA | 21348–21343 | 21324 | 21325 | 52, 73 |
| ORF-K5 | – | Y | 26483–25713 | AUUAAA | 25574–25569 | 25545 | 25538 | 74 |
| ORF-K6 | – | Y | 27424–27137 | AAUAAA | 26920–26915 | 26895 | This study | |
| ORF-K7 | + | Y | 28622–29002 | AAUAAA | 29719–29724 | 29743 | This study | |
| ORF16 | + | Y | 30145–30672 | AUUAAA/AAUGAA | 30732–30737/30736-30741 | 30753 | This study | |
| ORF17 | – | Y | 32482–30821 | AAUAAA | 30765–30760 | 30744 | This study | |
| ORF18 | + | Y | 32424–33197 | AUUAAA | 33431–33436 | 33450 | This study | |
| ORF19 | – | Y | 34843–33194 | AUUAAA | 32873–32868 | 32862 | This study | |
| ORF20 | – | N | 35573–34611 | Not mapped | ||||
| ORF21 | + | Y | 35383–37125 | AAUAAA | 39312–39317 | 39328 | This study | |
| ORF22 | + | Y | 37113–39305 | AAUAAA | 39312–39317 | 39334 | This study | |
| ORF23 | – | Y | 40516–39302 | AAGAAA | 39278–39273 | 39250 | This study | |
| ORF24 | – | Y | 42778–40520 | AAUAAA | 39251–39246 | 39227 | This study | |
| ORF25 | + | Y | 42777–46907 | AAUAAA | 48741–48746 | 48789 | This study | |
| ORF26 | + | Y | 46933–47850 | AAUAAA | 48741–48746 | 48769 | This study | |
| ORF27 | + | Y | 47873–48745 | AUUAAA | 4877–48775 | 48780 | This study | |
| ORF28 | + | N | 48991–49299 | Not mapped | ||||
| ORF29b | – | Y | 50417–49362 | AAUAAA | 49366–49361 | 49342 | 49347 | 75 |
| ORF30 | + | Y | 50623–50856 | AAUAAA | 54072–54077 | 54099 | 53 | |
| ORF31 | + | Y | 50763–51437 | AAUAAA | 54072–54077 | 54099 | 53 | |
| ORF32 | + | Y | 51404–52768 | AAUAAA | 54072–54077 | 54099 | 53 | |
| ORF33 | + | Y | 52761–53699 | AAUAAA | 54072–54077 | 54100 | 53 | |
| ORF29a | – | N | 54676–53738 | Not mapped | ||||
| ORF34 | + | Y | 54675–55658 | AAUAAA | 58853–58858 | 58878 | This study | |
| ORF35 | + | Y | 55639–56091 | AAUAAA | 58853–58858 | 58878 | 58881 | 76 |
| ORF36 | + | Y | 55976–57301 | AAUAAA | 58853–58858 | 58878 | This study | |
| ORF37 | + | Y | 57273–58733 | AAUAAA | 58853–58858 | 58878 | This study | |
| ORF38 | + | Y | 58688–58873 | Not mapped | ||||
| ORF39 | – | Y | 60175–58976 | AAUAAA | 58906–58901 | 58885 | This study | |
| ORF40 | + | Y | 60308–61681 | AAUAAA | 62540–62545 | 62561, 62559 | This study | |
| ORF41 | + | Y | 61827–62444 | AAUAAA | 62540–62545 | 62559 | This study | |
| ORF42 | – | Y | 63272–62436 | AAUAAA | 62440–62435 | 62416 | This study | |
| ORF43 | – | Y | 64953–63136 | AAUAAA | 62440–62435 | 62416 | This study | |
| ORF44 | + | Y | 64892–67258 | AAUAAA | 67295–67300 | 67319 | This study | |
| ORF45 | – | Y | 68576–67353 | AAUAAA | 67350–67345 | 67323 | 67329 | 52 |
| ORF46 | – | Y | 69404–68637 | AAUAAA | 67350–67345 | 67323 | This study | |
| ORF47 | – | Y | 69915–69412 | AAUAAA | 67350–67345 | 67328 | This study | |
| ORF48 | – | Y | 71381–70173 | AAUAAA | 67350–67345 | 67333 | This study | |
| ORF49 | – | Y | 72538–71630 | AAUAAA | 71634–71629 | 71615 | 71618 | 77 |
| ORF50 | + | Y | 72734–74629 | AAUAAA | 76714–76719 | 76737 | 76737 | 52 |
| ORF-K8 | + | Y | 74850–75569 | AAUAAA | 76714–76719 | 76737 | 76737 | 52 |
| ORF-K8.1 | + | N | 75785–76690 | AAUAAA | 76714–76719 | 76737 | 76737 | 52 |
| ORF52 | – | Y | 77197–76802 | AAUAAA | 76729–76724 | 76706 | This study | |
| ORF53 | – | Y | 77665–77333 | AAUAAA | 76729–76724 | 76706 | This study | |
| ORF54 | + | Y | 77667–78623 | AAUAAA | 78753–78778 | 78778 | This study | |
| ORF55 | – | Y | 79448–78765 | AAUAAA | 78724–78719 | 78706 | This study | |
| ORF56 | + | Y | 79436–81967 | AAUAAA | 83608–83613 | 83631 | 83628, 83631, 82634 | 48 |
| ORF57 | + | Y | 83717–83544 | AAUAAA | 83608–83613 | 83630 | 83628, 83631, 82634 | 48 |
| ORF-K9 | – | Y | 85209–83860 | AAUAAA | 83808–83803 | 83789 | 83787, 83788, 83789 | 47 |
| ORF-K10 | – | Y | 88164–86074 | AAUAAA | 86030–86025 | 86004 | 86006, 86007, 86008 | 47 |
| ORF-K10.5 | – | Y | 91394–89600 | AAUAAA | 89392–89387 | 89372 | 89373 | 47 |
| ORF-K11 | – | N | 93367–91964 | AAUAAA | 91753–91748 | 91751, 91756 | 47 | |
| ORF58 | – | Y | 95544–94471 | AAUAAA | 94488–94483 | 94466 | 94469, 94477 | 48 |
| ORF59 | – | Y | 96939–95549 | AAUAAA | 94492–94487 | 94466 | 94469, 94477 | 48 |
| ORF60 | – | Y | 97787–96870 | AAUAAA | 94492–94487 | 94466 | This study | |
| ORF61 | – | Y | 100194–97816 | AAUAAA | 94492–94487 | 94471 | This study | |
| ORF62 | – | Y | 101194–100199 | AAUAAA | 98298–98293 | 98723 | This study | |
| ORF63 | + | N | 01208–103994 | Not mapped | ||||
| ORF64 | + | Y | 104000–111907 | AAUAAA | 111886–111891 | 111912 | This study | |
| ORF65 | – | Y | 112443–111931 | AAUAAA | 111826–111807 | 111807 | This study | |
| ORF66 | – | Y | 113759–112470 | AAUAAA | 111826–111807 | 111809 | This study | |
| ORF67 | – | Y | 114508–113693 | AAUAAA | 111826–111807 | 111809 | This study | |
| ORF68 | + | N | 114768–116405 | Not mapped | ||||
| ORF69 | + | Y | 116669–117346 | AAUAAA | 117403–117408 | 117424 | 117425 | 78 |
| ORF-K12 | – | Y | 118101–117919 | AAUAAA | 117447–117442 | 117432 | 117432 | 79 |
| ORF71 | – | Y | 122710–122145 | AAUAAA | 122094–122089 | 122068 | 122070 | 50 |
| ORF72 | – | Y | 123566–122793 | AAUAAA | 122094–122089 | 122068 | 122070 | 50 |
| ORF73 | – | Y | 127296–123808 | AAUAAA | 122094–122089 | 122065 | 122070 | 50 |
| ORF-K14 | + | Y | 127883–128929 | AUUAAA | 130518–130523 | 130547 | 130545 | 80 |
| ORF74 | + | Y | 129371–130399 | AUUAAA | 130518–130523 | 130547 | 130545 | 80 |
| ORF75 | – | Y | 134440–130550 | AUUAAA | 130518–130523 | 130499 | This study | |
| ORF-K15 | – | N | 136279–135997 | Not mapped |
Pol, the direction of RNA Pol II transcription; PAS, poly(A) signal.; TTS, transcription termination site.
FIG 2.
Annotation of 3′UTRs in the KSHV genome. 3′UTRs of KSHV genes were mapped using uninduced and TPA-induced BCBL-1 cells as described in Materials and Methods. Genes are labeled in blue with arrowheads indicating the transcription orientation. Intergenic regions are labeled in gray. 3′UTRs of viral transcripts are indicated as purple lines.
3′UTR analyses of KSHV genes.
Poly(A) signals are a defining feature of eukaryotic protein-coding genes (46). Analysis of KSHV 3′UTR sequences revealed the presence of poly(A) signals in all transcripts. The position of a number of poly(A) sites differ from the published data by ca. 1 to 3 nucleotides (Table 4). Such variations have been described in previous studies (47, 48). This is likely due to the flexibility on the selection of the cleavage sites (49).
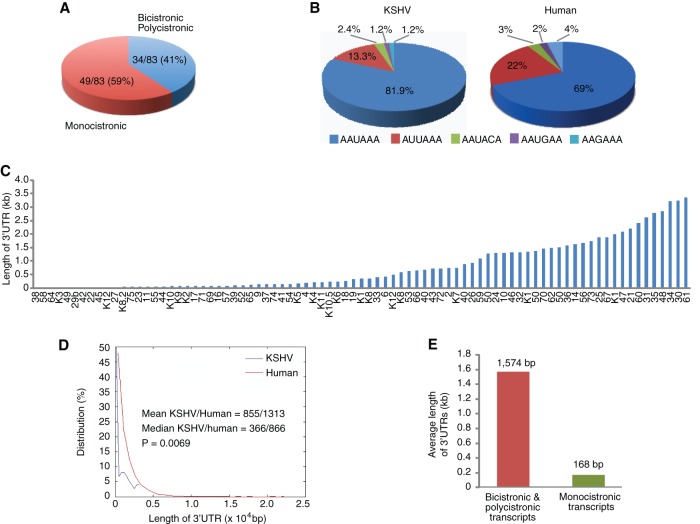
In eukaryotes, a transcript usually encodes a single polypeptide. KSHV genome encodes a large number of genes clustered in specific genomic loci with overlapping coding or short intergenic sequences. These loci have the potential to encode bicistronic and polycistronic transcripts that often share the same poly(A) signals. Indeed, bicistronic and polycistronic transcripts have previously been identified at several KSHV genomic loci, such as the ORF71-73, ORF50-K8, ORF34-37, and ORF30-33 loci (50–53). Of 83 3′UTRs identified in the present study, 34 (41%) were from viral genes with bicistronic or polycistronic transcripts, all of which contained two or more ORFs, respectively, based on the sequenced 3′UTR clones (Fig. 3A). Of 83 3′UTRs, 68 (81.9%) used the conventional “AAUAAA” as the PAS sequence (Fig. 3B). However, we also identified 15 (19.1%) other PAS sequences. Thus, similar to human transcripts, KSHV uses a variant poly(A) signal (PAS) for gene transcription. The pattern of distribution of KSHV PAS sequences was similar to that of human transcripts, although KSHV had fewer PAS variants (Fig. 3B).
FIG 3.
Features of KSHV 3′UTRs. (A) Distribution of KSHV genes with monocistronic transcripts and bicistronic and polycistronic transcripts. (B) Distribution of poly(A) signal sequences of KSHV and human transcripts. (C) Distribution of KSHV 3′UTRs by length. (D) Distribution of different lengths of KSHV and human 3′UTRs by percentage. (E) Average lengths of 3′UTRs of KSHV genes with monocistronic transcripts and bicistronic and polycistronic transcripts.
For bicistronic and polycistronic transcripts, protein translation is usually only initiated with the 5′-proximal ORFs, whereas other 5′-distal ORFs and intergenic regions are considered 3′UTR sequences (1, 2). Because KSHV encodes a large number of bicistronic and polycistronic transcripts, we observed a wide range of 3′UTRs, with sizes varying from 5 bp to >3,000 bp (Fig. 3C). However, compared to human 3′UTRs, the lengths of KSHV 3′UTRs are shorter than those of human (mean 855 bp versus 1,313 bp, median 366 bp versus 866 bp, P = 0.0069) (Fig. 3D). Among the KSHV transcripts, the average length for the 3′UTRs of viral genes with bicistronic and polycistronic transcripts is substantially longer than that for monocistronic transcripts (1,574 bp versus 168 bp) (Fig. 3E).
Screening of 3′UTR reporters for novel viral targets of KSHV miRNAs.
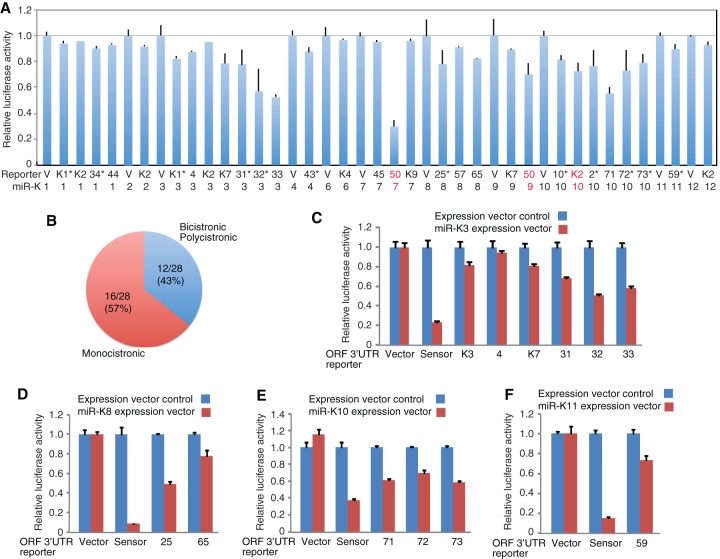
The relative long 3′UTRs of KSHV genes suggest that they might be more likely regulated by miRNAs. We performed screening of the 3′UTR reporters for their responsiveness to KSHV miRNAs. Each 3′UTR reporter, as well as the 3′UTR reporter control vector, were examined for their responsiveness to the individual expression constructs of the 12 KSHV pre-miRNAs or the miRNA expression vector control. Reporters with two repeats of perfect matching sequences of the respective miRNAs, termed “sensor reporters,” were used to monitor the expression of the miRNAs (45). In total, three rounds of independent screening were performed, each with three repeats. The results of the three rounds of screenings were combined and analyzed by using the values of miRNA expression vector control as “1”.
Previous studies have shown that RTA (ORF50) 3′UTR is targeted by miR-K7-5p and miR-K9* (19, 34). Indeed, both pairs were identified in our screening, albeit only weak repression activity was observed for miR-K9 construct expressing both miR-K9 and -K9* (Fig. 4A). In total, we identified 28 new 3′UTR-miRNA pairs with repression levels ranging from 5 to 45% (Fig. 4A), 12 (43%) of which were 3′UTRs from viral genes with bicistronic and polycistronic transcripts (Fig. 4B). Multiple 3′UTR reporters from the ORF71-73 locus or the ORF31-33 locus were repressed by the same miRNA, respectively, indicating that the repression effects were likely mediated by the same targeting site(s). We further confirmed the responsiveness of the 3′UTR reporters to miR-K3 (ORF-K3, ORF4, ORF-K7, ORF31, ORF32, and ORF33) (Fig. 4C), miR-K8 (ORF25 and ORF65) (Fig. 4D), miR-K10 (ORF71, ORF72, and ORF73) (Fig. 4E), and miR11 (ORF59) (Fig. 4F).
FIG 4.
Newly identified viral genes targeted by KSHV miRNAs. (A) Repression of 3′UTRs by KSHV miRNAs detected by reporter assays. 3′UTR reporters from KSHV genes with bicistronic and polycistronic transcripts are labeled with an asterisk (*). The miR-K10/ORF-K2 pair, which was independently identified in a HITS-CLIP screening (40), is labeled in red type. The miR-K7/ORF50 and miR-K9/ORF50 pairs identified in previous studies are shown in “red” and used as positive controls (19, 34). (B) Distribution of 3′UTRs from KSHV genes with monocistronic transcripts, and bicistronic and polycistronic transcripts repressed by KSHV miRNAs. (C to F) Confirmation of repression of 3′UTRs by KSHV miRNAs. 3′UTRs were targeted by miR-K3 (C) miR-K8 (D), miR-K10 (E), and miR-K11 (F). 293T cells were cotransfected with 3′UTR reporter plasmids or a reporter vector control and the respective miRNA expression plasmids or a miRNA expression vector control, together with a pSV-β-galactosidase plasmid for 48 h, and the relative luciferase activities were measured after normalization. In the initial screening experiments (A), three rounds of independent screening were performed, each with three repeats. The results of the three rounds of screenings were combined and analyzed. All other experiments (C to F) were repeated at least three times. The results presented as averages and STD by setting the values of miRNA expression vector control as “1”.
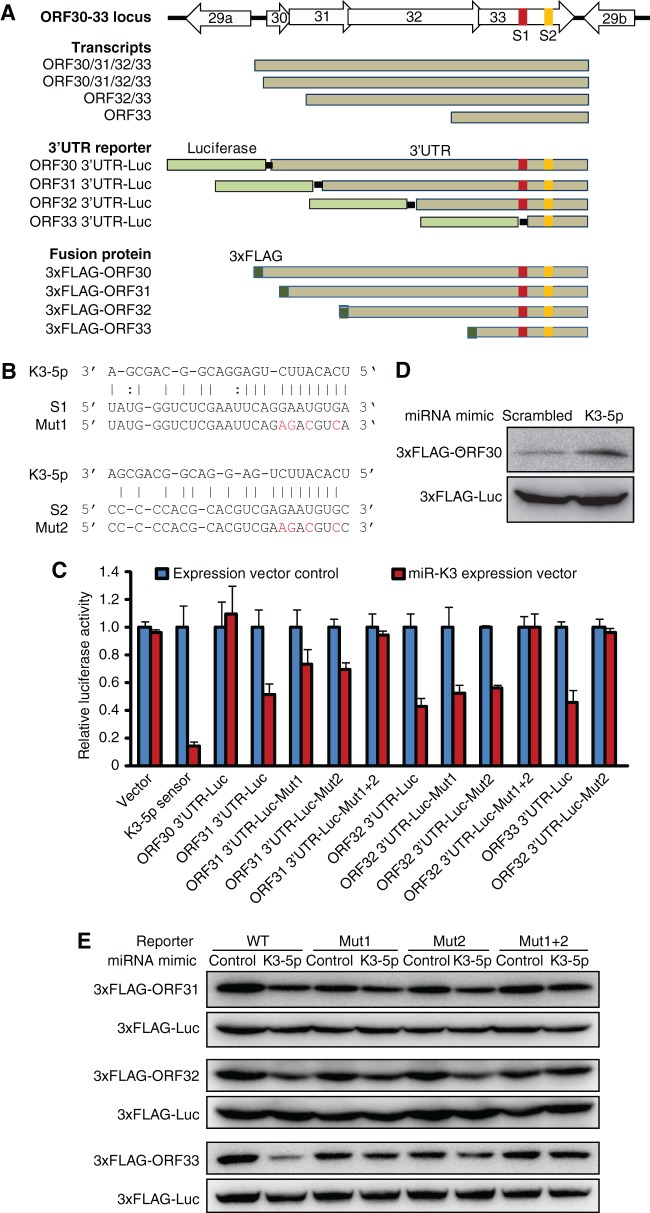
miR-K3-5p targets ORF31, ORF32, and ORF33 transcripts.
KSHV ORF30-33 locus, which encodes lytic genes ORF30, ORF31, ORF32, and ORF33, has not been extensively studied thus far. In MHV-68, these genes are involved in the late stage of viral replication (54–56). In particular, ORF33 is a tegument protein associated with virion maturation (57, 58). Transcription mapping of this gene locus identified four transcripts with the same poly(A) site (53). We further examined miR-K3 targeting of the transcripts from this locus. Bioinformatics analysis predicted two putative miR-K3-5p binding sites in the coding region of ORF33 (Fig. 5A). Interestingly, our initial screening only observed repression of 3′UTRs of ORF31, ORF32, and ORF33 by miR-K3 (Fig. 4A and C). To confirm that the two putative sites indeed mediated miR-K3 targeting of these 3′UTRs, we generated reporters with single and double mutations in these sites (Fig. 5B). Reexamination confirmed miR-K3 repression of the 3′UTRs of ORF31, ORF32, and ORF33 but not that of ORF30 (Fig. 5C). Mutation of site 1 or 2 alone only partially relieved miR-K3 repression of ORF31 and ORF32 3′UTR reporters (Fig. 5C). However, mutation of both sites completely relieved miR-K3 repression of these two 3′UTRs. The ORF33 3′UTR reporter contained only site 2 (Fig. 5A). Mutation of this site alone completely relieved miR-K3-5p repression of this 3′UTR (Fig. 5C). These results indicate that both predicted sites are functional. We further determined whether miR-K3 could inhibit the protein expression of ORF31, ORF32, and ORF33. Because there is no antibody available for these proteins, we generated 3×FLAG-tagged fusion protein expression plasmids under the control of their respective 3′UTRs with or without mutations of the binding sites (Fig. 5A). Note that both sites were present in the ORF33 fusion construct. Consistent with the 3′UTR reporter results, miR-K3-5p mimic did not repress the expression of ORF30 fusion protein (Fig. 5D). In fact, the expression of ORF30 fusion protein appeared to be slightly increased in the presence of miR-K3-5p. In contrast, miR-K3-5p mimic repressed the expression of ORF31, ORF32, and ORF33 fusion proteins (Fig. 5E). Mutation of a single site only partially relieved the repression effect of miR-K3-5p. However, mutation of both sites completely abolished the repression effect of miR-K3-5p on the expression of these proteins (Fig. 5E). Together, these results indicate that sites 1 and 2 mediate the miR-K3-5p repression of ORF31, ORF32, and ORF33 3′UTRs, resulting in the inhibition of their protein expression.
FIG 5.
miR-K3 targets 3′UTRs of ORF31, ORF32, and ORF33 through binding sites in the ORF33 coding region. (A) Schematic illustration of the ORF30-33 gene locus, their transcripts, 3′UTR luciferase reporters, and constructs expressing 3×FLAG fusion proteins. S1 and S2 are predicted as miR-K3-5p binding sites located in the coding region of ORF33. For the 3×FLAG fusion protein constructs, each ORF, including the downstream 3′UTR sequence, is fused in frame with the N terminus of the 3×FLAG tag. (B) Alignment of miR-K3-5p with predicted binding sites in ORF30-33 transcripts. Mutated nucleotides in the binding sites are shown in red type. Solid lines indicate the Watson-Crick base pairing, and dotted lines indicate wobble base pairing. (C) miR-K3 represses ORF31, ORF32, and ORF33 but not ORF30 3′UTRs by targeting two binding sites (S1 and S2) residing in the ORF33 coding region. miRNA expression plasmid or a miRNA expression control vector was cotransfected with the wild-type 3′UTR reporter, mutant 3′UTR reporter, or a reporter control vector into 293T cells, together with a pSV-β-galactosidase plasmid for 48 h, and the relative luciferase activities were examined and normalized for β-galactosidase activity. Experiments were repeated three times, and the results are presented as averages and STD by setting the values of miRNA expression vector control as “1”. (D and E) The mimic of miR-K3-5p does not repress the expression of fusion protein from the 3×FLAG-ORF30 construct (D). The mimic, but not the scrambled control mimic, of miR-K3-5p inhibits the expression of fusion proteins of ORF31, ORF32, and ORF33 by targeting S1 and S2 binding sites (E). Western blot analyses of 3×FLAG-ORF30 fusion protein (D) and of 3×FLAG-ORF31, -ORF32, and -ORF33 fusion proteins (E) in 293T cells were performed. The mimic of miR-K3-5p or a scrambled control mimic was cotransfected with each FLAG-tagged protein expression construct or its mutants with mutation in S1, S2, or both binding sites into 293T cells for 48 h, and the expression of the fusion protein was detected by Western blotting. A 3×FLAG-tagged firefly luciferase construct was also cotransfected as an internal control.
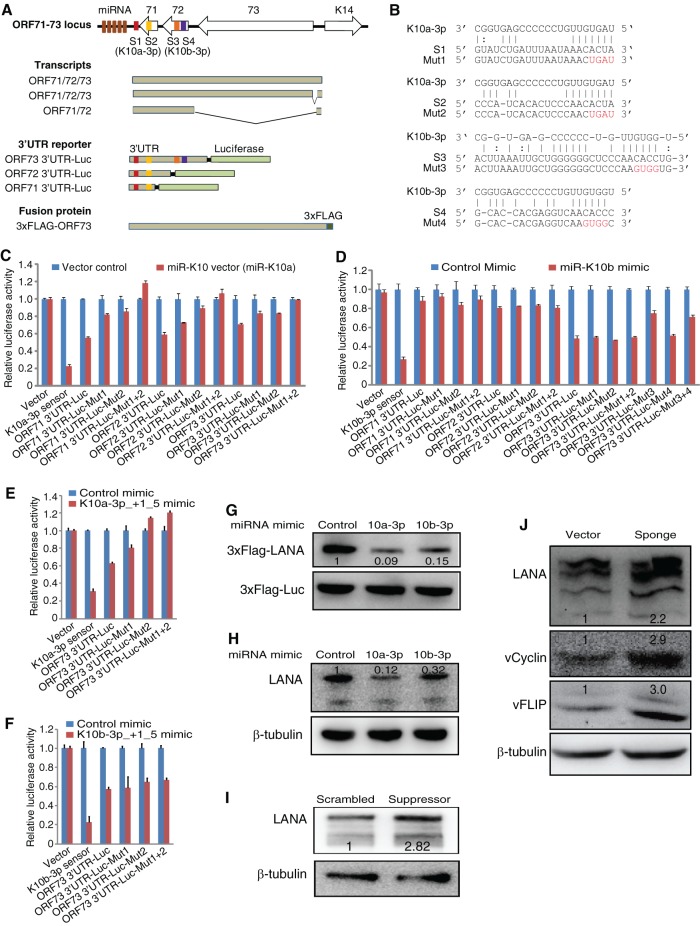
miR-K10a-3p and miR-K10b-3p targets ORF71, ORF72, and ORF73 transcripts.
KSHV pre-miR-K10 is derived from the coding region of ORF-K12 (59). Although there are several functional variants, the predominant mature miRNA in latent KSHV cells is miR-10a-3p, which can be switched to miR-10b-3p with a single point mutation during lytic replication through RNA editing (13, 32, 38, 60, 61). The ORF71-73 gene locus encodes three viral latent proteins, including vFLIP (ORF71), vCyclin (ORF72), and LANA (ORF73), that have diverse functions during viral latency. Consistent with the screening results, bioinformatics analysis predicted two putative miR-K10a-3p targeting sites (S1 and S2) located in the downstream sequence and coding sequence of ORF71, respectively, and two putative miR-K10b-3p targeting sites within the coding sequence of ORF72 (Fig. 6A and B). To determine whether sites S1 and S2 mediated miR-K10a-3p targeting of ORF71, ORF72, and ORF73 3′UTRs, we cotransfected the 3′UTR reporters with a pre-miR-K10 expression plasmid, which only expressed miR-K10a (38). miR-K10a repressed ORF71, ORF72, and ORF73 3′UTR reporters by 45, 42, and 30%, respectively (Fig. 6C). Mutation of either the S1 or the S2 site partially relieved miR-K10a-3p repression of all three reporters, while mutation of both S1 and S2 sites fully restored the reporter activities (Fig. 6C), indicating that both sites were functional.
FIG 6.
miR-K10 and variants target 3′UTRs of the ORF71-ORF73 cluster through distinct binding sites. (A) Schematic illustration of the ORF71-73 gene locus, their transcripts, 3′UTR luciferase reporters, and a 3×FLAG-ORF73 construct. S1 and S2 are predicted miR-10a-3p binding sites, while S3 and S4 are predicted miR-10b-3p binding sites. For the 3×FLAG-ORF73 construct, ORF73 and the downstream 3′UTR sequence is fused in frame with the N terminus of the 3×FLAG tag. (B) Alignment of miR-K10a-3p and miR-K10b-3p with the predicted binding sites. Mutated nucleotides in the binding sites are marked in red type. Solid lines indicate the Watson-Crick base paring, and dotted lines indicate wobble base pairing. (C) miR-K10a represses ORF71, ORF72, and ORF73 3′UTRs by targeting binding sites S1 and S2 residing in the downstream sequence and ORF71 coding region, respectively. miR-K10 expression plasmid, which expresses only miR-K10a or a miRNA expression vector control, was cotransfected with wild-type 3′UTR reporter, mutant 3′UTR reporter, or a reporter control vector into 293T cells, together with a pSV-β-galactosidase plasmid for 48 h, and the relative luciferase activities were examined and normalized for β-galactosidase activity. Experiments were repeated three times, and the results are presented as averages and STD by setting the values of miRNA expression vector control as “1”. (D) Repression of ORF71, ORF72, and ORF73 3′UTRs by miR-K10b-3p. miR-K10b-3p represses ORF73 3′UTR by targeting the S3 and possibly an unidentified site but not the S4 site. miR-K10b also weakly represses ORF71, ORF72, and ORF73 3′UTRs by targeting the unidentified binding site. Mimic of miR-K10b-3p or scrambled control mimic was transfected into 293T cells for 16 h, followed by cotransfection of the 3′UTR reporter or a reporter control vector with pSV-β-galactosidase plasmid for 48 h, and the relative luciferase activities were examined as described in panel C. (E and F) Repression of ORF73 3′UTR by miR-K10 variants. miR-K10a-3p variant miR-K10a-3p_+1_5 represses the ORF73 3′UTR by targeting both S1 and S2 sites (E), but these two sites do not mediate miR-K10b-3p variant miR-K10b-3p_+1_5 repression of the ORF73 3′UTR (F). Mimic of miR-K10a-3p_+1_5 or miR-K10b-3p_+1_5 or scrambled control mimic was transfected into 293T cells for 16 h, followed by cotransfection of the 3′UTR reporter or a reporter control vector with pSV-β-galactosidase plasmid for 48 h, and the relative luciferase activities were examined as described in panel C. (G) Mimic of miR-K10a-3p or miR-K10b-3p represses the expression of LANA from the 3×FLAG-ORF73 construct. Western blot analyses of 3×FLAG-ORF73 fusion protein in 293T cells. Mimic of miR-K10a-3p or miR-K10b-3p or scrambled control mimic was cotransfected with 3×FLAG-ORF73 expression construct into 293T cells for 48 h. A 3×FLAG-tagged firefly luciferase construct was also cotransfected as an internal control. (H) Mimic of miR-K10a-3p or miR-K10b-3p represses the expression of LANA in 293T-BAC36 cells. Mimic of miR-K10a-3p or miR-K10b-3p or scrambled control mimic was transfected into 293T-BAC36 cells for 48 h, and the expression of LANA was examined by Western blotting. β-Tubulin was used as a loading control. (I) LNA-based miR-K10-3p suppressor upregulates the endogenous protein expression of ORF73 in BCP-1 cells. BCP-1 cells transfected with LNA suppressor or scrambled control suppressor for 48 h were examined for the expression of LANA by Western blotting. β-Tubulin was used as a loading control. (J) Specific miR-K10a-3p and miR-K10b-3p sponges increase the expression of LANA, vCyclin, and vFLIP proteins in BC-3 cells. BC-3 cells transduced with lentiviral viruses expressing a miR-K10-3p sponge or a vector control for 48 h were examined for the expression of LANA, vCyclin, and vFLIP proteins by Western blotting. β-Tubulin was used as a loading control.
To determine whether sites S3 and S4 mediated miR-K10b-3p targeting of ORF71, ORF72, and ORF73 3′UTRs, we cotransfected the 3′UTR reporters with a miR-K10b mimic (Fig. 6D). miR-K10b repressed ORF73 3′UTR reporter by 52% (Fig. 6D). As expected, mutation of S1, S2, or both had no effect on miR-K10b repression of the reporter. Mutation of site 3 relieved miR-K10b's repression effect to 24%, while mutation of site 4 had no effect, indicating that site 3 but not site 4 was functional, and there was potentially another unidentified unconventional miR-K10b site(s). Although sites S3 and S4 were absent in both ORF71 and ORF72 3′UTR reporters, miR-K10b repressed their activities by 13 and 20%, respectively, indicating that the unidentified site could be resided in the 3′UTR of ORF73. As expected, mutation of both S1 and S2 sites alone or together did not relieve the repression effect of miR-K10b. These results were consistent with the results of ORF73 3′UTR reporter (Fig. 6D), further indicating that there could be likely an unidentified miR-K10b site(s) in these 3′UTRs. Nevertheless, it is possible that the observed weak repression is due to other unknown indirect effect.
Recent studies indicate that both miR-K10a-3p and miR-10b-3p have minor variants with a single extended nucleotide at the 5′ end, termed miR-K10a_+1_5 and miR-K10b_+1_5, respectively (32, 60). Moreover, human miR-142-3p_-1_5 shares the same 7mer seed region with that of miR-K10a-3p_+1_5 (32, 38, 60). Both miR-K10a-3p_+1_5 and miR-10b-3p_+1_5 mimics repressed the expression of ORF73 3′UTR reporter (Fig. 6E and F). Mutation of S1 partially relieved the repression effect of miR-K10a-3p_+1_5 on the reporter, while repression of S2 or both sites completely relieved the repression effect (Fig. 6E) indicating that S2 played a dominant role in mediating miR-K10a-3p_+1_5 targeting of the ORF73 3′UTR. Nevertheless, both S1 and S2 appeared to be functional sites for miR-K10a-3p_+1_5. In contrast, mutation of S1, S2, or both did not relieve any repression effect of miR-K10b-3p_+1_5 (Fig. 6F), indicating that, similar to miR-K10b-3p, S1 and S2 did not mediate miR-K10b-3p_+1_5 targeting of the ORF73 3′UTR. To our surprise, miR-142-3p_-1_5 had no effect on ORF73 3′UTR reporter (data not shown). Together, these results indicated that miR-K10a-3p and miR-K10a-3p_+1_5 targeted the 3′UTRs of ORF71, ORF72, and ORF73 through both sites S1 and S2. miR-K10b-3p and miR-K10b-3p_+1_5 targeted the 3′UTR of ORF73 primarily through site S3 and a putative identified site resided in the downstream and coding regions of ORF71. miR-K10b-3p and miR-K10b-3p_+1_5 targeted ORF71 and ORF72 3′UTRs through this unidentified site, but the effect was minimal.
To confirm the results of 3′UTR reporters, we examined the effects of miR-K10a-3p and miR-K10b-3p on the expression of LANA, vCyclin, and vFLIP proteins. Cotransfection of a 3×FLAG-tagged ORF73 expression plasmid and a 3×FLAG-tagged luciferase expression plasmid as a transfection control, together with miR-K10a-3p and miR-K10b-3p mimics, into 293T cells repressed the LANA protein level by 91 and 85%, respectively (Fig. 6G), whereas transfection of miR-K10a-3p and miR-K10b-3p mimics into KSHV-infected cells 293T-BAC36 repressed LANA protein levels by 88 and 68%, respectively (Fig. 6H). Consistent with these results, inhibition of miR-K10a-3p and miR-K10b-3p in BCP-1 cells with a locked nucleic acid (LNA)-based suppressor increased LANA protein level by 2.82-fold (Fig. 6I). Furthermore, inhibition of miR-K10a-3p and miR-K10b-3p with an miRNA sponge in BC-3 cells increased the protein levels of LANA, vCyclin, and vFLIP by 2.2-, 2.9-, and 3.0-fold, respectively (Fig. 6J). Together, these results indicated that both miR-K10a-3p and miR-K10b-3p indeed targeted the 3′UTRs of ORF71, ORF72, and ORF73 to repress the expression of these proteins.
DISCUSSION
It is estimated that each miRNA can have as many as several hundreds of cellular targets (10, 42). Therefore, it is not surprising that the complete functions of most miRNAs, including those that have been heavily scrutinized, remain to be revealed. Similar to most cellular miRNAs, the full functions of KSHV miRNAs remain to be determined, albeit a number of cellular and viral targets have been identified in the last few years (16–39). By using RNA-cross-linking immunoprecipitation approaches, several groups have recently identified a large number of potential targets of KSHV miRNAs (23, 32, 40), providing a roadmap for their further functional delineations. Nevertheless, among the putative targets identified by these genomic approaches, few are of viral origins. To dissect the roles of KSHV miRNAs in regulating the expression of viral genes, we sought to identify the viral targets of these miRNAs. Because miRNAs primarily target the 3′UTRs of gene transcripts, we postulated that a functional approach by screening a 3′UTR library of KSHV genes could lead to the identification of viral targets of KSHV miRNAs. This approach has recently been used to identify targets of miR-122 by screening a library of 3′UTR reporters of human genes (62). Of 142 predicted target genes, as many as 57 (40%) were significantly inhibited by miR-122 mimics, validating the effectiveness of this functional approach for the global identification of miRNA targets.
We carried out a genome-wide mapping for the 3′UTRs of KSHV genes by using 3′RACE and cloned 83 3′UTR reporters for 74 KSHV genes. Of the 3′UTRs of 74 viral genes, 43 are newly mapped. Further analyses of these 3′UTRs have revealed a distinct feature of KSHV genome, which is the presence of a large number of viral genes that are transcribed as bicistronic and polycistronic transcripts. Of 83 transcripts identified, as many as 34 (41%) of them are from viral genes with bicistronic or polycistronic transcripts. Although bicistronic and polycistronic transcripts have previously been identified in several KSHV loci (5), our results indicate that these transcripts are widely spread across the entire viral genome.
In mammalian cells, it is usually the 5′-proximal ORFs that are translated into proteins by cap-dependent mechanism in bicistronic and polycistronic transcripts (1, 2). Although the 5′-distal ORFs have the potentials for encoding proteins, alternative mechanism other than the cap-dependent initiation for protein translation is required (1, 2). Surprisingly, such an alternative mechanism has thus far only been identified for two of these 5′-distal KSHV ORFs (6, 7). ORF71 positioning as a 5′-distal ORF in either the ORF71-72 transcript or the ORF71-73 transcript is translated through initiation at an internal ribosomal entry site residing in ORF72 (6). ORF36, which is only transcribed as a 5′-distal ORF in the ORF35-37 transcript, is translated through a termination-reinitiation mechanism (7). It is unclear whether any other 5′-distal ORFs in KSHV bicistronic and polycistronic transcripts is also translated into proteins. Nevertheless, it is clear that the presence of 5′-distal ORF(s) in these bicistronic and polycistronic transcripts results in significant increase of the 3′UTR lengths of their 5′-proximal ORFs (Fig. 3E). These extended 3′UTRs could serve as regulatory targets of other cellular and viral factors, such as RNA-binding proteins and miRNAs. Indeed, results from the screening of 3′UTR reporters obtained here have revealed a number of 5′-proximal ORFs that are regulated by KSHV miRNAs through their 3′UTRs containing the 5′-distal ORFs, including ORF25 (miR-K8), ORF31 (miR-K3), ORF32 (miR-K3), ORF43 (miR-K4), ORF59 (miR-K11), ORF72 (miR-K10), and ORF73 (miR-K10) (Fig. 4). Mutagenesis analyses have confirmed the regulation of ORF31, ORF32, ORF72, and ORF73 by the respective miRNAs (Fig. 5 and 6). A recent study has also identified target sites of miR-K5 and miR-K6-3p in the ORF57 coding region, which is part of the ORF56 3′UTR of a bicistronic transcript (41). Nevertheless, to what extent that KSHV miRNAs regulate viral gene expression through the extended 3′UTR regions remains to be determined.
Among the KSHV genes with potential transcripts identified here, the transcript of ORF62, which encodes a capsid protein, has previously been shown to be coterminal with that of ORF58, ORF59, ORF60, and ORF61 (48). However, it has been mapped to the coding region of the downstream ORF61 with a typical “AAUAAA” PAS signal sequence in the present study. PCR analysis with RT primers has confirmed the presence of the poly(A) sequence (data not shown). It is possible that ORF62 has two transcripts, with one presenting as a shorter and minor transcript that has escaped detection by Northern blot analysis thus far, while the second one presents as the longer and major transcript previously identified by Northern blot analysis but was missed by 3′RACE here because of the preferential amplification of shorter products in PCR. Similar to ORF62, the poly(A) site of the transcript of another KSHV gene ORF6 is also mapped to the coding region of ORF7 (Fig. 2 and see Fig. S1 in the supplemental material).
ORF-K1 is the first gene located in the KSHV genome. The intergenic region between ORF-K1 and the adjacent ORF4 is 168 nucleotides. ORF-K1 3′RACE analysis did not identify any poly(A) site in the intergenic region but did reveal two 3′UTRs coterminal with that of the ORF4 transcript (see Fig. S2 in the supplemental material). One of the 3′UTRs spans the intergenic region and the entire ORF4 coding region, while the second one is a spliced 3′UTR derived from sequence immediately after the stop codon to the poly(A) tail, which has been previously described (63, 64). Thus, ORF-K1 has long and short 3′UTRs, which might subject them to differential regulation by miRNAs or RNA-binding proteins in different stages of the KSHV life cycle.
By screening 3′UTR reporters of KSHV genes, we have identified 28 new miRNA-target pairs, which cover 22 KSHV genes from all four gene classes, including ORF-K1, ORF4, ORF10, ORF-K2, ORF2, ORF-K4, ORF-K7, ORF25, ORF31, ORF32, ORF33, ORF34, ORF43, ORF44, ORF45, ORF57, ORF-K9, ORF59, ORF65, ORF71, ORF72, and ORF73 (Fig. 4A). Previous studies have already identified several KSHV genes that are targeted by KSHV miRNAs (19, 28, 34, 40, 41). ORF50, which has been identified as the target of miR-K9* and miR-K7-5p (19, 34), has also been confirmed in our experiments (Fig. 4A). As indicated above, ORF56 is targeted by both miR-K5 and miR-K6-3p through binding in the 3′UTR of a bicistronic transcript (41). The same study has also shown that ORF57 is targeted by miR-K5 in the protein-coding region of an ORF57 monocistronic transcript. In our screening, we failed to identify ORF56 as the target of miR-K5 and miR-K6-3p, and ORF57 as the target of miR-K5. Examination of our results indicated that both miRNAs had low repression efficiencies for their respective “sensor reporters,” suggesting poor expression of these miRNAs in the screening assays. Similarly, we have noticed low repression efficiencies (<50%) of “sensor reporters” by miR-K2, -K7, -K9, and -K12, suggesting that some targets of these miRNAs could be missed in our screening. A number of the newly identified targets were only weakly repressed by the respective miRNAs (<10%). Although it is possible that this could also be due to the poor expression of these miRNAs, a recent study using a comprehensive proteomics approach has shown that the repression effects of miRNAs on most of their targets are modest (65). Thus, it might be worthwhile to further confirm the biological significance of such modest repression.
Most the 3′UTR reporters used here contained portions of the ORFs. Our results showed that they did not interfere with the screening of miRNA target sites in the 3′UTRs. For any identified putative sites, further mutagenesis analysis can confirm whether they are in the 3′UTR or ORF sequences, as illustrated for ORF71-73 and ORF31-33 3′UTR reporters (Fig. 5 and 6). Because some ORFs can be targeted by miRNAs (41), any targets identified in our screening can also be resided in the ORF sequences. However, our reporters lack a full coverage of the ORFs. Thus, our screening for target sites in the ORFs was far from exhaustive. Although it was not the primary goal of the study, any sites identified in the ORFs could also be valuable for understanding the functions of KSHV miRNAs once they were validated and their functions further defined.
Four lytic transcripts are transcribed from the ORF30-33 locus in the KSHV genome (53). Both ORF30 and ORF31 are present in two polycistronic transcripts containing all four ORFs. However, only ORF30 is a 5′-proximal ORF, which can be translated by cap-dependent mechanism. Our previous study failed to identify any functional internal ribosomal entry site that can be used for the translational initiation of ORF31 (53). Thus, it is unclear whether ORF31 is translated in KSHV-infected cells. Interestingly, all of the 3′UTR reporters from the ORF30-33 locus were repressed by miR-K3-5p except the ORF30 construct in the 3′UTR reporter assays, despite the translation of a luciferase reporter gene designed to function in cap-dependent fashion for all of the constructs. Sequencing of the cDNA revealed that the full transcript of the ORF30 3′UTR reporter was expressed, indicating that the lack of repression was not due to truncation of the targeting sites in the transcript (data not shown). Because both ORF30 and ORF31 reporters share most of the 3′UTR regions, including those that are targeted by miR-K3-5p, it remains to be determined whether the additional ORF31 sequence in the ORF30 3′UTR reporter prevents miR-K3-5p repression of the reporter.
Recent RNA cross-linking immunoprecipitation analyses have revealed a limited number of enriched KSHV gene transcripts (23, 32, 40). Among them ORF-K2 (vIL6) has been confirmed as a miR-K10a-3p target (40), a finding which is consistent with our screening results. Transcripts from the ORF71-73 latent locus were also enriched in the RNA cross-linking immunoprecipitation analyses (32, 40). Indeed, we demonstrated that the 3′UTRs of these transcripts were robustly repressed by miR-K10 in our 3′UTR screening. One reason RNA cross-linking immunoprecipitation studies have identified few viral targets is the use of latent uninduced KSHV-infected cells. It would be interesting to carry out similar analyses with lytically induced KSHV-infected cells, which are likely to reveal additional viral transcripts. Cross-validation of the results from the RNA cross-linking immunoprecipitation and 3′UTR screening approaches would provide insightful information into how KSHV miRNAs might regulate the viral gene expression program.
Our mutagenesis analyses have revealed that miR-K10a-3p and miR-K10b-3p, as well as their variants, directly target ORF71-73 transcripts through distinct sites (Fig. 6). Extensive studies have shown that the products encoded by these transcripts vFLIP, vCyclin, and LANA regulate KSHV latency and promote cell growth and survival (8). Nonetheless, excessive expression of these genes could also have detrimental effects on the cells. It has been shown that overexpression of vFLIP or vCyclin retards cell proliferation (66). Thus, maintaining the appropriate expression levels of KSHV latent proteins is likely essential for the survival of latent KSHV-infected cells. In this case, viral miRNAs might play a role in fine-tuning the expression of KSHV latent genes. A similar mechanism has also been described for Epstein-Barr virus (EBV) latent gene LMP1, which is regulated by EBV-encoded BART miRNAs (67). Furthermore, our previous study showed that miR-K10a-3p and miR-K10b-3p and their variants inhibit the transforming growth factor β (TGF-β) pathway by targeting TGF-β type II receptor to promote cell survival (38). Taken together, these viral miRNAs are likely to have a critical role in KSHV latency by maintaining the cell homeostasis and promoting the cell survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the S.-J.G. laboratory for technical assistance and helpful discussion.
This study was supported by grants from the National Institutes of Health (NIH; CA096512, CA124332, CA132637, and CA177377) to S.-J.G., from the NIH (CA180779, CA082057, CA31363, CA115284, CA180779, AI105809, and AI073099), the Hastings Foundation, and the Fletcher Jones Foundation to J.U.J., and from the National Natural Science Foundation of China (81171552, 81361120387, and 31270199) to C.L.
ADDENDUM IN PROOF
While the present study was under review, a study describing the mapping of KSHV 3′UTRs was published. Most of the 3′UTRs mapped in these two studies are consistent [68].
Footnotes
Published ahead of print 23 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02689-13.
REFERENCES
- 1.Pi H, Lee LW, Lo SJ. 2009. New insights into polycistronic transcripts in eukaryotes. Chang Gung Med. J. 32:494–498 [PubMed] [Google Scholar]
- 2.Tautz D. 2009. Polycistronic peptide coding genes in eukaryotes—how widespread are they? Brief Funct. Genomic Proteomic 8:68–74. 10.1093/bfgp/eln054 [DOI] [PubMed] [Google Scholar]
- 3.Ryabova LA, Pooggin MM, Hohn T. 2002. Viral strategies of translation initiation: ribosomal shunt and reinitiation. Prog. Nucleic Acids Res. Mol. Biol. 72:1–39. 10.1016/S0079-6603(02)72066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye F, Lei X, Gao SJ. 2011. Mechanisms of Kaposi's sarcoma-associated herpesvirus latency and reactivation. Adv. Virol. 2011:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng ZM. 2010. Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses. Int. J. Biol. Sci. 6:730–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundhoff A, Ganem D. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857–1863. 10.1128/JVI.75.4.1857-1863.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. 2013. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog. 9:e1003156. 10.1371/journal.ppat.1003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao SJ. 2007. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 133:69–127. 10.1007/978-0-387-46816-7_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. 2001. MicroRNAs: tiny regulators with great potential. Cell 107:823–826. 10.1016/S0092-8674(01)00616-X [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 12.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570–5575. 10.1073/pnas.0408192102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276. 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- 14.Samols MA, Hu J, Skalsky RL, Renne R. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301–9305. 10.1128/JVI.79.14.9301-9305.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundhoff A, Sullivan CS, Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gammaherpesviruses. RNA 12:733–750. 10.1261/rna.2326106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099. 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. 10.1371/journal.ppat.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845. 10.1128/JVI.01804-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellare P, Ganem D. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570–575. 10.1016/j.chom.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376–385. 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 21.Ziegelbauer JM, Sullivan CS, Ganem D. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41:130–134. 10.1038/ng.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abend JR, Uldrick T, Ziegelbauer JM. 2010. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol. 84:12139–12151. 10.1128/JVI.00884-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M, Lieber D, Bailer SM, Hoffmann R, Ruzsics Z, Kremmer E, Pfeffer S, Zimmer R, Koszinowski UH, Grasser F, Meister G, Haas J. 2010. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gammaherpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7:324–334. 10.1016/j.chom.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 24.Gottwein E, Cullen BR. 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 84:5229–5237. 10.1128/JVI.00202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, Kellam P, Boshoff C. 2010. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24:195–205. 10.1101/gad.553410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. 2010. Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat. Cell Biol. 12:193–199. 10.1038/ncb2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu CC, Li Z, Chu CY, Feng J, Sun R, Rana TM. 2010. MicroRNAs encoded by Kaposi's sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 11:784–790. 10.1038/embor.2010.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697–2706. 10.1128/JVI.01997-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin Z, Freitas E, Sullivan R, Mohan S, Bacelieri R, Branch D, Romano M, Kearney P, Oates J, Plaisance K, Renne R, Kaleeba J, Parsons C. 2010. Upregulation of xCT by KSHV-encoded microRNAs facilitates KSHV dissemination and persistence in an environment of oxidative stress. PLoS Pathog. 6:e1000742. 10.1371/journal.ppat.1000742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin Z, Kearney P, Plaisance K, Parsons CH. 2010. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J. Leukoc. Biol. 87:25–34. 10.1189/jlb.0409251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R. 2011. A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rγ-null mice. J. Virol. 85:9877–9886. 10.1128/JVI.05558-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526. 10.1016/j.chom.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang D, Gao Y, Lin X, He Z, Zhao Q, Deng Q, Lan K. 2011. A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKε. Cell Res. 21:793–806. 10.1038/cr.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K. 2011. miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA. PLoS One 6:e16224. 10.1371/journal.pone.0016224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suffert G, Malterer G, Hausser J, Viiliainen J, Fender A, Contrant M, Ivacevic T, Benes V, Gros F, Voinnet O, Zavolan M, Ojala PM, Haas JG, Pfeffer S. 2011. Kaposi's sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 7:e1002405. 10.1371/journal.ppat.1002405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. 2012. Kaposi's sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the Toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J. Virol. 86:11663–11674. 10.1128/JVI.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlke C, Maul K, Christalla T, Walz N, Schult P, Stocking C, Grundhoff A. 2012. A microRNA encoded by Kaposi's sarcoma-associated herpesvirus promotes B-cell expansion in vivo. PLoS One 7:e49435. 10.1371/journal.pone.0049435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei X, Zhu Y, Jones T, Bai Z, Huang Y, Gao SJ. 2012. A Kaposi's sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor beta pathway to promote cell survival. J. Virol. 86:11698–11711. 10.1128/JVI.06855-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Sun R, Lin X, Liang D, Deng Q, Lan K. 2012. Kaposi's sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5. J. Virol. 86:1372–1381. 10.1128/JVI.06245-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. 2012. Ago HITS-CLIP expands understanding of Kaposi's sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 8:e1002884. 10.1371/journal.ppat.1002884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin HR, Ganem D. 2011. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc. Natl. Acad. Sci. U. S. A. 108:5148–5153. 10.1073/pnas.1102033108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196. 10.1128/JVI.76.12.6185-6196.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Ye F, Xie J, Kuhne K, Gao SJ. 2009. Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290–302. 10.1016/j.virol.2009.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottwein E, Cai X, Cullen BR. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 80:5321–5326. 10.1128/JVI.02734-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev. 25:1770–1782. 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham C, Barnard S, Blackbourn DJ, Davison AJ. 2003. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J. Gen. Virol. 84:1471–1483. 10.1099/vir.0.19015-0 [DOI] [PubMed] [Google Scholar]
- 48.Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 80:11968–11981. 10.1128/JVI.01394-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 29:1690–1694. 10.1093/nar/29.8.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haque M, Wang V, Davis DA, Zheng ZM, Yarchoan R. 2006. Genetic organization and hypoxic activation of the Kaposi's sarcoma-associated herpesvirus ORF34-37 gene cluster. J. Virol. 80:7037–7051. 10.1128/JVI.00553-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai Z, Zhou F, Lei X, Ma X, Lu C, Gao SJ. 2012. A cluster of transcripts encoded by KSHV ORF30-33 gene locus. Virus Genes 44:225–236. 10.1007/s11262-011-0698-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol. 83:10582–10595. 10.1128/JVI.00497-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J. Virol. 78:6610–6620. 10.1128/JVI.78.12.6610-6620.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu TT, Park T, Kim H, Tran T, Tong L, Martinez-Guzman D, Reyes N, Deng H, Sun R. 2009. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J. Virol. 83:2265–2273. 10.1128/JVI.01785-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952–4964. 10.1128/JVI.79.8.4952-4964.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800–811. 10.1128/JVI.79.2.800-811.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894. 10.1101/gad.17352611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695–703. 10.1128/JVI.02013-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gandy SZ, Linnstaedt SD, Muralidhar S, Cashman KA, Rosenthal LJ, Casey JL. 2007. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 81:13544–13551. 10.1128/JVI.01521-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collins P, Rose M, Aldred SF, Trinklein N. 2009. High-throughput microRNA target screening: miR-122 case study. Biotechniques 47:883 [Google Scholar]
- 63.Spiller OB, Robinson M, O'Donnell E, Milligan S, Morgan BP, Davison AJ, Blackbourn DJ. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 77:592–599. 10.1128/JVI.77.1.592-599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandriani S, Ganem D. 2010. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 84:5565–5573. 10.1128/JVI.02723-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455:64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang YF, Wang LY, Li YL, Shyu HW, Chiou YH, Chou MC, Lin KH, Tai MH, Chen CY. 2010. Human herpesvirus 8 viral FLICE-inhibitory protein retards cell proliferation via downregulation of Id2 and Id3 expression. Mol. Cell. Biochem. 343:83–89. 10.1007/s11010-010-0501-y [DOI] [PubMed] [Google Scholar]
- 67.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. U. S. A. 104:16164–16169. 10.1073/pnas.0702896104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McClure LV, Kincaid RP, Burke JM, Grundhoff A, Sullivan CS. 2013. Comprehensive mapping and analysis of KSHV 3′ UTRs identifies differential post-transcriptional control of gene expression in lytic versus latent infection. J. Virol. 87:12838–12849. 10.1128/JVI.02374-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L, Park MS. 2009. Identification and characterization of the promoter region of Kaposi's sarcoma-associated herpesvirus ORF11. Virus Res. 142:160–168. 10.1016/j.virusres.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 70.Deng H, Song MJ, Chu JT, Sun R. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252–8264. 10.1128/JVI.76.16.8252-8264.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rimessi P, Bonaccorsi A, Sturzl M, Fabris M, Brocca-Cofano E, Caputo A, Melucci-Vigo G, Falchi M, Cafaro A, Cassai E, Ensoli B, Monini P. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161–7174. 10.1128/JVI.75.15.7161-7174.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicholas J, Ruvolo VR, Burns WH, Sandford G, Wan X, Ciufo D, Hendrickson SB, Guo HG, Hayward GS, Reitz MS. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287–292. 10.1038/nm0397-287 [DOI] [PubMed] [Google Scholar]
- 73.Taylor JL, Bennett HN, Snyder BA, Moore PS, Chang Y. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol. 79:15099–15106. 10.1128/JVI.79.24.15099-15106.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867–2875. 10.1128/JVI.74.6.2867-2875.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saveliev A, Zhu F, Yuan Y. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301–314. 10.1006/viro.2002.1561 [DOI] [PubMed] [Google Scholar]
- 76.Masa SR, Lando R, Sarid R. 2008. Transcriptional regulation of the open reading frame 35 encoded by Kaposi's sarcoma-associated herpesvirus. Virology 371:14–31. 10.1016/j.virol.2007.08.023 [DOI] [PubMed] [Google Scholar]
- 77.Gonzalez CM, Wong EL, Bowser BS, Hong GK, Kenney S, Damania B. 2006. Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 80:3062–3070. 10.1128/JVI.80.6.3062-3070.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santarelli R, Farina A, Granato M, Gonnella R, Raffa S, Leone L, Bei R, Modesti A, Frati L, Torrisi MR, Faggioni A. 2008. Identification and characterization of the product encoded by ORF69 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4562–4572. 10.1128/JVI.02400-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pearce M, Matsumura S, Wilson AC. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 79:14457–14464. 10.1128/JVI.79.22.14457-14464.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nador RG, Milligan LL, Flore O, Wang X, Arvanitakis L, Knowles DM, Cesarman E. 2001. Expression of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor monocistronic and bicistronic transcripts in primary effusion lymphomas. Virology 287:62–70. 10.1006/viro.2001.1016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.