Abstract
Measles virus (MV) lacking expression of C protein (CKO) is a potent activator of the double-stranded RNA (dsRNA)-dependent protein kinase (PKR), whereas the isogenic parental virus expressing C protein is not. Here, we demonstrate that significant amounts of dsRNA accumulate during CKO mutant infection but not following parental virus infection. dsRNA accumulated during late stages of infection and localized with virus replication sites containing N and P proteins. PKR autophosphorylation and stress granule formation correlated with the timing of dsRNA appearance. Phospho-PKR localized to dsRNA-containing structures as revealed by immunofluorescence. Production of dsRNA was sensitive to cycloheximide but resistant to actinomycin D, suggesting that dsRNA is a viral product. Quantitative PCR (qPCR) analyses revealed reduced viral RNA synthesis and a steepened transcription gradient in CKO virus-infected cells compared to those in parental virus-infected cells. The observed alterations were further reflected in lower viral protein expression levels and reduced CKO virus infectious yield. RNA deep sequencing confirmed the viral RNA expression profile differences seen by qPCR between CKO mutant and parental viruses. After one subsequent passage of the CKO virus, defective interfering RNA (DI-RNA) with a duplex structure was obtained that was not seen with the parental virus. We conclude that in the absence of C protein, the amount of PKR activator RNA, including DI-RNA, is increased, thereby triggering innate immune responses leading to impaired MV growth.
INTRODUCTION
Measles virus (MV), a member of the Paramyxoviridae family, is an important human pathogen and a model virus of the Morbillivirus genus. Belonging to the order Mononegavirales, MV possesses a nonsegmented, single-stranded RNA genome with negative polarity (−ssRNA) of 15,894 nucleotides (nt) that carries six individual genes (1, 2). These genes encode nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin (H), and the viral RNP-dependent RNA polymerase (or large protein [L]), in this order. The P gene also encodes two additional proteins, the nonstructural V and C proteins (3, 4), which play important roles in controlling both the induction of interferon (5–9) and interferon signaling (10–14), two major arms of antiviral innate immunity (15, 16).
We previously established that a recombinant version of the MV Moraten vaccine strain unable to express the C protein [MVvac-CKO(GFP) (where GFP is green fluorescent protein), also designated CKO virus] induces double-stranded RNA (dsRNA)-dependent innate immune responses via the pattern recognition receptor retinoic acid-inducible gene I (RIG-I) and protein kinase R (PKR) (17–19). RIG-I activation leads to the induction of interferon β (IFN-β) expression via activation of interferon regulatory factor 3 (IRF3) (20, 21). PKR activation triggers a cellular stress response, which includes eukaryotic translation initiation factor 2α (eIF2α)-mediated translational arrest and formation of stress granules (SGs) (22). In contrast to the CKO virus, isogenic parental virus [MVvac-WT(GFP), also designated wild-type (WT) virus] impairs the induction of the innate immune response (17–19, 21), suggesting that one function of the C protein is to control dsRNA production. Earlier studies described the MV C protein as a virulence factor (23) that might have a regulatory effect on the viral RNA-dependent RNA polymerase, modulating its engagement in transcription or replication (24, 25). In addition, the C protein is able to prevent induction of IFN-β by a mechanism that involves its nuclear localization (9). However, in the case of PKR activation, direct inhibitory effects of C protein on the signaling pathways were excluded (17, 26), leading to speculation that C protein might prevent the accumulation of RNA that otherwise would act as a trigger for PKR activation.
Negative-strand viruses, such as MV or influenza A virus, produce little if any dsRNA during their infectious cycle (27), in contrast to dsRNA viruses or even positive-strand ssRNA (+ssRNA) viruses or dsDNA viruses. However, recent studies suggested that Sendai virus (SENV) and parainfluenza virus type 1 mutants unable to express C protein produce significant amounts of dsRNA that trigger MDA5- and PKR-mediated innate immune responses (26, 28). Similarly, we found enhanced activation of the dsRNA-dependent PKR and phosphorylation of IRF3 in CKO mutant virus-infected cells (17, 21). The source of the putative activator dsRNA in paramyxovirus-infected cells remains to be elucidated. Possibilities include RNA of viral origin, such as a dsRNA product of aberrant viral replication, or a structured ssRNA product whose spatial localization is altered in CKO virus-infected cells. Alternatively, the RNA could be a cellular product that is upregulated during viral infection, which acts as a trigger for pathogen recognition receptors (PRRs).
Here, we show that formation of dsRNA is the trigger for PKR activation during infection with recombinant CKO mutant virus and that the RNA responsible for this event is most likely of viral origin, as measured by intracellular localization, kinetics of accumulation, and drug sensitivity tests. In addition, we found activated phospho-PKR (pPKR) accumulation at sites of dsRNA accumulation. Quantification of viral protein and RNA expression revealed reduced replication of the CKO mutant compared to that of the parental virus as well as a steeper transcription gradient. We identify and characterize copyback defective RNAs amplified during CKO virus infection but not during infection with parental virus.
MATERIALS AND METHODS
Cells and viruses.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Life Technologies) with 5% (vol/vol) fetal bovine serum (FBS; HyClone; Thermo Scientific), penicillin (100 μg/ml; Gibco), and streptomycin (100 U/ml; Gibco). Generation of HeLa cells with stable knockdown for PKR (PKRkd), ADAR1 (ADAR1kd), or a nonspecific control (CONkd) has been described previously (29, 30). Cells were maintained under selection in the presence of puromycin (1 μg/ml; Sigma-Aldrich), but during experiments, puromycin was omitted. Virus stock production and titer determination were performed with low-passage-number inoculum at a multiplicity of infection (MOI) of 0.01 on Vero cells as previously described (31). Recombinant MV expressing GFP from an additional transcription unit downstream of the H gene has been described previously (31). Parental virus MVvac-WT(GFP) and CKO mutant virus MVvac-CKO(GFP) are derived from the Moraten vaccine strain of MV.
Virus infections.
Cells were seeded into 12-well plates (1 × 105 cells/well) 1 day before infection. For immunofluorescence analyses, cells were seeded on glass coverslips at a lower density (approximately 5 × 104 cells/well). Virus infections were carried out with an MOI of 0.1 unless otherwise stated. Virus was diluted with Opti-MEM to obtain a final inoculum volume of 300 μl/well. Cells were incubated with virus for 2 h at 37°C with gentle rocking every 15 min. Virus-containing Opti-MEM was then replaced with DMEM containing 5% FBS, and the cells were incubated at 37°C until they were harvested or fixed.
Antibodies.
We used rabbit antisera directed against MV-N(505), MV-P(254), MV-V(C-terminal), MV-C(2), MV-M(81), MV-F(cyt), and MV-H(cyt) as described previously (17, 31, 32). Rabbit polyclonal antibodies were used to detect GFP (Molecular Probes, Life Technologies), PKR (clone D7F7; Cell Signaling), and G3BP (Sigma-Aldrich). A rabbit monoclonal antibody was used to detect phospho-PKR(T446) (Epitomics). Mouse monoclonal antibodies were used to detect tubulin (Sigma-Aldrich) and dsRNA (clone J2; English & Scientific Consulting). A chicken polyclonal antiserum was used to detect GFP by immunofluorescence (Molecular Probes). Secondary antibodies for Western blot detection were anti-rabbit IRDye800 and anti-mouse IRDye680 (both from LI-COR). Secondary antibodies for immunofluorescence were anti-rabbit Alexa Fluor 350, anti-mouse Alexa Fluor 594, and anti-chicken Alexa Fluor 488 (all from Molecular Probes).
Immunoblot analysis.
Protein lysates were prepared as described previously (17) and stored at −80°C. Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad) and calculated on the basis of a bovine serum albumin (BSA) standard. Denaturing SDS-PAGE and Western blotting were performed with 25 μg total protein/lane as described previously (17). Membranes were blocked with 5% (wt/vol) nonfat dry milk in phosphate-buffered saline (PBS) or with 5% (wt/vol) BSA in Tris-buffered saline (TBS) for 60 min and incubated with antibodies diluted in PBS containing 3% (wt/vol) nonfat dry milk and 0.5% Tween 20 or in TBS containing 3% (wt/vol) BSA and 0.5% Tween 20 overnight at 4°C. Membranes were washed 3 times for 5 min with PBS or TBS containing 0.5% Tween 20, incubated for 1 h with LI-COR IRDye-conjugated secondary antibody diluted 1:5,000 in LI-COR blocking buffer containing 0.1% (vol/vol) Tween 20 and 0.01% (wt/vol) SDS, and washed again 3 times for 5 min. Membranes were scanned using the LI-COR Odyssey FX imaging system and quantified using the Odyssey image processing software (version 3.0; LI-COR). Images were further processed using GIMP (version 2.8.2).
Immunofluorescence analysis.
Cells grown on coverslips, either infected or not, were washed once with PBS and fixed by incubation with 3% paraformaldehyde in PBS for 20 min followed by PBS supplemented with 50 mM NH4Cl for 10 min at room temperature. Cells were permeabilized with PBS containing 0.5% (vol/vol) Triton X-100 for 5 min and blocked in PBS containing 2.5% (wt/vol) nonfat dry milk and 0.1% (vol/vol) Triton X-100 for 30 min. Coverslips were incubated with primary antibodies diluted in PBS containing 0.1% Triton X-100 for 2 h at room temperature, followed by being washed 3 times with PBS and incubation with Alexa Fluor-labeled secondary antibodies (Molecular Probes) for 1.5 h. Finally, coverslips were washed 3 times with PBS and 3 times with Nanopure H2O, before being mounted using ProLong Gold (Molecular Probes). Nuclear staining was carried out using DAPI (4′,6-diamidino-2-phenylindole; Invitrogen, Life Technologies) in PBS for 5 min between the PBS and H2O washing steps. Slides were analyzed using an IX71 fluorescence microscope (Olympus), and images were captured with a Retiga-2000R camera and QCapture Pro software (version 6.0; both QImaging). Images were processed using IrfanView (version 4.33) and GIMP (version 2.8.2).
RNA isolation and qPCR analysis.
Total RNA was isolated using the RNeasy minikit and protocol (Qiagen) and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). RNAs were stored at −80°C until used. cDNA was prepared using 1 μg total RNA and either random hexamer or oligo(dT)15 primers (Promega) and the Superscript II reverse transcription (RT)-PCR kit (Invitrogen) in a total reaction volume of 10 μl. The resultant cDNA product was then diluted 10-fold, and 1 μl was then used for PCR quantification carried out using the iQ SYBR green supermix (Bio-Rad) and specific primer pairs (Table 1). Quantitative PCRs (qPCRs) were performed using a MyiQ single-color optical detection system and software (version 1.0, Bio-Rad). The qPCR program included 45 cycles with real-time quantitation at each cycle as well as a melting curve analysis at the end to verify the uniformity of the PCR products in each individual sample. To calculate absolute copy numbers, a standard 10-fold dilution series (from 1 ng [∼4.7 × 107 copies] to 0.1 pg [∼4.7 × 103 copies]) of the MV full-length cDNA encoding plasmid pB(+)MVvac2(GFP)H (33) was analyzed in parallel. Copy numbers for each gene were calculated, including PCR efficiency and background corrections using Microsoft Excel 2010 (Microsoft).
TABLE 1.
MV-specific primers for qPCR analysis
| Primer name | Sequence (5′→3′) | Binding sitea |
|---|---|---|
| Le-N fwd | CCAAACAAAGTTGGGTAAGG | 2 |
| Le-N rev | ACCGGATCCTGATGTAATGG | 182r |
| N fwd | ATTGACACTGCAACGGAGTC | 1530 |
| N rev | GCCTTGTTCTTCCGAGATTC | 1634r |
| N-P fwd | AACAACATCCGCCTACCATC | 1707 |
| N-P rev | TTCTGACCATGCTGCCATAG | 1920r |
| P fwd | GTCGGGTTTGTTCCTGACAC | 3154 |
| P rev | AGGTAACGCTTCCGATCCTC | 3248r |
| P-M fwd | GCCAGTCGACCCACCTAGTA | 3360 |
| P-M rev | CTTCCTGTCGCCTAGACCAG | 3575r |
| M fwd | AACGCAAACCAAGTGT | 3843 |
| M rev | TGAAGGCCACTGCATT | 4002r |
| M-F fwd | CCCCCTCTTCCTCAACACAAG | 4755 |
| M-F rev | TGGGTTCTGTTGGGTGTGTATG | 4908r |
| F fwd | TGATTGCAGTGTGTCTTGGAG | 6953 |
| F rev | CCCGTAAGATCAGGCTTTAGG | 7079r |
| F-H fwd | CTTCGTCATCAAGCAACCAC | 7153 |
| F-H rev | GCATTTATCCGGTCTCGTTG | 7299r |
| H fwd | AACTCTGGTGCCGTCACTTC | 8997 |
| H rev | CCATCCCAGAGTGAGTGATATG | 9068r |
| H-GFP fwd | CCCACTAGCCTACCCTCCAT | (9172) |
| H-GFP rev | GAACTTCAGGGTCAGCTTGC | (9401r) |
| GFP fwd | AGAACGGCATCAAGGTGAAC | (9736) |
| GFP rev | TGCTCAGGTAGTGGTTGTCG | (9870r) |
| GFP-L fwd | ACATGGTCCTGCTGGAGTTC | (9913) |
| GFP-L rev | AGTCCATAACGGGGAACCAC | 9240r (10074r) |
| L fwd | AATCTCAAGTCCGGCTATCTG | 15600 (16434) |
| L rev | CCATTCTTTGGTCTCCTTGAC | 15746r (16580r) |
| L-Tr fwd | TGAACTCCGGAACCCTAATC | 15791 (16625) |
| L-Tr rev | AAAGCTGGGAATAGAAACTTCG | 15887r (16721r) |
Binding site are as in the Moraten vaccine strain reference sequence (GenBank accession number AF266287.1) and/or in the MVvac(GFP)H sequence (parentheses) for sequences downstream of the H gene.
RNA sequencing.
Aliquots (10 μg) of total RNA from infected CONkd cells were depleted of rRNA using the RiboMinus eukaryote kit (Ambion, Life Technologies), and RNA sequencing libraries were generated using the ion total RNA-Seq kit, version 2 (Ion Torrent; Life Technologies). Sequencing was carried out on an Ion Torrent PGM instrument. Alignment to a combined Homo sapiens hg19 (Build 37.2)-MVvac2(GFP)H genome was accomplished with a two-stage process. Sequences were first aligned using TopHat2 (version 2.06) (34). Reads that failed to map with TopHat2 were then aligned with TMAP using 5′ and 3′ soft clipping (Life Technologies). The two mappings were merged, and the results were visualized and quantified using Partek Genomics Suite (Partek). Identification and quantification of single nucleotide polymorphisms were performed using SAMTools Pileup (35). Only base reads with Q values of >17 were considered.
DI-RNA detection.
Confluent 100-mm dishes of Vero cells were infected with either parental MVvac-WT(GFP) or mutant MVvac-CKO(GFP) at an MOI of 0.1. Cells were scraped into 4 ml Opti-MEM at the time of maximum GFP expression (∼48 h after infection), subjected to one round of freeze (−80°C) and thaw (ice), cleared of cellular debris by centrifugation (1,600 rpm, 4°C, 10 min), and stored at −80°C, yielding passage P0.
CONkd cells were infected either with original virus stock (MOI of 0.1) or with 100 μl of P0 inoculum. Cells were harvested 48 h after infection, and total RNA was isolated using TRIzol reagent (Ambion, Life Technologies). cDNA was generated from 1 μg total RNA using Superscript II (Invitrogen, Life Technologies) and two DI-specific primers (A1, TCT GGT GTA AGT CTA GTA TCA GA; and A2, AAA GCT GGG AAT AGA AAC TTC G) (36) in a total volume of 20 μl, using a modified protocol (98° for 10 min; 4°C for 10 min; 42°C for 50 min; 72°C for 15 min). A total of 1 μl of the resultant cDNA was then amplified with primers A1 and A2 using GoTaq DNA polymerase (Promega) in a total volume of 50 μl, and the products were analyzed on a 2% agarose gel with an exACTGene 100-bp PCR DNA ladder (Fisher Scientific) as the size standard. The ∼220-bp product obtained from CKO-P1 was gel purified and sequenced from both ends using primers A1 and A2 (Genewiz). A control PCR for standard full-length RNA was performed using primer A2 in combination with primer B1 (ATG ACA GAT CTC AAG GCT AAC) (36).
RESULTS
Detection of dsRNA during C protein-deficient MV infection.
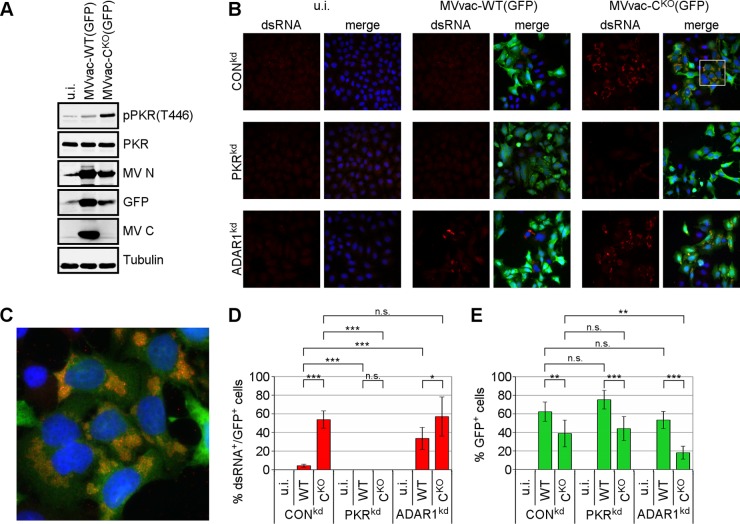
CKO mutant MV, in contrast to parental virus, is a potent activator of PKR in infected cells, as shown by immunoblotting with a phospho(T446)-specific PKR antibody (Fig. 1A) (17, 21). Since RNA with double-stranded character is the activating pathogen-associated molecular pattern for PKR, we tested whether dsRNA formation can be monitored in MV-infected cells by immunostaining with an antibody against dsRNA (37). We used HeLa clones stably knocked down for either PKR (PKRkd) or ADAR1 (ADAR1kd) or a nonspecific control knockdown (CONkd). These cells were infected with either parental MVvac-WT(GFP) or the isogenic MVvac-CKO(GFP) mutant virus. As shown in Fig. 1B and C, we detected dsRNA in CONkd and ADAR1kd cells infected with CKO virus but not in uninfected cells. The dsRNA signal appeared as a punctate and compartmentalized staining pattern within the cytoplasm of infected cells (Fig. 1C). Quantification revealed detectable dsRNA in approximately 50 to 60% of CKO virus-infected (GFP-positive) cells (Fig. 1D); dsRNA expression was much lower in cells infected with the parental WT virus than in those infected with the CKO mutant virus. ADAR1kd cells were also tested for dsRNA, because the dsRNA adenosine deaminase is known to destabilize dsRNA structures (19, 38), and hence cells deficient in ADAR1 might permit detection of lower levels of dsRNA that otherwise would not be seen in ADAR1-sufficient CONkd cells. Indeed, the abundance of dsRNA in parental virus-infected cells was significantly increased in ADAR1kd cells compared to in CONkd cells, consistent with the earlier observations that parental virus activates PKR in ADAR1kd cells but not in CONkd cells (21, 30). Surprisingly, neither parental nor mutant CKO virus infection resulted in detectable amounts of dsRNA in PKRkd cells (Fig. 1B). Higher levels of dsRNA in CKO virus-infected cells inversely correlated with lower infectivity, as quantified by virus-dependent expression of GFP (Fig. 1E). However, increased generation of dsRNA in WT virus-infected ADAR1kd cells led only to a modest reduction of GFP expression (Fig. 1E).
FIG 1.
MV deficient in C protein expression causes increased phosphorylation of PKR and production of dsRNA. (A) Immunoblot of whole-cell lysates against indicated proteins expressed in HeLa cells infected with MVvac-WT(GFP) or MVvac-CKO(GFP) or left uninfected (u.i.) for 48 h. (B) Immunofluorescence staining against dsRNA (red), GFP (green), and nuclei (DAPI; blue) in HeLa cells stably expressing short hairpin RNA (shRNA) directed against PKR (PKRkd), ADAR1 (ADAR1kd), or an unspecific control (CONkd) and infected for 48 h or left uninfected (u.i.). The merge shows combination of all three channels. Images were taken at ×40 magnification. (C) Magnification of the framed section in panel B. (D) Quantification of dsRNA-expressing cells as a percentage of WT or CKO mutant virus-infected (GFP-positive) cells. (E) Quantification of infection as the percentage of GFP-positive cells. Averages and standard deviations were determined from six fields from two independent experiments with ∼100 cells/field for each condition. Statistical significance was determined by Student's t test (two-tailed, two-sample test with equal variance) and P values are marked: *, P < 0.05; **, P < 0.01; ***, P < 0.005. n.s., not significant.
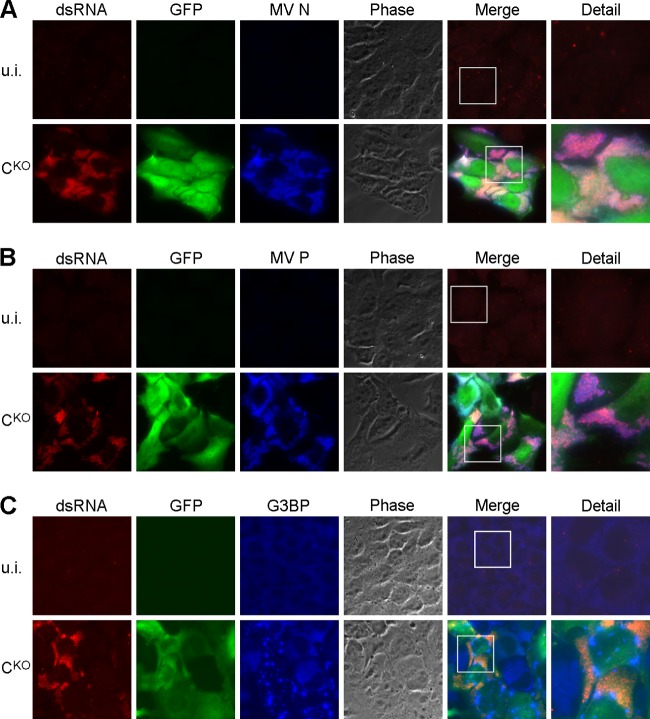
dsRNA localizes to sites of viral replication but not to stress granules.
Next, we assessed the subcellular localization of the dsRNA in CKO-infected cells. We performed coimmunostaining of dsRNA in CKO mutant-infected CONkd cells with that of viral nucleoprotein (N), phosphoprotein (P), and the cellular stress granule marker G3BP (Fig. 2). Both N and P colocalize in viral “inclusion bodies,” presumably the sites of MV replication (39). Indeed, we observed that the localization of the dsRNA signal and the N and P signals largely overlapped (Fig. 2A and B). In contrast, cellular stress granules, which form following CKO virus infection (19), did not colocalize with dsRNA (Fig. 2C). However, the stress granule structures formed in close proximity to the sites of viral replication.
FIG 2.
dsRNA localizes to sites of viral replication but not to stress granules. HeLa CONkd cells were infected with CKO mutant virus or left uninfected (u.i.) and fixed 48 h after infection. Immunofluorescence was performed against dsRNA (red), GFP (green), and viral N protein (A; blue), viral P protein (B; blue), or the cellular stress granule marker G3BP (C; blue). The merge column shows the combination of all three colors. The detail column shows the expanded framed regions in the merge column. Images were taken at ×64 magnification and are representative of three independent experiments.
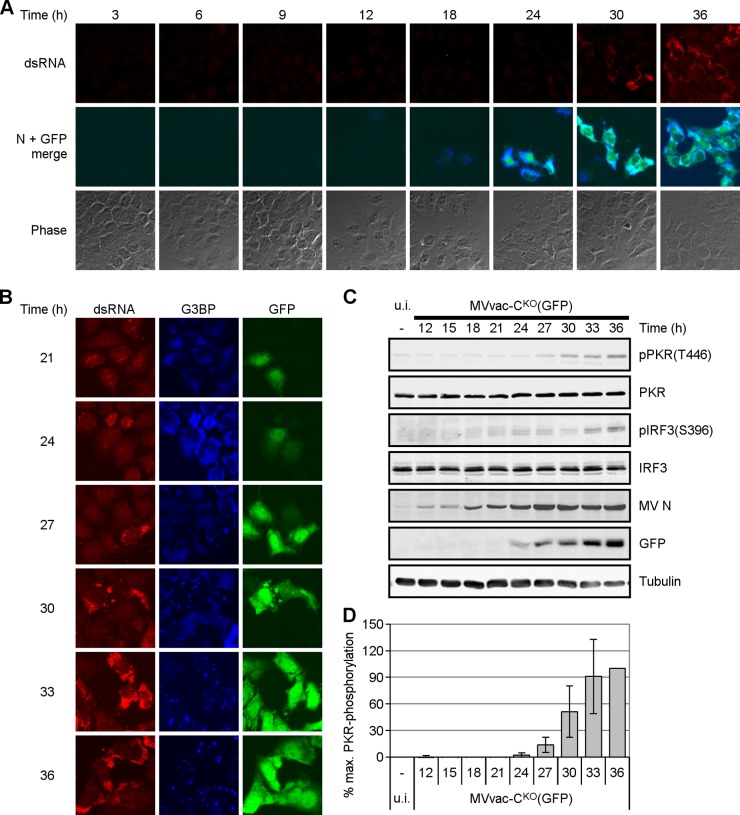
dsRNA accumulates with kinetics similar to PKR phosphorylation and stress granule formation.
We performed time course experiments to determine the kinetics of dsRNA appearance relative to mutant CKO virus gene expression. HeLa CONkd cells were fixed for immunostaining at different times after infection. dsRNA was detected at 24 h postinfection (p.i.), after which the signal intensity and percentage of cells showing dsRNA increased (Fig. 3A). Viral N protein was detected at 12 h p.i., which likely correlates with primary transcription. Robust N expression as well as GFP reporter expression was seen at 24 h p.i. and thereafter, likely reflecting secondary transcription. Detection of dsRNA during a phase of strong viral gene expression, together with the fact that dsRNA colocalized with N and P proteins (Fig. 2A and B), led us to consider that the dsRNA observed during CKO mutant infection is a viral replication product.
FIG 3.
dsRNA accumulates with kinetics similar to PKR activation and stress granule formation. HeLa CONkd cells were infected with MVvac-CKO(GFP) mutant virus and fixed at the indicated times (h) after infection. (A) Cells were simultaneously stained with antibodies against dsRNA (red), GFP (green), and viral N protein (blue). The first row shows dsRNA, the second row shows the merged GFP and N signals, and the third row shows phase contrast. Images were taken at ×64 magnification. (B) Immunofluorescence was performed using antibodies directed against dsRNA (red; left column), G3BP (blue; middle column), and GFP (green; right column). Images were taken at ×64 magnification. (C) Immunoblot analysis of the indicated proteins. Blots are representative of three independent experiments. (D) Quantification of pPKR levels. Levels are averages and standard deviations from two independent experiments. They were normalized to the intensity of the corresponding tubulin band in each lane and are expressed as the percentage of the value at 36 h.
We also analyzed the formation of stress granules, which depends on PKR and the phosphorylation of eIF2α (19, 22). Coimmunostaining of dsRNA and the stress granule marker G3BP indicated similar kinetics of appearance (Fig. 3B): the G3BP signal (blue) was diffuse and cytoplasmic both in uninfected cells and in infected cells at early stages of the replication cycle, but at later times G3BP was recruited to stress granules. Next, we analyzed by immunoblot analysis the phosphorylation status of PKR and IRF3. Both phospho-PKR(T446) and phospho-IRF3(S396) were observed at 24 h after infection at a low MOI of 0.1 (Fig. 3C and D). N protein was detected as early as 12 h after infection, but robust viral protein expression was observed after 24 h of infection, correlating with the expression of dsRNA detected by immunofluorescence (Fig. 3A and B). Thus, the kinetics of dsRNA production and PKR and IRF3 phosphorylation correlate.
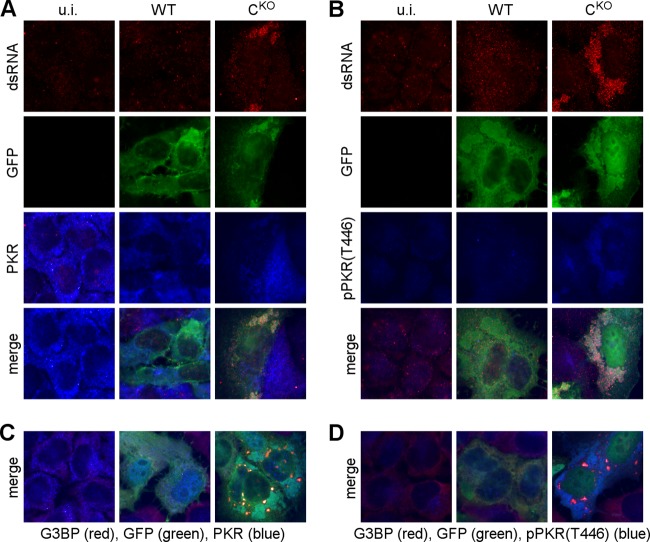
Phospho-PKR localizes at sites of dsRNA accumulation.
Coimmunostaining of dsRNA with either PKR (Fig. 4A) or phospho-PKR (Fig. 4B) revealed that PKR relocalized to the sites of dsRNA accumulation in CKO virus-infected cells. In contrast, PKR showed a more uniform cytoplasmic staining in uninfected cells and in cells infected with parental virus (Fig. 4A, left and middle columns). Under these conditions, no phospho-specific PKR signal was detected (Fig. 4B, left and middle columns). In addition, costaining with G3BP showed that the PKR-derived signal localized to sites between stress granules in CKO-infected cells (Fig. 4C and D). Thus, dsRNA may be the activator of PKR during CKO virus infection.
FIG 4.
Phospho-PKR localizes to sites of dsRNA accumulation. HeLa CONkd cells were infected with parental MVvac-WT(GFP) or mutant MVvac-CKO(GFP) virus and fixed 48 h after infection. (A) Immunofluorescence against dsRNA (red), GFP (green), and total PKR (blue). (B) Immunofluorescence against dsRNA (red), GFP (green), and pPKR(T446) (blue). (C) Immunofluorescence against G3BP (red), GFP (green), and total PKR (blue). (D) Immunofluorescence against G3BP (red), GFP (green), and pPKR(T446) (blue). The merge row shows the combination of all three colors.
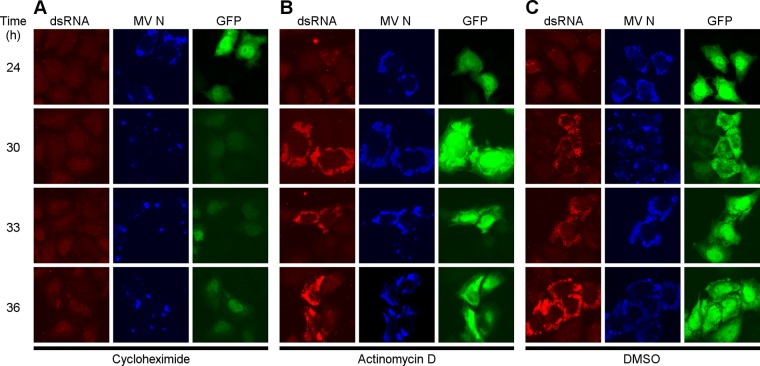
dsRNA production is inhibited by cycloheximide but not actinomycin D.
To further investigate the hypothesis that the dsRNA is a product of viral replication, we examined the production of dsRNA in infected cells in the presence and absence of actinomycin D. This drug inhibits RNA synthesis by dsDNA-dependent RNA polymerases but not by the MV RNA-dependent RNA polymerase. As a control, we treated infected cells with cycloheximide, an inhibitor of protein synthesis. Cycloheximide inhibited the synthesis of both the viral N protein and dsRNA as illustrated by the weak intensity staining (Fig. 5A) compared to that of DMSO vehicle alone. In contrast, actinomycin D had no appreciable effect on the synthesis of either N protein or dsRNA (Fig. 5B). DMSO vehicle likewise did not affect dsRNA production (compare Fig. 5C to Fig. 5A and B). The cycloheximide treatment led to a strong reduction in the dsRNA-specific signal at all times examined (Fig. 5A). Notably, viral gene expression was strongly inhibited as well (compare N [blue] and GFP [green] signal intensities and size of viral replication sites in Fig. 5). We conclude that the dsRNA observed during CKO virus infection is likely of viral origin. This experiment further suggests that viral secondary gene expression, which is protein synthesis dependent (40), is necessary for the accumulation of dsRNA.
FIG 5.
dsRNA is a viral product. HeLa CONkd cells infected with MVvac-CKO(GFP) mutant virus and treated with 10 μg/ml cycloheximide (A), 10 μg/ml actinomycin D (B), or DMSO (C) at 24 h after infection. Cells were fixed at the indicated time points, and immunofluorescence was performed in parallel with antibodies directed against dsRNA (red; left column), viral N protein (blue; middle column), or GFP (green; right column). Images were taken at ×64 magnification and are representative of three independent experiments.
Deletion of C causes less efficient transcription and a steeper transcription gradient.
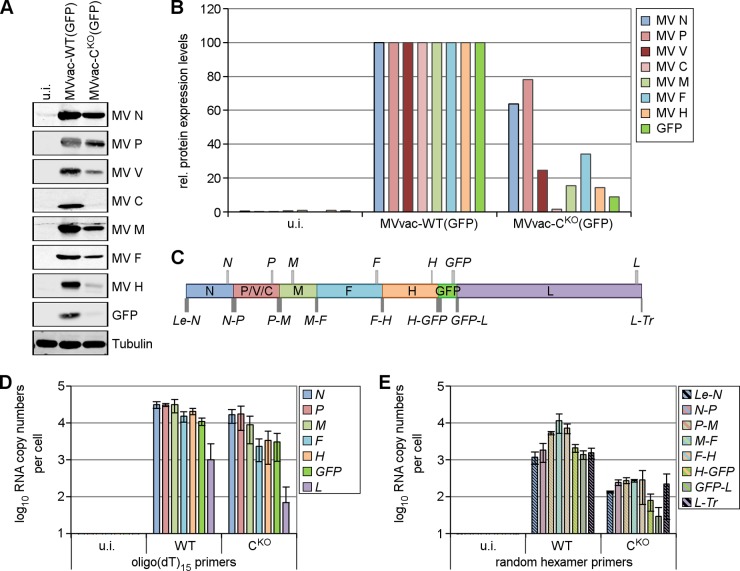
Since the C protein may regulate the balance of viral transcription and replication (24, 25, 41), we quantified viral protein and RNA expression of both the parental and CKO viruses (Fig. 6). Viral protein expression was reduced in CKO virus-infected cells compared to that in parental virus-infected cells (Fig. 6A); furthermore, while levels of N (64%) and P (78%) proteins were slightly reduced, the levels of the downstream-encoded proteins M, F, H, and GFP were strongly reduced (16%, 34%, 14%, and 9%, respectively) (Fig. 6B).
FIG 6.
Knockout of C protein slows viral macromolecular synthesis and results in a steeper transcription gradient. HeLa CONkd cells were infected with parental MVvac-WT(GFP) or mutant MVvac-CKO(GFP) virus or left uninfected (u.i.). Cells were harvested at 48 h after infection. (A) Immunoblot analysis with antibodies directed against the indicated viral and cellular proteins. Blots are representative of three independent experiments. (B) Quantification of the proteins from panel A. Protein levels were normalized to the corresponding tubulin levels in each lane and are relative to the levels of MVvac-WT(GFP)-infected cells, which were set as 100% for each protein. (C) Schematic representation of the genome organization of MVvac-WT(GFP). The relative size and location of the viral gene-specific (light-gray boxes) and intercistronic (dark-gray boxes) qPCR products are shown. (D) qPCR analysis of viral mRNA from infected cells using cDNA prepared with oligo(dT)15 primers. (E) qPCR analysis of intercistronic viral sequences from infected cells using cDNA prepared with random hexamer primers. The resulting cDNAs were analyzed for product copy number as described in Materials and Methods. Data shown are average numbers ± standard deviations from three independent experiments.
We next examined the question of how protein expression levels correlate with the viral RNA expression pattern using qPCR analysis with probe sets corresponding to each viral gene, the individual gene borders, and the leader-N (Le-N) and the L-trailer (L-Tr) regions (Fig. 6C and Table 1). We generated cDNA libraries from RNA of infected HeLa CONkd cells using either oligo(dT)15 primers or random hexamer primers. The former library was used to quantify viral mRNA levels with intragenic primer pairs (Fig. 6D and Table 2). Although generally reflecting the transcription gradient for nonsegmented −ssRNA viruses (42), the intergenic attenuation was more pronounced for the CKO virus, resulting in a steeper transcription gradient.
TABLE 2.
Expression of MV-specific transcriptsa
| oligo(dT)15 | MVvac-WT(GFP) |
MVvac-CKO(GFP) |
P value (WT vs CKO)b |
|||
|---|---|---|---|---|---|---|
| Copy no. | % of N mRNA | Copy no. | % of N mRNA | Copy no. | % of N mRNA | |
| N | 31,000 ± 6,600 | 100 ± 0 | 17,000 ± 6,700 | 100 ± 0 | 0.054 (NS) | ND |
| P | 31,000 ± 2,900 | 100 ± 21 | 17,000 ± 11,000 | 97 ± 38 | 0.120 (NS) | 0.888 (NS) |
| M | 31,000 ± 12,000 | 107 ± 53 | 9,000 ± 6,300 | 48 ± 24 | 0.049* | 0.151 (NS) |
| F | 15,000 ± 4,900 | 49 ± 8 | 2,300 ± 1,400 | 13 ± 5 | 0.011* | 0.002** |
| H | 21,000 ± 3,900 | 67 ± 12 | 3,400 ± 2,600 | 18 ± 11 | 0.003** | 0.006** |
| GFP | 11,000 ± 2,600 | 37 ± 10 | 3,100 ± 2,100 | 16 ± 8 | 0.014* | 0.056 (NS) |
| L | 1,000 ± 1,700 | 3 ± 5 | 700 ± 120 | 1 ± 1 | 0.404 (NS) | 0.530 (NS) |
Oligo(dT)15-primed cDNA obtained from 10 ng of total RNA was used for qPCR quantification. Absolute copy numbers were calculated using an MV plasmid reference standard as described in Materials and Methods. Results are average values ± standard deviations from three independent experiments.
P values were calculated using a two-tailed, two-sample Student t test, with equal variance. ND, not determined; NS, not significant; *, P ≤ 0.05; **, P ≤ 0.01.
Analysis of intercistronic regions in randomly primed cDNA inferred readthrough transcripts for most genes (Fig. 6E). Quantitation of the Le-N and GFP-L intergene borders of parental virus (WT) showed approximately 103 copies per cell, which correlates well with quantitative Northern blot analyses (43). Intercistronic N-P, P-M, M-F, and F-H sequences were 5 to 10 times more greatly expressed than the other intercistronic regions, possibly due to high levels of bicistronic mRNAs (43). As expected, products obtained with intragenic primer pairs that also detect monocistronic mRNAs were most abundant, about 104 copies for the N, P, M, F, and H mRNAs (Fig. 6D). CKO virus exhibited a steeper transcription profile with greater attenuation at gene junctions than the parental virus. RNA quantities were reduced about 10-fold more in cells infected with CKO virus compared to those in the parental virus. However, the L-Tr region was overrepresented in CKO virus-infected cells but not in cells infected with parental virus (Fig. 6E, compare GFP-L and L-Tr in WT and CKO), perhaps due to selective amplification (see below). We conclude that, in CKO-infected cells in the absence of C protein, the viral polymerase may be less processive than in parental virus-infected cells where C protein is present.
RNA deep sequencing confirms a steeper transcription gradient in CKO virus-infected cells.
We then analyzed RNA prepared from CKO mutant and parental virus-infected cells by RNA deep sequencing. Total RNA samples depleted of rRNA were sequenced using the Ion Torrent platform, and sequences that aligned to the viral genome were identified and quantified (Fig. 7). In addition to obtaining single nucleotide resolution, RNA-Seq permitted us to discriminate between viral sequences derived from either +RNA (mRNA/antigenome; shown in blue) or −RNA (genomic RNA; shown in red). The relative abundance of viral transcript RNAs determined by RNA-Seq (Fig. 7) was in good agreement with the relative abundance of gene-specific transcripts determined by qPCR analysis (Fig. 6D and E). The number of +RNA reads obtained for CKO virus was about 10-fold less than those obtained for parental virus infection (Fig. 7, compare y axes in top and bottom panels). N and P mRNA transcripts showed the highest abundance by RNA-Seq for both viruses, and the GFP and L transcript levels were the lowest. In contrast, −RNA sequence reads were rare and distributed comparably along the whole MV reference sequence. The RNA-Seq results also confirmed that the transcription gradient was steeper for the CKO mutant virus than for the parental virus. The uneven distribution of reads within individual genes, which is a common feature of short-read sequencing that likely arises from sequence-dependent differences in sequencing efficiencies, was not further analyzed. Finally, the RNA-Seq nucleotide analyses revealed similar mutation frequencies with measured error rates of about 0.1% for both viruses (parental virus, 4,979,339 sequenced viral nucleotides/4,247 observed mutations/0.1% mutation rate; CKO virus, 529,760/449/0.1%). Population-wide changes other than the engineered mutations to suppress C protein translation (U1830C and G1845A on the plus strand) (31) were not observed.
FIG 7.
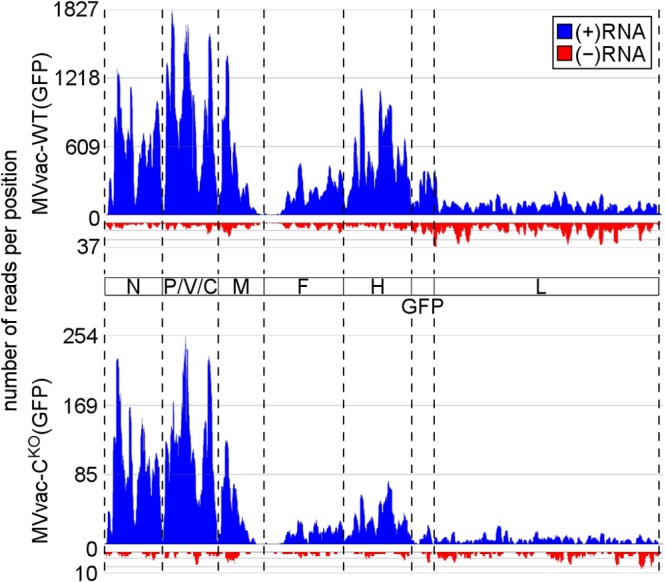
RNA-Seq analyses of RNA from CKO mutant and parental virus-infected cells. cDNA libraries prepared using 10 μg total RNA from virus-infected cells depleted of ribosomal RNAs were analyzed by Ion Torrent sequencing as described in Materials and Methods. Diagrams show the alignment of reads to the viral genome sequence for parental MVvac-WT(GFP)-infected cells (top) and mutant MVvac-CKO(GFP)-infected cells (bottom). Reads are separated corresponding to their polarity with +RNA shown in blue (mRNA, antigenome) and −RNA (genome) shown in red. Dashed lines correspond to gene borders.
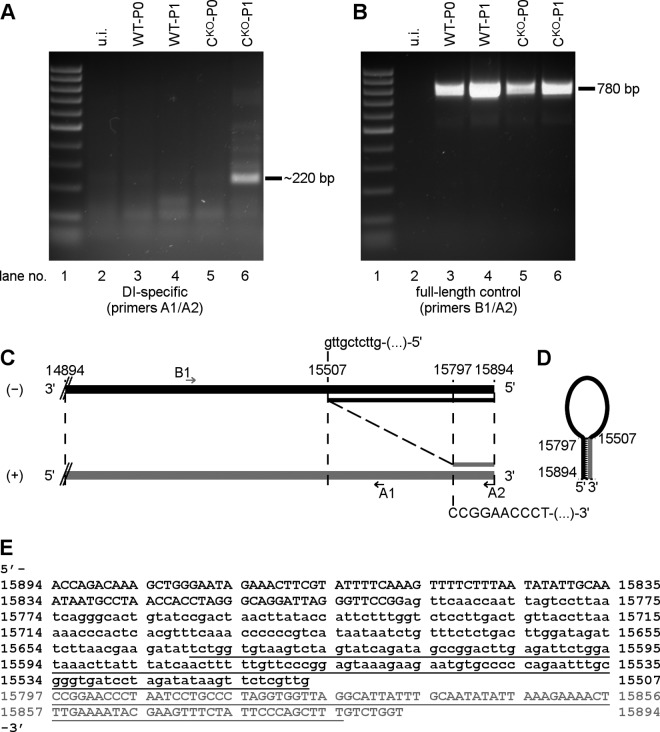
CKO virus generates DI-RNAs.
The above-described sequencing data did not reveal MV-specific RNA species with new junctions in CKO-infected cells that may have been generated by replication errors, such as DI-RNAs. Because DI-RNAs can have long complementary sequences prone to form dsRNA (44), and because copyback DI-RNAs have been described in MV infections (36, 45), we tested for them using a specific PCR approach based on two staggered primers, both of negative polarity (36). We analyzed total RNA from parental and CKO virus-infected CONkd cells (passage P0) and from cells that were infected with an inoculum generated after one high MOI passage on Vero cells (passage P1). With parental (WT-P0) and CKO (CKO-P0) viruses that had not been passed, specific PCR products were not observed beyond the low background level (Fig. 8A, lanes 3 and 5). However, using CKO virus passed once at high MOI (CKO-P1), several product bands in the size range of ∼100 to ∼1,000 bp were detected (Fig. 8A, lane 6). An ∼220-bp product was most abundant. The parental virus (WT-P1) after high MOI passage did not show these products (Fig. 8A, lane 4). A control PCR was carried out using a primer pair of opposite polarity to test amplification of the standard MV genome. This control reaction yielded a 780-bp fragment for all four conditions of infection as expected but not from the uninfected cells (Fig. 8B). Direct sequence analysis of the CKO-P1-derived 220-bp PCR product identified the breakpoint at genome position 15507 of Moraten vaccine strain (numbering as in GenBank accession number AF266287.1) and the reinitiation site (nt 15797), predicting a 486-nt copyback DI-RNA with a 98-nt terminal stem and a 290-nt internal loop (Fig. 8C to E). Thus, C deletion may favor amplification of copyback DI-RNAs.
FIG 8.
CKO mutant virus generates copyback DI-RNA. cDNA generated from HeLa CONkd cells infected with original virus stocks of WT or CKO mutant viruses (P0) or with viruses that have been passaged once on Vero cells (P1) were analyzed for copyback DI-RNAs by PCR. (A) DI-specific PCR using primers A1 and A2, both of which are negative polarity with respect to virus genome sequence. (B) Control PCR using primers B1 (positive polarity) and A2 (negative polarity). (C) Schematic diagram of the sequence obtained for the 220-bp fragment observed with CKO-P1. Breakpoint (genome position −15507) and reinitiation point (genome position +15797) of copyback DI are indicated, as well as relative binding sites of primers A1, A2, and B1. (D) Schematic diagram of the DI-RNA stem-loop model with the predicted 98-bp stem. (E) Sequence of the detected copyback DI-RNA. Complementary ends are shown in uppercase font, and the loop sequence of the DI-RNA is shown in lowercase font. The PCR fragment that was sequenced is shown in underlined font. Antigenomic +RNA is shown in gray; −RNA (genomic RNA) is shown in black in panels C, D, and E. Nucleotide positions correspond to the Moraten vaccine strain of MV (GenBank accession number AF266287).
DISCUSSION
We previously reported that an MV mutant lacking the expression of the nonstructural C protein (CKO) is a potent activator both of PKR (17) and PKR-mediated antiviral responses (19, 21, 46). Similar observations have been made by others with paramyxoviruses lacking C protein expression, including MV (6, 25) and human parainfluenza virus 1 (28). We show herein that in contrast to parental MV, the amplification of a DI-RNA forming a dsRNA structure was readily demonstrable in CKO mutant-infected cells. This DI-RNA may account for the dsRNA formation detected immunochemically that correlated both with activation of PKR kinase autophosphorylation and formation of stress granules. Furthermore, the suppression of ADAR1 expression in HeLa cells led to increased formation of dsRNA in parental virus-infected cells but had no significant effect on the already high dsRNA level seen in cells infected with CKO virus. This observation is consistent with earlier studies describing a proviral role for ADAR1 by counteracting PKR activation (19, 21, 30). ADAR1, an A-to-I dsRNA editing enzyme, deaminates adenosine in dsRNA structures, thereby destabilizing dsRNA because I·U base pairs are less stable than A·U pairs (38, 47). ADAR1 is known to compete with and suppress PKR activity (38, 48). Thus, generation of dsRNA is not restricted to the CKO virus but also appears in parental virus-infected ADAR1kd cells. The levels of dsRNA generated during replication of parental virus, however, are apparently low enough to become functionally destabilized in ADAR1-sufficient cells. Remarkably, we did not detect dsRNA in HeLa cells deficient in PKR expression. Since ADAR1 is expressed at comparable levels in HeLa PKRkd cells and parental cells (30), we suggest that the lack of a dsRNA signal in PKRkd cells may be due to the destabilizing effect of ADAR1 on these structures. PKR conceivably competes with ADAR1 for binding these dsRNAs, leading to their stabilization.
We observed the dsRNA expressed in CKO-infected cells in close proximity to the sites of MV replication (39). These sites contained high levels of viral N and P proteins, and in the case of parental virus, they also contained C protein (data not shown) (6). In contrast, most of the dsRNA did not localize with the SG protein marker G3BP. Stress granules form in response to CKO virus infection but not in response to parental virus infection (19). Interaction and exchange of molecular components between cytoplasmic SG bodies and the sites of MV replication seem plausible.
Formation of SG and autophosphorylation of PKR occurred exclusively in cells showing dsRNA immunostaining and with kinetics that correlated with the appearance of the dsRNA. Formation of SG is PKR dependent in a number of virus-infected cell systems (49), including MV (19), hepatitis C virus (50), respiratory syncytial virus (51), and West Nile virus (52). Subcellular localization, kinetics of accumulation, and sensitivity to cycloheximide, a potent inhibitor of MV replication (40), suggest that the RNA responsible for the dsRNA signal observed in CKO mutant virus-infected cells is of viral and not of cellular origin. Phospho-PKR staining was observed in all infected cells that showed detectable dsRNA staining, although only about 50 to 60% of the infected cells measured by GFP or N protein expression were positive for dsRNA. This ratio was consistently observed and possibly reflects a higher concentration threshold required for dsRNA detection by the J2 monoclonal antibody than for GFP or N detection in the immunochemical assays.
We carried out an extensive analysis of viral RNA and protein expression profiles to gain insights into the mechanisms promoting dsRNA formation and PKR activation seen in cells infected with CKO virus. A steep transcription gradient observed for CKO-infected cells compared to parental virus-infected cells suggests that the C protein may enhance the processivity of the viral polymerase at gene junctions. This effect appears largely independent of the cellular interferon response, because we also observed a stronger transcription gradient for the CKO virus than for the parental virus in IFN-deficient Vero cells (data not shown). We also found an ∼10-fold reduction of viral mRNAs in cells infected with CKO virus compared to parental virus-infected cells. This ratio is further reflected in the ∼10-fold or greater reduction of final infectious titers seen for the CKO virus than for the parental virus (17, 25, 53). The reduction of viral genomic RNA seen in CKO-infected cells by RNA-Seq may also have a direct impact on the mRNA copy numbers, since fewer template molecules for viral transcription are present. A strong overrepresentation of the L-Tr sequence in CKO virus-infected cells was observed, which is suggestive of the possible generation of defective genomes during replication.
RNA-Seq allowed us to analyze the specific nucleotide sequence of expressed viral RNAs. We did not observe an increased rate of viral mutations in cells infected with the CKO virus. Therefore, the C protein is not necessary for polymerase fidelity nor does it affect subsequent editing of the product RNA, for example, A-to-I editing by an ADAR, at least during a single cycle of infection.
We were unable to detect DI-RNA in CKO-infected cells by whole-genome analysis during single-cycle infection. However, after further passage of the inoculum, we detected copyback DI-RNAs, which are known to efficiently trigger innate immune responses: in the case of vesicular stomatitis virus (VSV) DI-011, just a single particle is sufficient to induce a quantum yield of interferon (54, 55). DI-RNAs have been observed not only with VSV, SENV, and parainfluenza virus 5 (PIV5) (54, 56, 57) but also for MV (36, 45), where a vaccine strain-derived inoculum able to express the C protein produced high quantities of DI-RNAs after high MOI passage in cell culture. Herein, we show that a single round of CKO passage on Vero cells led to amplification of the DI-RNA. In contrast, no copyback DI-RNAs were amplified from cells infected with parental virus.
We identified the breakpoint site of polymerase dissociation from the template strand and the reinitiation point on the newly synthesized strand. This then allowed us to deduce the complete 486-nt-long sequence of the DI-RNA. This DI-RNA with self-complementary ends can form a 98-bp dsRNA stem, which is a sufficient dsRNA structure to trigger activation of PKR (15, 58). Several additional higher-molecular-weight products were also observed, suggesting that additional copyback DI-RNAs with different breakpoints and reinitiation sites are likely also generated during CKO virus infection. dsRNA binding by PKR occurs at a repeated domain in its N-terminal region that in turn leads to autophosphorylation, including at Thr446 (47, 59). RNA binding by PKR is not sequence specific but is RNA structure dependent. Kinase activation is seen with ∼30 to 50 bp of dsRNA and is optimal with ∼80 bp, although PKR interacts with as little as 11 bp of dsRNA (15, 58).
At least two functions have been proposed for the MV C protein: modulation of the innate immune response, including PKR activation and RIG-I signaling (25, 53), and downregulation of viral RNA synthesis (24, 41). Our findings are consistent with the notion that C protein may stabilize the RNP-polymerase complex, and its absence results in more frequent chain termination occurring both during transcription (steeper mRNA gradient) and during replication (DI-RNA generation) modes of expression. Whether the C protein functions through direct interaction with the viral RNP-polymerase complex or indirectly through host proteins, including SHCBP1 (60, 61), or perhaps through both, is not clear. The C protein may interfere with PKR activation by preventing the generation of dsRNA rather than by sequestering dsRNA or by directly inhibiting the signal transduction cascade. This hypothesis is supported by the observation that the C protein, unlike the influenza virus NS1 protein (25), does not sequester activator dsRNA in coinfections with MV and ΔE3L vaccinia virus (17) or SENV and Newcastle disease virus (26). Likewise, MV C protein does not detectably bind to PKR (17). It is possible that the C protein might have a supporting role in proper RNA encapsidation. Knockout of C would increase the frequency with which DI-RNAs are generated that subsequently form dsRNA structures that activate PKR and PKR-mediated cellular responses, including SG formation. The inhibitory effects of these responses on viral replication would account for the decreased viral RNA production and reduced infectious virus yields seen in Moraten and IC-B CKO infections (17, 25, 53).
While we observed the amplification of DI-RNA in CKO but not parental virus-infected cells following one passage, additional mechanisms of dsRNA-mediated activation of PKR that are independent of copyback DI-RNAs remain conceivable. We cannot eliminate the possibility of other potential contributing mechanisms of dsRNA-mediated activation of PKR that are dependent upon spacial and temporal differences in subcellular architecture that may occur between CKO and parental virus-infected cells. For example, a viral ssRNA may activate PKR following annealing with a complementary strand that normally is efficiently encapsidated or otherwise shielded in parental virus-infected cells but is not in CKO virus infections. It is known that, in addition to synthetic and natural duplex dsRNA effectors of PKR, naturally occurring viral ssRNAs with dsRNA character indeed can either activate, as exemplified by reovirus s1 mRNA, or antagonize, as exemplified by adenovirus VA RNA (15, 62).
In conclusion, our results link the CKO mutation of MV with a propensity to generate elevated levels of dsRNA, including copyback DI-RNAs. This provides an explanation as to why the CKO virus is such a potent activator not only of PKR phosphorylation and stress granule formation (19) but also of RIG-I activation and signaling triggered by 5′-triphosphate-containing dsRNAs (18, 63).
ACKNOWLEDGMENTS
This work was supported in part by research grants AI-12520 and AI-20611 (to C.E.S.) and AI-63476 (to R.C.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Public Health Service, and by research fellowship PF791/1-1 from the Deutsche Forschungsgemeinschaft (to C.K.P.).
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Griffin DE. 2007. Measles virus, p 1551–1585 In Knipe DM, Howley PM. (ed), Fields virology, 5 ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Cattaneo R, McChesney M. 2008. Measles virus, p 285–291 In Mahy BWJ, van Regenmortel MHV. (ed), Encyclopedia of virology, 3rd ed. Academic Press, Oxford, United Kingdom [Google Scholar]
- 3.Cattaneo R, Kaelin K, Baczko K, Billeter MA. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759–764. 10.1016/0092-8674(89)90679-X [DOI] [PubMed] [Google Scholar]
- 4.Bellini WJ, Englund G, Rozenblatt S, Arnheiter H, Richardson CD. 1985. Measles virus P gene codes for two proteins. J. Virol. 53:908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escoffier C, Manie S, Vincent S, Muller CP, Billeter M, Gerlier D. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsu Y, Takeda M, Ohno S, Shirogane Y, Iwasaki M, Yanagi Y. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296–8306. 10.1128/JVI.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaller CK, Conzelmann KK. 2008. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365–12373. 10.1128/JVI.01321-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuhmann KM, Pfaller CK, Conzelmann KK. 2011. The measles virus V protein binds to p65 (RelA) to suppress NF-kappaB activity. J. Virol. 85:3162–3171. 10.1128/JVI.02342-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparrer KM, Pfaller CK, Conzelmann KK. 2012. Measles virus C protein interferes with beta interferon transcription in the nucleus. J. Virol. 86:796–805. 10.1128/JVI.05899-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaffer JA, Bellini WJ, Rota PA. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389–397. 10.1016/S0042-6822(03)00537-3 [DOI] [PubMed] [Google Scholar]
- 11.Caignard G, Guerbois M, Labernardiere JL, Jacob Y, Jones LM, Wild F, Tangy F, Vidalain PO. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368:351–362. 10.1016/j.virol.2007.06.037 [DOI] [PubMed] [Google Scholar]
- 12.Fontana JM, Bankamp B, Bellini WJ, Rota PA. 2008. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 374:71–81. 10.1016/j.virol.2007.12.031 [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran A, Parisien JP, Horvath CM. 2008. STAT2 is a primary target for measles virus V protein-mediated alpha/beta interferon signaling inhibition. J. Virol. 82:8330–8338. 10.1128/JVI.00831-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caignard G, Bourai M, Jacob Y, Tangy F, Vidalain PO. 2009. Inhibition of IFN-alpha/beta signaling by two discrete peptides within measles virus V protein that specifically bind STAT1 and STAT2. Virology 383:112–120. 10.1016/j.virol.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 15.Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809. 10.1128/CMR.14.4.778-809.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47. 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- 17.Toth AM, Devaux P, Cattaneo R, Samuel CE. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83:961–968. 10.1128/JVI.01669-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J. Virol. 84:380–386. 10.1128/JVI.02630-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okonski KM, Samuel CE. 2013. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 87:756–766. 10.1128/JVI.02270-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Okonski KM, Samuel CE. 2012. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J. Virol. 86:3787–3794. 10.1128/JVI.06307-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 23:573–582. 10.1016/j.coi.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devaux P, Hodge G, McChesney MB, Cattaneo R. 2008. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 82:5359–5367. 10.1128/JVI.00169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankamp B, Wilson J, Bellini WJ, Rota PA. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336:120–129. 10.1016/j.virol.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 25.Nakatsu Y, Takeda M, Ohno S, Koga R, Yanagi Y. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J. Virol. 80:11861–11867. 10.1128/JVI.00751-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 82:10102–10110. 10.1128/JVI.00599-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boonyaratanakornkit J, Bartlett E, Schomacker H, Surman S, Akira S, Bae YS, Collins P, Murphy B, Schmidt A. 2011. The C proteins of human parainfluenza virus type 1 limit double-stranded RNA accumulation that would otherwise trigger activation of MDA5 and protein kinase R. J. Virol. 85:1495–1506. 10.1128/JVI.01297-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Samuel CE. 2007. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J. Virol. 81:8192–8200. 10.1128/JVI.00426-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toth AM, Li Z, Cattaneo R, Samuel CE. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284:29350–29356. 10.1074/jbc.M109.045146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devaux P, von Messling V, Songsungthong W, Springfeld C, Cattaneo R. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 360:72–83. 10.1016/j.virol.2006.09.049 [DOI] [PubMed] [Google Scholar]
- 32.Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Valle JR, Devaux P, Hodge G, Wegner NJ, McChesney MB, Cattaneo R. 2007. A vectored measles virus induces hepatitis B surface antigen antibodies while protecting macaques against measles virus challenge. J. Virol. 81:10597–10605. 10.1128/JVI.00923-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calain P, Curran J, Kolakofsky D, Roux L. 1992. Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology 191:62–71. 10.1016/0042-6822(92)90166-M [DOI] [PubMed] [Google Scholar]
- 37.Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19:2993–3000. 10.1093/nar/19.11.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuel CE. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411:180–193. 10.1016/j.virol.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai M, Imagawa DT. 1969. Electron microscopy of measles virus replication. J. Virol. 3:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall WW, Genius D, ter Meulen V. 1977. The effect of cycloheximide on the replication of measles virus. J. Gen. Virol. 35:579–582. 10.1099/0022-1317-35-3-579 [DOI] [PubMed] [Google Scholar]
- 41.Reutter GL, Cortese-Grogan C, Wilson J, Moyer SA. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100–109. 10.1006/viro.2001.0962 [DOI] [PubMed] [Google Scholar]
- 42.Rima BK, Duprex WP. 2009. The measles virus replication cycle. Curr. Top. Microbiol. Immunol. 329:77–102. 10.1007/978-3-540-70523-9_5 [DOI] [PubMed] [Google Scholar]
- 43.Cattaneo R, Rebmann G, Schmid A, Baczko K, ter Meulen V, Billeter MA. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Hara PJ, Nichol ST, Horodyski FM, Holland JJ. 1984. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36:915–924. 10.1016/0092-8674(84)90041-2 [DOI] [PubMed] [Google Scholar]
- 45.Calain P, Roux L. 1988. Generation of measles virus defective interfering particles and their presence in a preparation of attenuated live-virus vaccine. J. Virol. 62:2859–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McAllister CS, Samuel CE. 2009. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J. Biol. Chem. 284:1644–1651. 10.1074/jbc.M807888200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth AM, Zhang P, Das S, George CX, Samuel CE. 2006. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 81:369–434. 10.1016/S0079-6603(06)81010-X [DOI] [PubMed] [Google Scholar]
- 48.Gelinas JF, Clerzius G, Shaw E, Gatignol A. 2011. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 85:8460–8466. 10.1128/JVI.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20:175–183. 10.1016/j.tim.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85. 10.1016/j.chom.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindquist ME, Mainou BA, Dermody TS, Crowe JE., Jr 2011. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 413:103–110. 10.1016/j.virol.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courtney SC, Scherbik SV, Stockman BM, Brinton MA. 2012. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J. Virol. 86:3647–3657. 10.1128/JVI.06549-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devaux P, Cattaneo R. 2004. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J. Virol. 78:11632–11640. 10.1128/JVI.78.21.11632-11640.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815–819. 10.1038/266815a0 [DOI] [PubMed] [Google Scholar]
- 55.Schubert M, Lazzarini RA. 1981. Structure and origin of a snapback defective interfering particle RNA of vesicular stomatitis virus. J. Virol. 37:661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308. 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, Goodbourn S, Randall RE. 2013. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J. Virol. 87:4798–4807. 10.1128/JVI.03383-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nallagatla SR, Toroney R, Bevilacqua PC. 2011. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 21:119–127. 10.1016/j.sbi.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCormack SJ, Thomis DC, Samuel CE. 1992. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology 188:47–56. 10.1016/0042-6822(92)90733-6 [DOI] [PubMed] [Google Scholar]
- 60.Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y. 2009. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 83:10374–10383. 10.1128/JVI.01056-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito M, Iwasaki M, Takeda M, Nakamura T, Yanagi Y, Ohno S. 2013. Measles virus nonstructural C protein modulates viral RNA polymerase activity by interacting with host protein SHCBP1. J. Virol. 87:9633–9642. 10.1128/JVI.00714-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujita T. 2009. A nonself RNA pattern: tri-p to panhandle. Immunity 31:4–5. 10.1016/j.immuni.2009.06.014 [DOI] [PubMed] [Google Scholar]