Abstract
Technologies for engineering synthetic transcription factors have enabled many advances in medicine and science. In contrast to existing methods based on engineering of new DNA-binding proteins, we created a Cas9-based transactivator that is targeted to DNA sequences by guide RNA molecules. Co-expression of this transactivator and combinations of guide RNAs in human cells induced specific expression of endogenous target genes, demonstrating a simple and versatile approach for RNA-guided gene activation.
Synthetic transcription factors have been engineered to control gene expression for many different medical and scientific applications in mammalian systems, including stimulating tissue regeneration1, compensating for genetic defects2, activating silenced tumor suppressors3, controlling stem cell differentiation4, 5, performing genetic screens6, 7, and creating synthetic gene circuits8, 9. These transcription factors can target promoters or enhancers of endogenous genes10, or be purposefully designed to recognize sequences orthogonal to mammalian genomes for transgene regulation11. To date, the most common strategies for engineering novel transcription factors targeted to user-defined sequences have been based on the programmable DNA-binding domains of zinc finger proteins10, 12 and transcription-activator like effectors (TALEs)13, 14. Both of these approaches involve applying the known principles of protein-DNA interactions of these domains to engineer new proteins with unique DNA-binding specificity. Although these methods have been widely successful for many applications, the protein engineering protocols can be laborious and require specialized expertise. Additionally, these new proteins are not always effective. The reasons for this are not yet known but may be related to context-dependent protein-DNA interactions or the effects of epigenetic modifications and chromatin state on protein binding to the genomic target site.
Recently, an engineered form of the clustered, regularly interspaced, short palindromic repeats (CRISPR)-CRISPR-associated (Cas) protein system15 of Streptococcus pyogenes was shown to function in human cells for genome engineering16-19. In this system, the type II CRISPR protein Cas9 is directed to genomic target sites by short RNAs, where it functions as an endonuclease. In the naturally occurring system, Cas9 is directed to its DNA target site by two noncoding CRISPR RNAs (crRNAs), including a trans-activating crRNA (tracrRNA) and a precursor crRNA (pre-crRNA)15. In the synthetically reconstituted system, these two short RNAs can be fused into a single chimeric guide RNA (gRNA)15. A Cas9 mutant with undetectable endonuclease activity (dCas9)15 has been recently targeted to genes in bacteria, yeast, and human cells by gRNAs to silence gene expression through steric hindrance20. In this study, we demonstrate a novel strategy to activate the expression of endogenous human genes by targeting a fusion protein of dCas9 and a transactivation domain to human promoters via combinations of gRNAs.
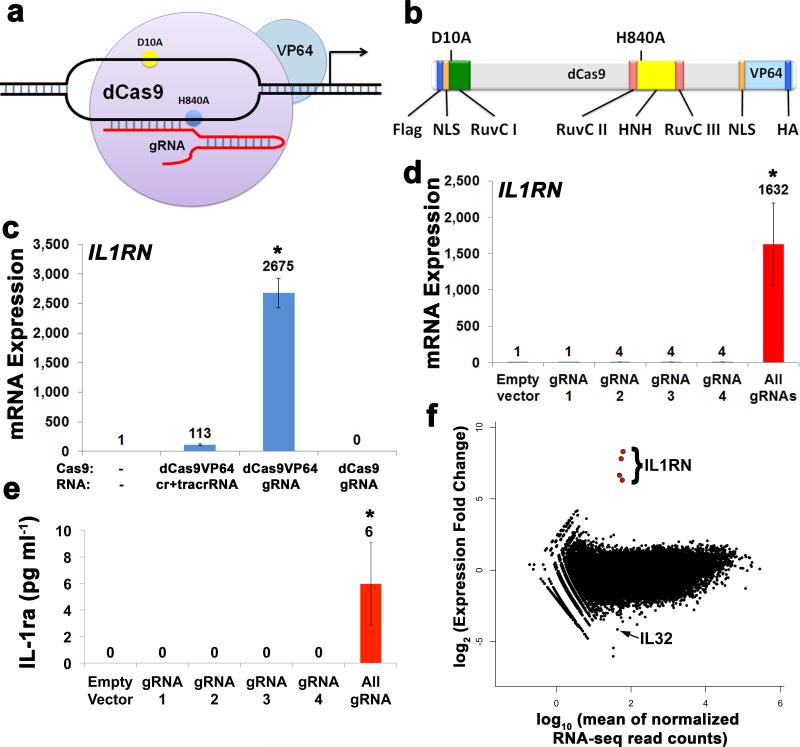
To create a CRISPR/Cas9-based transcriptional activation system, we mutated the endonuclease catalytic residues of Cas9 (D10A, H840A) to create dCas915, 20 and genetically fused it with a C-terminal VP64 acidic transactivation domain (Fig. 1a,b)10. We observed robust expression of dCas9-VP64 from the transfected plasmid in human embryonic kidney (HEK) 293T cells by western blot of the N-terminal Flag epitope tag (Supplementary Fig. 1). We and others have recently shown that combinations of synthetic transcription factors targeted to endogenous human promoters result in synergistic and robust activation of gene expression21, 22. Therefore we identified four gRNA target sites of 20 bp followed by the NGG PAM sequence15-20 in the promoter of the IL1RN gene (Supplementary Fig. 2, Supplementary Table 1). To compare crRNA- and gRNA-based targeting strategies, we introduced the four target site sequences into crRNA and gRNA expression plasmids17 and co-transfected them with the dCas9-VP64 expression plasmid into HEK293T cells. Although we observed substantial induction of IL1RN expression by qRT-PCR in samples treated with the combination of crRNAs, much higher levels were achieved with the combination of gRNAs (Fig. 1c). There were no changes to IL1RN gene expression in cells treated with gRNAs and an expression plasmid for dCas9 without VP64, demonstrating the critical role of the activation domain in modulating gene expression (Fig. 1c). We also confirmed that nuclease activity at these target sites had been abrogated in the dCas9-VP64 system by performing the Surveyor assay to detect DNA repair events in samples treated with dCas9-VP64 and wild-type Cas9 (Supplementary Fig. 3). By transfecting each of the four gRNAs individually or in combination, we demonstrated that targeting multiple sites in the promoter with combinations of gRNAs was necessary for robust increases in gene expression (Fig. 1d), as we have seen with other classes of engineered transcription factors21, 22. Similarly, production of the IL-1 receptor antagonist (IL-1ra) protein, encoded by the IL1RN gene, was observed in three of the six samples treated with the combination of gRNAs across three different experiments, whereas it was never detected in samples treated with single gRNAs or control plasmid (Fig. 1e). To examine the specificity of gene activation by dCas9-VP64, we assessed global gene expression of HEK293T cells treated with the combination of four gRNAs by RNA-seq (Fig. 1f). Notably, the only genes with significantly increased expression relative to control (false discovery rate ≤ 3 × 10−4) were the four isoforms expressed from the IL1RN locus (Supplementary Fig. 2), indicating a high level of specificity of gene activation.
Figure 1.
RNA-guided activation of the human IL1RN gene by dCas9-VP64. (a) An RNA-guided transcriptional activator was created by fusing dCas9 (D10A/H840A) to the VP64 transactivation domain. dCas9-VP64 recognizes genomic target sites through the hybridization of a guide RNA (gRNA) to a 20 bp target sequence. (b) The structure of dCas9-VP64 is shown, including Flag and HA epitope tags, two nuclear localization signals (NLS), and the positions of the RuvC- and HNH-like endonuclease domains that target the noncomplementary and complementary DNA strands, respectively, if not inactivated by the mutations D10A and H840A.15(c) Expression plasmids for four gRNAs or crRNA/tracrRNAs targeted to sequences in the IL1RN promoter were co-transfected with the dCas9-VP64 expression plasmid into HEK293T cells. Activation of IL1RN expression was assessed by qRT-PCR. (d) The four gRNA expression plasmids were co-transfected with dCas9-VP64 individually or in combination. Robust gene activation was observed by qRT-PCR only in response to the combination of gRNAs. (e) Activation of IL1RN expression was confirmed by assessing secretion of the IL-1ra gene product into the media by ELISA. IL-1ra was only detectable above background in three of the six samples treated with the combination of gRNAs. For (c-e), data are shown as the mean ± s.e.m (n = 3 independent experiments). Treatment with the combination of gRNAs was statistically different than all other treatments (*P ≤ 0.05) by Tukey’s test. (f) RNA-seq was performed on samples treated with empty expression vector (n = 2) or co-transfected with the expression plasmids for dCas9-VP64 and the four gRNAs targeting IL1RN (n = 2). The only statistically significant changes in gene expression between these treatments were an increase in the four IL1RN isoforms (false discovery rate ≤ 3 × 10−4) and a decrease in IL32 (false discovery rate = 0.03).
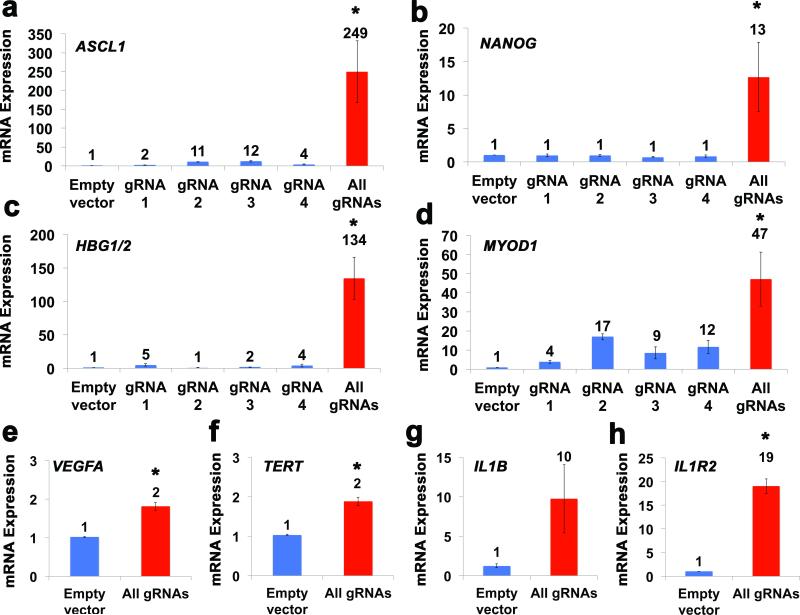
To demonstrate the general applicability of this system, we designed four gRNAs targeted to each of the promoters of eight other genes relevant to medicine and biotechnology, including ASCL1, NANOG, HBG1/2, MYOD1, VEGFA, TERT, IL1B, and IL1R2 (Supplementary Fig. 2, Supplementary Table 1). Expression of each of these genes was enhanced by co-transfection of expression plasmids for dCas9-VP64 and the four gRNAs into HEK293T cells, as determined by qRT-PCR (Fig. 2). In some cases expression of a single gRNA was sufficient to induce gene expression, but in all cases co-transfection of the four gRNAs led to synergistic effects (Fig. 2a-d). Notably, chromatin accessibility, as determined by DNase-seq, was not a predictor of successful gene activation (Supplementary Fig. 2), consistent with our previous observations21. RNA-seq was performed on cells transfected with dCas9-VP64 and the four gRNAs targeting HBG1, three of which also perfectly target HBG2. This revealed specific and reproducible increases in expression of both HBG1 and HBG2, which cannot be distinguished by RNA-seq, although statistical significance was not achieved due to low total expression levels (Supplementary Fig. 4). Increases in protein expression of Ascl1 and γ-globin following treatment with dCas9-VP64 and the four gRNAs were detected by western blot (Supplementary Fig. 5), corroborating higher mRNA levels observed by qRT-PCR (Fig. 2). Low baseline levels of Ascl1 and γ-globin protein expression were detectable in empty vector controls. As evidence that the dCas9-VP64/gRNA system can activate gene expression in other cell types, we transfected expression plasmids for dCas9-VP64 and the four gRNAs targeting ASCL1 into murine embryonic fibroblasts (MEFs) (Supplementary Fig. 6). Because the gRNA target sites are conserved in the human and mouse ASCL1 promoters (Supplementary Fig. 6a), activation of ASCL1 mRNA expression was also observed four days post-transfection in MEFs treated with plasmids encoding dCas9-VP64 and the four gRNAs (Supplementary Fig. 6b).
Figure 2.
RNA-guided activation of human genes relevant to cell and gene therapy, genetic reprogramming, and regenerative medicine. HEK293T cells were transfected with the dCas9-VP64 expression plasmid and four gRNAs individually or in combination. Target gene expression was measured by qRT-PCR and normalized to GAPDH mRNA levels. Data are shown as the mean ± s.e.m (n = 3 independent experiments). Treatment with the combination of gRNAs was statistically different than all other treatments (*P < 0.05) by Tukey’s test.
This study provides a new mechanism for activating the expression of endogenous mammalian genes based on targeting a transcriptional activator to promoters via RNA. This is fundamentally different from previously described methods based on engineering sequence-specific DNA-binding proteins and therefore may provide new opportunities for targeted gene regulation. Because the generation of new gRNA expression plasmids simply involves synthesizing two short custom oligonucleotides and one cloning step, it is possible to generate many new gene activators quickly and economically. The gRNAs can also be transfected directly to cells following in vitro transcription.18 Here we show multiple gRNAs targeted to single promoters, but simultaneous targeting of multiple promoters could also be envisioned, similar to recent methods for CRISPR-based multiplexed gene editing16. It is likely that future efforts for combining multiple gRNA expression cassettes into single delivery vectors will facilitate this type of approach. Recognition of genomic target sites with RNAs, rather than proteins, may also circumvent limitations of targeting epigenetically modified sites, such as methylated DNA. Notably, the activation of IL1RN by dCas9-VP64 in this study was weaker than what we have reported for TALE-based transcription factors targeting the same promoter21, and we have observed a similar trend for other targets, but additionally studies are necessary to understand the differences in these nascent technologies.
Using RNA-seq, we show that targeted gene activation was exquisitely specific with no detectable off-target gene activation (Fig. 1f, Supplementary Fig. 4). We specifically chose IL1RN and HBG1/2 for this specificity analysis as we expected the gene products, IL-1ra and γ-globin, would not generate secondary effects on gene expression in HEK293T cells. We hypothesize that exploiting the synergistic activity of multiple weak transcriptional activators as we have done here, in contrast to using a single strong activator, may increase specific gene regulation since it is unlikely that multiple adjacent off-target sites would exist at another locus. Although these results show that gene activation by dCas9-VP64 combinations can be very specific, this is not necessarily a surrogate for DNA-binding and additional studies are necessary to define the role of the gRNA target sequence in determining genome-wide Cas9 activity in mammalian cells. Interestingly, the RNA-seq results for both IL1RN and HBG1/2 showed moderate downregulation of the IL32 gene (false discovery rate < 0.03) in the samples treated with dCas9-VP64 and gRNAs compared to control samples treated with only an empty expression plasmid (Fig. 1f, Supplementary Fig. 4). Because both the IL1RN and HBG1/2-targeted samples were similarly affected, it is unlikely that this is the result of off-target dCas9-VP64 activity related to the identity of the target sequences. These results were confirmed by qRT-PCR for IL32 which showed that this downregulation is a general response to dCas9-VP64, even in the absence of gRNAs (Supplementary Fig. 7). Additional studies are necessary to determine the mechanism of IL32 downregulation in response to dCas9-VP64.
Potential applications of RNA-guided transcription factors are diverse across many areas of science and biotechnology. Here we provide examples of activation of genes related to cell and gene therapy, genetic reprogramming, and regenerative medicine, including the regulators of cell lineage specification ASCL1, NANOG, and MYOD1 (Fig. 2). Although we did not observe robust conversion of MEFs into a neuronal phenotype following ASCL1 activation, in contrast to direct sustained overexpression of Ascl123, there are many ways by which this method could be improved. For example, repeated transfections of dCas9-VP64/gRNA combinations, stable expression of these factors, and targeting multiple genes, such as Brn2 and Myt1l in addition to Ascl1,23 may facilitate more efficient activation of cell lineage specification networks. We hypothesize that with continued optimization, the activation of endogenous genes encoding the key regulators of cell fate, rather than forced overexpression of these factors, could potentially lead to more rapid, efficient, stable, or specific methods for genetic reprogramming and transdifferentiation. Finally, dCas9 fusions to other domains, including repressive and epigenetic-modifying domains,24 could provide a greater diversity of RNA-guided transcriptional regulators to complement other RNA-based tools for mammalian cell engineering.25
Online Methods
Cell culture and transfection
HEK293T cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facilities and were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2. HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Transfection efficiencies were routinely higher than 80% as determined by fluorescence microscopy following delivery of a control eGFP expression plasmid. dCas9-VP64 expression plasmid was transfected at a mass ratio of 3:1 to either the individual gRNA expression plasmids or the identical amount of gRNA expression plasmid consisting of a mixture of equal amounts of the four gRNAs.
Primary mouse embryonic fibroblasts (PMEF-HL, Millipore, Billerica, MA) were seeded (75,000 per well) in 24-well TCPS plates (BD, Franklin Lakes, NJ) and maintained at 37°C and 5% CO2 in complete MEF medium consisting of high glucose DMEM supplemented with 10% Premium Select FBS (Atlanta Biologicals, Lawrenceville, GA), 25 μg mL−1 gentamicin (Invitrogen), 1X GlutaMAX, non-essential amino acids, sodium pyruvate, and β-mercaptoethanol (Invitrogen). MEF transfections were performed with a single 1 μg cm−2 dose of total plasmid DNA, delivered as cationic nanocomplexes following electrostatic condensation with poly(CBA-ABOL) in serum- and antibiotic-free OptiMEM, as described previously26. OptiMEM was replaced with complete MEF medium four hours after transfection. MEFs were processed for qRT-PCR four days after transfection. A GFP reporter vector (pmax-GFP, 3486 bp, Amaxa, Cologne, Germany) was used to optimize transfection conditions. dCas9-VP64 expression plasmid was transfected at a mass ratio of 3:1 or 1:1 to an equal mixture of four gRNA expression plasmids.
Plasmids
The plasmids encoding wild-type and H840A Cas9 were obtained from Addgene (Plasmid #39312 and Plasmid #39316).15 H840A Cas9 was cloned into the vector pcDNA3.1 in frame with a FLAG epitope tag and a NLS at the N-terminus with a primer pair that introduced the D10A mutation. The VP64 domain,10, 27 an NLS, and an HA epitope tag were cloned in frame with the Cas9 ORF at the C-terminus (Fig. 1a, Supplementary Fig. 8). The tracrRNA and crRNA expression cassettes were ordered as gBlocks (IDT) based on published sequences16 and cloned into a pZDonor plasmid (Sigma) with KpnI and SacII sites. A previously described chimeric guide RNA expression cassette17 was also ordered as gBlocks with modifications to include a BbsI restriction site16 to facilitate rapid cloning of new guide RNA spacer sequences (Supplementary Fig. 9). The oligonucleotides containing the target sequences were obtained from IDT, hybridized, phosphorylated, and cloned in the appropriate plasmids using BbsI sites. The target sequences and positions relative to the transcriptional start site are provided in Supplementary Table 1.
Western blot
Cells were lysed in 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, and 0.1% SDS. Lysates were mixed with loading buffer, boiled for 5 min, and equal volumes of protein were run in NuPAGE® Novex 4-12% or 10% Bis-Tris Gel polyacrylamide gels and transferred to nitrocellulose membranes. Non-specific antibody binding was blocked with 50 mM Tris/150 mM NaCl/0.1% Tween-20 (TBS-T) with 5% nonfat milk for 30 min. The membranes were incubated with primary antibodies (HRP-conjugated anti-Flag (Cell Signaling, Cat#2044) in 5% BSA in TBS-T diluted 1:1000 overnight; anti-GAPDH (Cell Signaling, clone 14C10) in 5% milk in TBS-T diluted 1:5000 for 30 min; anti-ASCL1 (Santa Cruz, clone sc-48449) in 5% BSA diluted 1:500; or anti-g-globin (Santa Cruz, clone 51-7) in 5% milk diluted 1:500 and the membranes were washed with TBS-T for 30 min. Membranes labeled with primary antibodies were incubated with anti-rabbit HRP-conjugated antibody (Sigma-Aldrich) diluted 1:5000 for 30 min, anti-goat (1:3000) or anti-mouse (1:5000) and washed with TBS-T for 30 minutes. Membranes were visualized using the Immun-Star WesternC™ Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc™ XRS+ System and processed using ImageLab software (Bio-Rad).
ELISA
Serum-free culture media (OPTI-MEM) was collected and frozen at −80°C. Human IL-1ra secretion into culture media was quantified via enzyme-linked immunosorbent assay (ELISA), according to the manufacturer’s protocols (R&D Systems, Cat. No. DY280). The standard curve was prepared by diluting recombinant human IL-1ra in OPTI-MEM and the IL-1ra in culture media was measured undiluted. The samples were concentrated ~8 fold via centrifugation through 3 kDa MWCO filters for 20 minutes (Amicon Ultra, Cat # UFC500396). Reported values were corrected by the concentration factor for each sample.
Optical density was measured at 450 nm, with a wavelength correction at 540 nm. Each standard and sample was assayed in duplicate. The duplicate readings were averaged and normalized by subtracting the average zero standard optical density. A standard curve was generated by log-transforming the data and performing a linear regression of the IL-1ra concentration versus the optical density. Reported values are the mean and standard error of the mean from three independent experiments (n = 3) that were performed on different days with technical duplicates that were averaged for each experiment.
qRT-PCR
Total RNA was isolated using the RNeasy Plus RNA isolation kit (Qiagen). cDNA synthesis was performed using the SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen). Real-time PCR using PerfeCTa® SYBR® Green FastMix was performed with the CFX96 Real-Time PCR Detection System (Bio-Rad) with oligonucleotide primers reported in Supplementary Table 2 that were designed using Primer3Plus software and purchased from IDT. Primer specificity was confirmed by agarose gel electrophoresis and melting curve analysis. Reaction efficiencies over the appropriate dynamic range were calculated to ensure linearity of the standard curve (Supplementary Fig. 10). The results are expressed as fold-increase mRNA expression of the gene of interest normalized to GAPDH expression by the ΔΔCT method. Reported values are the mean and standard error of the mean from three independent experiments performed on different days (n = 3) with technical duplicates that were averaged for each experiment.
RNA-Seq
RNA seq libraries were constructed as previously described28. Briefly, first strand cDNA was synthesized from oligo dT Dynabead® (Invitrogen) captured mRNA using SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen). Second strand cDNA was synthesized using DNA Polymerase I ew England Biolabs). cDNA was purified using Agencourt AMPure XP beads (Beckman Coulter) and Nextera transposase (Illumina; 5 minutes at 55°C) was used to simultaneously fragment and insert sequencing primers into the double-stranded cDNA. Transposition reactions were halted using QG buffer (Qiagen) and fragmented cDNA was purified on AMPure XP beads. Indexed sequencing libraries were generated by 6 cycles of PCR.
Libraries were sequenced using 50-bp single end reads on two lanes of an Illumina HiSeq 2000 instrument, generating between 29 million and 74 million reads per library. Reads were aligned to human RefSeq transcripts using Bowtie29. The statistical significance of differential expression, including correction for multiple hypothesis testing, was calculated using DESeq30. Raw RNA-seq reads and the number of reads aligned to each RefSeq transcript have been deposited for public access in the Gene Expression Omnibus (GEO) (accession number GSE47114).
Statistics
Statistical analysis was performed by Tukey’s test with alpha equal to 0.05 in JMP 10 Pro.
Supplementary Material
Acknowledgments
This work was supported by a US National Institutes of Health (NIH) Director’s New Innovator Award (DP2OD008586), National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035), NIH R03AR061042, and an American Heart Association Scientist Development Grant (10SDG3060033) to C.A.G, and grants from the NIH to G.E.C. (U54HG004563), K.W.L. (EB015300 and HL109442), and F.G. (R01AR48852). K.A.G. and P.I.T. were supported by NSF Graduate Research Fellowships. L.R.P. was supported by an NIH Biotechnology Training Grant to the Duke Center for Biomolecular and Tissue Engineering (T32GM008555). D.G.O was supported by a predoctoral fellowship from the American Heart Association. C. Grigsby (Duke University) provided the pABOL polymer used in MEF transfections.
Footnotes
Author Contributions
P.P., C.M.V., A.F.A., D.G.O, G.E.C., T.E.R. and C.A.G. designed experiments. P.P., D.D.K., C.M.V., A.F.A., A.M.K., L.R.P., P.I.T., K.A.G., and D.G.O. performed the experiments. P.P., D.D.K., C.M.V., A.F.A., A.M.K., L.R.P., P.I.T., K.A.G., D.G.O., K.W.L., F.G., G.E.C., T.E.R. and C.A.G. analyzed the data. P.P. and C.A.G. wrote the manuscript.
COMPETINGFINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Rebar EJ, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 2.Graslund T, Li X, Magnenat L, Popkov M, Barbas CF., 3rd Exploring strategies for the design of artificial transcription factors: targeting sites proximal to known regulatory regions for the induction of gamma-globin expression and the treatment of sickle cell disease. J Biol Chem. 2005;280:3707–3714. doi: 10.1074/jbc.M406809200. [DOI] [PubMed] [Google Scholar]
- 3.Beltran A, et al. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene. 2007;26:2791–2798. doi: 10.1038/sj.onc.1210072. [DOI] [PubMed] [Google Scholar]
- 4.Bartsevich VV, Miller JC, Case CC, Pabo CO. Engineered zinc finger proteins for controlling stem cell fate. Stem Cells. 2003;21:632–637. doi: 10.1634/stemcells.21-6-632. [DOI] [PubMed] [Google Scholar]
- 5.Bultmann S, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blancafort P, Magnenat L, Barbas CF., 3rd Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–274. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 7.Park KS, et al. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- 8.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Scientific reports. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 15.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 19.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Pinera P, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeder ML, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell. 2011;43:915–926. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler AF, et al. Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells. Molecular therapy. Nucleic acids. 2012;1:e32. doi: 10.1038/mtna.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gertz J, et al. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22:134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.