Abstract
This study establishes that sparse canonical correlation analysis (SCCAN) identifies generalizable, structural MRI-derived cortical networks that relate to five distinct categories of cognition. We obtain multivariate psychometrics from the domain-specific sub-scales of the Philadelphia Brief Assessment of Cognition (PBAC). By using a training and separate testing stage, we find that PBAC-defined cognitive domains of language, visuospatial functioning, episodic memory, executive control, and social functioning correlate with unique and distributed areas of gray matter (GM). In contrast, a parallel univariate framework fails to identify, from the training data, regions that are also significant in the left-out test dataset. The cohort includes164 patients with Alzheimer’s disease, behavioral-variant frontotemporal dementia, semantic variant primary progressive aphasia, nonfluent/agrammatic primary progressive aphasia, or corticobasal syndrome. The analysis is implemented with open-source software for which we provide examples in the text. In conclusion, we show that multivariate techniques identify biologically-plausible brain regions supporting specific cognitive domains. The findings are identified in training data and confirmed in test data.
Keywords: Alzheimer disease, Frontotemporal lobar degeneration, Philadelphia Brief Assessment of Cognition, PBAC, MRI, Sparse canonical correlation analysis
Introduction
Multivariate methods have advantages over univariate methods in genomics (Hibar et al., 2011; Le Floch et al., 2012; Parkhomenko et al., 2009), pattern recognition (Bishop, 1995; Roberts, 1997; Tipping, 2001) and neuroimaging (De Martino et al., 2008; Fan et al., 2008; McIntosh et al., 1996; Shamy et al., 2011; Tosun et al., 2012) due to the high dimensionality and latent structure within these types of datasets. Various forms of multivariate pattern analysis (MVPA) (Habeck et al., 2008; Hanke et al., 2009; Kloeppel et al., 2008; Norman et al., 2006; Stonnington et al., 2010) are frequently used in (often functional) magnetic resonance imaging (MRI) studies to increase detection power (McIntosh et al., 1996; Norman et al., 2006; O’Toole et al., 2007; Yamashita et al., 2008). Recently, multivariate analysis of structural MRI has gained more attention (Grosenick et al., 2013; Ryali et al., 2010; Sabuncu and Van Leemput, 2011). The large majority of these techniques relate a multivariate pattern to a univariate outcome.
Modern datasets allow the opportunity to relate two independent multivariate patterns. Neuroimaging and psychometric batteries describe cognition and the brain itself, respectively, with a matrix of quantitative measurements. These types of datasets may be analyzed with methods such as canonical correlation analysis (CCA) (Cherry, 1996) which is closely related to multivariate regression and partial least squares (Sun et al., 2009). Partial least squares (PLS), without sparseness, has been used for several years in multivariate brain mapping studies (Addis et al., 2004; Chen et al., 2009; Leibovitch et al., 1999; Lin et al., 2003; McIntosh et al., 1996). Ridge and related penalties allow these methods to be applied even when the number of subjects is far fewer than the number of measurements (Nestor et al., 2002b). However, a caveat of these approaches is that the resulting solution vectors have global extent i.e. cover the entire brain with basis vectors that are non-zero and may have both positive and negative values. Traditional approaches are more clearly directional: a long neurological history is founded on relating behavioral deficits (losses) associated with destruction of brain tissue by stroke or related disorders. Perhaps the most famous example is H.M. This epilepsy patient lacked the ability to form new memories after anterior temporal lobe resection. That is, loss of a specific part of the brain resulted in a specific deficit.
Tools such as independent component analysis and principal components analysis (PCA) (Borroni et al., 2012; Comon, 1994; Mansfield et al., 1977; Shamy et al., 2011; Yeung and Ruzzo, 2001) increase power by efficiently describing data. However, PCA solutions provide signed basis vectors with global support and therefore lose the specificity of classical region of interest approaches or lesion studies. Sparse multivariate methods have advantages of interpretability (Lee and Seung, 1999; Suykens et al., 2002) and, potentially, improved generalizability (Elad, 2006; Ryali et al., 2010; Yamashita et al., 2008; Zhang, 2008; Zibulevsky and Elad, 2010). In this paper, we use the cognitive variance induced by a spectrum of neurodegenerative conditions to examine how new, sparse multivariate analysis techniques more powerfully reveal relationships between brain and behavior. At the same time, sparse methods achieve a degree of specificity that cannot naturally be obtained by dimensionality reduction tools such as PCA (Lee and Seung, 1999). Here, we apply sparse multivariate methods to find cortical networks that vary with cognition in a mixed group composed of controls and phenotypes related to Alzheimer’s disease (AD) and frontotemporal lobar degeneration (FTLD) pathology. An example of the difference between sparse solutions and more traditional approaches appears in Fig. 1.
Fig. 1.

Sparse canonical correlation analysis solution vectors are overlaid on a slice of the brain where the brightness of the red-hued overlay is related to the solution’s weighting at the local voxel. A traditional canonical correlation analysis produces component vectors with global extent (to reader’s far left). Sparse solutions (increasingly sparse to the reader’s right) seek to extract controllably focal information thereby, in the context of this paper, isolating “networks” of voxels that collectively relate to cognition. This enables component vectors to be more easily interpreted in terms of traditional neuroscientific coordinate systems.
Like AD, FTLD is a progressive neurodegenerative condition that is accompanied by changes in behavior. Unlike AD, which typically presents atrophy in the precuneus and temporal lobes, FTLD’s pathology occurs more frequently in frontal and temporal lobes (Rabinovici et al., 2007; Whitwell et al., 2007). FTLD phenotypes include patients with a disorder of social comportment and executive functioning (bvFTD); a non-fluent/agrammatic variant of primary progressive aphasia (naPPA), also known as progressive nonfluent aphasia; a semantic variant of primary progressive aphasia (svPPA), also known as semantic dementia; and corticobasal syndrome (CBS). A common test for cognitive deficits in dementia is the Mini-Mental State Examination (Hill and Baeckman, 1995). However, the MMSE does not assess the behavioral and cognitive deficits associated with FTLD (Hutchinson and Mathias, 2007). Other tests have been developed to screen and compare patients with dementia syndromes, including the Frontal Assessment Battery (Dubois et al., 2000) (FAB); the Addenbrooke Cognitive Examination (Galton et al., 2005) (ACE); and the Montreal Cognitive Assessment (MoCA) (Nasreddine et al., 2005).
The Philadelphia Brief Assessment of Cognition (Libon et al., 2007, 2011) (PBAC) provides an economical means to screen and assess important domains of cognitive and behavioral impairment associated with AD and FTLD spectrum phenotypes. The PBAC requires about 12 min for administration and scoring. An important component of the PBAC is the construction of sub-scales designed to assess specific cognitive and behavioral/comportment deficits that typify AD and FTLD syndromes, including executive/working memory, language, visuospatial/constructional skills, verbal/visual episodic memory, and behavior/social comportment. Dementia severity is assessed by summing all PBAC sub-scales. Recent research with the PBAC has demonstrated that AD and FTLD patients present with specific areas of impairment on sub-scales that correspond to phenotypic syndromes (Libon et al., 2011) i.e. clinical diagnosis.
The current study extends previous research with the PBAC (Libon et al., 2011) by examining the gray matter neuroimaging correlates of PBAC’s cognitive and social measurements in a large number of AD and FTLD patients. From a neurological perspective, the purpose, here, is to use the variance within these patients to assess brain and behavior relationships across multiple behavioral loci, as opposed to diagnosis. From a technical perspective, the goal is to contrast univariate and multivariate techniques. To test the hypothesis that PBAC indirectly measures the integrity of different cortical networks (versus individual voxels), we employ a new data-driven machine learning technique, sparse canonical correlation analysis for neuroimaging (SCCAN), to associate high-dimensional imaging measurements with the full information provided by a multivariate psychometric battery such as PBAC. Specifically, this approach allows an optimal weighting of psychometric sub-scales (as opposed to averaging their values) such that the relationship with neuroimaging is maximized. At the same time, SCCAN optimizes and selects regions of gray matter (GM) to maximize correlation with psychometrics. This results in a set of gray matter regions that may be interpreted as the network most-associated with the given psychometric domain. SCCAN previously identified covariation between GM and diffusion tensor imaging white matter (WM) changes that optimally discriminate between CSF- and autopsy-defined patients with AD and FTLD (Avants et al., 2010b). The purpose of the current research is to test the hypothesis that SCCAN may employ individual PBAC sub-scales to extract GM networks that are reproducibly associated with variation in cognition. This would provide additional criterion validity for both the PBAC and multivariate techniques such as SCCAN, in contrast to univariate techniques, and establish a novel strategy for performing multivariate analyses of brain and behavior.
Methods
An overview of our study is in Fig. 2. We first discuss the core dataset and measurements. We then discuss the PBAC and SCCAN methods. We proceed with an evaluation framework, including a comparison against a univariate approach.
Fig. 2.
Diagram of the study. The data splitting in step 1 happens only once. We perform stages 2 and 3 for each of the five PBAC sub-scales.
Patients
Individuals participating in the current research were drawn from a corpus of 270 patients, as previously described (Libon et al., 2011). Dementia patients were evaluated by experienced behavioral neurologists (AC, HBC, RGG, MG) and classified clinically on the basis of previously published criteria (Gorno-Tempini et al., 2011; McKhann et al., 2001; Rascovsky et al., 2011). A research diagnosis was made on the basis of an independent review of a semi-structured history obtained from patients and their families and a detailed neurologic examination. At least two trained reviewers from a consensus committee (inter-rater reliability, r=0.91, p < 0.001) confirmed patients’ clinical diagnosis and the presence of a specific dementia syndrome involving AD or FTLD. Discrepancies were resolved based on group discussion and follow-up assessment. The PBAC was not used for the initial diagnosis of research participants.
The clinical diagnosis of dementia was consistent with serum studies, clinical studies of cerebrospinal fluid (when available), clinical imaging studies such as MRI or CT, and functional neuroimaging studies such as SPECT or PET (these studies were not available to the consensus committee). Exclusion criteria included the presence of other neurologic conditions such as stroke or hydrocephalus, primary psychiatric disorders (e.g., major depression, psychosis), or a systemic illness that can interfere with cognitive functioning. Some patients were taking a cholinesterase inhibitor (e.g. donepezil, galantamine), memantine, or a non-sedating anti-depressant (e.g., serotonin-specific re-uptake inhibitors such as sertraline), or an atypical neuroleptic agent (e.g., quetiapine) consistent with clinical care; however, no patient demonstrated evidence of sedation. The current research examined patients with AD (n=17), behavioral variant-FTD (bvFTD; n = 41), semantic variant-primary progressive aphasia (svPPA; n = 14), non-fluent/agrammatic-primary progressive aphasia (nfaPPA; n = 15) and corticobasal syndrome (CBS; n = 24). The imaging analysis also included elderly controls (n=56) who were living independently in the community and not taking psychoactive medications. Normal control participants presented with no cognitive complaints or impaired instrumental activities of daily living. Table 1 summarizes participant demographic features. This research was approved by the University of Pennsylvania Institutional Review Board and informed consent was obtained consistent with the Declaration of Helsinki.
Table 1.
The demographics for this study of 164 subjects are listed as mean/standard deviation in each column. For the testing and training split, subjects are matched on age, education and MMSE. MMSE = Mini-Mental State Examination, AD = Alzheimer’s disease, bvFTD=behavioral variant of frontotemporal dementia, nfaPPA=non-fluent agrammatic variant of primary progressive aphasia; svPPA = semantic variant of primary progressive aphasia, CBS = corticobasal syndrome, NC = normal control.
| DX | N | Age | Education | MMSE | |
|---|---|---|---|---|---|
| 1 | AD | 17.00 | 70.88/7.79 | 14.82/3.17 | 21.94/4.75 |
| 2 | bvFTD | 41.00 | 61.51/9.55 | 16.05/3.66 | 25.44/4.85 |
| 3 | nfaPPA | 15.00 | 66.2/10.64 | 14.27/2.25 | 20.73/7.94 |
| 4 | svPPA | 14.00 | 63.21/5.85 | 17.64/2.34 | 19.14/10.16 |
| 5 | CBS | 24.00 | 62.42/8.52 | 14.33/2.46 | 22.29/3.94 |
| 6 | NC | 56.00 | 64.88/8.86 | 15.38/2.76 | 20.55/8.33 |
The Philadelphia Brief Assessment of Cognition (PBAC)
Full details regarding the rationale and construction of the PBAC can be found elsewhere (Libon et al., 2011). The PBAC consists of 20 variables grouped into 5 domain-specific rating scales. These variables are grouped into five sub-scales measuring: working memory/executive control, language, visuospatial/constructional ability, verbal/visual episodic memory, and behavior/social comportment. The total PBAC score ranges between 0 and 93. The executive scale includes measurements of fluency, digits backward and digits forward. The language scale measures naming, speech, reading, writing and semantic ability. The memory scale quantifies delayed free recall, recognition and Rey recall. The visuo-spatial scale measures judgment of line orientation (JOLO) and the Rey copy test. The behavioral scale includes subjective measurements of apathy, disinhibition, social comportment, agitation, empathy and ritual. The correlations between these different sub-scales are shown in Fig. 3.
Fig. 3.
We visualize, with a heatmap, the correlations between the different PBAC individual scales which are clustered together to form the sub-scales studied here. The total PBAC is an average of the 5 sub-scale scores. The sub-scales provide a reasonable separation of measurements.
Image acquisition
All images were acquired with a Siemens Trio 3.0Tesla MRI scanner. Following a rapid sagittal T1-weighted scan to determine patient position, a T1-weighted structural image was acquired with TR = 1620 ms, TE = 3 ms, slice thickness = 1 mm, in-plane resolution = 0.9766 mm × 0.9766 mm, and FOV = 256 × 192.
Image processing
The imaging analysis is based on the publicly available and open-source Advanced Normalization Tools (ANTs, http://stnava.github.io/ANTs/) and the associated pipelining framework PipeDream (http://neuropipedream.sourceforge.net). PipeDream automates and quality-assures ANTs processing via a single parameter file and data organization hierarchy. Each patient’s T1 imaging data are inhomogeneity corrected via the N4 bias correction algorithm (Tustison et al., 2010). PipeDream then performs diffeomorphic normalization via the top-performing symmetric normalization methodology available in ANTs (Avants et al., 2008, 2011a, 2012; Tustison et al., in press) to map each subject to a population-specific template built from the same scanner and imaging parameters. The template contains prior labeling and probability maps that are used to guide both brain extraction and neuroanatomical segmentation. Segmentation is performed with a Markov Random Field approach (Avants et al., 2011b) implemented in the ANTs toolkit which has been validated on publicly-available datasets. GM probability maps are then smoothed by a 2 mm Gaussian kernel, mapped to the template space, and down-sampled to 2mm resolution. These normalized GM probability maps are used for subsequent multivariate correlation with PBAC.
Dimensionality reduction/statistical analysis
There are two primary reasons that univariate approaches lack power. First, power is compromised because the same test is repeated at each measurement site leading to an often severe multiple comparisons problem. Second, univariate methods do not exploit the latent signal in the data that is spread across measurement sites. In this study, rather than perform voxel-wise testing, we employ a dimensionality reduction method implemented in SCCAN. Briefly, this is an imaging-specific extension of sparse canonical correlation analysis (Avants et al., 2010a; Cao et al., 2009; Witten and Tibshirani, 2009; Witten et al., 2009) that is itself a sparse extension to Hotelling’s seminal canonical correlation analysis (CCA) (Hotelling, 1935, 1936). CCA, in turn, is a multi-modality extension to principal component analysis.
Classical CCA may be used to compute a multivariate association between two different views of a dataset. One of Hotelling’s original examples associated measurements of height and weight to measurements of cognition. More recently, sparse versions of CCA have been developed to increase the interpretability of the output where we take motivation for sparse methods from our introductory material. Sparse CCA methods, like classical CCA, compute eigenvectors (in actuality, pseudo-eigenvectors) that maximize the Pearson correlation between the input modalities.1 In this work, we use sparse CCA in a similar way to Hotelling’s classic study, i.e. to associate two different types of measurements, one anatomical and one psychometric, in a population.
We employed the SCCAN implementation of sparse CCA to directly associate the five PBAC sub-scales described above with GM measures taken from T1-weighted MR images. In general, SCCAN elucidates the relationship between two sets of measurements taken across a population and is thus well-suited to multivariate neuroimaging data. The input to SCCAN is two matrices X and Y. The size of X is n × p where n is the number of subjects and p is the number of voxels from the cortical gray matter. The matrix thus collects all cortical imaging data for each subject. The size of Y is n × q where there are q psychometric measurements in the given PBAC sub-scale i.e. q=3 for executive, 5 for language, 3 for memory, 2 for visuospatial and 6 for behavioral scales.2 SCCAN maximizes the Pearson correlation, in a rotated space, between nonnegatively weighted columns of these matrices. More formally, SCCAN, like classic CCA, introduces new unknown solution vectors, x (p × 1) and y (q × 1) that act as weight functions on columns of X and Y. The SCCAN optimization criterion is:
| (1) |
where x* and y* are the optimal solution vectors, xj denotes the jth entry of x, s determines the sparseness level and ||·||1 denotes the ℓ1 norm. The ℓ1 norm forces the solution x to be sparse i.e. have zero value over the majority of the brain. In this application, we do not enforce sparseness on the y vector as it is relatively small, i.e. q≪n. SCCAN’s objective function can be optimized even when the matrices are fat i.e. p≫n, often the case in neuroimaging studies. Due to the non-linear (even np-hard) nature of subset selection (sparse optimization) from a large matrix, optimizing for a single canonical variate pair, x, y, involves a nonlinear gradient descent on the objective function above. The analytical gradient of the objective function (ignoring the ℓ1 and positivity constraints) w.r.t. x is:
| (2) |
Following this gradient will cause the candidate solution x to leave the permissible solution space and, as such, we follow the gradient update step with a projection step as in Polak (1997) and Schwartz and Polak (1997). The projection step involves a standard soft-thresholding operation used in ℓ1 optimization which is easily modified to include positivity constraints (Donoho, 1995; Elad, 2006; Tibshirani, 1996; Witten and Tibshirani, 2009). The gradient for y is obtained at the same time as that for x with a simple switching of variables. Following, we refer to x* as x and y* as y interchangeably.
SCCAN therefore produces a sparse projection vector acting on GM voxels that, taken as a set, maximally correlate with the user-selected PBAC domain of interest. Here we use the working memory/executive, language, visuospatial, memory, and social/behavioral PBAC sub-scales, thus requiring only five direct multivariate tests, i.e., one for each cognitive domain. The sparse eigenvectors that emerge from SCCAN identify the brain regions supporting the specific PBAC domain that was passed to the algorithm as the second view. In this study, we restrict the SCCAN eigenvectors to be positive. Thus, each eigenvector can be reinterpreted as a weighted average of values over a restricted region of GM, like a region of interest (Chen et al., 2010; Poldrack, 2007; Rasmussen et al., 2012; Zhou et al., 2011). This may be achieved in a post-processing step that sets the sum of the solution vector, x or y, to equal one. The implementation details are available in the Advanced Normalization Toolkit’s sccan.cxx program which contains the SCCAN source code. The significance of SCCAN results may be tested by permuting one of the two views over many different simulations. One then compares the correlation value produced by the original ordering of the data to the correlations produced over the N permutations. Large N (typically >1000) is needed to provide a reasonable sampling of the empirical null distribution. In this analysis, we use a training and testing paradigm3 to avoid permutation testing and to allow us to perform parameter selection (for s) on the training dataset in a manner that is independent of the test dataset. Finally, we note that ANTsR example code for SCCAN section highlights a few of the key steps in this analysis as implemented with a pre-release of the ANTsR software.
Parameter selection
Image processing
Our studies used reproducible research practices by employing open methods, standardized parameter sets and version control of both parameters and code via git. We employed standard parameters in PipeDream and ANTs for template construction, normalization, segmentation and GM estimation. These analysis protocols are described elsewhere (Avants et al., 2011a, b; Tustison et al., 2010, in press).
Statistical parameters
SCCAN was used to extract the single most dominant features associating GM and a PBAC sub-scale in a training dataset. Our study design involves setting only a single parameter, s, which controls the sparseness of the imaging space solution vector, x. We chose to restrict x to also be nonnegative such that the solutions can be interpreted with clear directionality i.e. as a weighted average. To identify the parameter s, we run SCCAN with a range of scandidate values (s = 0.005, 0.01, …, 0.5) and store the resulting SCCAN correlation for each trial. We then fit a model, SCCAN-correlation ≈β1scandidate + β2(scandidate)2 + β3(scandidate)3 + β4(scandidate)4, to the function mapping s → SCCAN-correlation. We set the final parameter s to correspond to the value of our model that maximizes the dependent SCCAN-correlation value. If there are ties, we take the first from the left (maximally sparse) solution. We then compute the final solutions x*, y* that will be evaluated for reproducibility in our test dataset. We did not employ sparseness on the 2nd view (the PBAC sub-scale scores). We need only to perform 5 tests in total as opposed to 5p tests required by a univariate setting. Here, p = 90,084 and 5p = 450,420. Parameter selection results are in Fig. 4.
Fig. 4.

A one parameter search over sparseness, in the training dataset, allows us to identify the optimal sparseness parameter for each cognitive domain. The network variables x* and y* that arise from SCCAN computed at the optimal sparseness level will be evaluated in the test dataset for reproducibility.
Comparison of univariate to multivariate feature selection
This study employs a training and testing paradigm. Therefore, SCCAN may be viewed as a tool that generates “feature vectors” (x, y) that are optimized in a multivariate manner to associate GM and PBAC. Using our existing terminology, these maximize Corr(Xx, Yy) where Corr denotes Pearson correlation and X and Y correspond to the training dataset matrices. These feature vectors may be used as “hypotheses” and applied to a new testing dataset, Xtest and Ytest, to determine if the patterns extracted from training data are reliable in test data. In this analysis, we use the correlation in test data as our outcome measurement which is computed as OutcomeMultivariate=Corr(Xtestx, Ytesty) where x and y are derived from training data.
We can also compute a parallel univariate outcome, Outcomeunivariate, via a similar methodology based on univariate feature selection. We use Occam’s razor to decide upon a univariate feature selector i.e. we do something simple-minded yet akin to several other studies (Chen et al., 2010; Dickerson and Wolk, 2012). We compute the univariate p-values of a model associating the PBAC sub-scale summary measurement (an average of the Z transformations of the individual measurements within the sub-scale) with each voxel in the cortex. This univariate model may be written as PBAC-sub-scale ≈ vi where vi is a vector containing the subject GM values at a given voxel (so i takes values 1…p). The p-value associated with vi is then denoted as pi. Now, define u as a p-length weight vector similar to the weight vector x. However, the entry at ui is zero if pi is > 0.01 (unadjusted) and 1 otherwise. This finally allows us to define the univariate outcome as Outcomeunivariate = Corr(Xtestu, PBAC-sub-scaletest).
This protocol allows us to compare a joint multivariate feature selector that identifies weights on PBAC sub-scale values (y) and the brain (x) with a univariate feature selector acting only on the brain (u). While the univariate approach does not weight the PBAC-sub-scale, it does use a standard average of the sub-scales commonly used in PBAC assessment. Thus, we compare a new, fully multivariate approach, to an existing standard approach. One may argue that the threshold selected to binarize the u vector is arbitrary. However, this is a common/standard style of analysis employed in univariate methods, i.e. select a significance threshold and accept the results as the truth. The univariate solutions do not survive multiple comparison correction so we had no choice but to use uncorrected p-values as feature selectors.
Results
Univariate: PBAC sub-scales and GM density
No univariate outcome measurement achieves significance when relating brain and behavior in the test data. That is, the correlation between PBAC-sub-scaletest with Xtestu is weak in each of the five PBAC sub-scales (p-values: exec 0.93, language 0.078, memory 0.85, visuospatial 0.66, behavioral 0.78). However, the relationship between PBAC-sub-scaletrain with Xtrainu is strong, as expected (all p-value<0.01). This indicates that, at least in this dataset, naive univariate feature selection overfits to the training data.
Multivariate: PBAC sub-scales and GM density
Based on training data, we found, in testing data, significant associations between GM density and each of the five PBAC sub-scales consistent with putative neuroanatomical substrates at the Bonferroni-corrected p < 0.01 level. Scatterplots are in Fig. 6 where raw p-values are also reported (rounded below 0.0001 to 0). Fig. 5 shows the x function for each sub-scale plotted on the brain. Note these are sparse functions i.e. no additional thresholding is performed to generate these overlays. Fig. 6 shows the test data relationship between brain and behavior, as well as a visualization (in heatmap form) of each PBAC sub-scale.
Fig. 6.
We visualize the correlation between Xtestx* and Ytesty* for each of the five PBAC sub-scales. We also show the PBAC sub-scales and their corresponding putative support regions in the cortex, as identified by SCCAN and verified in testing data. Each row, from the top, contains the results for the behavioral/social comportment scale, the executive/working memory scale, the language scale, the episodic memory scale and the visuospatial scale.
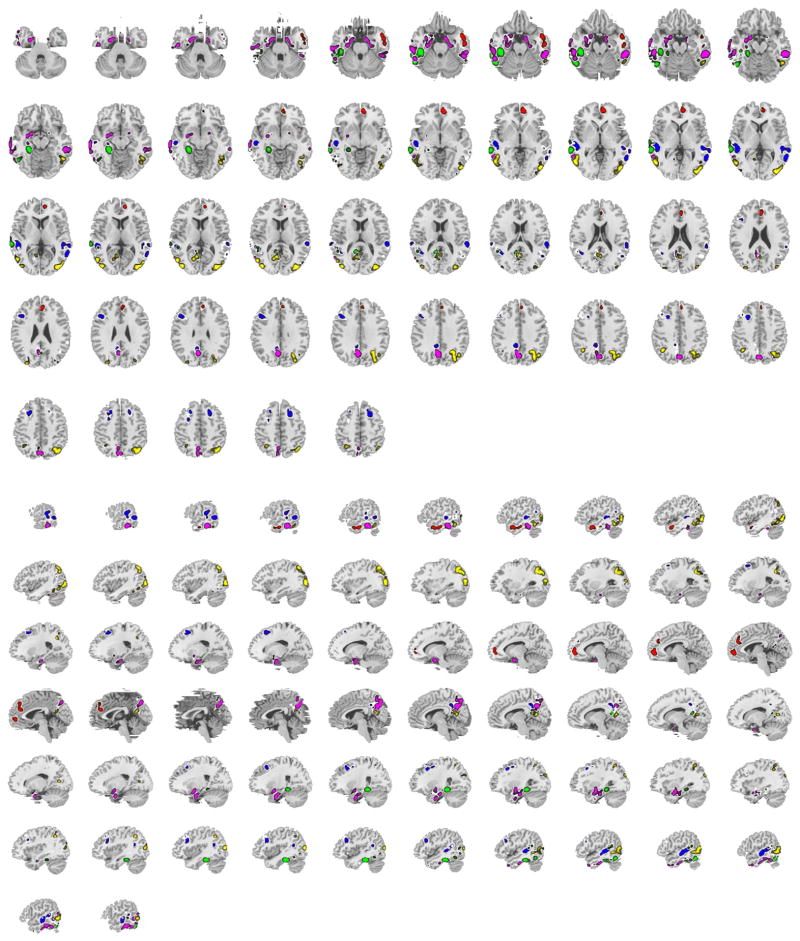
Fig. 5.
All of the x* solution vectors are combined in axial and sagittal views of the brain. Red is behavior, blue executive, green language, magenta episodic memory and yellow visuospatial. The left hemisphere of the brain is on the reader’s left in the axial view.
Reporting multivariate results
We display SCCAN pseudo-eigenvectors on the brain in a manner that is similar to traditional voxel-based analysis, as in Fig. 5. As mentioned in Methods section, these weight vectors are similar to weighted averages in that the difference between these vectors and a traditional weighted average is only a scalar multiplication. Therefore, the effect on a correlation is null. We also tabulate results in Talairach coordinates similar to traditional methods (e.g. Table 2). However, instead of plotting coordinates of peak p-values, we annotate the Talairach coordinate of each spatial component of the pseudo-eigenvector such that we gain a degree of spatial resolution in the interpretation of SCCAN results. Despite using a familiar reporting system, the key point is that p-values are assessed over the relationships computed from the entire operation on the multivariate dataset (e.g. Corr(Xtestx*, y*)). Thus, we are reporting the collection of neuroanatomical data points that contribute to the relationship with cognition as opposed to a single point. We estimate Brodmann areas and AAL labels from the label sets included with mricron (Rorden et al., 2007). Fig. 6 displays results in a glass brain where we draw connections between the major sub-regions of the pseudo-eigenvectors. We also show, via a heatmap, the distribution of cognitive scores for each PBAC sub-scale. Most importantly, we display the distribution of the test dataset gray matter against cognition.
Table 2.
The approximate Talairach coordinates and AAL labels for network behav with significance 1.31e–06 and sparseness 0.034. Weights on the multivariate PBAC sub-scale (the y) are 0.029, 0.027, 0.028, 0.017, 0.017, and 0.016.
| x | y | z | t | Label | Brodmann | AAL | |
|---|---|---|---|---|---|---|---|
| 1 | 52.00 | −6.00 | −21.00 | 0 | 1 | 21 | Temporal_Mid_R |
| 2 | 8.00 | 50.00 | −5.00 | 0 | 2 | 10 | Frontal_Med_Orb_R |
| 3 | 4.00 | 39.00 | 23.00 | 0 | 3 | 32 | Cingulum_Ant_R |
As summarized in Fig. 6 and Table 2, PBAC-defined behavior/social comportment focuses on medial and inferior prefrontal cortex. The memory sub-scales (Table 5) relates to precuneus, hippocampus, and bilateral posterior temporal atrophy more prominently on the left than the right. Significant visuospatial/constructional impairment (Table 6) relates to bilateral posterior temporal–occipital and bilateral parietal–occipital lobe. The language sub-scale (Table 4) was related to left temporal and left temporal–parietal gray matter. The executive network involves a distributed temporal and frontal network and is described in Table 3.
Table 5.
The approximate Talairach coordinates and AAL labels for the memory network with significance 1.03e–05 and sparseness 0.173. Weights on the multivariate PBAC sub-scale (the y) are 0.064, 0.061, and −0.004. Left hippocampus is clustered together with parahippocampal gyrus.
| x | y | z | t | Label | Brodmann | AAL | |
|---|---|---|---|---|---|---|---|
| 1 | −3.00 | −65.00 | 34.00 | 0 | 1 | 0 | Precuneus_L |
| 2 | 57.00 | −38.00 | −16.00 | 0 | 2 | 20 | Temporal_Inf_R |
| 3 | −57.00 | −30.00 | −21.00 | 0 | 3 | 20 | Temporal_Inf_L |
| 4 | −22.00 | −11.00 | −27.00 | 0 | 4 | 36 | ParaHippocampal_L |
| 5 | −26.00 | −5.00 | −16.00 | 0 | 5 | 34 | Amygdala_L |
| 6 | 16.00 | −9.00 | −22.00 | 0 | 6 | 28 | ParaHippocampal_R |
| 7 | −68.00 | −28.00 | −11.00 | 0 | 7 | 21 | Temporal_Mid_L |
| 8 | −57.00 | −54.00 | −0.00 | 0 | 8 | 37 | Temporal_Mid_L |
| 9 | −56.00 | −50.00 | −19.00 | 0 | 9 | 37 | Temporal_Inf_L |
| 10 | −66.00 | −14.00 | −17.00 | 0 | 10 | 21 | Temporal_Mid_L |
| 11 | −4.00 | −59.00 | 47.00 | 0 | 11 | 7 | Precuneus_L |
| 12 | −57.00 | −66.00 | 10.00 | 0 | 12 | 37 | Temporal_Mid_L |
| 13 | −66.00 | −44.00 | −11.00 | 0 | 13 | 20 |
Table 6.
The approximate Talairach coordinates and AAL labels for network vs with significance 0.0007 and sparseness 0.13. Weights on the multivariate PBAC sub-scale (the y) are 0.065 and 0.054.
| x | y | z | t | Label | Brodmann | AAL | |
|---|---|---|---|---|---|---|---|
| 1 | 34.00 | −64.00 | 39.00 | 0 | 1 | 7 | Angular_R |
| 2 | 41.00 | −81.00 | 8.00 | 0 | 2 | 19 | Occipital_Mid_R |
| 3 | −52.00 | −65.00 | 3.00 | 0 | 3 | 37 | Temporal_Mid_L |
| 4 | −40.00 | −83.00 | 11.00 | 0 | 4 | 19 | Occipital_Mid_L |
| 5 | 50.00 | −55.00 | −10.00 | 0 | 5 | 37 | Temporal_Inf_R |
| 6 | −10.00 | −64.00 | 11.00 | 0 | 6 | 17 | Calcarine_L |
| 7 | −37.00 | −55.00 | 39.00 | 0 | 7 | 39 | Angular_L |
| 8 | 46.00 | −65.00 | −7.00 | 0 | 8 | 37 | Temporal_Inf_R |
| 9 | −29.00 | −56.00 | 49.00 | 0 | 9 | 7 | Parietal_Sup_L |
| 10 | 2.00 | −60.00 | 19.00 | 0 | 10 | 23 | Calcarine_R |
| 11 | −30.00 | −86.00 | 26.00 | 0 | 11 | 19 | Occipital_Mid_L |
Table 4.
The approximate Talairach coordinates and AAL labels for network lang with significance 3.26e–10 and sparseness 0.109. Weights on the multivariate PBAC sub-scale (the y) are 0.051, 0.006, 0.035, 0.03, and 0.013.
| x | y | z | t | Label | Brodmann | AAL | |
|---|---|---|---|---|---|---|---|
| 1 | −56.00 | −39.00 | −13.00 | 0 | 1 | 20 | Temporal_Inf_L |
| 2 | −26.00 | −38.00 | −11.00 | 0 | 2 | 30 | ParaHippocampal_L |
| 3 | −55.00 | −67.00 | 1.00 | 0 | 3 | 37 | Temporal_Mid_L |
| 4 | −6.00 | −58.00 | 18.00 | 0 | 4 | 23 | Precuneus_L |
| 5 | −54.00 | −60.00 | −16.00 | 0 | 5 | 37 | Temporal_Inf_L |
Table 3.
The approximate Talairach coordinates and AAL labels for network exec with significance 9.31e–07 and sparseness 0.159. Weights on the multivariate PBAC sub-scale (the y) are 0.031, 0.054, and 0.038.
| x | y | z | t | Label | Brodmann | AAL | |
|---|---|---|---|---|---|---|---|
| 1 | −51.00 | −35.00 | 3.00 | 0 | 1 | 22 | Temporal_Mid_L |
| 2 | −64.00 | −41.00 | −15.00 | 0 | 2 | 20 | Temporal_Inf_L |
| 3 | 60.00 | −54.00 | 4.00 | 0 | 3 | 37 | Temporal_Mid_R |
| 4 | 20.00 | 19.00 | 46.00 | 0 | 4 | 0 | Frontal_Sup_R |
| 5 | 61.00 | −32.00 | 15.00 | 0 | 5 | 42 | Temporal_Sup_R |
| 6 | −43.00 | 19.00 | 24.00 | 0 | 6 | 48 | Frontal_Inf_Tri_L |
| 7 | −25.00 | 19.00 | 39.00 | 0 | 7 | 0 | Frontal_Mid_L |
| 8 | −7.00 | −54.00 | 27.00 | 0 | 8 | 0 | Precuneus_L |
| 9 | 51.00 | −33.00 | 2.00 | 0 | 9 | 21 | Temporal_Mid_R |
| 10 | 65.00 | −37.00 | 4.00 | 0 | 10 | 22 | Temporal_Mid_R |
| 11 | −9.00 | −45.00 | 35.00 | 0 | 11 | 0 | Cingulum_Mid_L |
Discussion
This study establishes, for the first time, that sparse canonical correlation analysis finds repeatable relationships between multivariate psychometric batteries and network level gray matter density measurements. Our analysis restricts the signs of the gray matter density solution vector, x*, to be sparse and positive enabling directional relationships to be established (i.e. low gray matter, low cognitive score). The SCCAN multivariate analysis framework shows improved generaliz-ability and power over the univariate method against which it is compared.
This study also represents the first comparison of PBAC sub-scales to structural neuroimaging. The PBAC was designed as a screening instrument to assess overall dementia severity and to differentiate between neurodegenerative syndromes within the AD and FTLD spectrum. Previous research with the PBAC found a significant correlation between the total PBAC score and the MMSE (Libon et al., 2007, 2011), thus demonstrating that performance on the instrument reasonably captures overall dementia severity. The psychometric qualities of the PBAC include good internal consistency among the tests within each PBAC sub-scale and highly significant intra-class correlations between PBAC sub-scales and standard neuropsychological tests tetlibon-Philadelphia-2011. PBAC sub-scales also show good clinical utility in distinguishing between AD and FTLD, and between FTLD-related phenotypes using sub-scale cut scores, although diagnosis is not the focus here.
The current research used novel imaging methods to provide further evidence for the criterion validity of the PBAC by associating PBAC sub-scale test performance with corresponding areas of GM atrophy. The heterogeneity of this population (which includes controls and five different clinical phenotypes) drives these relationships. For instance, patients with AD are known to present with striking memory impairment as well as atrophy involving the hippocampus, precuneus and related anatomic structures. In the current research, the PBAC memory sub-scale was associated with GM involving left hippocampus mid-body, anterior hippocampus and precuneus. Bilateral inferior temporal cortex and right parahippocampal gyrus are also involved. Hippocampal and precuneus involvement in clinically-defined patients with anterograde amnesia and AD is well known (Nestor et al., 2002a; Pengas et al., 2010). Moreover, prior research with AD and amnesic mild cognitive impairment suggests that these areas are part of a neurocognitive network for memory (Delano-Wood et al., 2012; Gardini et al., 2011). Table 5 shows all regions associated by SCCAN to the verbal and visual episodic memory sub-scale.
The behavioral variant of FTLD induces variance in the behavioral sub-scale. bvFTLD presents with alterations in social comportment (Rascovsky et al., 2011; Shany-Ur and Rankin, 2011), including apathy, disinhibition, and lack of empathy. In the current study, the PBAC-behavior/social scale is related to striking bilateral medial and ventral frontal GM, perhaps with greater right-sided involvement. Previous research has associated these behavioral abnormalities with bilateral ventral and medial frontal atrophy (Massimo et al., 2009; Rosen et al., 2010).
Similarly, CBS is associated with significant impairment in visuospatial and visuoconstructional tests, as well as some executive deficits (Libon et al., 2007, 2009). The current research associates the PBAC-visuospatial/constructional sub-scale with right posterior GM atrophy including parietal and occipital lobes. Prior neuroimaging research with CBS patients is controversial, with some researchers suggesting both parietal and frontal atrophy (Grossman and Ash, 2004) and others only frontal involvement (Hassan et al., 2011). In the present study, atrophy in primarily right visual association cortex was related to the visuospatial/constructional sub-scale which, in turn, was designed to identify individuals with the clinical diagnosis of CBS. Table 6 highlights the visuospatial network defined by SCCAN.
In contrast to other domains, the executive/working memory sub-scale is not typically associated with a single diagnosis but instead is associated with several diagnostic subgroups, including CBS, bvFTLD and AD. Prior research with the PBAC has shown that patients with naPPA score lower on the PBAC-working memory/executive sub-scale (Libon et al., 2007, 2011) confirming executive and working memory deficits in these patients and reflecting that their disease centered primarily in the frontal lobe (Gunawardena et al., 2010; Rogalski et al., 2011). The executive network highlights diverse regions (see Table 3) including superior frontal and middle frontal gyrus (a classic working memory region) and superior temporal gyrus which has been previously implicated as a neural substrate for the digits backward test (Li et al., 2012). Inspecting Fig. 5 shows the executive scale extracts several brain regions that form key points in the default mode network such as precuneus, superior prefrontal cortex, lateral temporal lobe and lateral superior parietal cortex (Buckner et al., 2008). The PBAC-working memory/executive sub-scale consists of verbally-mediated tests and, as such, several language regions are involved as well.
The PBAC language sub-scale is strongly related to left temporal GM density, primarily with prominent posterior and inferior temporal GM. These data are consistent with prior research demonstrating left temporal atrophy in patients with svPPA who have language-related cognitive impairment (Bonner et al., 2009; Williams et al., 2005). The measures on the PBAC language sub-scale focus mostly on object comprehension, single word expression, reading and writing, with little emphasis on the characteristics important for identifying naPPA (Gorno-Tempini et al., 2011). Future modifications of the PBAC may include measures to improve detection of regions that would be specifically impaired in the naPPA subgroup. Such focused tests are not currently included in PBAC.
Taken together, these data extend past research to show that PBAC sub-scales are associated with MRI-defined anatomical substrates. The identified relationships between neurocognitive GM networks and psychometrics underscore the power of SCCAN over traditional univariate approaches to imaging analysis. By using SCCAN, we obtain massive dimensionality reduction which significantly enhances the empirical ability to detect these behavioral and neuroimaging relationships in both training and testing data. Prior work demonstrated the advantages of SCCAN in combining GM and diffusion tensor imaging data to distinguish between autopsy- and CSF-defined cases of AD and FTLD (Avants et al., 2010b). The results of the current study reveal that SCCAN is capable of associating multi-dimensional psychometric and neuroimaging data to reveal the large-scale neural networks that are degraded when cognition is compromised in neurodegenerative conditions like AD and FTLD.
There are several caveats to this study. A primary technical issue is whether the network structure (i.e. distributed sets of voxels) that SCCAN extracts is either a minimal or maximal reliable network supporting the selected cognitive domain. Indeed, it is likely that subsets of the voxels added to or subtracted from the SCCAN solutions may not alter predictive accuracy to a strong degree. This hypothesis is strengthened by the fact that our sparseness versus correlation curves (Fig. 4) is relatively smooth. However, identifying an optimally predictive set of high-dimensional variables across arbitrary subsets of a population is a generally unsolved problem. We elected to address this by defining our networks based on a tractable parameter search strategy that maximizes the SCCAN correlation in training data. A secondary issue is that our comparison with the univariate strategy may not be ideal. There are several alternative approaches for univariate feature selection that may be employed with better results, for instance optimizing the recently proposed cluster-level FDR (Chumbley and Friston, 2009). We leave this question to future work by ourselves or others and also note that this approach would remain univariate in the dependent variable. SCCAN, on the other hand, uses a multivariate treatment of both datasets, cognition and neuroimaging.
The clinical aspects of this research also have limitations. First, the PBAC was not designed nor should it be used as a substitute for a comprehensive neuropsychological evaluation. This is reflected most clearly in the fact that PBAC sub-scales appear to extract primarily posterior temporal regions, medial frontal regions, medial posterior regions and parietal lobe. There is little representation of motor regions and anterior temporal regions. Second, the sample size for several groups was relatively modest which makes evaluating diagnostic power very challenging due to the large number of diverse phenotypes, i.e. 5 neurodegenerative and one control group. The results of the present study will require confirmation with larger groups of patients. Further evaluation of SCCAN as a tool to aid diagnosis and prognosis is also required. Third, virtually all work with the PBAC has been confined to differentiating AD from FTLD and to distinguishing between FTLD phenotypes. The utility of the PBAC in other populations such as Parkinson’s disease, amyotrophic lateral sclerosis, and Mild Cognitive Impairment requires further examination. Finally, SCCAN associated some unexpected brain regions with a specific cognitive domain. Additional work is needed to understand the basis for these associations and whether these additional voxels are part of a minimal or maximal predictive network. With these limitations in mind, the current study suggests the power of a multivariate analytic approach such as SCCAN to interrogate multivariate brain and behavior relationships.
Acknowledgments
Dr. Grossman receives support from the National Institutes of Health (AG32953, AG17586, AG15116, NS44266, and NS53488) and the Wyncote Foundation, and is a consultant for Bristol Meyers Squibb and Allon Therapeutics. Dr. Chatterjee receives support from National Institutes of Health (RO1 DC008779, RO1 HD050199) and the National Science Foundation (SBE0541957). Dr. McMillan receives support from NIH HD0406. Ms. Massimo receives support from National Institutes of Health (F31NR013306) and John A. Hartford Foundation’s Building Academic Geriatric Nursing Capacity Award Program. Dr. Libon receives support for Bayer Pharmaceuticals.
Appendix A. ANTsR example code for SCCAN
We first load the relevant libraries for this analysis.
suppressPackageStartupMessages(library(ANTsR)) library(xtable) library(abind) library(grDevices) library(visreg) library(vegan)
Several organizational steps are not included. The key steps of univariate and multivariate feature selection, along with assessment on testing data, are shown below.
# do the univariate test to get the sparseness param
print(“UNIVARIATE”)
voxside <- “~ vox”
if (nam == “exec”)
myform <- as.formula(paste(unames[l], voxside))
if (nam == “lang”)
myform <- as.formula(paste(unames[2], voxside))
if (nam == “vs”)
myform <- as.formula(paste(unames[3], voxside))
if (nam == “mem”)
myform <- as.formula(paste(unames[5], voxside))
if (nam == “behav”)
myform <- as.formula(paste(unames[6], voxside))
print(myform)
bynum <- 1
ss <- seq(l, ncol(trainmat), by = bynum)
ntests <- length(ss)
progress <- txtProgressBar(min = 0, max = ntests, style = 3)
upvs <- rep(1, ncol(trainmat))
for (i in ss) {
vox <- trainmat[, i]
fit <- lm(myform, data = demog[permutesubs, ])
upvs[i] <- coefficients(summary(fit))[2, 4]
if (i%%1000 == 0) {
setTxtProgressBar(progress, i)
}
}
sigthresh <- 0.01
upvs[is.na(upvs)] <- 1
qvsm <- upvs
qvsm[upvs < sigthresh] <- 1
qvsm[upvs >= sigthresh] <- 0
qvsm <- qvsm/sum(qvsm)
qvsm <- matrix(qvsm, nrow = 1)
vox <- testmat %*% t(qvsm)
fit <- lm(myform, data = demog[permutesubs2, ])
print(summary(fit))
uvimg <- antsImageClone(maskimg)
uvimg[mask] <- (1 - upvs)
antsImageWrite(uvimg, paste(DIR, nam, “_uv.nii.gz”, sep =“ ”))
resUV <- lappend(resUV, c(nam, coefficients(summary(fit))[2, 4]))
# now do the multi-variate equivalent
print(paste(“MULTIVARIATE”))
if (length(sparMV) < 5) {
sparlist <- (seq(1, 50, by = 4)/100)
corrlist <- list()
for (sigfrac in sparlist) {
print (paste(“SEARCH:”, sigfrac))
ff <- sparseDecom2(inmatrix = list(trainmat, cogmat[permutesubs, nw]),
inmask = c(maskimg, NA), sparseness = c(sigfrac, -1), nvecs = 1,
its = 15)
myprojla <- trainmat %*% t(imageListToMatrix(ff$eigl, maskimg))
myprojlb <- cogmat [permutesubs, nw] %*% as.matrix(ff$eig2)
corrlist <- lappend (corrlist, cor.test(myprojla, myprojlb)$est)
}
corrlist <- unlist(unlist(corrlist))
fit <- lmCcorrlist ~ sparlist + I(sparlist^2) + I(sparlist^3) + I(sparlist^4))
pdf(paste(“figs/sccan_param_”, nam, “.pdf”, sep = “ ”))
mytitle <- paste(“Correlation v. Sparseness”, nam, sep = “ ”)
visreg(fit, main = mytitle, xlab = “Sparseness”, ylab = “SCCAN correlation”,
cex.main = 2, cex.lab = 2)
dev.off()
sigfrac <- sparlist[which(corrlist == max(corrlist))]
sp <- data.frame(sparlist = (c(1:200)/1000))
pc <- predict(fit, newdata = sp)
sigfrac <- sp$sparlist[which.max(pc)]
sparMV <- lappend(sparMV, sigfrac)
} else sigfrac <- sparMV[[opt]]
ff <- sparseDecom2(inmatrix = list(trainmat, cogmat[permutesubs, nw]),
inmask = c(maskimg, NA), sparseness = cCsigfrac, -1),
nvecs = 1, its = 15)
myproj1a <- trainmat %*% t(imageListToMatrix(ff$eig1, maskimg))
myproj1b <- cogmat [permutesubs, nw] %*% as.matrix(ff$eig2)
myproj2a <- testmat %*% t(imageListToMatrix(ff$eig1, maskimg))
myproj2b <- cogmat[permutesubs2, nw] %*% as.matrix(ff$eig2)
cogweights <- c(round(ff$eig2 * 1000)/1000)
print(cogweights)
imaging <- myproj2a[, 1]
cognition <- myproj2b[, 1]
myform <- as.formula(“cognition ~ imaging”)
fit <- lm(myform, data = demog[permutesubs2, ])
mypv <- coefficients(summary(fit))[2, 4]
print(summary(fit))
Footnotes
Pseudo-eigenvectors are based on optimizing an objective function that is related to the eigenvalue problem but may not strictly satisfy the eigenvalue problem and related constraints.
One column of the behavioral scales (self insight) was excluded due to lack of variance.
As recommended by reviewers.
Conflict of interest
Drs. Rascovsky and Avants, and Ms. Boller report no disclosures.
References
- Addis Donna Rose, McIntosh Anthony R, Moscovitch Morris, Crawley Adrian P, Pat McAndrews Mary. Characterizing spatial and temporal features of autobiographical memory retrieval networks: a partial least squares approach. Neuroimage. 2004;23(4):1460–1471. doi: 10.1016/j.neuroimage.2004.08.007. http://dx.doi.org/10.1016/j.neuroimage.2004.08.007 (Dec. URL http://dx.doi.org/10.1016/j.neuroimage.2004.08.007) [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008 Feb;12(1):26–41. doi: 10.1016/j.media.2007.06.004. http://dx.doi.org/10.1016/j.media.2007.06.004. URL http://dx.doi.org/10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants Brian, Cook Philip A, McMillan Corey, Grossman Murray, Tustison Nicholas J, Zheng Yuanjie, Gee James C. Sparse unbiased analysis of anatomical variance in longitudinal imaging. Med Image Comput Comput Assist Interv. 2010a;13(Pt 1):324–331. doi: 10.1007/978-3-642-15705-9_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants Brian B, Yushkevich Paul, Pluta John, Minkoff David, Korczykowski Marc, Detre John, Gee James C. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010b Feb;49(3):2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. http://dx.doi.org/10.1016/j.neuroimage.2009.09.062. URL http://dx.doi.org/10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants Brian B, Tustison Nicholas J, Song Gang, Cook Philip A, Klein Arno, Gee James C. A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage. 2011a Feb;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. http://dx.doi.org/10.1016/j.neuroimage.2010.09.025. URL http://dx.doi.org/10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants Brian B, Tustison Nicholas J, Wu Jue, Cook Philip A, Gee James C. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics. 2011b Dec;9(4):381–400. doi: 10.1007/s12021-011-9109-y. http://dx.doi.org/10.1007/s12021-011-9109-y. URL http://dx.doi.org/10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants Brian B, Tustison Nicholas J, Song Gang, Wu Baohua, Stauffer Michael, McCormick Matthew, Johnson Hans J, Gee James C. A unified image registration framework for ITK. WBIR. 2012:266–275. [Google Scholar]
- Bishop Christopher M. Neural Networks for Pattern Recognition. Oxford University Press; 1995. [Google Scholar]
- Bonner Michael F, Vesely Luisa, Price Catherine, Anderson Chivon, Richmond Lauren, Farag Christine, Avants Brian, Grossman Murray. Reversal of the concreteness effect in semantic dementia. Cogn Neuropsychol. 2009 Sep;26(6):568–579. doi: 10.1080/02643290903512305. http://dx.doi.org/10.1080/02643290903512305. URL http://dx.doi.org/10.1080/026432909203512305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni Barbara, Grassi Mario, Premi Enrico, Alberici Antonella, Cosseddu Maura, Cancelli Vanessa, Caobelli Federico, Paghera Barbara, Padovani Alessandro. Is long-term prognosis of frontotemporal lobar degeneration predictable by neuroimaging? Evidence from a single-subject functional brain study. J Alzheimers Dis. 2012;29(4):883–890. doi: 10.3233/JAD-2012-112078. http://dx.doi.org/10.3233/JAD-2012-112078 (URL http://dx.doi.org/10.3233/JAD-2012-112078) [DOI] [PubMed] [Google Scholar]
- Buckner Randy L, Andrews-Hanna Jessica R, Schacter Daniel L. The brain’s default network. Ann N Y Acad Sci. 2008;1124(1):1–38. doi: 10.1196/annals.1440.011. http://dx.doi.org/10.1196/annals.1440.011/full. [DOI] [PubMed] [Google Scholar]
- Cao Kim-Anh L, Martin Pascal GP, Robert-GraniŽ Christ le, Besse Philippe. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinforma. 2009;10:34. doi: 10.1186/1471-2105-10-34. http://dx.doi.org/10.1186/1471-2105-10-34 (URL http://dx.doi.org/10.1186/1471-2105-10-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Kewei, Reiman Eric M, Huan Zhongdan, Caselli Richard J, Bandy Daniel, Ayutyanont Napatkamon, Alexander Gene E. Linking functional and structural brain images with multivariate network analyses: a novel application of the partial least square method. Neuroimage. 2009 Aug;47(2):602–610. doi: 10.1016/j.neuroimage.2009.04.053. http://dx.doi.org/10.1016/j.neuroimage.2009.04.053. URL http://dx.doi.org/10.1016/j.neuroimage.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Kewei, Langbaum Jessica BS, Fleisher Adam S, Ayutyanont Napatkamon, Reschke Cole, Lee Wendy, Liu Xiaofen, Bandy Dan, Alexander Gene E, Thompson Paul M, Foster Norman L, Harvey Danielle J, de Leon Mony J, Koeppe Robert A, Jagust William J, Weiner Michael W, Reiman Eric M Alzheimer’s Disease Neuroimaging Initiative. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: findings from the Alzheimer’s disease neuroimaging initiative. Neuroimage. 2010 Jun;51(2):654–664. doi: 10.1016/j.neuroimage.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry Steve. Singular value decomposition analysis and canonical correlation analysis. J Clim. 1996;9:2003–2009. [Google Scholar]
- Chumbley Justin R, Friston Karl J. False discovery rate revisited: FDR and topological inference using Gaussian random fields. Neuroimage. 2009;44(1):62–70. doi: 10.1016/j.neuroimage.2008.05.021. ( http://www.sciencedirect.com/science/article/pii/S1053811908006472) [DOI] [PubMed] [Google Scholar]
- Comon P. Independent component analysis, a new concept? Signal Process. 1994;36(3):287–314. [Google Scholar]
- De Martino Federico, Valente Giancarlo, Staeren Noël, Ashburner John, Goebel Rainer, Formisano Elia. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. Neuroimage. 2008;43(1):44–58. doi: 10.1016/j.neuroimage.2008.06.037. ( http://www.sciencedirect.com/science/article/pii/S1053811908007854) [DOI] [PubMed] [Google Scholar]
- Delano-Wood Lisa, Stricker Nikki H, Sorg Scott F, Nation Daniel A, Jak Amy J, Woods Steven P, Libon David J, Delis Dean C, Frank Lawrence R, Bondi Mark W. Posterior cingulum white matter disruption and its associations with verbal memory and stroke risk in mild cognitive impairment. J Alzheimers Dis. 2012;29(3):589–603. doi: 10.3233/JAD-2012-102103. http://dx.doi.org/10.3233/JAD-2012-102103 (URL http://dx.doi.org/10.3233/JAD-2012-102103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson Bradford C, Wolk David A. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology. 2012 Jan;78(2):84–90. doi: 10.1212/WNL.0b013e31823efc6c. http://dx.doi.org/10.1212/WNL.0b013e31823efc6c. URL http://www.ncbi.nlm.nih.gov/pubmed/22189451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho David L. De-noising by soft-thresholding. IEEE Trans Inf Theory. 1995;41(3):613–627. [Google Scholar]
- Dubois B, Slachevsky A, Litvan I, Pillon B. The FAB: a frontal assessment battery at bedside. Neurology. 2000 Dec;55(11):1621–1626. doi: 10.1212/wnl.55.11.1621. [DOI] [PubMed] [Google Scholar]
- Elad M. Why simple shrinkage is still relevant for redundant representations? IEEE Trans Inf Theory. 2006 Dec;52(12):5559–5569. http://dx.doi.org/10.1109/TIT.2006.885522. [Google Scholar]
- Fan Yong, Batmanghelich Nematollah, Clark Chris M, Davatzikos Christos Alzheimer’s Disease Neuroimaging Initiative. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008 Feb;39(4):1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galton Clare J, Erzin lioglu Sharon, Sahakian Barbara J, Antoun Nagui, Hodges John R. A comparison of the Addenbrooke’s Cognitive Examination (ACE), conventional neuropsychological assessment, and simple MRI-based medial temporal lobe evaluation in the early diagnosis of Alzheimer’s disease. Cogn Behav Neurol. 2005 Sep;18(3):144–150. doi: 10.1097/01.wnn.0000182831.47073.e9. [DOI] [PubMed] [Google Scholar]
- Gardini Simona, Concari Letizia, Pagliara Salvatrice, Ghetti Caterina, Venneri Annalena, Caffarra Paolo. Visuo-spatial imagery impairment in posterior cortical atrophy: a cognitive and spect study. Behav Neurol. 2011;24(2):123–132. doi: 10.3233/BEN-2011-0279. http://dx.doi.org/10.3233/BEN-2011-0279 (URL http://dx.doi.org/10.3233/BEN-2011-0279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, Ogar JM, Rohrer JD, Black S, Boeve BF, Manes F, Dronkers NF, Vandenberghe R, Rascovsky K, Patterson K, Miller BL, Knopman DS, Hodges JR, Mesulam MM, Grossman M. Classification of primary progressive aphasia and its variants. Neurology. 2011;76(11):1006–1014. doi: 10.1212/WNL.0b013e31821103e6. (cited By (since 1996) 115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenick Logan, Klingenberg Brad, Katovich Kiefer, Knutson Brian, Taylor Jonathan E. Interpretable whole-brain prediction analysis with GraphNet. Neuroimage. 2013 Jan; doi: 10.1016/j.neuroimage.2012.12.062. http://dx.doi.org/10.1016/j.neuroimage.2012.12.062. [DOI] [PubMed]
- Grossman Murray, Ash Sharon. Primary progressive aphasia: a review. Neurocase. 2004 Feb;10(1):3–18. doi: 10.1080/13554790490960440. http://dx.doi.org/10.1080/13554790490960440. URL http://dx.doi.org/10.1080/13554790490960440. [DOI] [PubMed] [Google Scholar]
- Gunawardena D, Ash S, McMillan C, Avants B, Gee J, Grossman M. Why are patients with progressive nonfluent aphasia nonfluent? Neurology. 2010 Aug;75(7):588–594. doi: 10.1212/WNL.0b013e3181ed9c7d. http://dx.doi.org/10.1212/WNL.0b013e3181ed9c7d. URL http://dx.doi.org/10.1212/WNL.0b013e3181ed9c7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck Christian, Foster Norman L, Perneczky Robert, Kurz Alexander, Alexopoulos Panagiotis, Koeppe Robert A, Drzezga Alexander, Stern Yaakov. Multivariate and univariate neuroimaging biomarkers of Alzheimer’s disease. Neuroimage. 2008 May;40(4):1503–1515. doi: 10.1016/j.neuroimage.2008.01.056. http://dx.doi.org/10.1016/j.neuroimage.2008.01.056. URL http://dx.doi.org/10.1016/j.neuroimage.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke Michael, Halchenko Yaroslav O, Sederberg Per B, José Hanson Stephen, Haxby James V, Pollmann Stefan. PyMVPA: a python toolbox for multivariate pattern analysis of fmri data. Neuroinformatics. 2009;7(1):37–53. doi: 10.1007/s12021-008-9041-y. http://dx.doi.org/10.1007/s12021-008-9041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan Anhar, Whitwell Jennifer L, Josephs Keith A. The corticobasal syndrome–Alzheimer’s disease conundrum. Expert Rev Neurother. 2011 Nov;11(11):1569–1578. doi: 10.1586/ern.11.153. http://dx.doi.org/10.1586/ern.11.153. URL http://dx.doi.org/10.1586/ern.11.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar Derrek P, Stein Jason L, Kohannim Omid, Jahanshad Neda, Saykin Andrew J, Shen Li, Kim Sungeun, Pankratz Nathan, Foroud Tatiana, Huentelman Matthew J, Potkin Steven G, Jack Clifford R, Jr, Weiner Michael W, Toga Arthur W, Thompson Paul M Alzheimer’s Disease Neuroimaging Initiative. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011 Jun;56(4):1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RD, Baeckman L. The relationship between the mini-mental state examination and cognitive functioning in normal elderly adults: a componential analysis. Age Ageing. 1995 Sep;24(5):440–446. doi: 10.1093/ageing/24.5.440. [DOI] [PubMed] [Google Scholar]
- Hotelling H. Canonical Correlation Analysis (CCA) J Educ Psychol 1935 [Google Scholar]
- Hotelling H. Relations between two sets of variants. Biometrika. 1936:321–377. [Google Scholar]
- Hutchinson AD, Mathias JL. Neuropsychological deficits in frontotemporal dementia and Alzheimer’s disease: a meta-analytic review. J Neurol Neurosurg Psychiatry. 2007 Sep;78(9):917–928. doi: 10.1136/jnnp.2006.100669. http://dx.doi.org/10.1136/jnnp.2006.100669. URL http://dx.doi.org/10.1136/jnnp.2006.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloeppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. (cited By (since 1996) 221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floch Edith, Guillemot Vincent, Frouin Vincent, Pinel Philippe, Lalanne Christophe, Trinchera Laura, Tenenhaus Arthur, Moreno Antonio, Zilbovicius Monica, Bourgeron Thomas, Dehaene Stanislas, Thirion Bertrand, Poline Jean-Baptiste, Duchesnay Edouard. Significant correlation between a set of genetic polymorphisms and a functional brain network revealed by feature selection and sparse partial least squares. Neuroimage. 2012 Oct;63(1):11–24. doi: 10.1016/j.neuroimage.2012.06.061. http://dx.doi.org/10.1016/j.neuroimage.2012.06.061. URL http://dx.doi.org/10.1016/j.neuroimage.2012.06.061. [DOI] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999 Oct;401(6755):788–791. doi: 10.1038/44565. http://dx.doi.org/10.1038/44565. URL http://www.ncbi.nlm.nih.gov/pubmed/10548103. [DOI] [PubMed] [Google Scholar]
- Leibovitch FS, Black SE, Caldwell CB, McIntosh AR, Szalai JP, et al. Brain spect imaging and left hemispatial neglect covaried using partial least squares: the Sunnybrook Stroke Study. Hum Brain Mapp. 1999;7(4):244–253. doi: 10.1002/(SICI)1097-0193(1999)7:4<244::AID-HBM3>3.0.CO;2-K. http://dx.doi.org/10.1002/(SICI)1097-0193(1999)7:4%3C244 (AID-HBM3%3E3.0.CO;2-K/full) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Rui, Qin Wen, Zhang Yunting, Jiang Tianzi, Yu Chunshui. The neuronal correlates of digits backward are revealed by voxel-based morphometry and resting-state functional connectivity analyses. PLoS One. 2012;7(2):e31877. doi: 10.1371/journal.pone.0031877. http://dx.doi.org/10.1371/journal.pone.0031877 (URL http://dx.doi.org/10.1371/journal.pone.0031877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libon DJ, Xie SX, Moore P, Farmer J, Antani S, McCawley G, Cross K, Grossman M. Patterns of neuropsychological impairment in frontotemporal dementia. Neurology. 2007;78:369–375. doi: 10.1212/01.wnl.0000252820.81313.9b. [DOI] [PubMed] [Google Scholar]
- Libon DJ, McMillan C, Gunawardena D, Powers C, Massimo L, Khan A, Morgan B, Farag C, Richmond L, Weinstein J, Moore P, Coslett HB, Chatterjee A, Aguirre G, Grossman M. Neurocognitive contributions to verbal fluency deficits in frontotemporal lobar degeneration. Neurology. 2009 Aug;73(7):535–542. doi: 10.1212/WNL.0b013e3181b2a4f5. http://dx.doi.org/10.1212/WNL.0b013e3181b2a4f5. URL http://dx.doi.org/10.1212/WNL.0b013e3181b2a4f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libon David J, Rascovsky Katya, Gross Rachel G, White Matthew T, Xie Sharon X, Dreyfuss Michael, Boller Ashley, Massimo Lauren, Moore Peachie, Kitain Jessica, Branch Coslett H, Chatterjee Anjan, Grossman Murray. The Philadelphia brief assessment of cognition (PBAC): a validated screening measure for dementia. Clin Neuropsychol. 2011 Nov;25(8):1314–1330. doi: 10.1080/13854046.2011.631585. http://dx.doi.org/10.1080/13854046.2011.631585. URL http://www.ncbi.nlm.nih.gov/pubmed/22084867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Fa-Hsuan, McIntosh Anthony R, Agnew John A, Eden Guinevere F, Zeffiro Thomas A, Belliveau John W. Multivariate analysis of neuronal interactions in the generalized partial least squares framework: simulations and empirical studies. Neuroimage. 2003 Oct;20(2):625–642. doi: 10.1016/S1053-8119(03)00333-1. http://dx.doi.org/10.1016/S1053-8119(03)00333-1. URL http://dx.doi.org/10.1016/S1053-8119(03)00333-1) [DOI] [PubMed] [Google Scholar]
- Mansfield ER, Webster JT, Gunst RF. An analytic variable selection technique for principal component regression. J R Stat Soc: Ser C: Appl Stat. 1977 Jan;26(1):34–40. http://dx.doi.org/10.2307/2346865. URL http://www.jstor.org/stable/2346865. ArticleType: research-article/Full publication date: 1977/Copyright © 1977 Royal Statistical Society) [Google Scholar]
- Massimo Lauren, Powers Chivon, Moore Peachie, Vesely Luisa, Avants Brian, Gee James, Libon David J, Grossman Murray. Neuroanatomy of apathy and disinhibition in frontotemporal lobar degeneration. Dement Geriatr Cogn Disord. 2009;27(1):96–104. doi: 10.1159/000194658. http://dx.doi.org/10.1159/000194658 (URL http://dx.doi.org/10.1159/000194658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AR, Bookstein FL, Haxby JV, Grady CL. Spatial pattern analysis of functional brain images using partial least squares. Neuroimage. 1996 Jun;3(3 Pt 1):143–157. doi: 10.1006/nimg.1996.0016. http://dx.doi.org/10.1006/nimg.1996.0016. URL http://dx.doi.org/10.1006/nimg.1996.0016. [DOI] [PubMed] [Google Scholar]
- McKhann G, Trojanowski JQ, Grossman M, Miller BL, Dickson D, Albert M. Clinical and pathological diagnosis of frontotemporal dementia: report of a work group on frontotemporal dementia and Pick’s disease. Arch Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Nasreddine Ziad S, Phillips Natalie A, BŽdirian ValŽrie, Charbonneau Simon, Whitehead Victor, Collin Isabelle, Cummings Jeffrey L, Chertkow Howard. The Montreal cognitive assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005 Apr;53(4):695–699. doi: 10.1111/j.1532-5415.2005.53221.x. http://dx.doi.org/10.1111/j.1532-5415.2005.53221.x. URL http://dx.doi.org/10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- Nestor PJ, Graham KS, Bozeat S, Simons JS, Hodges JR. Memory consolidation and the hippocampus: further evidence from studies of autobiographical memory in semantic dementia and frontal variant frontotemporal dementia. Neuropsychologia. 2002a;40(6):633–654. doi: 10.1016/s0028-3932(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nestor Paul G, O’Donnell Brian F, McCarley Robert W, Niznikiewicz Margaret, Barnard John, Jen Shen Zi, Bookstein Fred L, Shenton Martha E. A new statistical method for testing hypotheses of neuropsychological/MRI relationships in schizophrenia: partial least squares analysis. Schizophr Res. 2002b Jan;56(1–2):57–66. doi: 10.1016/s0920-9964(00)00171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman Kenneth A, Polyn Sean M, Detre Greg J, Haxby James V. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn Sci. 2006 Sep;10(9):424–430. doi: 10.1016/j.tics.2006.07.005. http://dx.doi.org/10.1016/j.tics.2006.07.005. URL http://dx.doi.org/10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- O’Toole Alice J, Jiang Fang, Abdi Hervé, Pénard Nils, Dunlop Joseph P, Parent Marc A. Theoretical, statistical, and practical perspectives on pattern-based classification approaches to the analysis of functional neuroimaging data. J Cogn Neurosci. 2007;19(11):1735–1752. doi: 10.1162/jocn.2007.19.11.1735. http://dx.doi.org/10.1162/jocn.2007.19.11.1735. [DOI] [PubMed] [Google Scholar]
- Parkhomenko Elena, Tritchler David, Beyene Joseph. Sparse canonical correlation analysis with application to genomic data integration. Stat Appl Genet Mol Biol. 2009 Jan;8(1):Article 1. doi: 10.2202/1544-6115.1406. http://dx.doi.org/10.2202/1544-6115.1406. URL http://dx.doi.org/10.2202/1544-6115.1406. [DOI] [PubMed] [Google Scholar]
- Pengas George, Hodges John R, Watson Peter, Nestor Peter J. Focal posterior cingulate atrophy in incipient Alzheimer’s disease. Neurobiol Aging. 2010 Jan;31(1):25–33. doi: 10.1016/j.neurobiolaging.2008.03.014. http://dx.doi.org/10.1016/j.neurobiolaging.2008.03.014. URL http://dx.doi.org/10.1016/j.neurobiolaging.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Polak Elijah. Optimization: Algorithms and Consistent Approximations. Springer; 1997. [Google Scholar]
- Poldrack Russell A. Region of interest analysis for fMRI. Soc Cogn Affect Neurosci. 2007 Mar;2(1):67–70. doi: 10.1093/scan/nsm006. http://dx.doi.org/10.1093/scan/nsm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici GD, Seeley WW, Kim EJ, Gorno-Tempini ML, Rascovsky K, Pagliaro TA, Allison SC, Halabi C, Kramer JH, Johnson JK, Weiner MW, Forman MS, Trojanowski JQ, Dearmond SJ, Miller BL, Rosen HJ. Distinct MRI atrophy patterns in autopsy-proven Alzheimer’s disease and frontotemporal lobar degeneration. Am J Alzheimers Dis Other Demen. 2007;22(6):474–488. doi: 10.1177/1533317507308779. http://dx.doi.org/10.1177/1533317507308779 (URL http://dx.doi.org/10.1177/1533317507308779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascovsky Katya, Hodges John R, Knopman David, Mendez Mario F, Kramer Joel H, Neuhaus John, van Swieten John C, Seelaar Harro, Dopper Elise GP, Onyike Chiadi U, Hillis Argye E, Josephs Keith A, Boeve Bradley F, Kertesz Andrew, Seeley William W, Rankin Katherine P, Johnson Julene K, Gorno-Tempini Maria-Luisa, Rosen Howard, Prioleau-Latham Caroline E, Lee Albert, Kipps Christopher M, Lillo Patricia, Piguet Olivier, Rohrer Jonathan D, Rossor Martin N, Warren Jason D, Fox Nick C, Galasko Douglas, Salmon David P, Black Sandra E, Mesulam Marsel, Weintraub Sandra, Dickerson Brad C, Diehl-Schmid Janine, Pasquier Florence, Deramecourt Vincent, Lebert Florence, Pijnenburg Yolande, Chow Tiffany W, Manes Facundo, Grafman Jordan, Cappa Stefano F, Freedman Morris, Grossman Murray, Miller Bruce L. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011 Sep;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. http://dx.doi.org/10.1093/brain/awr179. URL http://dx.doi.org/10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen Jerod M, Lakatos Anita, van Erp Theo GM, Kruggel Frithjof, Keator David B, Fallon James T, Macciardi Fabio, Potkin Steven G Alzheimer’s Disease Neuroimaging Initiative. Empirical derivation of the reference region for computing diagnostic sensitive “18fluorodeoxyglucose ratios in Alzheimer’s disease based on the ADNI sample. Biochim Biophys Acta. 2012 Mar;1822(3):457–466. doi: 10.1016/j.bbadis.2011.09.008. http://dx.doi.org/10.1016/j.bbadis.2011.09.008. URL http://dx.doi.org/10.1016/j.bbadis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts Stephen J. Parametric and non-parametric unsupervised cluster analysis. Pattern Recogn. 1997 Feb;30(2):261–272. http://dx.doi.org/10.1016/S0031-3203(96)00079-9. URL http://www.sciencedirect.com/science/article/pii/S0031320396000799. [Google Scholar]
- Rogalski E, Cobia D, Harrison TM, Wieneke C, Weintraub S, Mesulam M-M. Progression of language decline and cortical atrophy in subtypes of primary progressive aphasia. Neurology. 2011 May;76(21):1804–1810. doi: 10.1212/WNL.0b013e31821ccd3c. http://dx.doi.org/10.1212/WNL.0b013e31821ccd3c. URL http://dx.doi.org/10.1212/WNL.0b013e31821ccd3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden Chris, Karnath Hans-Otto, Bonilha Leonardo. Improving lesion–symptom mapping. J Cogn Neurosci. 2007 Jul;19(7):1081–1088. doi: 10.1162/jocn.2007.19.7.1081. http://dx.doi.org/10.1162/jocn.2007.19.7.1081. URL http://dx.doi.org/10.1162/jocn.2007.19.7.1081. [DOI] [PubMed] [Google Scholar]
- Rosen Howard J, Alcantar Oscar, Rothlind Johannes, Sturm Virginia, Kramer Joel H, Weiner Michael, Miller Bruce L. Neuroanatomical correlates of cognitive self-appraisal in neurodegenerative disease. Neuroimage. 2010 Feb;49(4):3358–3364. doi: 10.1016/j.neuroimage.2009.11.041. http://dx.doi.org/10.1016/j.neuroimage.2009.11.041. URL http://dx.doi.org/10.1016/j.neuroimage.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryali Srikanth, Supekar Kaustubh, Abrams Daniel A, Menon Vinod. Sparse logistic regression for whole-brain classification of fMRI data. Neuroimage. 2010;51(2):752–764. doi: 10.1016/j.neuroimage.2010.02.040. (URL http://www.sciencedirect.com/science/article/pii/S1053811910002089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu Mert R, Van Leemput Koen. The relevance voxel machine (RVoxM): a Bayesian method for image-based prediction. Med Image Comput Comput Assist Interv. 2011;14(Pt 3):99–106. doi: 10.1007/978-3-642-23626-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A, Polak E. Family of projected descent methods for optimization problems with simple bounds. J Optim Theory Appl. 1997 Jan;92(1):1–31. http://dx.doi.org/10.1023/A:1022690711754. URL http://link.springer.com/article/10.1023/A%3A1022690711754. [Google Scholar]
- Shamy Jul Lea, Habeck Christian, Hof Patrick R, Amaral David G, Fong Sania G, Buonocore Michael H, Stern Yaakov, Barnes Carol A, Rapp Peter R. Volumetric correlates of spatiotemporal working and recognition memory impairment in aged rhesus monkeys. Cereb Cortex. 2011 Jul;21(7):1559–1573. doi: 10.1093/cercor/bhq210. http://dx.doi.org/10.1093/cercor/bhq210. URL http://dx.doi.org/10.1093/cercor/bhq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shany-Ur Tal, Rankin Katherine P. Personality and social cognition in neurodegenerative disease. Curr Opin Neurol. 2011 Dec;24(6):550–555. doi: 10.1097/WCO.0b013e32834cd42a. http://dx.doi.org/10.1097/WCO.0b013e32834cd42a. URL http://dx.doi.org/10.1097/WCO.0b013e32834cd42a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington Cynthia M, Chu Carlton, Klšppel Stefan, Jack Clifford R, Jr, Ashburner John, Frackowiak Richard SJ Alzheimer Disease Neuroimaging Initiative. Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage. 2010 Jul;51(4):1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Liang, Ji Shuiwang, Yu Shipeng, Ye Jieping. On the equivalence between canonical correlation analysis and orthonormalized partial least squares. IJCAI. 2009:1230–1235. [Google Scholar]
- Suykens Johan AK, De Brabanter Jos, Lukas Lukas, Vandewalle Joos. Weighted least squares support vector machines: robustness and sparse approximation. Neurocomputing. 2002;48(1):85–105. (URL http://www.sciencedirect.com/science/article/pii/S0925231201006440) [Google Scholar]
- Tibshirani Robert. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Methodol. 1996 Jan;58(1):267–288. URL http://www.jstor.org/stable/2346178. ArticleType: research-article/Full publication date: 1996/Copyright © 1996 Royal Statistical Society) [Google Scholar]
- Tipping Michael E. Sparse Bayesian learning and the relevance vector machine. J Mach Learn Res. 2001;1:211–244. (URL http://dl.acm.org/citation.cfm?id=944741) [Google Scholar]
- Tosun Duygu, Rosen Howard, Miller Bruce L, Weiner Michael W, Schuff Norbert. MRI patterns of atrophy and hypoperfusion associations across brain regions in frontotemporal dementia. Neuroimage. 2012 Feb;59(3):2098–2109. doi: 10.1016/j.neuroimage.2011.10.031. http://dx.doi.org/10.1016/j.neuroimage.2011.10.031. URL http://dx.doi.org/10.1016/j.neuroimage.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison Nicholas J, Avants Brian B, Cook Philip A, Zheng Yuanjie, Egan Alexander, Yushkevich Paul A, Gee James C. N4itk: improved n3 bias correction. IEEE Trans Med Imaging. 2010 Jun;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. http://dx.doi.org/10.1109/TMI.2010.2046908. URL http://dx.doi.org/10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison Nicholas J, Avants Brian B, Cook Philip A, Kim Junghoon, Whyte John, Gee James C, Stone James R. Logical circularity in voxel-based analysis: normalization strategy may induce statistical bias. Hum Brain. 2013 doi: 10.1002/hbm.22211. http://www.ncbi.nlm.nih.gov/pubmed/23151955 (in press) [DOI] [PMC free article] [PubMed]
- Whitwell Jennifer L, Jack Clifford R, Parisi Joseph E, Knopman David S, Boeve Bradley F, Petersen Ronald C, Ferman Tanis J, Dickson Dennis W, Josephs Keith A. Rates of cerebral atrophy differ in different degenerative pathologies. Brain. 2007 Apr;130(Pt 4):1148–1158. doi: 10.1093/brain/awm021. http://dx.doi.org/10.1093/brain/awm021. URL http://dx.doi.org/10.1093/brain/awm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GB, Nestor PJ, Hodges JR. Neural correlates of semantic and behavioural deficits in frontotemporal dementia. Neuroimage. 2005;24(4):1042–1051. doi: 10.1016/j.neuroimage.2004.10.023. (cited By (since 1996) 94) [DOI] [PubMed] [Google Scholar]
- Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8(1) doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten Daniela M, Tibshirani Robert, Hastie Trevor. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009 Jul;10(3):515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Okito, Sato Masa-aki, Yoshioka Taku, Tong Frank, Kamitani Yukiyasu. Sparse estimation automatically selects voxels relevant for the decoding of fMRI activity patterns. Neuroimage. 2008;42(4):1414–1429. doi: 10.1016/j.neuroimage.2008.05.050. ( http://www.sciencedirect.com/science/article/pii/S1053811908006940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17(9):763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- Zhang Tong. Adaptive forward–backward greedy algorithm for sparse learning with linear models. Advances in Neural Information Processing Systems. 2008:1921–1928. [Google Scholar]
- Zhou Luping, Wang Yaping, Li Yang, Yap Pew-Thian, Shen Dinggang Alzheimer’s Disease Neuroimaging Initiative (ADNI) Hierarchical anatomical brain networks for MCI prediction: revisiting volumetric measures. PLoS ONE. 2011 Jul;6(7):e21935. doi: 10.1371/journal.pone.0021935. http://dx.doi.org/10.1371/journal.pone.0021935. http://dx.doi.org/10.1371/journal.pone.0021935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibulevsky M, Elad M. L1–L2 optimization in signal and image processing. IEEE Signal Process Mag. 2010 May;27(3):76–88. [Google Scholar]