Abstract
Leveraging TCGA’s multi-dimensional data in glioblastoma (GBM), we inferred the putative regulatory network between microRNA and mRNA using the Context-Likelihood-Relatedness (1) modeling algorithm. Interrogation of the network in context of defined molecular subtypes identified 8 microRNAs with a strong discriminatory potential between proneural and mesenchymal subtypes. Integrative in silico analyses, functional genetic screen and experimental validation identified miR-34a as a tumor suppressor in proneural subtype GBM. Mechanistically, in addition to its direct regulation of PDGFRA, promoter enrichment analysis of CLR-inferred mRNA nodes established miR-34a as a novel regulator of a Smad4 transcriptional network. Clinically, miR-34a expression level is shown to be prognostic, where miR-34a low-expressing GBMs exhibited better overall survival. This work illustrates the potential of comprehensive multi-dimensional cancer genomic data combined with computational and experimental models in enabling mechanistic exploration of relationships among different genetic elements across the genome space in cancer.
Keywords: Brain/central nervous system cancers, new algorithms
INTRODUCTION
Glioblastoma (GBM; World Health Organization IV) is the most common primary brain tumors in adults. Patients with newly diagnosed GBM have a median survival of 12 months with generally poor responses to chemo-radiotherapy (2). Recent genome-wide profiling studies have shown extensive genetic heterogeneity among GBM samples with distinct molecular subtypes, that these transcriptomic subtypes reflect distinct underlying biology is supported by observed difference in clinical outcome of the patients (3, 4), enrichment of different genomic and epigenetic alterations within each subtypes and differential activation of major signaling pathways (5, 6).
Non-coding RNAs have emerged as an important class of regulatory molecules in both normal and neoplastic development. MicroRNAs (or miRs) are a class of non-coding small RNAs produced by RNA polymerase II as hairpins of longer precursor RNAs that are subsequently processed to approximately 22-nt-long fragments by RNase III enzymes, Drosha and Dicer. Mature miRs regulate gene expression by promoting mRNA degradation or by inhibiting mRNA translation (7, 8). The connection between miRs and cancer was first implicated by their genomic alteration and dysregulated expression in various human tumors (9–13). Multiple miRs have since been identified to promote or suppress oncogenesis in various tumors, presumably by modulating gene expression in the oncogenic and tumor suppressor networks. Additionally, recent studies have proposed new mechanisms of miR/mRNA regulation such as modulation of mRNA with competitive miR binding sites (sponge interactions) or mRNA that affect constituents of the miR regulatory machinery (non-sponge interactions) (14–16).
Global views of the relationship between miR and mRNA expression has been reported. For instance, Su et al. used integrative genomics and genetic techniques to characterize the roles of mouse miRs within the mouse liver miR-mRNA network (17); Dong et al. deciphered the pathway connecting mutations under the GBM miR-mRNA expression network (18); Mestdagh et al. established the miR body map online resource to dissect miR function through integrative genomics (19); Grigoryev et al. presented the genome-wide miR regulation of T lymphocyte activation through the mapped miR, mRNA and protein networks (20); Sharbati et al. studied the macrophages infections via an integrated miR-mRNA network (21). In these global integrative miR-mRNA network analyses, either general correlation coefficient methods (17–19) or putative miR target prediction methods (20, 21) have been used to construct or map miR-mRNA connections. These approaches preferentially quantify linear dependencies between pair-wise variables.
Recognizing that the functional relationship between two variables in cancer is not necessarily linear, we explored a Mutual Information (MI) based approach in this work that scores miR/mRNA interaction strength based on relevant expression contexts. This MI-based approach has been used recently by Sumazin et al. to uncover a novel class of modulators of miR/mRNA interactions (14). Here, we applied Context Likelihood of Relatedness (CLR) network modeling algorithm to generate pair-wise measures of associations based on MI through calculation of the entropy. At the heart of the CLR algorithm is a unique statistical background correction test which utilizes the full set of MI values to estimate a significance value for each miR-mRNA pair under a given observed network context (see Supplementary Information). The algorithm evaluates the MI value of a miR:mRNA pair against the background MI distribution of all mRNAs in the data set with the miR, as well as with the distribution of MI values of all miRs with the mRNA under consideration. A combined Z-score summarizing these two comparisons is generated and the list of all such pairwise Z-scores is subsequently used to generate p-values by comparing to the normal distribution. A stringent FDR of 5% is finally applied to identify putative miR:mRNA regulatory edges. Interactions whose MI values are outliers in the right tails of the pertinent context background distributions of MI scores have the greatest likelihood of being identified as significant. This background correction method allows the CLR algorithm to filter out those edges between miRs and mRNA that have spurious similarities with large numbers of other gene/miR species.
The Cancer Genome Atlas (TCGA) has characterized the genomes of GBM on multiple dimensions including coding and non-coding RNAs (22). We applied the CLR modeling algorithm to this multi-dimensional dataset to infer putative regulatory relationships (edges) between miRs and mRNAs in GBM. Specifically, we were interested in directional miR:mRNA interactions where the miR downregulates mRNA expression either directly through binding or indirectly through intermediary effectors. Against this global network, we explored the functional relationship between miRs and mRNAs in gliomagenesis.
RESULTS
Global miR and mRNA regulatory network in GBM
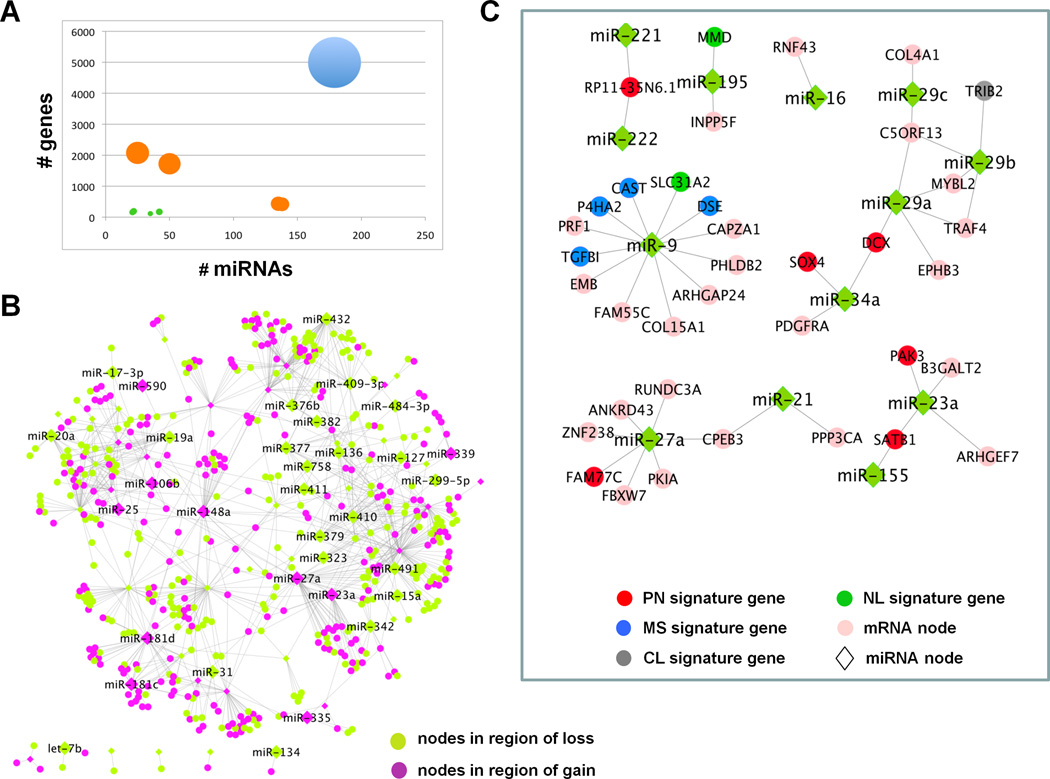
To explore the relationship between miRs and mRNAs, we applied the CLR network algorithm to miR and mRNA transcriptome data from 290 GBM samples from TCGA (Supp. Table 1). A total of 26,297 edges between 254 miR and 6,152 mRNA nodes were defined (Supp Table 2). Next, we integrated genome-wide copy number profiles with the inferred edges of the network with the assumption that biologically relevant miRs or mRNAs are likely to show additional levels of dysregulation across the samples. Here, we found that a third (34.1%) of the miR nodes or mRNA nodes in the CLR network resided in regions of copy number aberration in GBM, and 3.9% involved both miR and mRNA that are localized in regions of genomic alterations (Figure 1A–B). This integrative analysis thus prioritized 1,018 edges involving 69 miRs and 467 mRNAs nodes as candidates with likely biological importance.
Figure 1. Integrative analyses of CLR network in context of copy number alterations and direct target prediction.
(A) CLR network grouped by connectivity among miRs and mRNAs in regions of copy number aberration. Size of the node represents the number of edges between the miR and mRNAs in each group. There are 9 sub-networks represented. Edges in the network where both participating miR and mRNA are in regions of copy number alterations account for less than 3.9% of the total connectivity (green circles). Edges where either the participating miR or the mRNA is in region of gain or loss, but not both, account for 34.1% of the network (orange circles). Approximately 62% of the network involves miRs and mRNAs without evidence of genomic alterations in GBM (blue circle). (B) Network of CLR edges where both miR (diamonds) and mRNA (circles) are in regions of copy number gain (purple nodes) or loss (green nodes). (C) Putative direct target subnetworks among CLR-identified edges in GBM. miRs are represented as green diamonds and genes as circles; classical signature genes (grey), neural signature genes (dark green), proneural signature genes (red), mesenchymal signature genes (blue), genes not in any of the molecular subtype signatures (pink). Abbreviations: PN= Proneural, MS=Mesenchymal, CL=Classical, NL=Neural.
To further rank the above nodes, we next applied a putative-direct-target filter. Specifically, based on previous findings showing that miRs down-regulate their target mRNAs by binding to their 3’ UTR regions (7), we defined a subset of mRNA nodes as putative direct targets of their miR nodes based on the following parameters: i) a significant negative correlation (Pearson correlation coefficient ≤ −0.3) between expression of miR and mRNA; and ii) sequence-based prediction of interaction in all three sequence-based prediction databases, namely Pictar, TargetScan and Miranda (23–27). This in silico analysis identified 3 edges involving 6 of the 536 prioritized nodes as representing putative direct interactions, namely miR-34a:PDGFRA; mir-27a:CPEB3; miR-23a:ARHGEF7). The same trend is observed with the global CLR network edges, where 45 (0.17%) of the 26,297 CLR-inferred edges were predicted computationally to be direct (Figure 1C, Supp. Table 3). This suggests that a significant proportion of the putative miR-mRNA relationships may be indirect, possibly mediated via intermediaries such as transcription factors (see below).
Functional analyses of subtype-discriminant miR nodes
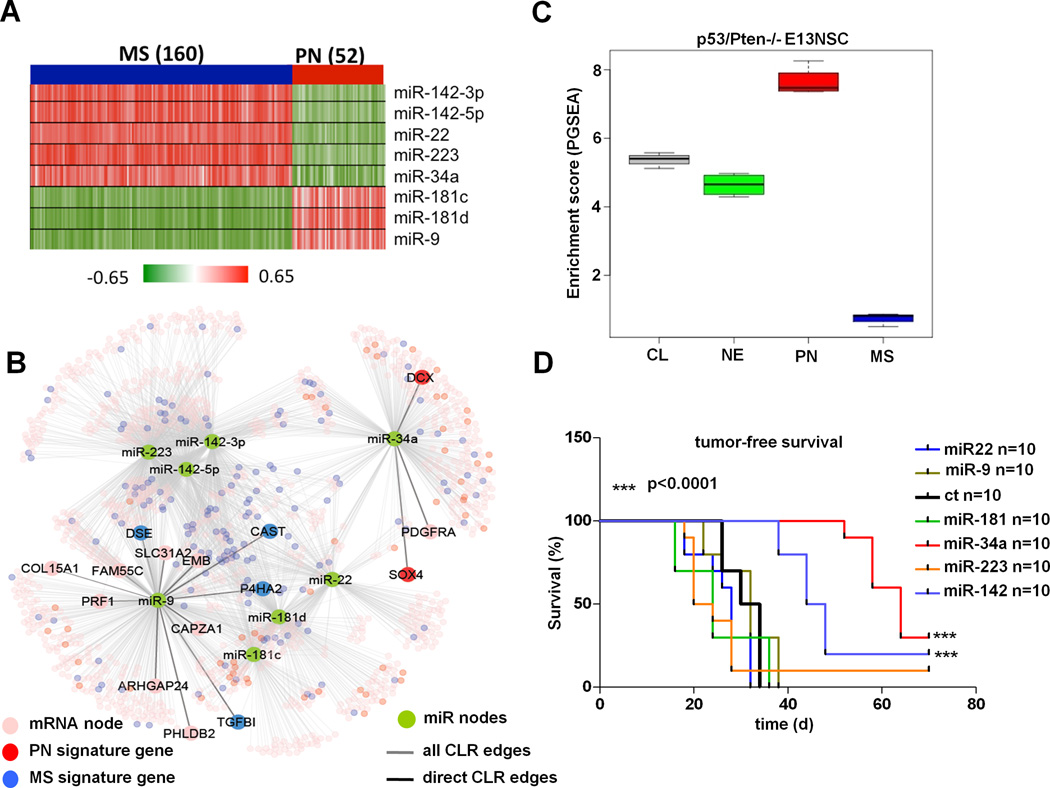
Next, we asked whether the CLR-inferred global network captures the salient transcriptomic features of the four molecular subtypes of GBM. Here, we looked for edges that are unique to each molecular subtype (Supp. Table 4) as well as differential expression of the miR and mRNA nodes between any two molecular subtypes. We found that the variability among molecular subtypes appeared to be the predominant driver of relationships defined by CLR (Supp. Figure 1). For example, 67% (n=17,934) of the network edges involved miR and mRNA that are differentially expressed (p<0.001) between two molecular subtypes. While the difference in expression between the subtype signature genes is not surprising, it is striking that the CLR-identified miRs associated with these genes should show a reciprocal and opposite change of expression along with their mRNA nodes. In particular, the greatest transcriptomic shift was observed between proneural (PN) and mesenchymal (MS) subtypes with nearly half of the edges in the global network (or 12,673 edges, 48%) marked by expression differences between them. This observation suggested that miR regulation of mRNA may play a role in defining the molecular signatures of these two subtypes. To this end, we looked for CLR-inferred edges among the 685 significantly overexpressed genes used by Verhaak et al. for subtype pathway/GO enrichment analysis (3). Of these 685 genes, 506 of them (73%) have inferred edges to a miR node in the global CLR network. Conversely, of the 2,984 inferred edges to these 506 subtype-classifier genes, a disproportionate number (70%) are part of either the PN or MS subtype signatures (e.g. 328 to classical, 560 to neural, 858 to PN and 1,238 to MS signature genes), suggesting that miR-mRNA regulation may contribute to gene signatures underlying these two molecular subtypes. Indeed, eight miR nodes (p<0.001) were found to be highly discriminatory between PN and MS subtypes (Figure 2A) (see Supp. Methods). Five of these miR nodes (miR-22; miR-34a; miR-223; miR-142-3p and miR-142-5p) are under-expressed in PN-subtype GBMs, harboring inferred negative edges with PN signature genes; conversely, three of them (miR-9, miR-181c and miR-181d) are under-expressed in MS-subtype GBM with inferred negative edges with MS signature genes (Figure 2B). When integrated with copy number and expression profiles as well as putative direct-target prediction as above (Supp. Table 5), we found that miR-34a is the only one that also resides in a region of frequent loss and harbors a putative direct CLR-edge to PDGFRA, a well-known proneural signature gene that is also a target of genomic amplification.
Figure 2. Discriminatory miRs between PN and MS subtypes.
(A) Heatmap of correlations between 8 most discriminative miRs with PN and MS signature genes in the CLR network. Each cell in the heatmap is the correlation between the corresponding miR and signature gene across samples. (B) CLR network around the 8 miRs with putative direct targets highlighted. (C) Gene set enrichment analysis of pre-malignant spheroid cultures derived from p53/Pten−/− E13 mouse embryos (p53L/L;PtenL/L;Gfap-Cre). For any subtype signature gene list, the enrichment of the subtype in a given sample is calculated by comparing the average expression of the signature genes to the overall mean using a t-statistic. Each box-plot in the figure encapsulates the distribution of these subtype enrichment scores across all samples for the individual subtypes. A clear enrichment is observed for PN signature genes (p= 3.66 × 10−12, n=4). (D) Kaplan-Meier survival analysis of Ncr/nude mice transplanted sub-cutaneously with pre-malignant PN-like p53/Pten−/− neural stem cells transduced with 6 pre-miRs corresponding to 8 discriminant miRs or the GFP control (n=10 per group). miR-34a and miR-142 potently impaired tumorigenesis when compared with GFP control (*** p<0.0001). Abbreviations: PN=Proneural, MS=Mesenchymal, CL=Classical, NL=Neural, NSC=neural stem cell.
Next, we sought evidence that these 8 miRs are functionally active in a proneural context. Recognizing the limitation of established cell systems in modeling PN molecular subtype, we first investigated whether a genetically-engineered mouse (GEM) model of GBM constructed with concomitant p53 and Pten deletion in neural stem cells and neural progenitors (Gfap-Cre;p53L/L;PtenL/+), can be considered an appropriate model of PN GBM (28). Here, we performed gene set enrichment-analysis (PGSEA) (29) of the transcriptomes of pre-malignant p53/Pten double-null E13 embryonic neural stem cells (NSC) (Gfap-Cre;p53L/L;PtenL/L) and tumor spheroids (TS) isolated from malignant gliomas developed in the context of p53 and Pten deficiency (Gfap-Cre;p53L/L;PtenL/+) (Supp. Figure 2A). As shown in Figure 2C and Supp. Figure 2B, significant enrichments for the PN signature genes were observed in both pre-malignant embryonic neural stem cells and in malignant tumor spheroids (p= 3.66 × 10−12 and p=2.2 × 10−16, respectively), thus substantiating the p53/Pten double-null model as a PN model. Next, we enforced expression of the miR precursors corresponding to the 8 subtype discriminant miRs in pre-malignant p53/Pten−/−NSC (Supp. Figure 3). As summarized in Figure 2D, both miR-34a and miR-142 significantly inhibited tumorigenesis of p53/Pten−/−E13 NSC in vivo, suggesting tumor suppressive activity in this context. In summary, subtype-discriminant miRs are functionally active in proneural GBM context.
miR-34a expression is prognostic in GBM
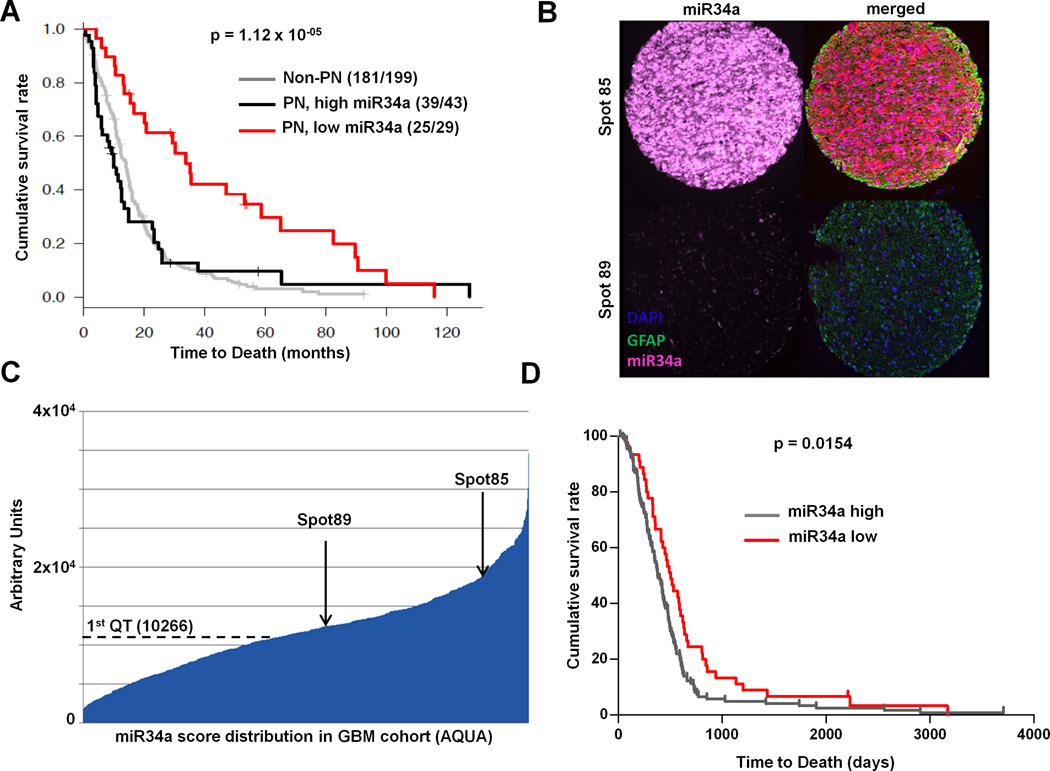
To explore the potential clinical relevance of subtype-discriminant miRs in human GBM, we asked whether their expression tracks with any clinical parameters, particularly prognosis (see Methods). As summarized in Supplementary Table 6, when dichotomized, miR-34a expression was the only one of the 8 miRs that showed significant prognostic correlation (Bonferroni-adjusted p value = 0.0047) in the TCGA dataset. Specifically, patients with miR-34a low-expressing GBMs exhibited an overall improved survival (Figure3A). Moreover, miR-34a low-expression did not simply identify the proneural-subtype which is known to have better outcome (3, 4). Instead, within the PN-subtype, low miR-34a expression stratifies a subgroup of PN GBM patients with improved overall survival (Hazard Ratio = 2.2, p= 2.2E-05), compared to miR-34a high-expressing PN patients or other non-PN patients (Supp. Table 6); moreover the prognostic significance of miR-34a was independent from other clinical variables such as therapy and gender or batches in multivariate analyses (Supp. Table 7).
Figure 3. miR-34a levels are prognostic in Glioblastoma.
(A) Kaplan-Meier survival analysis of patients in the TCGA dataset (n=271). Low expression of miR-34a stratifies a subgroup of PN GBM patients with a significant survival advantage compared to others in TCGA cohort (log-rank p=1.12 × 10−5). (B) GBM Tissue Microarray (TMA). Representative spots showing high and low expressors. A LNA modified probe for miR-34a and and an antibody specific for Glial Fibrillar Acidic Protein (GFAP) were hybridized to the arrays. (C) AQUA score distribution across the GBM cohort. Arrows indicate the AQUA score for the samples shown in figure 3B. (D) Kaplan-Meier survival analysis of patients in the independent training cohort (n=220). Subjects were stratified in high and low miR-34a expressors based on the quantile expression assessed by the AQUA score. 1st (lower) quartile of the AQUA score distribution was used as cut-off to assign patients to the low or high expressors groups. “miR-34a low” patients show a significant better outcome when compared to “high expressors” (p=0.0154).
Importantly, this prognostic correlation of miR-34a was recapitulated in an independent cohort of human GBM. Briefly, we performed quantitative in situ hybridization using miR-34a as probe on a Tissue Microarrays (TMA) from an independent cohort of human gliomas (Figure 3B–C, Supp. Figure 4, Supp. Table 8–9) and showed that low miR-34a expressors were indeed associated with better overall survival (p = 0.0154) (Figure 3D).
miR-34a is a tumor suppressor in proneural GBM
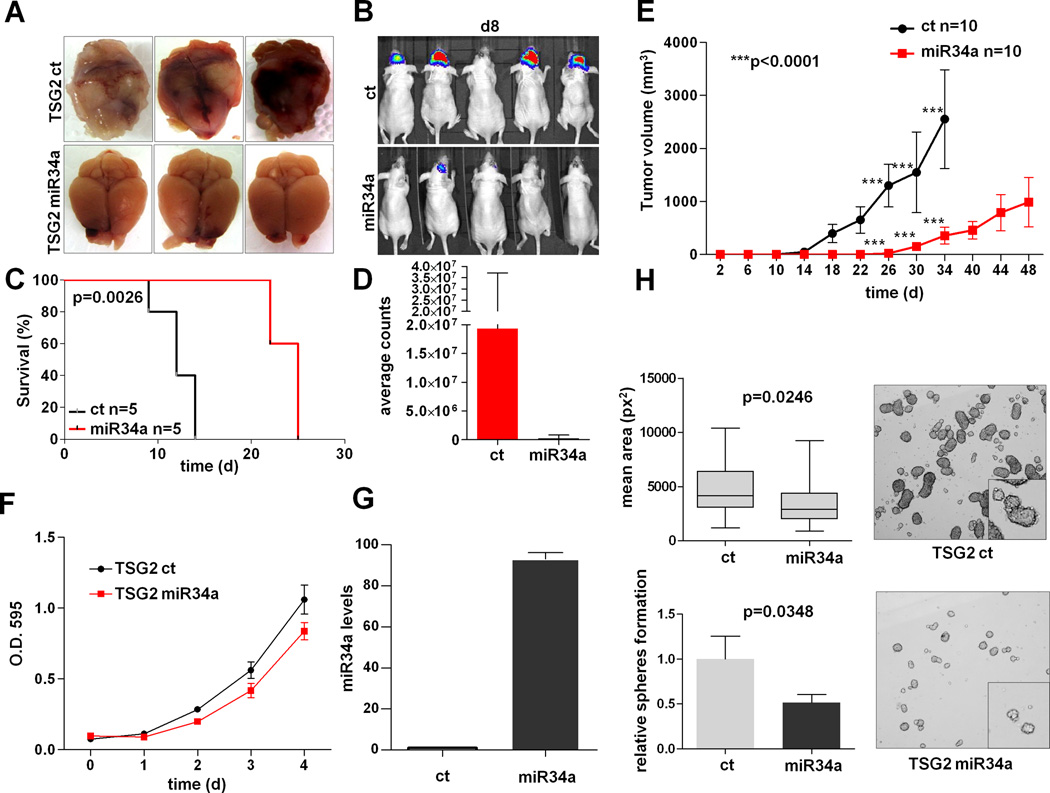
Its potent tumor suppressive activity in vivo, evidence of genomic alteration and its putative direct mRNA target PDGFRA, coupled with its prognostic significance, together strongly nominated miR-34a for mechanistic studies. Although it has been shown to be a tumor suppressor in multiple human cancer types including GBM (30, 31), a specific role in proneural GBM has not been fully demonstrated. Similar to the pattern of lower expression in human PN subtype GBM in human (p=1.92e-19) (Supp. Figure 5A), miR-34a expression was lower in p53/Pten double-null TS or NSC compared to normal neural stem cells (p=0.0034 and p=0.0036 respectively) (Supp. Figure 5B), moreover the acute deletion of p53 and Pten by ex-vivo CRE expression in early passage primary astrocytes results in significant down-regulation of endogenous miR-34a (p=0.002) (Supp. Figure 5C–D). Functionally, in TSG2, a tumor spheroid culture derived from a murine p53/Pten−/− GBM, miR-34a expression significantly inhibited in vivo tumorigenesis in both orthotopic xenografts (p=0.0026) and in subcutaneous transplants (p<0.0001) (Figure 4A–E), resulting in prolonged survival of tumor-bearing mice and a less aggressive growth pattern (Supp. Figure 6A). While potently inhibiting tumorigenesis in vivo, it was interesting that miR-34a over-expression did not impact significantly on cell proliferation in 2D culture experiments (Figure 4F–G). While it is possible that the lack of growth phenotype may reflect the possibility that cells under the selective pressure of miR-34a overexpression may undergo negative selection in vitro, as observed in vivo in tumors arising from cells stably transduced with a miR-34a lentivector (Supp. Figure 6B), the phenotype of significant impairment of spherogenic potency in both human and mouse tumor neurospheres implicates renewal potential as phenotype impacted by miR34a expression. In particular, spherogenic renewal of murine TSG2 cultured in neural stem cell medium in presence of FGF and EGF was inhibited by miR34a expression (Figure 4H). Similarly, this phenotype was recapitulated in TS543, a human proneural-like GBM spheroid showing concomitant amplification and over-expression of PDGFRA, upon miR-34a over-expression (Supp. Figure 7A–B).
Figure 4. miR-34a regulates self-renewal and tumorigenesis in PN GBM.
TSG2 cells, a PN-like malignant cell line derived from a p53L/L;PtenL/+;Gfap-Cre mouse glioma, were co-infected with a luciferase reporter and with lentiviral particles harboring the miR-34a precursor or the control vector and injected orthotopically (frontal lobe) in Ncr/nude mice. (A) Macroscopic appearance of mouse brains injected with p53/Pten−/− GBM cells harbouring miR-34a or the vector control isolated from terminally ill mice. Re-introduction of miR-34a results in poorly invasive tumors confined to the frontal lobe (lower panels). (B) Bioluminescence images of Ncr-nude mice transplanted orthotopically with p53/Pten−/− murine GBM cells harboring a luciferase construct along with miR-34a or the vector control. Images taken 8 days after surgery show a significantly higher tumor burden in control mice. (C) Kaplan-Meier survival analysis of Ncr/nude mice transplanted orthotopically (frontal lobe) with p53/Pten−/− GBM cells harboring miR-34a or the vector control. (D) Quantification of bioluminescence imaging. Mice were imaged 8 days after surgery, the plots shows a significant difference in tumor burden between control and miR-34a tumors as assessed by photon counts. Error bars represent Standard Deviation of 5 experimental replicates. (E) miR-34a impairs in vivo the tumorigenic potential of PN-like TSG2 cells transplanted subcutaneously into the flanks of Ncr/nude mice. Average tumor volumes are plotted. Error bars represent the Standard Deviation of experimental replicates. (F) miR-34a does not affect cell proliferation in 2D culture. Cells were stained with Crystal Violet and assessed for their ability to grow in vitro. Error bars represent the Standard Deviation of experimental replicates. (G) Expression levels of miR-34a in murine PN-like TSG2 malignant glioma cells by RT-qPCR 72 hours after infection with lentivirus harboring a miR-34a precursor or the control vector. Error bars represent Standard Deviation of replicates. (H) Re-introduction of miR-34a in TSG2 spheroids decreases their self-renewal potential. Error bars represent extreme values (upper graph) and Standard Deviation of experimental triplicates (lower graph).
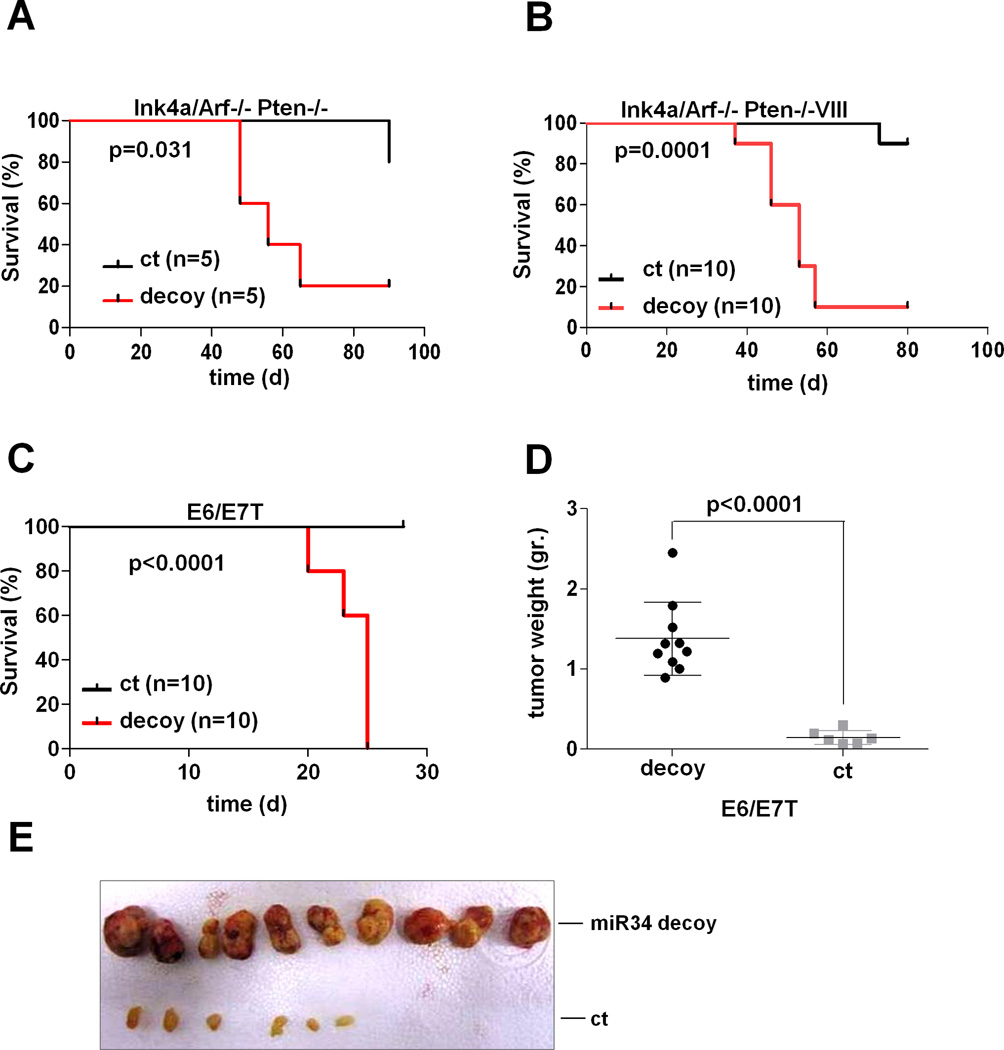
Conversely, to demonstrate a gain-of-oncogenic function in the context of miR-34a loss, we employed a decoy 3’UTR system to functionally knock-down miR-34a as previously described (32). Indeed, stable decoy expression resulted in enhanced tumorigenicity in vivo in human E6/E7T astrocytes and in mouse Ink4a/Arf−/−;Pten−/− and Ink4a/Arf−/−Pten−/−;EgfrVIII/+ astrocytes resulting in decreased tumor-free survival (Figure 5A–E and Supp. Figure 8A–B). Taken together, these in vitro and in vivo functional data in mouse and human systems, both by exogenous genetic perturbation as well as by endogenous down-regulation of miR-34a, unequivocally prove that miR-34a is a GBM tumor suppressor in PN subtype.
Figure 5. miR-34a knock-down transforms genetically engineered mouse and human astrocytes.
(A–B) Knockdown by miR-34a decoy enhances the tumorigenic potential of Ink4a/Arf−/−;Pten−/− and Ink4a/Arf−/−;Pten−/−;EgfrVIII/+ adult mouse astrocytes in vivo. Kaplan-Meier analysis of tumor-free survival of Ncr/nude mice transplanted sub-cutaneously with decoy and control adult mouse astrocytes. (C) Kaplan-Meier analysis of tumor-free survival of Ncr/nude mice transplanted sub-cutaneously with decoy and control E6/E7T human astrocytes. (D) Average weight of tumors generated by decoy and ct E6/E7T cells. Error bars represent Standard Deviation of experimental replicates. (E) Macroscopic appearance of tumors generated by decoy and ct E6/E7T astrocytes.
miR-34a directly regulates PDGFRA
To understand its mechanism of action in proneural GBM, we explored the molecular basis of miR-34a action within the context of the established CLR network. First, we focused on PDGFRA as the predicted direct target of miR-34a since they represent the two nodes of a CLR-defined edge and both are subjected to genomic alterations, in addition to PDGFRA being a known signature oncogene in proneural subtype of GBM (3). Specifically, we looked to confirm direct binding of PDGFRA by miR-34a utilizing a 3’UTR luciferase reporter assay. Indeed, the direct nature of endogenous miR-34a – PDGFRA interaction was validated by this assay in E6/E7T human astrocytes (Supp. Figure 9A). Supporting the biological relevance of this interaction, PDGFRA expression was down-regulated by enforced expression of miR-34a in 2 independent mouse (TSG1 and TSG2) and human (TS543) PN tumor spheroid cultures (Figure 6A) as well as in established human and mouse GBM cells (Supp. Figure 9B). Conversely, decoy-based miR-34a knockdown in primary mouse astrocytes induced expression of PDGFRA (Figure 6A) and reintroduction of PDGFRA open-reading frame was able to reverse the inhibition of tumorigenesis by miR-34a in the TSG2 system (Figure 6B) as well as the spherogenic potential of mouse and human PN malignant spheroids (Supp. Figure 10A and Supp. Figure 10B–C, respectively). Together, these experimental data confirm that PDGFRA is a direct target of miR-34a that is functionally linked to proneural GBM biology, consistent with recent report (33).
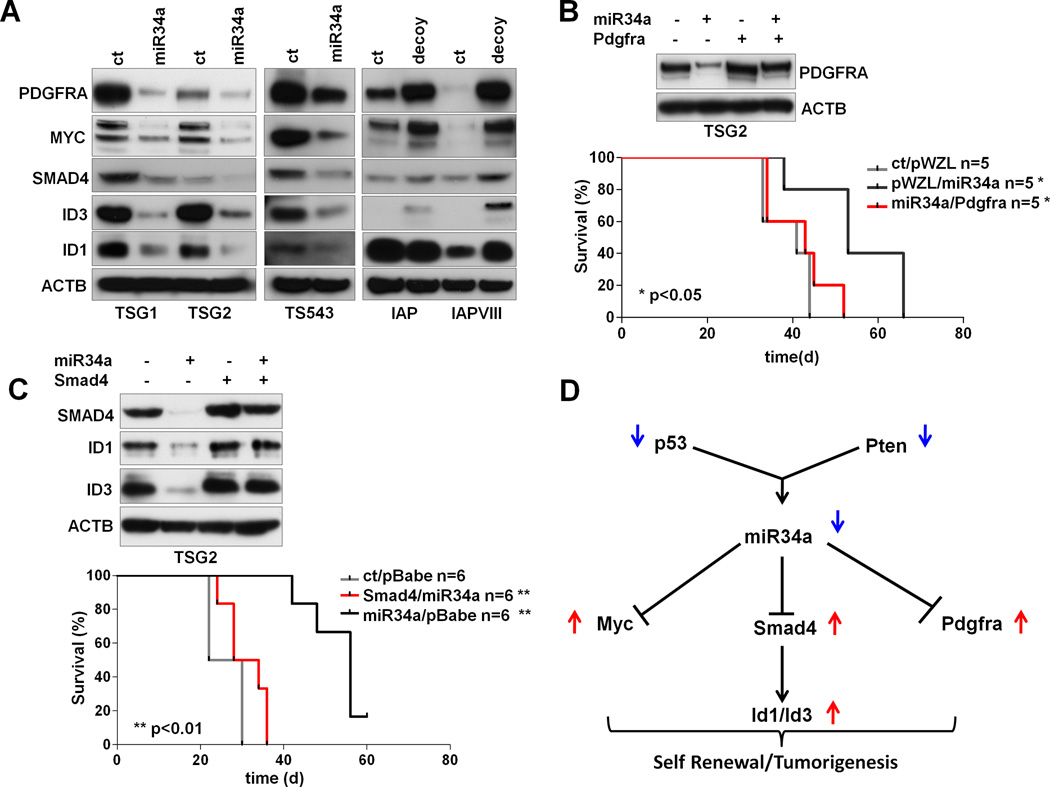
Figure 6. miR-34a functions through PDGFRA and ID proteins in PN Glioblastomas.
(A) Expression levels of PDGFRA, MYC, SMAD4, ID1 and ID3 in response to miR-34a modulation, in mouse (TSG1/2) and human (TS543) PN GBM systems and in genetically engineered, immortalized mouse astrocytes (IAP and IAPVIII). (B) In-vivo rescue experiments. Kaplan-Meier analysis of survival shows that PDGFRA rescues the anti-tumorigenic effects of miR-34a in p53/Pten−/− murine GBM (TSG2) transplanted sub-cutaneously in Ncr/nude mice. Western Blot analysis of protein expression in cells transduced with miR-34a, PDGFRA and the appropriate controls. Protein levels were measured 72 hours after infection (upper panel). (C) In-vivo rescue experiments. Kaplan-Meier analysis of survival shows that Smad4 rescues the anti-tumorigenic effects of miR-34a in p53/Pten−/− murine GBM (TSG2) transplanted sub-cutaneously in Ncr/nude mice. Western Blot analysis of protein expression in cells transduced with miR-34a, Smad4 and the appropriate controls. Protein levels were measured 72 hours after the infection (upper panel). (D) A model showing a hierarchical sub-network involving miR-34a and its upstream and downstream oncogenic nodes in proneural GBM. Abbreviations: IAP=Ink4a/Arf−/−;Pten−/−, IAPVIII=Ink4a/Arf−/−;Pten−/−;EgfrVIII/+.
miR-34a regulates TGFβ signaling via a Smad4 transcriptional network
In line with the observation in the global CLR network, less than 1% of the miR-34a sub-network (three of the 342 nodes or 0.88%) was computationally predicted to be direct, suggesting that indirect mechanisms such as transcriptional regulation mediated through intermediate regulators, e.g. transcription factors, may be at play (34). To identify such intermediates in the network, we performed an in silico enrichment analysis for transcription factor (TF) binding sites in promoter regions of mRNA nodes that are (i) connected to miR-34a and (ii) defined as part of the PN or MS signature(3) (see Methods). Since miR-34a is strongly under-expressed in PN samples, TFs that were (i) overrepresented in the binding sites of these PN signature genes linked to miR-34a, (ii) overexpressed in PN samples with (iii) a statistically significant negative correlation with miR-34a in the PN subtype were defined as potential intermediaries through which miR-34a may act. This in silico analysis identified 3 transcription factors, PBX1, Smad4 and Myc, whose binding sites were significantly enriched (p<0.05; Supp. Table 10). While PBX1 was not expressed in either our human or mouse GBM systems (data not shown), Myc has been shown to be a target regulated by miR-34a (35) (data not shown), and is known to play a pivotal role in glioma-genesis in the p53/Pten GEM model of GBM (28), thus providing support that the computational approach taken to identify transcription factor intermediate is biologically sound. With that, we next explored the putative relationship between miR-34a and Smad4, a connection that has not been reported in any tumor type including GBM.
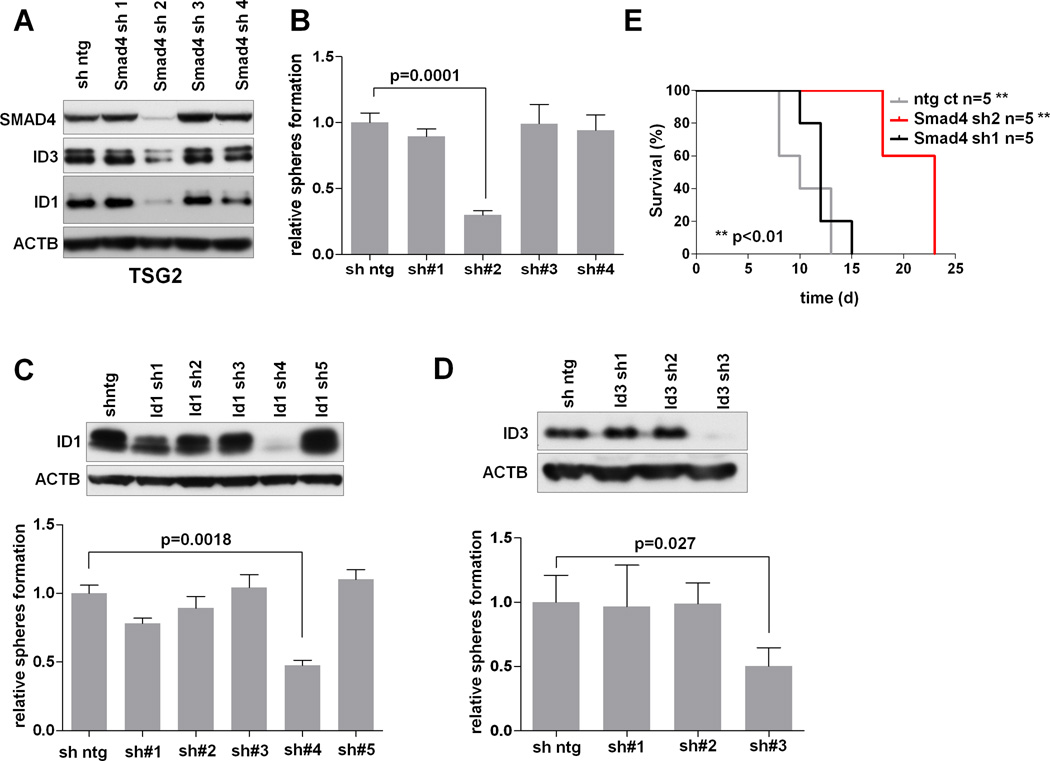
Smad4 is a co-Smad required for R-Smad mediated activation of TGFβ signaling. A large body of literature has implicated critical roles for TGFβ signaling in cancers (36). In GBM, TGFβ pathway has been shown to act as an oncogenic factor (37) and TGFβ signaling can enhance self-renewal capacity of tumor-derived spheroids in vitro, an effect that is dependent on Id1 and Id3 (38). This gains significance in view of the observed effect of miR-34a on the spherogenic potency in vitro. Against this backdrop, the bioinformatic prediction of Smad4 as a putative TF intermediate in the CLR network suggests that miR-34a may be a regulator of the TGFβ-Smad-Id signaling pathway. Consistent with this is the observation of a strong negative correlation between expression of miR-34a and Smad4 in the TCGA dataset (Supp. Figure 11). To functionally test this hypothesis, we examined the expression of Smad4 and the Id family proteins in independent mouse (TSG1 and TSG2) and human (TS453) PN-like GBM tumor spheroid cultures in response to miR-34a. As shown in Figure 6A, miR-34a down-regulated Smad4 and Id1/3 expression, and such regulation was through direct binding to a conserved consensus region in the 3’UTR of Smad4 based on the 3’UTR luciferase reporter assay (Supp. Figure 12). Conversely, in primary astrocytes, decoy sponging of endogenous miR-34a increased Smad4 and Id1/3 expression (Figure 6A). Next, to demonstrate a functional role of Smad4 in the proneural system, we co-transduced p53/Pten−/− TS cells with miR-34a and a Smad4 expression vector to show that Smad4 expression was sufficient to reverse the effect of miR-34a on Id1 and Id3 expression (Figure 6C), and rescues its anti-tumorigenic activity both in vitro and in vivo (Figure 6C and Supp. Figure 13). Overall our functional rescue studies suggest that, in proneural GBM, both PDGFRA and Smad4 are key effectors downstream of miR-34a (Figure 6D). Further, since TGFβ’s activity on renewal of GBM spheroid cultures has been reported to depend on Id1/Id3 expression (38), we showed that lentiviral-based hairpin knockdown of Smad4, Id1or Id3 in the mouse PN-like system significantly reduced the number of spheroids when compared to the non-targeting control (Figure 7A–D). Finally, we found that the stable knock-down of Smad4 impairs tumorigenesis in vivo in intracranial transplants (p<0.01) (Figure 7E), suggesting that the Smad4/Id1/Id3 axis is crucial in regulating the homeostasis of malignant glioma cells in mouse PN-like GBM. In summary, mechanistic exploration guided by integrative analyses of TF binding site enrichment in context of this CLR inferred network established a novel mechanism of actions by miR-34a whereby it exerts its diverse transcriptomic influences through the modulation of the Tgfβ transcriptomic network through the direct binding of Smad4 3’UTR.
Figure 7. The functional inactivation of Smad4 and Id proteins impairs tumorigenesis and self-renewal in PN Glioblastoma.
(A) Western blot analysis of SMAD4, ID1 and ID3 proteins 72 hours after infecting p53/Pten−/−glioma cells (TSG2) with shRNA constructs specific for murine Smad4. (B) Stable knock-down of Smad4 by lentiviral shRNA impairs the self-renewal capacity of mouse PN spheroids (TSG2) in vitro. Error bars represent Standard Deviation of experimental triplicates. (C) Stable knock-down of Id1 by lentiviral shRNA impairs the self-renewal capacity of mouse PN spheroids (TSG2) in vitro. Protein levels were measured 72 hours after the infection (upper panel). Error bars represent Standard Deviation of experimental triplicates. (D) Stable knock-down of Id3 by lentiviral shRNA impairs the self-renewal capacity of mouse PN-like spheroids (TSG2) in vitro. Protein levels were measured 72 hours after the infection (upper panel). Error bars represent Standard Deviation of experimental triplicates. (E) Kaplan-Meier survival analysis of Ncr/nude mice transplanted orthotopically with p53/Pten−/− murine GBM cells harboring hairpins specific for murine Smad4 (sh1 and sh2) or the vector control (ntg ct).
DISCUSSION
GBM is a heterogeneous disease characterized by distinct molecular subtypes underlying different biological behaviors and response to therapies. Leveraging the multi-dimensional TCGA dataset, reverse-engineering with the CLR algorithm has provided an inferred map of the putative miR-mRNA regulatory network in GBM. Integrating this network model with molecular subtype definition and functional genomic screen, as well as in silico sequence-based target prediction and promoter analysis, we prioritized miR-34a for downstream mechanistic studies. These studies uncovered a novel regulatory network emanating from miR-34a which acts as a tumor suppressor in PN-like GBM in part through direct action on PDGFRA as recently shown by Silber et al. (33) in addition to commandeering of the Smad4 transcriptomic network to regulate Id1 and Id3 levels. That the majority of mRNAs computed to link to a miR node appears to be regulated indirectly through transcription factors, such as Smad4 (this study) and Myc (35,39), likely serves as an amplifier of a miR’s effect on the global transcriptome. This finding thus provides a rationale for an alternative approach to inhibit TFs activity, through modulating its upstream miR regulatory node. In sum, this work illustrates not only the power of comprehensive cancer genomic datasets such as that of TCGA and the importance of mining and interpreting such datasets in the context of cancer biology, but also the value of computational and experimental models in enabling an understanding of the underlying complexity of the disease.
Although miR-34a has been implicated in multiple cancer types (40), its roles in the regulation of Smad4/Id1/Id3 have not been previously suggested. While the relationship between PDGFRα and miR-34a is predicted by sequence-based algorithms, relationship between miR-34a and Smad4 or its downstream Id1 and Id3 are not; therefore, the hypothesis that they could be mediators of miR-34a activity came only through unbiased analysis for TF binding site enrichment in the promoters of CLR-inferred mRNA nodes linked to miR-34a. This reinforces the power of global system-biology approaches in generating unanticipated hypotheses.
Equally important are appropriate experimental models that enable genetic (or pharmacological) perturbations to examine the validity of network connections inferred by the computational models. The importance of the GEM model of PN-like GBM is of particular relevance as most of the well-established human GBM cell models or human tumor spheroid cultures do not represent the PN subtype; indeed, using this system, we show that miR-34a exerts its tumor suppressive function by impinging on self-renewal through the Tgfβ/Smad pathway (38). The ability to demonstrate the relationship between miR-34a with PDGFRA and Smad4/Id nuclear oncoproteins in spontaneous de novo GBM in p53/Pten−/− GEM model, and the effects of the experimental perturbation of these oncogenic axes on cancer cell homeostasis in this system, provide strong evidence that the predicted regulatory relationships are relevant and operative in vivo in an intact microenvironment.
Finally, our findings in this study have potential clinical application, as miR-34a expression level is shown in two independent cohorts of GBM to stratify patients into good and poor prognosis subgroups with significant difference in overall survival. Further, within the TCGA cohort, we found that miR-34a carries significant overlap in prognostic significance with G-CIMP status (6) (data not shown), suggesting a possible mechanistic relationship between miR-34a and G-CIMP, although elucidation of the molecular basis for this relationship will require further studies.
Previous studies have reported miR-34a expression level as a prognostic parameter. For instance, in Pancreatic Ductal Adenocarcinoma (PDAC), miR-34a loss (i.e. low to no expression) is associated with a decreased survival probability (41); in other words, miR-34a expressing PDAC have a relatively better survival. In breast cancer, although high miR-34a expression is correlated with poor prognosis factors including positive nodal status, high tumor grade, ER-negativity, HER2-positivity and high proliferation rate, after adjusted for these known prognostic parameters in multi-variant analysis, high miR-34a expression is in fact associated with a lower risk of recurrence or death from breast cancer (42), indicating that high levels of miR-34a are a good prognostic factor. In contrast to these previous studies, our analyses of two independent cohorts of GBM showed that miR-34a low-expressing GBMs have better outcome with longer overall survival. In other words, GBM tumors driven by inactivation of miR-34a are less aggressive compared to GBM that evolve through deregulation of other genetic elements. The differences in prognostic significance of miR-34a loss in different tumor types likely reflect the modulatory effects of pre-existing genetic alterations and the specific susceptibility of different cell types to the aberrant activation of any given pathway. This is not dissimilar to the case of another glioma gene, IDH1, whose specific point mutation affecting a key residue in the protein (IDH1R132) has been shown to be oncogenic. Interestingly, gliomas carrying this mutation in IDH1 as well as analogous mutations affecting IDH2, have a significant better prognosis (43–46). On the other hand, in cytogenetically normal AML carrying NPM1 mutations, IDH1 mutations at the same residue are a poor prognostic factor as patients with IDH1 mutations do worse (47,48). In summary, we illustrate here that computational network modeling of the complex inter-relationships among diverse genetic elements can generate a logical framework in which to explore and understand the genetics and biology of cancers, and when integrated with disease knowledge and clinical annotation, can lead to discovery of new pathogenetic insights in addition to potential prognostic biomarkers or therapeutic targets. In this regard, we believe that the results from this study should motivate future efforts to explore the therapeutic implication of miR-34a re-constitution. The potent tumor suppressive activity in our preclinical models would suggest possible therapeutic benefit of miR-34a re-constitution by tumor-targeted delivery in low miR-34a expressing GBM. In view of its mechanism of action through PDGFRA, Myc and Smad4, one may further speculate that re-constitution of miR-34a could represent an attractive strategy to deliver combination therapy against multiple bona fide cancer gene targets.
METHODS SUMMARY
Bioinformatic analysis
Network inference was performed, both globally and within each of the four molecular subtype sample sets, using the CLR algorithm on 290 matched miR and mRNA expression profiles from TCGA. Copy number analysis was performed on Level 3 segmented data by the CN Tools Bioconductor package using 90th percentile Segment Gain or Loss (SGOL) values as thresholds. Gene weight analysis was done to test for correlation of expression and copy number change for each miR and mRNA species. Transcription factor motifs were identified based on coincidental prediction of binding sites by the CisGenome and MotifScanner programs in promoter regions (−8kb, +2kb of TSS).
Mouse and human proneural Glioblastoma cell lines
p53L/L;PtenL/+;Gfap-Cre and p53L/L;PtenL/L; Gfap-Cre, mice have been previously described (28). Primary murine astrocytes were isolated from Gfap-Cre;Ink4a/ArfL/L;PtenL/L and Gfap-Cre;Ink4a/ArfL/L;PtenL/L;EgfrLSLVIII/+. GEMM models were provided by Ronald A. Depinho. Murine cell lines were created by enzymatic and mechanical dissociation of individual samples, all genotypes were verified by PCR amplification of genomic DNA and gel electrophoresis. The Harvard Institutional Animal Care and Use Committee approved all animal studies and procedures. TS543 human malignant spheroids are short term cultures derived from a primary proneural GBM kindly provided by Dr. Cameron Brennan. PDGFRA expression was validated by Western Blot.
Human astrocytes immortalized with the E6/E7 oncoproteins and Tert were provided by Ronald A. Depinho.
An extensive description of the materials and methods is provided in the Supplementary Information.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
We illustrate here that network modeling of complex multi-dimensional cancer genomic data can generate a framework in which to explore the biology of cancers, leading to discovery of new pathogenetic insights as well as potential prognostic biomarkers. Specifically in GBM, within the context of the global network, promoter enrichment analysis of network edges uncovered a novel regulation of TGFβ signaling via a Smad4 transcriptomic network by miR-34a.
ACKNOWLEDGEMENTS
Authors wish to acknowledge the support of the Belfer Institute for Applied Cancer Science for computational analyses, and thank Drs. Ronald A. Depinho, Giulio Draetta, and members of the Chin laboratories, Alessandro Sgambato and Stefano Sioletic for comments and reagents. We acknowledge Joshua Mendell for the MSCV-PIG miR-34a construct. We thank Zhou Shan, Jiang Shan and Carla Bianchi for mouse husbandry and colony care. All the expression clones were obtained through the Dana Farber/Harvard Plasmid Core Facility. The pHage vector was originally developed by Darrel Kotton. Human glioma spheroids were provided by Cameron Brennan. GEMM models and immortalized human astrocytes were provided by Ronald A. Depinho.
Financial support: This work is supported by funding from the NIH (PO1 CA095616, U01 CA141508 and U24 CA143845) and from the Ben and Catherine Ivy Foundation to L.C., J.J.C. is a HHMI investigator, G.G. was supported by the AIRC “Associazione Italiana per la Ricerca sul Cancro” fellowship, S.A.S. was supported by The Robert A. and Renee E. Belfer foundation and by the GDAC grant (NIH U24 CA143845) P.G. was supported by a NIH F32 fellowship, S.N.Q. was supported by a fellowship from the Canadian Institutes of Health Research, K.R. was supported by a NIH K99/R00 Pathway to Independence Award (1K99CA160578-01) and Charles A King Trust post-doctoral Fellowship, K.D. was supported by the Terry Fox Foundation (#2009-700118), S.S. was supported by a fellowship from the Ovarian Cancer Research Foundation, Cameron Brennen is supported by the NIH P01 CA95616 grant.
Abbreviations list
- GBM
Glioblastoma
- CLR
Context-Likelihood-Relatedness
- PN
Proneural
- MS
Mesenchymal
- CL
Classical
- NL
Neural
- NSC
neural stem cells
- TS
tumor spheroids
- GSEA
gene set enrichment analysis
- IAP
Ink4a/Arf−/−;Pten−/−
- IAPVIII
Ink4a/Arf−/−;Pten−/−;EgfrVIII/+
Footnotes
Conflict of interest disclosure:
David L. Rimm is a consultant to and stockholder in HistoRx, the exclusive licensee of the AQUA patent, the other authors have no conflict of interest to disclose.
SUPPLEMENTARY INFORMATION is linked to the online version of the paper at http://cancerdiscovery.aacrjournals.org/
AUTHORS’ CONTRIBUTIONS
A.E., S.A.S. formulated overall approaches for analysis and identification of most discriminatory miRs. A.E. conducted the network inference modeling and integration with copy number data; S.A.S. performed gene set enrichment, direct target prediction results and promoter analyses. J.Z. and H.Z performed correlation of copy number with expression analysis. J.H., B.C. and D.R. generate and analyzed the TMA data. C.J.W. performed the survival analysis. G.G., P.G., H.Y. performed in-vivo experiments; G.G. and S.C. established the lentiviral vector systems. S.Q. performed luciferase reporter assays, B.C., K.R., S.S. and K.D. performed in-vitro studies. S.C., H.Z., and C.B. provided reagents, W.R.W. performed signaling analysis and L.K. analyzed microarray data. G.G., A.E., S.A.S. and L.C. wrote the paper with comments from all authors. J.C.C. and L.C. supervised all aspects of the project.
REFERENCES
- 1.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An Extensive MicroRNA-Mediated Network of RNA-RNA Interactions Regulates Established Oncogenic Pathways in Glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-Independent Regulation of the Tumor Suppressor PTEN by Competing Endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su WL, Kleinhanz RR, Schadt EE. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol. 7:490. doi: 10.1038/msb.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong H, Luo L, Hong S, Siu H, Xiao Y, Jin L, et al. Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst Biol. 4:163. doi: 10.1186/1752-0509-4-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mestdagh P, Lefever S, Pattyn F, Ridzon D, Fredlund E, Fieuw A, et al. The microRNA body map: dissecting microRNA function through integrative genomics. Nucleic Acids Res. 2011;39:e136. doi: 10.1093/nar/gkr646. (database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigoryev YA, Kurian SM, Hart T, Nakorchevsky AA, Chen C, Campbell D, et al. MicroRNA Regulation of Molecular Networks Mapped by Global MicroRNA, mRNA, and Protein Expression in Activated T Lymphocytes. J Immunol. 187:2233–2243. doi: 10.4049/jimmunol.1101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. PMCID: 2671642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 24.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 33.Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C, et al. miR-34a Repression in Proneural Malignant Gliomas Upregulates Expression of Its Target PDGFRA and Promotes Tumorigenesis. PLoS One. 2012;7:e33844. doi: 10.1371/journal.pone.0033844. PMCID: 3313940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 36.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 37.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermeking H. The miR-34 family in cancer and apoptosis. Cell death and differentiation. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 41.Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter R, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathological criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18:534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 42.Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, et al. MiR-34a Expression Has an Effect for Lower Risk of Metastasis and Associates with Expression Patterns Predicting Clinical Outcome in Breast Cancer. PLoS One. 2011;6:e26122. doi: 10.1371/journal.pone.0026122. PMCID: 3213093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. PMCID: 2820389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. PMCID: 2820383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. PMCID: 2881719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.