Abstract
Vast amounts of molecular data characterizing the genome, epigenome and transcriptome are becoming available for a variety of cancers. The current challenge is to integrate these diverse layers of molecular biology information to create a more comprehensive view of key biological processes underlying cancer. We developed a biocomputational algorithm that integrates copy number, DNA methylation, and gene expression data to study master regulators of cancer and identify their targets. Our algorithm starts by generating a list of candidate driver genes based on the rationale that genes that are driven by multiple genomic events in a subset of samples are unlikely to be randomly deregulated. We then select the master regulators from the candidate driver and identify their targets by inferring the underlying regulatory network of gene expression. We applied our biocomputational algorithm to identify master regulators and their targets in glioblastoma multiforme (GBM) and serous ovarian cancer. Our results suggest that the expression of candidate drivers is more likely to be influenced by copy number variations than DNA methylation. Next, we selected the master regulators and identified their downstream targets using module networks analysis. As a proof-of-concept, we show that the GBM and ovarian cancer module networks recapitulate known processes in these cancers. In addition, we identify master regulators that have not been previously reported and suggest their likely role. In summary, focusing on genes whose expression can be explained by their genomic and epigenomic aberrations is a promising strategy to identify master regulators of cancer.
1. Introduction
Technologies exist to rapidly and affordably profile the genome, epigenome and transcriptome of cancer. For example, advances in high throughput analysis allow quantification of global DNA variation, DNA methylation or RNA expression of biological samples (1–4). The current challenge is to integrate these layers of complex molecular biology information to produce a more comprehensive view of cancer (5–9). Successfully dealing with this complexity will allow determining how much each of the different types of genomic variations, i.e. mutation, copy number alteration or DNA methylation affect gene expression of key cancer drivers. Answering this question should provide a deeper understanding of cancer and insights on its initiation, progression and treatment response. Previous integration efforts have focused on how to distinguish driver genes from passenger genes. For example, Ciriello et al. developed a method to identify driver genes in glioblastoma based on mutual exclusivity by modeling copy number and mutation data (10). Vandin et al. developed a method to identify driver genes in cancer by focusing on pathways with a significant enrichment of approximately mutually exclusive genes (11). Several other investigators have identified driver genes through network analysis, such as Akavia et al. who used copy number data to filter potential regulators in a Bayesian module network analysis (12).
To identify master regulators of cancer and their targets, we built further on the network approach by filtering the candidate driver through a method that integrates copy number, DNA methylation, mutation and gene expression data. Our approach starts by generating a list of candidate driver genes based on the rationale that genes that are driven by multiple genomic events in a significant subset of samples are unlikely to be randomly deregulated. Examples of such genomic events for tumor suppressors are deletions, hyper-methylation or nonsense mutations. In the case of oncogenes, possible genomic or epigenomic events are amplification, hypo-methylation or a fusion with an active promoter region. Instead of using a statistical test on each genomic aberration separately, we developed a linear model that tests for concordance with three different types of genomic alterations simultaneously. We define these genes as candidate drivers because their expression can be significantly explained by the key mechanisms that drive oncogenesis: mutation, copy number alteration or DNA methylation. The second step of our algorithm selects the master regulators from these candidates and identifies their targets. This step applies a modified module networks analysis to computationally dissect the gene expression data into gene modules of co-expressed genes and assigning a regulatory program to each module (12–14). Our strategy has the advantage of using an informative way of selecting potential drivers and then focuses on those drivers that are likely to effect downstream targets.
We applied our algorithm to identify master regulators and their targets in glioblastoma multiforme and serous ovarian tumors from The Cancer Genome Atlas (TCGA). We found that the expression of the selected cancer drivers are greatly influenced by their copy number and to a much lesser extent by DNA methylation. In addition, for some drivers, we show synergy between genomic and epigenomic events. The second step of our algorithm selects a small set of potential cancer drivers as master regulators that explain much of the global gene expression in the reconstructed module network. Our results show that using candidate drivers from the first step improves the predictive performance on an independent test set of our models developed in the second step. This indicates that focusing on genes that are explained by their genomic and epigenomic profiles is a promising strategy to select master regulators of cancer.
2. Methods
2.1. Algorithm
We developed a biocomputational approach to identify key genes that drive human cancer. Our approach involves generating a list of candidate drivers (Step 1), followed by selecting the master regulators from the candidate drivers and their downstream targets (Step 2).
2.1.1. Step 1: Identifying candidate drivers of cancer
For a given gene to be considered a candidate cancer driver, we require that its gene expression be explained by its own genomic alterations, measured by its copy number, CpG DNA methylation and/or mutational variation. Our rationale is that cancer drivers whose expression can be explained by multiple genomic events are unlikely to be randomly deregulated. We used generalized linear models to predict the expression of each gene in terms of its own copy number, DNA methylation and mutation status. Our algorithm is initiated with a quality filter that removes copy number probes that are negatively correlated with gene expression and DNA methylation probes that are positively correlated with gene expression data. We reasoned that these probes have a higher chance of being associated with technical problems than a true underlying biological event. Next, we built a linear model to capture the effect of copy number, DNA methylation and mutation status on the expression level of a gene:
| (1) |
with βi the coefficients of the three predictors (i.e. CGH, DNA methylation or mutation status). We used sequential feature selection when adding multiple predictors by including a predictor only when it increases the R-square statistic more than expected by chance, based on the chi-square distribution with one degree of freedom. This model building procedure was wrapped inside a 10-fold cross validation loop (10F-CV) to estimate the generalization performance of the model on unseen data. We required that a predictor – e.g. CGH status – was selected in all cross validation iterations. The performance of the model was estimated using the R-square statistic on unseen data in each cross validation loop. We used several thresholds on the R-square statistic ranging from 0.2 to 0.5 and evaluated the number of genes at each threshold. We focused on genes with high R-square values since for these genes the expression is significantly explained by their copy number, methylation or mutation status. Within this set of genes we identified genes that are identified as a transcription factor. We used several external sources of information to define a gene as a transcription factor such as HPRD, a census of human transcription factors (15) and Gene Ontology resulting in a final list of 3964 transcription factors. This results in a list of candidate drivers that will serve as input for step 2.
2.1.2. Step 2: Identifying master regulators and their targets
The second step of our algorithm involves identifying the master regulators (as a key subset of cancer drivers from step 1) and determining their downstream targets by reconstructing a regulatory module network. Our module network approach builds upon previous work (13, 14). The algorithm is initiated by clustering the gene expression data into gene modules of co-expressed genes and then assigns a regulatory program to each module. The regulatory program of each module is defined by a sparse linear combination of driver genes that predict the module’s mean expression and are chosen from the list of transcription factors among the candidate drivers. The sparseness of the regulatory program is induced using elastic net regularization. We extended the module network framework in three ways: (a) first, we developed an approach to deal with auto-regulation, which is a situation where a regulator is selected in the regulatory program and is also a member of the same module. We allow this event to occur but relearn the regulatory program after removing the gene from the cluster. The regulator only stays in the regulatory program when it is also selected after removal of its expression in the module. (b) Second, we add a 10F-CV strategy that determines the regularization parameter for each module through minimization of the error. (c) Third, we use an iterative algorithm when adding regulators to the regulatory program by using the LARS-EN algorithm which has the advantage that it updates the elastic net solution sequentially (16) and thereby allows to stop adding regulators early.
After initial clustering of the data, the module network algorithm is run iteratively by learning the regulatory program and re-assigning genes to modules based on the updated regulatory program. Genes are reassigned to the module that they are closest to, based on Pearson correlation. We used k-means clustering with 100 clusters as the initial clustering algorithm. Next, our algorithm is run untill convergence corresponding to less than 1% of the genes being assigned to new modules. The module network is then interpreted using enrichment analysis using a hyper-geometric test to check for enrichment of gene sets in the gene modules to identify the key biological processes that are driven by the regulators. We used several databases of gene sets from MSigDB (17), GeneSetDB (18) and manually curated gene sets.
2.2. Data
We used data from The Cancer Genome Atlas (TCGA) on glioblastoma and ovarian cancer (data downloaded in May 2011). Gene symbols were used to map different technologies. Normal samples were removed. We used Level 3 Agilent G4502A gene expression data and Level 2 27K CpG methylation data. CpG sites were mapped to its closest gene transcription start site. The methylation probe level data was used since bi-modal signals were found for genes where multiple probes were present. Because averaging all probes for such a gene removed signal from the data, we defined methylation clusters based on a minimum Pearson correlation of 0.4 within a cluster. For the CGH data, two different platforms were used for the glioblastoma and ovarian project. For the glioblastoma project the CGH data was produced by the Agilent 244A platform and for the ovarian project the Agilent 1×1M platform was used. In both cases, we used the Level 3 CGH data. In the glioblastoma dataset, 251 patients had gene expression, DNA methylation and CGH data; these datasets were available for 511 ovarian cancer patients. For a limited number of patients, duplicate data was available however no averaging was done for these cases. We arbitrarily picked one case. When missing values were present, we estimated the missing value using 15-KNN (19). In most data sets a significant batch effect was observed and batch correction was done for all data sources using Combat (20). Mutation data was present for 140 glioblastoma patients and 324 ovarian patients through exome sequencing and we extracted all novel non-silent mutations. Gene expression data was present for 426 glioblastoma and 560 ovarian patients and used to generate the modules of the regulatory network. For all genes that had gene expression data, 14041 had also copy number, 9987 had DNA methylation and 8619 had both for ovarian cancer. For glioblastoma the overlap with gene expression data resulted in 13113 genes with copy number data, 9107 genes with DNA methylation data and 7510 with both.
3. Results
We used a two-step algorithm to identify the master regulators of cancer and their targets. We applied this algorithm on multi-dimensional TCGA ovarian cancer and glioblastoma datasets.
3.1. Identifying candidate drivers for glioblastoma and ovarian cancer
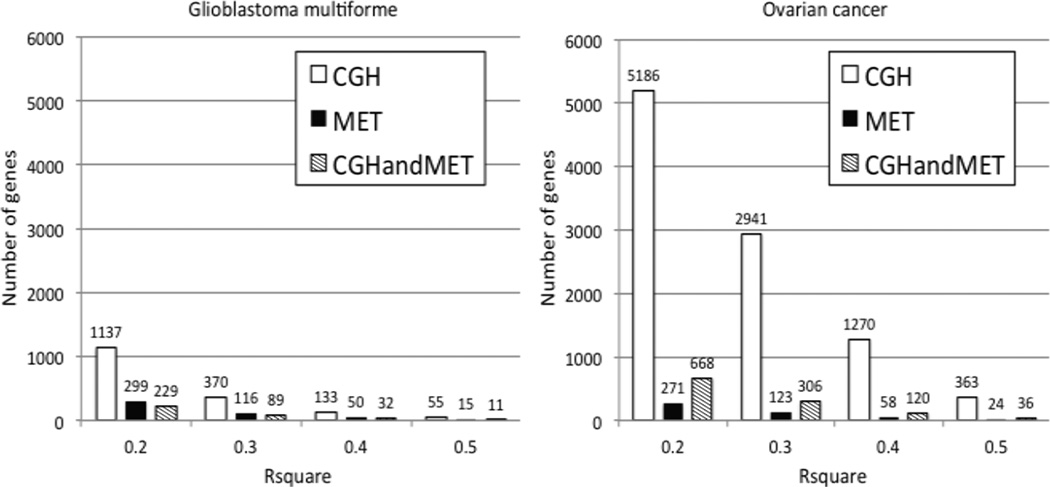
To identify candidate drivers of cancer, we developed a linear model to estimate the effect of copy number alterations, DNA methylation and mutation on gene expression levels (Step 1, Methods). Figure 1 shows the number of genes that is significantly explained by copy number, methylation or both at different R-square thresholds. More than 5000 genes have an R-square value for copy number alone of at least 0.20 in ovarian cancer compared to 1137 genes for glioblastoma reflecting the massive amount of copy number alterations that is present in serous ovarian cancer (21). Interestingly, DNA methylation is less informative when explaining gene expression data and much less genes are significant at each R-square threshold for both ovarian cancer and glioblastoma. For both glioblastoma and ovarian cancer, we found adding mutation data did not significantly change the results.
Figure 1.
Number of genes whose expression is significantly explained by its own copy number, DNA methylation or both.
3.1.1. Glioblastoma candidate drivers
When focusing on the genes with high R-square values in glioblastoma, we verified that our algorithm discovered a number of interesting genes previously reported and validated on TCGA glioblastoma (22). For example the gene PDGFRA, part of the platelet-derived growth factor receptor, has an R-square of 0.38 when considering only its copy number, 0.29 when considering only its DNA methylation profile and 0.60 when considering both. This indicates that 60% of the expression of PDGFRA is explained by synergy between its copy number and DNA methylation. PDGFRA is a receptor tyrosine kinase and an important part of the RAS pathway. In addition, PDGFRA is also mutated in 3 out of 140 patients (2%). Other interesting examples for glioblastoma include the genes MGMT and GLI1. MGMT, well known for its association with glioblastoma sensitivity to alkylating agents (23), has an R-square value of 0.46 and is significantly explained by its DNA methylation and copy number profile. Similarly, GLI1, glioma associated oncogene homolog 1, has an R-square value of 0.46 and is mutated in 1 patient.
3.1.2. Ovarian cancer candidate drivers
The ovarian cancer candidate drivers also included interesting genes with high R-square values. The gene BRCA1 is known to be associated with ovarian cancer due to mutations (21). In our analysis using copy number and DNA methylation data, we found that BRCA1 has an R-square of 0.10 when considering only its copy number, 0.41 when considering only its methylation profile and 0.49 when considering both. This indicates that besides mutation, DNA methylation is an important mechanism driving BRCA1 expression. This finding was also shown in the original TCGA ovarian results demonstrating that our method is able to recapitulate previous results (21). BRCA2 gene expression on the other hand is only explained by its copy number and is not significantly epigenetically regulated. Other interesting examples for ovarian cancer are KRAS, mutated in 2 out of 324 cases, with an R-square of 0.60 solely based on its copy number, and RAB25 with an R-square of 0.82 solely based on its DNA methylation. RAB25 was shown to be highest ranked gene epigenetically silenced in the original TCGA ovarian results and this is also the case using our model (21).
3.1.3. Gene set enrichment
We used several databases with gene sets to investigate the enrichment of known pathways and biological processes in the set of driver genes. We looked at gene set enrichment of the gene lists at an R-square of 0.3. The glioblastoma driver genes explained only by copy number were enriched in genes identified in the TCGA glioblastoma results as part of significant copy number changes (21). In addition, gene sets related to copy number changes in many other cancers were also in the top enriched gene sets (24–27). Interestingly the genes explained significantly by their DNA methylation at this R-square threshold were enriched in extracellular matrix genes and genes related to cell migration. For the ovarian cancer genes only explained by their copy number we observed enrichment of proliferation pathways, genes related to a BRCA1/CHEK1 network (28) and an ovarian cancer survival signature (29). Next, the top genes explained only by DNA methylation are enriched in genes affected by methylation in other cancers (30–32) validating our approach.
3.2. Identifying candidate master regulators of gene expression
To identify the master regulators of the network and their downstream targets, we apply a module network approach (Step 2, Methods). Our module network analysis is based on linear regression with elastic net regularization using the key transcription factors from the candidate drivers identified in Step 1 (13). For both glioblastoma and ovarian cancer we built a module network to associate transcriptional driver genes with their downstream targets. We used the gene expression data of 426 glioblastoma and 560 ovarian cancer patients but used only the top half most varying genes in all further analysis. As potential regulators for the modules, we selected genes with a high R-square as significantly regulated by a combination of copy number and DNA methylation and intersected this list with known transcription factors. This resulted in 431 and 469 genes for glioblastoma and ovarian cancer respectively that are defined as a transcription factor, show high variance and are regulated significantly by genomic alterations.
3.2.1. Glioblastoma module network
The master regulators in the glioblastoma network are TNFRSF1A, an important partner in the TNF and NF-kB pathway and ZNF300, both predicted as a regulator of 10 modules. We focused on the top DNA repair module in our network because this process is an important pathway in both glioblastoma and ovarian cancer. In the glioblastoma module network, module 22 is the top DNA repair module and is regulated by 6 regulators including DNMT1 and PARP1 and contains 81 genes. DNMT1 is a key player in regulation DNA methylation regulation and has been shown to be involved in inactivation of tumor suppressor genes and failure to maintain genomic stability (33). In addition PARP1 is known to regulate DNMT1 and forms a complex with DNMT1 (34, 35). Both are only explained by their copy number profile and are not driven by their own DNA methylation status.
3.2.2. Ovarian cancer module network
The master regulator for the ovarian cancer module network is BATF a transcription factor with unknown function. BATF is part of the regulatory program of 13 modules and its expression is significantly explained by its DNA methylation status. Other important regulators are NFKB1 and TGFB3. Similarly to the glioblastoma module network, we also focused on the most highly enriched DNA repair module for ovarian cancer: module 89. Module 89 contains 86 genes and has 10 regulators including EZH2, AURKA and CHAF1B. Interestingly CHAF1B was also predicted as a regulator of the top DNA repair module in glioblastoma. Other interesting regulators are CCNE1 and RAB25. CCNE was identified as a low frequency amplification in the original TCGA results and is predicted as the only regulator of a module enriched in the focal adhesion pathway. Next, RAB25 is the top methylated gene (21) and in our module network is part of the regulatory program together with MAML2, a member of the NOTCH pathway, a pathway also identified in the original ovarian TCGA results.
3.3. Algorithm Performance
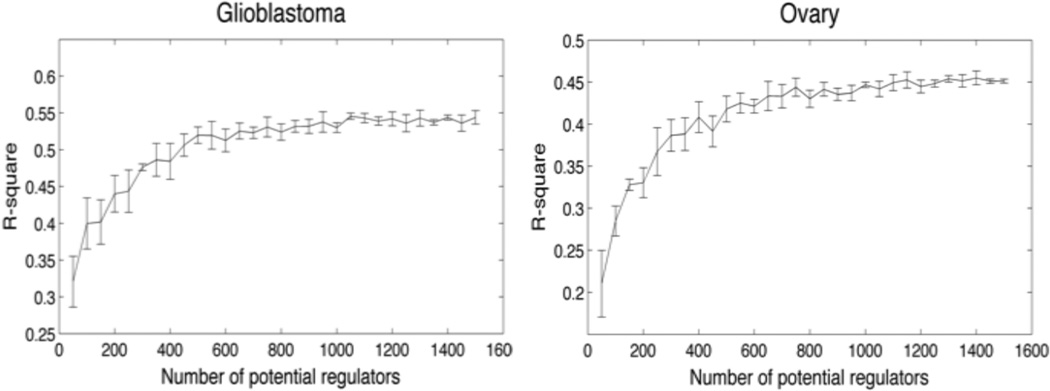
To evaluate the performance of our algorithm, we investigated how well our candidate drivers perform compared to random sets of transcription factors. We used an independent test set for both glioblastoma multiforme and ovarian cancer (9, 36) to estimate the generalized performance of each module on unseen data. To limit the computational power required and to facilitate comparison of the results, we ran the second step of our algorithm only once. This essentially corresponded to learning a regulatory program for the initial clustering. First, we established a baseline performance by incrementally and randomly adding transcription factors to the list of potential regulators. This was repeated 5 times for each number of potential transcription factors. Figure 2 shows how the performance evolves when adding more transcription factors. The performance is measured by averaging the R-square over all modules on the test set. Figure 2 shows that for glioblastoma and ovarian cancer, the performance plateaus after adding more than 600 potential regulators indicating that increasing the number of potential regulators beyond this point does not improve the predictive performance of the model on unseen data.
Figure 2.
Generalized performance of module networks generated from randomly selected candidate drivers.
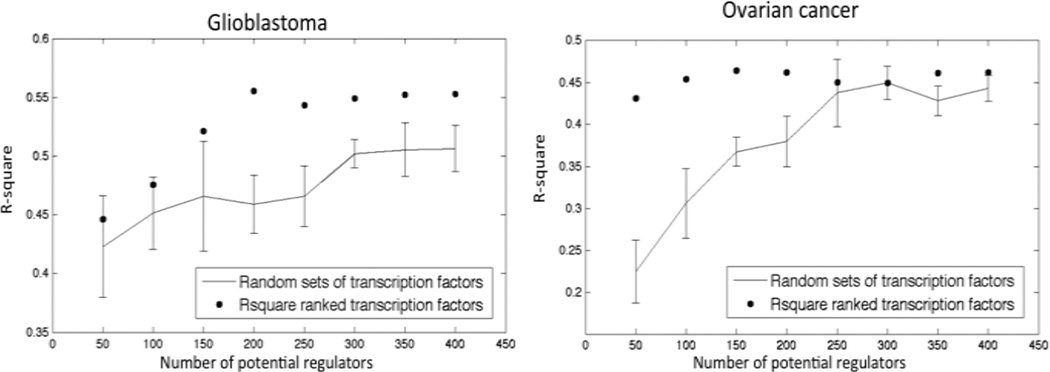
Finally, we investigated the performance of using transcription factors that are also driver genes. We reasoned that focusing only on candidate drivers as regulators would increase the performance of only a subset of modules and therefore focused on modules with a minimum R-square value on unseen data and compared the performance of these modules with random sets of transcription factors. For glioblastoma we saw an increase in performance independent of R-square threshold while for ovarian cancer the performance improved starting at a minimum R-square of 0.10. Figure 3 shows the average performance of modules with an R-square value of at least 0.20 on unseen data when adding incrementally regulators ranked by their own R-square value. Our results show for both glioblastoma and ovarian cancer that the generalization performance is comparable or better than random sets of transcription factors at several sizes of potential regulators (Figure 3).
Figure 3.
R-square performance of the module networks generated using candidate drivers vs. randomly drawn candidate drivers.
4. Discussion
To identify master regulators of gene regulation in cancer, we developed a biocomputational approach that first creates a select list of candidate cancer driver genes by integrating multiple genomic datasets. From this list, we select the master regulators and identify their targets when reconstructing a regulatory module network of cancer. For a gene to be on the list, we require that its expression be explained by known genomic and/or epigenomic aberrations, measured in terms of copy number variation, DNA methylation and mutational events. This requirement reduces the list of candidate drivers and improves the performance of the regulatory module network when applied to glioblastoma and ovarian cancer TCGA data.
For each candidate driver, we can determine which genomic aberration explains more of the gene expression. In the case of both GBM and ovarian cancer, the candidate drivers appear to be more influenced by their copy number variations than DNA methylation. DNA methylation appears to have a more subtle effect on gene expression. In addition, we identified many genes that showed synergy between their copy number and DNA methylation showing that a cancer cell can deregulate gene expression using both mechanisms. Besides copy number and DNA methylation, we also investigated the addition of mutation data to our linear model and investigated if mutation data has a significant effect on the amount of variance in gene expression that can be explained. As expected adding mutation data did not significantly change the results due to sparseness of mutation data. More importantly, only a subset of mutations will have an effect on gene expression because many missense mutations will not effect gene expression but may disrupt protein function. Determining this computationally requires dedicated methods that specifically model the mutation data and their impact on the final protein product to estimate which mutations have or do not have an effect on gene expression. For example, we observed that TP53 was not correlated with gene expression even though it is known to be an important tumor suppressor in ovarian cancer.
By focusing on candidate drivers as genes that are explained by their genomic and epigenomic profiles, we can identify more likely master regulators in the module networks analysis. We found that using transcription factors whose expression is determined by copy number or DNA methylation profile, had favorable performance on unseen data. In the context of the module networks generated from random sets of transcription factors, which were shown to plateau after adding more than 600 potential regulators, while our method provides a way of intelligently selecting regulators in module networks.
By the virtue of applying module network analysis, the master regulators are associated with downstream targets. The master regulators in both the glioblastoma and ovarian cancer network belong to known pathways affected in these cancers. In addition, we found several unknown genes that are important regulators in our module networks. For example, CHAF1B is predicted as a regulator for the top DNA repair module in both glioblastoma and ovarian cancer. CHAF1B is predicted to have a function in DNA repair and is part of a 4-gene signature predicting survival in glioma (37). Moreover, CHAF1B has been shown to be correlated to proliferation in several epithelial cancers (38). Interestingly we found that CHAF1B expression is dominated by its copy number in ovarian cancer and by DNA methylation in glioblastoma, showing the flexibility of our method. As more data comes available in the TCGA, such inter-cancer comparisons can be made with the potential to identify master regulators independent of cancer subtypes.
In summary, we developed a biocomputational approach for integrating multi-dimensional cancer data that allows to study how genomic and epigenomic features influence gene expression. Next, we used our method to identify master regulators of cancer and their downstream targets. Our approach has the potential to provide new insights in the molecular biology underlying cancer. Moreover, it associates drivers with their downstream targets, thereby enabling new insight into the biological mechanism underlying cancer progression.
Table 1.
Master regulators for the ovarian and GBM network. Genes highlighted in the main text are in bold.
| Glioblastoma network | Ovarian network | ||
|---|---|---|---|
| Regulators | Nr Modules | Regulators | Nr Modules |
| ZNF300 | 10 | BATF | 13 |
| TNFRSF1A | 10 | HTATIP2 | 9 |
| PTRF | 8 | PML | 9 |
| WWTR1 | 8 | NOD2 | 8 |
| MYT1 | 7 | JAK2 | 8 |
| PYCARD | 7 | HMGA2 | 7 |
| PATZ1 | 7 | TGFB3 | 7 |
| BASP1 | 6 | KLF12 | 7 |
| RAB32 | 6 | AKAP8L | 7 |
| SATB1 | 6 | YWHAH | 6 |
| ZMYND12 | 6 | HLA-DQB1 | 5 |
| CDC45 | 6 | JARID2 | 5 |
| ZNF217 | 6 | RNF19A | 5 |
| KCNIP3 | 6 | MORF4L1 | 5 |
| ARNT2 | 6 | SMAD4 | 5 |
| BTF3L4 | 6 | ZNF500 | 5 |
| POGZ | 6 | NFKB1 | 5 |
| TOB1 | 6 | TRIM29 | 4 |
| LGALS3 | 5 | SPDEF | 4 |
| KCNH8 | 5 | SREBF1 | 4 |
Acknowledgements
This research was supported by the Center for Cancer Systems Biology (CCSB) at Stanford (U54 CA149145). OG is a fellow of the Fund for Scientific Research Flanders (FWO-Vlaanderen), an Honorary Fulbright Scholar of the Commission for Educational Exchange between the United States of America, Belgium and Luxembourg, and a Henri Benedictus Fellow of the King Bauduin Foundation and the Belgian American Educational Foundation (BAEF).
Contributor Information
Olivier Gevaert, Radiology, Stanford, 1201 Welch Road, Stanford, CA 94305, olivier.gevaert@stanford.edu.
Sylvia Plevritis, Radiology, Stanford, 1201 Welch Road, Stanford, CA 94305, Sylvia.plevritis@stanford.edu.
References
- 1.Pao W, et al. Clin Cancer Res. 2009;15:5317. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C. Annals of Oncology. 2009;20:10. [Google Scholar]
- 3.Gevaert O, De Moor B. Expert Opinion on Medical Diagnostics. 2009;3:157. doi: 10.1517/17530050802680172. [DOI] [PubMed] [Google Scholar]
- 4.Gevaert O, Daemen A, De Moor B, Libbrecht L. BMC Med Genomics. 2009;2:69. doi: 10.1186/1755-8794-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin L, Hahn WC, Getz G, Meyerson M. Genes & development. 2011 Mar 15;25:534. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevaert O, Van Vooren S, De Moor B. Annals of the New York Academy of Sciences. 2007;1115:240. doi: 10.1196/annals.1407.002. [DOI] [PubMed] [Google Scholar]
- 7.Daemen A, Signoretto M, Gevaert O, Suykens JA, De Moor B. PLoS ONE. 2010;5:e10225. doi: 10.1371/journal.pone.0010225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leunen K, et al. Human mutation. 2009;30:1693. doi: 10.1002/humu.21135. [DOI] [PubMed] [Google Scholar]
- 9.Gravendeel L, et al. Cancer research. 2009;69:9065. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 10.Ciriello G, Cerami E, Sander C, Schultz N. Genome Res. 2011 Oct 12; doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandin F, Upfal E, Raphael BJ. Genome Res. 2011 Jul 11; doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akavia UD, et al. Cell. 2010 Dec 10;143:1005. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S-I, et al. PLoS genetics. 2009;5 doi: 10.1371/journal.pgen.1000358. e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal E, et al. Nature Genetics. 2003;34:166. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 15.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. Nat Rev Genet. 2009 Apr;10:252. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 16.Zou H, Hastie T. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2005;67:301. [Google Scholar]
- 17.Subramanian A, et al. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culhane AC, et al. Nucleic Acids Res. 2010 Jan;38:D716. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troyanskaya O, et al. Bioinformatics. 2001;17:520. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 20.Johnson WE, Li C, Rabinovic A. Biostatistics. 2007 Jan;8:118. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 21.Bell D, et al. Nature. 2011 Jun 30;474:609. [Google Scholar]
- 22.McLendon R, et al. Nature. 2008;455:1061. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteller M, et al. N Engl J Med. 2000 Nov 9;343:1350. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 24.Lin WM, et al. Cancer Res. 2008 Feb 1;68:664. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwood WW, et al. Oncogene. 2008 Jul 31;27:4615. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greshock J, et al. Cancer Res. 2007 Apr 15;67:3594. doi: 10.1158/0008-5472.CAN-06-3674. [DOI] [PubMed] [Google Scholar]
- 27.Osman I, et al. Clin Cancer Res. 2006 Jun 1;12:3374. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 28.Pujana M, et al. Nature Genetics. 2007;39:1338. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 29.Bonome T, et al. Cancer Res. 2008 Jul 1;68:5478. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heller G, et al. Cancer Res. 2008 Jan 1;68:44. doi: 10.1158/0008-5472.CAN-07-2531. [DOI] [PubMed] [Google Scholar]
- 31.Acevedo LG, Bieda M, Green R, Farnham PJ. Cancer Res. 2008 Apr 15;68:2641. doi: 10.1158/0008-5472.CAN-07-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato N, et al. Cancer Res. 2003 Jul 1;63:3735. [PubMed] [Google Scholar]
- 33.Rajendran G, et al. J Neurooncol. 2011 Sep;104:483. [Google Scholar]
- 34.Zampieri M, et al. Biochem J. 2012 Jan 15;441:645. doi: 10.1042/BJ20111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zampieri M, et al. PLoS ONE. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tothill RW, et al. Clin Cancer Res. 2008 Aug 15;14:5198. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 37.de Tayrac M, et al. Clin Cancer Res. 2011 Jan 15;17:317. doi: 10.1158/1078-0432.CCR-10-1126. [DOI] [PubMed] [Google Scholar]
- 38.Polo SE, et al. Histopathology. 2010 Nov;57:716. doi: 10.1111/j.1365-2559.2010.03681.x. [DOI] [PubMed] [Google Scholar]